Abstract
Huntington’s disease (HD) is a progressive neurodegenerative disorder primarily affecting the basal ganglia and is caused by expanded CAG repeats in the huntingtin gene. Except for CAG sizing, mitochondrial and nuclear DNA (mtDNA and nDNA) parameters have not yet proven to be representative biomarkers for disease and future therapy. Here, we identified a general suppression of genes associated with aerobic metabolism in peripheral blood mononuclear cells (PBMCs) from HD patients compared to controls. In HD, the complex II subunit SDHB was lowered although not sufficiently to affect complex II activity. Nevertheless, we found decreased level of factors associated with mitochondrial biogenesis and an associated dampening of the mitochondrial DNA damage frequency in HD, implying an early defect in mitochondrial activity. In contrast to mtDNA, nDNA from HD patients was four-fold more modified than controls and demonstrated that nDNA integrity is severely reduced in HD. Interestingly, the level of nDNA damage correlated inversely with the total functional capacity (TFC) score; an established functional score of HD. Our data show that PBMCs are a promising source to monitor HD progression and highlights nDNA damage and diverging mitochondrial and nuclear genome responses representing early cellular impairments in HD.
Introduction
Huntington’s disease is an autosomal dominant progressive neurodegenerative disorder caused by an inherited, pathogenic expansion of CAG repeats in exon 1 of the huntingtin gene (HTT). The mutant HTT (mHTT) protein manifests into HD when the CAG tracts expands beyond 36; while wild type HTT contains between 16 and 25 CAG repeats1. The size of the expansion is correlated with disease severity, with increasing CAG repeat lengths accelerating the age of onset. The HTT gene encodes a 350 kDa protein whose exact function is still not understood. The gene is essential for viability and is ubiquitously expressed2–4. The mHTT protein influences a plethora of cellular processes including transcription, axonal transport, cytoskeletal structure/function, signal transduction, and autophagy that ultimately result in cell death. HD predominantly affects the basal ganglia, leading to affective, cognitive, behavioural and motor decline, which then progresses to death. Medium GABAergic spiny neurons in the striatal part of the basal ganglia are particularly vulnerable. HD patients exhibit heterogeneous disease progression. The Unified Huntington’s Disease Rating Score (UHDRS) was established as a reliable hybrid scale to describe motor function, cognitive function, behaviour abnormalities, and total functional capacity (TFC)5, that together with molecular genetic testing (CAG sizing)6 provided tools to monitor disease. It has nevertheless been reported that loss of caudate volume exceeds 50% by the time of diagnosis7, and premanifest mHTT carriers display striatal atrophy 15–20 years before predicted disease onset8 The heterogeneity of the disease is further illustrated by that tests for behaviour, motor score, functional and cognitive decline do not overlap9.
Although neurological symptoms have been the main focus of the disease, mHTT exerts pathogenic effects in other tissues as well. Predisposition to diabetes and skeletal muscle wasting that precedes detectable neurodegeneration has been reported in patients, demonstrating that HD is a systemic disease10,11. Mouse models of HD demonstrate impaired glucose uptake and altered gene regulation12, as well as cardiac dysfunction13.
Mitochondrial alterations are well documented in HD. Immortalized lymphoblasts from HD patients are hypersensitive to mitochondrial inhibition14, and homozygous mHTT cells display mitochondrial aberrations15. Primary HD blood cells are characterized by increased levels of pro-apoptotic proteins and reactive oxygen species (ROS)16. The HD cellular phenotype is reminiscent of mitochondrial dysfunction. This association is emphasized by the HD-mimicking striatal degeneration by the mycotoxin 3-nitropropionic acid; a mitochondrial complex II inhibitor that was accidentally consumed as contamination of sugarcanes17. Subsequently, systemic exposure to 3-nitropropionic acid was introduced to investigate striatal neurodegeneration in rodents and verify the coupling between mitochondrial electron transport chain (ETC) complex II dysfunction and striatal degeneration.
Mitochondrial dysfunction is a general feature of mouse models of HD. The mHtt knock-in model carries an expanded CAG tract (150 CAGs) in the endogenous murine gene and represents a physiological relevant model to understand the initial phases of the disease18. The mHTT protein in this model influences nuclear gene expression including PGC-1α, the major regulator of mitochondrial biogenesis and result in reduced thermoregulation and reduced expression of the dismutases SOD1 and SOD219. Polyglutamines interfere with mitochondria and disturb intracellular trafficking and cause mitochondrial fragmentation20,21.
The extended CAG tract in the mHTT gene is genetically unstable, and increases with age in a tissue-specific manner, as shown in both patients and HD models. Studies from mouse models indicate that faulty DNA repair underlies somatic expansions in the polyQ tract, in particular the DNA base excision repair factors OGG1 and, NEIL122,23 and the DNA mismatch repair protein MSH224.
These observations imply that mitochondrial aberrations and/or DNA maintenance are key parameters underlying the disease. The abrogation of somatic expansions in MSH2 deficiency delayed nuclear mHTT aggregation and such implied that somatic expansion drives the progression of the disease25,26.
We hypothesized that mHTT would systemically affect mitochondrial function and that peripheral blood mononuclear cells (PBMCs) as post-mitotic (and peripheral) cells would serve as suitable and easy accessible material presenting HD. Parameters of oxidative metabolism, including mitochondrial ETC assessment were assessed to investigate cellular energy status. DNA damage was included as a quantitative measure of the intracellular redox state.
Results
Early mitochondrial aberrations in HD PBMCs
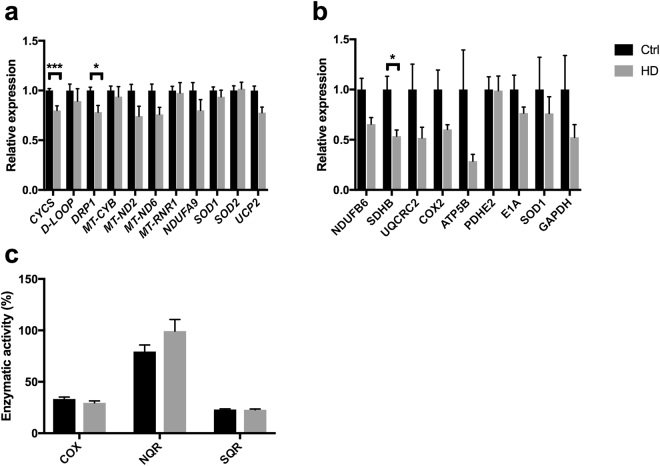
PBMCs were isolated from 36 patients and 49 healthy volunteers (controls). The patients were genetically tested for the CAG size. The sampling of PBMCs was paralleled by an assessment of the TFC, which was performed as part of the clinical follow-up. An overview of the included cohort is provided in Table 1. TFC was inversely correlated with duration of disease (R = −0.77, P < 0.001). There is partial correlation between onset of disease and TFC when age is corrected for (R = 0.61, P < 0.05).To investigate the influence of HD on mitochondrial function, we assessed the expression of mtDNA- and nDNA encoded genes related to mitochondrial pathways. We found significant downregulation of genes related to mitochondrial function in HD PBMCs. These are CYCS and DRP1 (P < 0.05) and UCP2 at the borderline after adjustment for multiple comparisons (Fig. 1a). The mtDNA-encoded genes MT-ND2, MT-ND6 genes and mitochondrial small ribosomal subunit (MT-RNR1) were unaffected. RNA prepared from 18 individual HD PBMCs were grouped into three categories according to CAG repeat length (low-CAG, mid-CAG and high-CAG), pooled in equimolar ratios in each category, and subjected to RNA sequencing analyses together with pooled RNA in equimolar ratios from 18 controls. The RNA sequencing analyses confirmed the suppression of genes related to mitochondrial regulation, aerobic metabolism and oxidative stress (Supplementary Table S2). We did not observe a significant reduced level of mitochondria-associated proteins in HD (Fig. 1b). However, the reduction of the iron-sulfur containing subunit B of succinate dehydrogenase (SDHB) reached statistical significance after correction for multiple testing. Importantly, the reduction in SDHB did not correlate with CAG, age or TFC. When enzymatic activities of the ETC complexes were assessed, it became clear that the SDHB reduction did not produce a net effect on complex II (SQR). Likewise, the average complex I (NQR) and complex IV (COX) activities were not different in HD compared to controls (Fig. 1c). The ETC complexes altogether contain up to 100 different subunits, and although only a minor proportion of these were assessed in Fig. 1b, the present results imply at least that other subunits that could be affected in HD, are not bottlenecks for the ETC mitochondrial biochemical complex activity.
Table 1.
Overview of included HD patients and age-matched controls.
| HD | Controls | ||||||
|---|---|---|---|---|---|---|---|
| Median | Range | (n) | Median | Range | (n) | ||
| Age (years) | All | 41.5 | [22–74] | 36 | 46 | [21–76] | 49 |
| Men | 40 | [31–73] | 19 | 41 | [21–76] | 25 | |
| Women | 46 | [22–74] | 17 | 47 | [24–66] | 24 | |
| CAG repeat size | All | 45 | [40–58] | 36 | N/A | ||
| Men | 45 | [41–53] | 19 | N/A | |||
| Women | 44 | [40–58] | 17 | N/A | |||
| TFC score | All | 7 | [0–13] | 36 | N/A | ||
| Men | 8 | [1–13] | 19 | N/A | |||
| Women | 6 | [0–11] | 17 | N/A | |||
Figure 1.
Early mitochondrial alterations in HD PBMCs. (a) Expression of mitochondria-related genes in HD (n = 12–18) relative to controls (n = 32–44). (b) Expression of proteins associated with mitochondrial function in HD (n = 17) and controls (n = 5). (c) Biochemical ETC complex activity in HD (n = 17) and control (n = 34) PBMCs. Data represent mean with error bars representing standard error mean. *P < 0.05., ***P < 0.001.
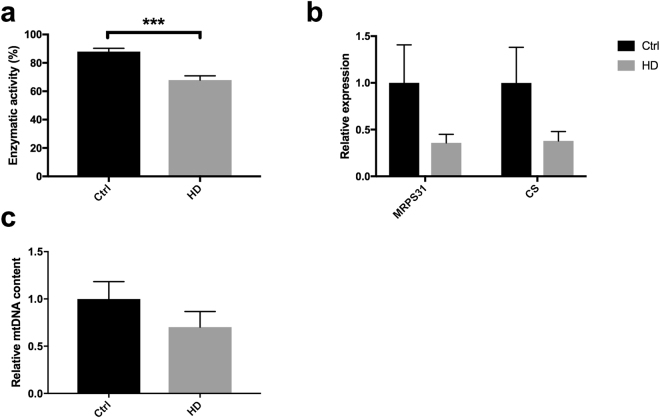
Despite ETC being unaffected in HD, the level and activity of specific mitochondrial proteins were assessed more in detail, as cellular ETC complex capacity by itself does not necessarily reflect aerobic activity27. The activity of citrate synthase, a commonly used marker for mitochondrial content, was indeed significantly reduced in HD (−25%; Fig. 2a). The corresponding levels of the citrate synthase protein was reduced as well, similarly as the small mitochondrial ribosomal subunit MRPS31, both with borderline significance (P = 0.05) (Fig. 2b). The reduction in the above parameters was however not coherent with the stable expression of MT-RNR1 (Fig. 1a) and might be indicative of an imbalanced composition of mitochondria in HD PBMCs. Interestingly, while there was a perfect symmetry between the activity of citrate synthase, complex I, II and IV in controls, the complex I activity did not correlate with any of the other activities in HD (Supplementary Table S2). The mtDNA copy number varies upon bioenergetic stimuli and oxidative stress. Upon its quantification, we found no difference between HD and controls (Fig. 2c).
Figure 2.
Mitochondrial biomarkers indicate reduced mitochondrial activity in HD. (a) Activity of citrate synthase in HD PBMCs (n = 17) relative to controls (n = 32). (b) Levels of ribosomal subunit MRPS31 and citrate synthase in HD (n = 17) and control (n = 5) PBMCs. (c) mtDNA copy number in HD and control PBMCs. ***P < 0.001.
Genome integrity is affected in HD
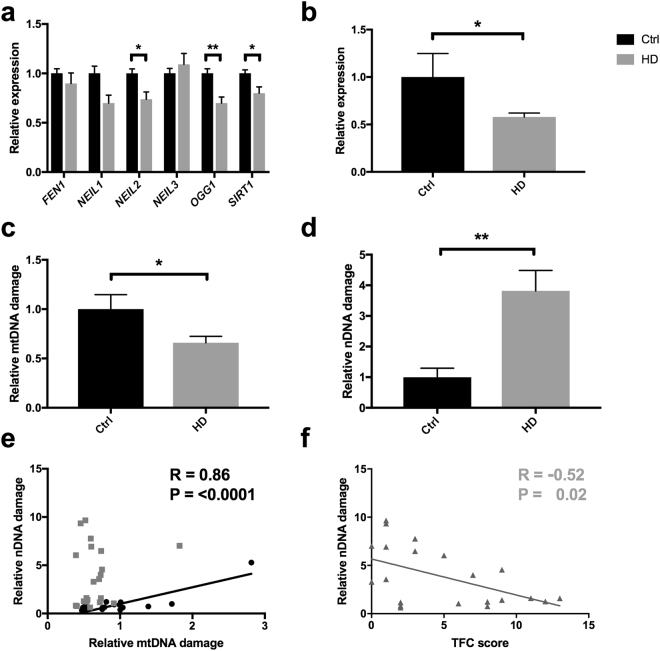
The overall mitochondrial aberration in HD PBMCs suggested that patient cells undergo metabolic alteration. We recently demonstrated that mtDNA responds to alterations in mitochondrial metabolism and that DNA repair factors influence the modification of mtDNA upon metabolic manipulations28. We therefore assessed the expression of genes associated to the base excision repair pathway, and found a tendency for general suppression of DNA repair capacity in HD (Fig. 3a). OGG1 was most significantly suppressed. We followed up by assessing the protein level of p53, whose role is implicated in DNA repair as well as mitochondrial function. In contrast to earlier studies29, we found p53 level to be reduced in HD PBMCs (Fig. 3b), which supported the lowering of mitochondrial parameters (Fig. 2). To assess DNA integrity, we used a qPCR-based method30, which detects any modification that inhibits the TaqαI restriction enzyme. In line with the tendency to lower mitochondrial activity, the mtDNA damage in HD PBMCs was reduced by one third compared to controls (Fig. 3c). The lower mtDNA damage level was not likely to be due to excessive/mitophagic removal of damaged molecules, as the copy number of mtDNA was similar in both genotypes (Fig. 2c).
Figure 3.
DNA integrity is impaired in HD PBMCs. (a) Expression of genes associated to base excision repair, in HD (n = 18) relative to controls (n = 44). (b) Expression of p53 protein level in HD (n = 17) and controls (n = 5). (c) mtDNA integrity and (d) nDNA integrity in HD (n = 21) and control (n = 16) PBMCs. (e) Correlation analyses between nDNA damage and mtDNA damage in HD and control PBMCs (n = 16). (f) Correlation analyses between nuclear DNA damage in PBMCs and TFC score in HD patients (n = 21) *P < 0.05; **P < 0.01. Pearson correlation with 95% confidence interval.
In contrast, a 4-fold (P < 0.01) increased level of nDNA damage was identified in HD PBMCs compared to healthy controls (Fig. 3d). We followed up by investigating the genomic DNA by mass spectrometry analyses. Despite the 4-fold increased nDNA damage level as measured by the qPCR-based method, we could not find any significant increase in any of the investigated base lesions/epigenetic marks in HD (Supplementary Fig. S1). Therefore, genomic alterations other than base lesions are likely causes of the elevated nDNA damage in HD.
These results suggest that mtDNA is relatively spared in HD compared to nDNA, and thus damaging events in the two cellular genomes are independent (Fig. 3c,d). To follow up on the latter point, we compared damage in mtDNA with that in nDNA. In control PBMCs, DNA damage level in the two genomes correlated strongly (R = 0.86, P < 0.0001), while there was no correlation between damage in mtDNA and nDNA in HD (Fig. 3e). Given the elevated nDNA damage in HD, we investigated potential correlation between nDNA damage and clinical symptoms, represented by the TFC score. Interestingly, TFC displayed an inverse correlation with the nDNA damage level in patient PBMCs (Fig. 3f). Additionally, the duration of disease (time from onset to PBMC collection) correlated with nDNA damage (R = 0.045, P < 0.05). These results demonstrate that nDNA damage emerges as a promising novel biomarker of systemic alterations in HD.
Discussion
In this study we have studied PBMCs from HD patients and healthy controls, and identified elevated nDNA damage and alterations in mitochondrial parameters in HD. Although mitochondrial aberrations previously have been confirmed in patient samples, this has not been established as suitable biomarkers for monitoring HD progression and effect of putative therapy. The correlation between the clinical score and DNA damage indicates that mHTT has a powerful systemic impact on DNA integrity in HD. Signs of early mitochondrial dysfunction was evident, but did not correlate with clinical score.
Associated side effects of mitochondrial dysfunction are oxidative stress and DNA damage. Urinary 8-oxoguanine is a marker of endogenous oxidative stress and was found to correlate with disease progression in the R6/2 HD model, as well as responding to therapy31. Impaired genome integrity has also been identified previously in caudate region of post-mortem HD brains32. Chen and co-workers demonstrated elevated genomic 8-oxoguanine in leukocytes from HD33, which is in contradiction to our data (Supplementary Fig. S1), as well as data from Borowsky et al.34. Although we don’t know the reason for the discrepancy, it is worth noting that neutrophils in general exhibit reduced genomic integrity compared to lymphocytes35. It may therefore be speculated that the neutrophile:lymphocyte composition vary in the two studies. Different leukocyte isolation procedures might also explain the lower mtDNA copy number in HD leukocytes found by Jędrak and co-workers36. Several observations herein are indicative of reduced mitochondrial activity in HD. The expression level of SIRT1, which supports mitochondrial biogenesis via deacetylating PGC-1α37, is lower in HD than in control PBMCs (Fig. 3a). This reduction is paralleled by reduced p53 levels, reduced citrate synthase and importantly, reduced level of mtDNA damage. One limitation of our study is that we did not include real-time analyses of PBMCs mitochondrial activity. However, Mejia and coworkers found strongly reduced mitochondrial activity in transformed lymphoblasts from HD patients, compared to controls. Despite this reduction the HD cells were proficient for ETC capacity, similarly as PBMCs in our study38.
We previously found that mitochondrial metabolism influences mtDNA integrity to the same extent as sub-lethal oxidative stress28. We do not believe that the epigenetic modifications contribute to the mtDNA damage signal39, and accordingly there was not an overall difference in epigenetic markers in genomic DNA from HD and controls (Supplementary Fig. S1). In view of these empirical data, we thus believe that the relatively spared mtDNA in HD is caused by an altered metabolic activity.
Interestingly, the strong correlation observed in control PBMCs between nDNA and mtDNA damage was abolished in HD, mainly due to strongly increased nDNA damage.
Different chromatin packing is one plausible explanation for the altered gene expression and sensitivity to nDNA damage in HD. HTT has been found to interact with and stimulate PRMT5, an arginine methyltransferase that affects chromatin packing via it’s methylation of arginine 3 of histone 2A and histone 4. This interaction was attenuated by the polyQ mutation, resulting in reduced dimethylation of H2A/H4R3 in HD patients and an HD mouse model40.
Reduced DNA repair activity in HD is implicated by the gene expression data (Fig. 3a) and could underlie the high nDNA damage level in HD. Emerging data infer that DNA repair factors reflect metabolism of the cell, OGG141, Neil142, p5343 are examples that respond to metabolic alterations in the cell. These alterations are interesting in view of the disproportional complex I in HD compared to controls (Supplementary Table S2).
Efficient ATP production depends on organization of ETC complexes into supercomplexes44, and altered stoichiometry of complex I relative to for example complex IV might have implications for intracellular ATP. The repair of nDNA was found to be relatively more dependent on intracellular ATP than repair of mtDNA45. Together, the indications of altered metabolism and disproportional ETC machinery are coherent with increased accumulation of nDNA damage in HD.
Earlier reports of ETC dysfunction in HD include reduction in complex III32, complex II, III and IV46, complex I47, as well as decreased COX activity48, which is in contrast to the normal ETC activities found here, beside a reduced level of SDHB (complex II) (Fig. 3b). This is indicative of a redundant capacity of ETC, where not all subunits are forming functional ETC complexes. A parallel phenomenon is illustrated by the strong increase (4-fold) in ETC activity upon cytomegalovirus infection that did not involve increased level of specific ETC subunits49. The ETC capacity as a good marker for mitochondrial activity is thus questionable. This is in contrast to the strong effect of ETC genes as shown by GO analyses (Supplementary Table S1), and implies that expression levels of ETC subunits rather than ETC capacity are early markers for mitochondrial dysfunction HD PBMC’s. We find it interesting that the complex II subunit SDHB is down-regulated in HD PBMCs. Systemic inhibition of complex II induces HD- like pathology17, and the SDHB (also known as Ip) is specifically targeted20. In particular, SDHB protects against 3-nitropropionic acid-induced striatal degeneration. Interestingly, SDHB is suppressed in monocytes exposed to inflammation/hypoxia in an APOBEC3-mediated manner50. GO analyses identified altered hypoxic signalling in the HD although not in the patients with highest CAG size (Supplementary Table S1).
The lack of correlation between SDHB and disease progression is interesting and motivates further evaluation of SDHB as a biomarker for the asymptomatic state of the disease. This is a task we are currently pursuing.
In summary, our results confirm other publications that peripheral cells are affected in HD. In addition to biomarkers found in cited reports above, mHTT protein fragments51 and inflammatory markers52 are identified in HD plasma. Importantly, we discover that nDNA damage in PBMCs correlates with duration of disease as well as TFC score. Consequently, future therapy may not necessarily need to target affected brain areas in HD. PBMCs provide an easy-accessible source that has a promising potential to be used in the future follow-up of patients, to monitor therapeutic interventions.
Methods
Patient material and sample collection
All work involving human samples was carried out in accordance with the World Medical Association Declaration of Helsinki and was approved by the Ethical Committee of the General University Hospital in Prague, approved project COST LD15099 (2015–2017) and Czech-Norwegian Research Programme (CZ09) 7F14308 (2014–2017). The written informed consent was obtained from patients as well as control subjects.
Transfer and analyses of human samples in Norway was approved by South-East Regional Ethical Committee (REK HSØ). Approved project no: REK 2015/1876 A.
PBMCs
Peripheral blood was taken from healthy controls and patients with confirmed HD. Within 1-2 h after collection, whole fraction of intact PBMCs was isolated from EDTA-blood by density medium centrifugation at 25 °C, using HistopaqueR1077 Hybri-Max TM (Sigma) following a standard protocol. Briefly, 6 ml of blood was layered on top of a 3 ml of Histopaque and centrifuged at 800 × g for 30 min at 25 °C. The separated mononuclear band was carefully collected (~1 ml), resuspended in 10 ml PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4.12H2O, 1.5 mM KH2PO4) and centrifuged again at 800 × g for 20 min. The pellet was washed twice in same conditions. Finally, the dry pellet was stored at −80 °C until use. For enzymatic and protein analyses samples from identical isolation of cells were used.
RNA isolation, real-time qPCR and gene expression analyses
Total RNA from samples was isolated using the RNeasy Kit (Qiagen) following manufacturer’s protocols with minor modifications, and evaluated by ND-1000 spectrophotometer (Nano-drop Technologies) and Agilent 2100 Bioanalyzer with Agilent RNA 6000 nano chips. cDNA was generated from at least 50 ng RNA in 20 μl total volume using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with the program: 10 minutes at 25 °C; 120 minutes at 37 °C; 5 minutes at 85 °C.
Quantitative real-time polymerase chain reaction (qPCR) was performed with a 7900HT Fast Real-Time PCR System (Applied Biosystems) using the Power SYBR green PCR Master mix (Applied Biosystems). qPCR was carried out with DNA/cDNA with a total volume of 10 μl containing 2 × Power SYBR Green PCR master Mix, 0.5 μM of each primer and 6 ng of mtDNA/30 ng nDNA/3 ng of cDNA. All primers were designed through Primer3 (see Supplementary Table S3 for primer sequences).
qPCR performance: 50 °C for 2 minutes, 95 °C for 10 minutes, 95 °C for 15 seconds, and 60 °C for 1 minute for 40 cycles. The ΔΔCT method was used to quantify the relative gene expression using the GAPDH gene as the internal reference gene. The baseline and threshold were adjusted for each target. RNA sequencing analyses were performed by pooling equimolar amounts of RNA from controls, and three groups of HD patients: with CAG size < 40 (low-CAG), 40–50 (mid-CAG) and >50 (high-CAG). RNA sequencing was performed by Novogene, Beijing. Gene onthology (GO) analyses on resulting Fastq data were performed by Novogene. GO with a corrected P-value of < 0.05 is significantly enriched in DEGs.
DNA isolation, DNA integrity and mtDNA copy number
Total DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen) following manufacturer’s protocols with minor modifications. mtDNA and nDNA damage was determined using the DNA damage assay described in Wang et al.30 by qPCR, using 6 and 30 ng DNA for mtDNA and nDNA analysis, respectively. Primers for MT-RNR1 and NDUFA9 were used to represent mtDNA and nDNA (see Supplementary Table S3 for primer sequences). CAG sizing was performed by PCR using primers (Supplementary Table S3) with a fluorescent tag designed to flank the region of the CAG repeats. A total volume of 20 μl containing 50 ng of total DNA, 5% DMSO, 1 × GeneAmp® PCR Buffer II, 1.25 mM MgCl2, 2.5 mM dNTP, 0.5 µM forward and reverse primers and 0.075 U/µl AmpliTaq Gold® DNA polymerase and the following program was used: 94 °C for 5 minutes then for 40 cycles, 94 °C for 30 seconds, 70 °C for 30 seconds, 72 °C for 30 seconds and finally 72 °C for 3 minutes. PCR products were run on a 1.2% agarose gel, the resulting band was cut out and separated by capillary electrophoresis in a POP-7 gel at 60 °C using LIZ 600 size standard. Length of the PCR products was determined by fragment analysis on an Applied Biosystems 3130 Genetic Analyzer. Data analysis was performed by GeneMapper® software V2.6.4.
mtDNA copy number was determined by real-time qPCR quantification of the nuclear (HBB) and mitochondrial (MT-RNR1) (see Supplementary Table S3) templates.
Western analyses
PBMC pellets (about 20 μl) were resuspended and incubated on ice in 2.5 × their volume of RIPA buffer (50 mm Tris/HCl (pH 7.4), 150 mm NaCl, 1 mm PMSF, 1 mm EDTA, 1% Triton X-100, 1% SDS (v/v) and 1% (v/v) Protease Inhibitor Cocktail (Sigma-Aldrich) for 20 minutes, vortexed every 5 minutes and then centrifuged for 20 min at 51,000 × g (4 °C). The protein concentration in supernatant was determined53 and proteins (20 μg per well) and a molecular weight marker (See Blue Plus2 Prestained Standard, Invitrogen) were separated by 12% SDS-PAGE54 (Mini-Protean System (Bio-Rad)). Protein was mixed with loading buffer (50 mm Tris/HCl (pH 6.8), 12% (v/v) glycerol, 4% SDS, 2% (v/v) 2-mercaptoethanol and 0.01% (w/v) Bromphenol Blue) for 30 minutes at 37 °C. Proteins were subsequently semidry electroblotted onto PVDF membranes (Merck), membranes air-dried overnight, rinsed with 100% methanol (v/v) and blocked in TBS (Tris-Buffered Saline) with 5% non-fat dried milk for 1 hour. Membranes were incubated with primary antibodies (see below) in TBS containing 0.1% (v/v) Tween-20 and 2% non-fat dried milk overnight at 4 °C. Secondary detection was carried out with peroxidase conjugated secondary antibodies (Sigma-Aldrich) in TBS containing 0.1% (v/v) Tween-20 and 2% non-fat dried milk for 1 hour. The proteins were then visualized with the Super Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific) using Syngene Imaging System and the intensity of signal was quantified with the Quantity One 1-D Analysis Software (Bio-Rad) for further analysis.
Primary antibodies: PDH antibody cocktail MSP02/C0611 (1:2000); Complex III subunit Core 2 monoclonal antibody MS304/D1129 (1:10000); Complex II subunit 30 kDa Ip monoclonal antibody MS203/D1205 (1:2000); Complex I subunit NDUFB6 monoclonal antibody MS108 (1:2000), all Mitosciences; Anti-ATPB antibody [3D5] - Mitochondrial Marker ab14730 (1:1000); Ms mAb to MTCO2 (12C4F12) ab10258 (1:10000); Anti-Superoxide Dismutase 1 antibody [SOD1] ab20926 (1:2000); Anti-GAPDH antibody [6C5] ab8245 (1:3333); Anti-MRPS31 antibody [EPR10707] ab167406 (1:3333); Anti-Citrate synthetase antibody [2H8BB6] ab128564 (1:2500) - all Abcam; p53 Antibody 9282(1:3333), Cell Signaling. Secondary antibodies: Anti-Mouse IgG (whole molecule)-Peroxidase antibody produced in goat A8924 (1:2500); Anti-Rabbit IgG (whole molecule)-Peroxidase antibody produced in goat A0545 (1:2500), Sigma-Aldrich.
Respiratory chain complexes and citrate synthase activity
Prior to measurements, dry pellets of isolated PBMCs were resuspended in PBS to concentration of protein approximately 3-4 mg/ml, determined as described in55. The specific activity of NADH-decylubiquinone oxidoreductase (NQR) (complex I), succinate decylubiquinone DCPIP reductase (SQR) (complex II) and cytochrome c oxidase (complex IV) were measured as was described previously56,57 using a Shimadzu 2401 UV-VIS Spectrophotometer. Briefly, to disrupt the mitochondrial membranes for complex I measurement 100 µg of the cell suspension was first lysed by 3 min incubation in distilled water. Rotenone-sensitive complex I activity was measured in assay medium with final volume of 1 ml (50 mM TRIS, pH 8.1, 2.5 mg/ml BSA, 0.3 mM KCN, 0.1 mM NADH, 50 µM decylubiquinone without and with 3 µM rotenone) and followed the decrease in absorbance at 340 nm due to the NADH oxidation (ε = 6.22 mM−1 cm−1)
Complex II activity was measured in 1 ml of assay medium containing 10 mM potassium phosphate pH 7.8, 2 mM EDTA, 1 mg/ml BSA, 200 µg of cell protein 0.3 mM KCN, 10 mM succinate, 3 µM rotenone, 0.2 mM ATP, 80uM DCPIP (2,6-dichlorophenolindophenol), 1 µM Antimycin, 50 µM decylubiquinone). The decrease in absorbance at 600 nm due to the oxidation of DCPIP at 600 nm (ε = 20.1 mM−1 cm−1) was recorded.
For determination of complex IV and CS enzyme activity, the cell suspension was solubilized by incubation with 1.5% n-dodecyl-β-D-maltoside for 15 minutes at 4 °C, then centrifuged for 10 minutes at 10,000 g and resulting supernatant used for activity assay.
Complex IV activity was measured by incubating of 100 µg of cell supernatant protein in 1 ml of assay medium (40 mM potassium phosphate, pH 7.0; 1 mg/ml BSA; 25 µM reduced cytochrome c) by following the oxidation of cytochrome c (II) at 550 nm (ε = 19.6 mM−1 cm−1). All assays were performed at 37 °C. Citrate synthase was measured with 100 µg of supernatant according to Srere58.
Statistics
All statistics were carried out in GraphPad Prism. Calculation of statistical significance was done using un-paired T-test without assuming consistent standard deviation. Multiple comparisons were corrected for using the Holm Sidak method with alpha = 0.05. Correlation analysis was calculated by Pearson method at 95% confidence interval. For enzymatic activity, all dependent variables were log-transformed (loge). Only in the case of COX we used y = −1/x.
For western blot analysis, relationships between dependent variables were examined using linear mixed effects models with interactions. Random effect of a set of measurements (of an experiment) was also considered. Dependent variables were transformed using Box-Cox transformations. For western blot analysis, box-cox power transformation was used with the following lambdas: NDUFB6:1 (identity), SDHB: 0.25, CORE2: 0.25, COX2: 2, ATPbeta: 0.25, PDH E2:0.5, E1a: 0 (loge), SOD1: −0.9, p53:1 (identity), MRPS31: 0.25, CS: −0.53, GAPDH: 0 (loge).
Data availability
Data that is approved for sharing to destined country is available from the corresponding author upon request.
Electronic supplementary material
Acknowledgements
The research leading to these results has received funding from the Norwegian Financial Mechanism 2009–2014 and the Ministry of Education, Youth and Sports under Project Contract no. MSMT-28477/2014, COST LD15099 and PROGRES Q26/LF1. JK receives support from PROGRES Q27/LF1. The PROMEC proteomics and metabolomics core facility is supported by the Liaison Committee between the Central Norway Regional Health Authority (RHA) and the Norwegian University of Science and Technology (NTNU) (grant no: 46040500).
Author Contributions
G.A., Z.D., M.R., A.K., G.N. performed the experiments. G.A., Z.D., M.R., I.L., M.B., A.K. H.H., L.E. designed the experiments and interpreted results. J.K. recruited patients, performed clinical evaluation and revised the manuscript. G.A. and L.E. wrote the manuscript and J.K., M.B., A.K. and H.H. revised it.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27985-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adegbuyiro A, Sedighi F, Pilkington AWT, Groover S, Legleiter J. Proteins Containing Expanded Polyglutamine Tracts and Neurodegenerative Disease. Biochemistry. 2017;56:1199–1217. doi: 10.1021/acs.biochem.6b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duyao MP, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 3.Nasir J, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 4.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 5.Unified Huntington’s Disease Rating Scale: reliability and consistency. Huntington Study Group. Mov Disord11, 136–142, 10.1002/mds.870110204 (1996). [DOI] [PubMed]
- 6.Losekoot M, et al. EMQN/CMGS best practice guidelines for the molecular genetic testing of Huntington disease. Eur J Hum Genet. 2013;21:480–486. doi: 10.1038/ejhg.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aylward, E. H. et al. Rate of caudate atrophy in presymptomatic and symptomatic stages of Huntington’s disease. Mov Disord15, 552–560, https://doi.org/10.1002/1531-8257(200005)15:3<552::AID-MDS1020>3.0.CO;2-P (2000). [DOI] [PubMed]
- 8.Paulsen JS, et al. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klempir J, Klempirova O, Spackova N, Zidovska J, Roth J. Unified Huntington’s disease rating scale: clinical practice and a critical approach. Funct Neurol. 2006;21:217–221. [PubMed] [Google Scholar]
- 10.Farrer LA. Diabetes mellitus in Huntington disease. Clin Genet. 1985;27:62–67. doi: 10.1111/j.1399-0004.1985.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 11.Busse ME, Hughes G, Wiles CM, Rosser AE. Use of hand-held dynamometry in the evaluation of lower limb muscle strength in people with Huntington’s disease. J Neurol. 2008;255:1534–1540. doi: 10.1007/s00415-008-0964-x. [DOI] [PubMed] [Google Scholar]
- 12.Borovecki F, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mihm MJ, et al. Cardiac dysfunction in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2007;25:297–308. doi: 10.1016/j.nbd.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawa A, et al. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999;5:1194–1198. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- 15.Squitieri F, et al. Severe ultrastructural mitochondrial changes in lymphoblasts homozygous for Huntington disease mutation. Mech Ageing Dev. 2006;127:217–220. doi: 10.1016/j.mad.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Almeida S, Sarmento-Ribeiro AB, Januario C, Rego AC, Oliveira CR. Evidence of apoptosis and mitochondrial abnormalities in peripheral blood cells of Huntington’s disease patients. Biochem Biophys Res Commun. 2008;374:599–603. doi: 10.1016/j.bbrc.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Luo X, Hu W. Studies on the epidemiology and etiology of moldy sugarcane poisoning in China. Biomed Environ Sci. 1992;5:161–177. [PubMed] [Google Scholar]
- 18.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 19.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Damiano M, et al. A role of mitochondrial complex II defects in genetic models of Huntington’s disease expressing N-terminal fragments of mutant huntingtin. Hum Mol Genet. 2013;22:3869–3882. doi: 10.1093/hmg/ddt242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 22.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mollersen L, et al. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum Mol Genet. 2012;21:4939–4947. doi: 10.1093/hmg/dds337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler VC, et al. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 26.Budworth H, et al. Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease. PLoS Genet. 2015;11:e1005267. doi: 10.1371/journal.pgen.1005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, et al. Mitochondrial DNA integrity is essential for mitochondrial maturation during differentiation of neural stem cells. Stem Cells. 2010;28:2195–2204. doi: 10.1002/stem.542. [DOI] [PubMed] [Google Scholar]
- 28.Pawar, T., Bjoras, M., Klungland, A. & Eide, L. Metabolism and DNA repair shape a specific modification pattern in mitochondrial DNA. Mitochondrion, 10.1016/j.mito.2017.09.002 (2017). [DOI] [PubMed]
- 29.Bae BI, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Scheffler K, Esbensen Y, Eide L. Quantification of DNA Damage by Real-Time qPCR. Methods Mol Biol. 2016;1351:27–32. doi: 10.1007/978-1-4939-3040-1_3. [DOI] [PubMed] [Google Scholar]
- 31.Disatnik MH, et al. Potential biomarkers to follow the progression and treatment response of Huntington’s disease. J Exp Med. 2016;213:2655–2669. doi: 10.1084/jem.20160776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browne SE, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 33.Chen CM, et al. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem Biophys Res Commun. 2007;359:335–340. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- 34.Borowsky B, et al. 8OHdG is not a biomarker for Huntington disease state or progression. Neurology. 2013;80:1934–1941. doi: 10.1212/WNL.0b013e318293e1a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levada-Pires AC, et al. The effect of an adventure race on lymphocyte and neutrophil death. Eur J Appl Physiol. 2010;109:447–453. doi: 10.1007/s00421-010-1363-4. [DOI] [PubMed] [Google Scholar]
- 36.Jedrak P, et al. Mitochondrial DNA levels in Huntington disease leukocytes and dermal fibroblasts. Metab Brain Dis. 2017;32:1237–1247. doi: 10.1007/s11011-017-0026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Mejia EM, Chau S, Sparagna GC, Sipione S, Hatch GM. Reduced Mitochondrial Function in Human Huntington Disease Lymphoblasts is Not Due to Alterations in Cardiolipin Metabolism or Mitochondrial Supercomplex Assembly. Lipids. 2016;51:561–569. doi: 10.1007/s11745-015-4110-0. [DOI] [PubMed] [Google Scholar]
- 39.Pawar T, Eide L. Pitfalls in mitochondrial epigenetics. Mitochondrial DNA A DNA Mapp Seq Anal. 2017;28:762–768. doi: 10.1080/24701394.2016.1180514. [DOI] [PubMed] [Google Scholar]
- 40.Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA. PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD) Cell Cycle. 2015;14:1716–1729. doi: 10.1080/15384101.2015.1033595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffler K, et al. 8-oxoguanine DNA glycosylase (Ogg1) controls hepatic gluconeogenesis. DNA Repair (Amst) 2018;61:56–62. doi: 10.1016/j.dnarep.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Vartanian V, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci USA. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prokesch A, et al. Liver p53 is stabilized upon starvation and required for amino acid catabolism and gluconeogenesis. FASEB J. 2017;31:732–742. doi: 10.1096/fj.201600845R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strand JM, Scheffler K, Bjoras M, Eide L. The distribution of DNA damage is defined by region-specific susceptibility to DNA damage formation rather than repair differences. DNA Repair (Amst) 2014;18:44–51. doi: 10.1016/j.dnarep.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Gu M, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 47.Arenas J, et al. Complex I defect in muscle from patients with Huntington’s disease. Ann Neurol. 1998;43:397–400. doi: 10.1002/ana.410430321. [DOI] [PubMed] [Google Scholar]
- 48.Tabrizi SJ, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann Neurol. 2000;47:80–86. doi: 10.1002/1531-8249(200001)47:1<80::AID-ANA13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 49.Kaarbo M, et al. Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion. 2011;11:935–945. doi: 10.1016/j.mito.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Sharma S, Wang J, Cortes Gomez E, Taggart RT, Baysal BE. Mitochondrial complex II regulates a distinct oxygen sensing mechanism in monocytes. Hum Mol Genet. 2017;26:1328–1339. doi: 10.1093/hmg/ddx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hensman Moss DJ, et al. Quantification of huntingtin protein species in Huntington’s disease patient leukocytes using optimised electrochemiluminescence immunoassays. PLoS One. 2017;12:e0189891. doi: 10.1371/journal.pone.0189891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang KH, Wu YR, Chen YC, Chen CM. Plasma inflammatory biomarkers for Huntington’s disease patients and mouse model. Brain Behav Immun. 2015;44:121–127. doi: 10.1016/j.bbi.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 54.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 55.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 56.Honzik T, et al. Activities of respiratory chain complexes and pyruvate dehydrogenase in isolated muscle mitochondria in premature neonates. Early Hum Dev. 2008;84:269–276. doi: 10.1016/j.earlhumdev.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Fornuskova D, et al. Novel insights into the assembly and function of human nuclear-encoded cytochrome c oxidase subunits 4, 5a, 6a, 7a and 7b. Biochem J. 2010;428:363–374. doi: 10.1042/BJ20091714. [DOI] [PubMed] [Google Scholar]
- 58.Srere PA, John ML. Citrate synthase: [EC 4.1.3.7. Citrate oxaloacetate-lyase (CoA-acetylating)] Methods in Enzymology. 1969;ume 13:8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that is approved for sharing to destined country is available from the corresponding author upon request.