Abstract
Corpora amylacea (CA) are spherical bodies mainly composed of polyglucans and, to a lesser extent, proteins. They are abundant in brains from patients with neurodegenerative diseases, particularly Alzheimer’s disease. Although CA were discovered many years ago, their precise origin and function remain obscure. CA from the insular cortex of two Alzheimer’s patients were purified and the protein composition was assessed by proteomic analysis. A number of microbial proteins were identified and fungal DNA was detected by nested PCR.A wide variety of human proteins form part of CA. In addition, we unequivocally demonstrated several fungal and bacterial proteins in purified CA. In addition to a variety of human proteins, CA also contain fungal and bacterial polypeptides.In conclusion, this paper suggests that the function of CA is to scavenge cellular debris provoked by microbial infections.
Introduction
Several different types of polyglucan bodies have been found in the central nervous system (CNS) of elderly people, as well as in patients with a variety of pathologies, including Alzheimer’s disease (AD), epilepsy, amyotrophic lateral sclerosis (ALS), multiple sclerosis (MS) and hippocampal sclerosis1,2. Among these polyglucan structures, Lafora bodies, Bielschowsky bodies and corpora amylacea (CA) have been identified based on their morphological characteristics3–5. Although CA were described early in the 18th century, their precise origin and potential function in normal or pathological conditions remains an enigma. The presence of CA in brain tissue was first described by Purkinje in 1837, and the Cajal school analyzed these structures, suggesting that they are formed by the microglia.
CA are amorphous rounded bodies approximately 10−50 μm in diameter. These glycoproteinaceous laminar structures accumulate in the brain during the course of normal aging and to a greater extent in the CNS of AD patients6–8. Of note, abundant CA are also found in some patients diagnosed with temporal epilepsy, where extensive deposits of CA have been observed in brain tissue9,10. Interestingly, CA have a perivascular distribution and are much more abundant in close proximity to blood vessels10–12. In addition to the CNS, CA are found in other organs and tissues, such as normal prostate, muscle, liver, lung, prostate tumours and other malignant tissues13–18. CA mostly contain polyglucans (over 85% are hexoses), with a minor component (4%) of proteins19. The rounded core is formed by different calcium salts, principally calcium phosphate and calcium oxalate depending on the bodies analyzed20–22. In general, a broad range of proteins can be found in CA and a number of them have been characterized using specific antibodies23. Accordingly, several blood proteins such as thrombospondin 1, some complement components, ubiquitin, heat-shock proteins and tau protein processed by caspase-3 have been detected in CA24–28. The protein composition of CA from prostate has been characterized in detail by proteomic analysis, showing that lactoferrin is the most abundant protein, together with myeloperoxidase, S100 calcium-binding proteins A8 and A9, which form the inflammatory protein calprotectin, and α-defensins, which form part of neutrophil granules29. Curiously, calprotectin is also present in CA from normal human brains30. The suggestion that CA are built up from breakdown products from neurons and oligodendroglial cells has been proposed23. Along this line, proteomic analysis of brain CA from patients with MS revealed the presence of cytoskeleton proteins and enzymes of the anaerobic glycolysis pathway . A number of microorganisms have also been suggested as the source of the chronic inflammation that triggers the formation of CA in prostate. Among these, several bacteria such as Chlamydia trachomatis, Escherichia coli and Pseudomonas spp., protozoa such as Trichomonas vaginalis and viruses known to contribute to different types of cancer, including human papillomavirus, have been considered29,31. Thus, the concept that CA represent the prostate response to a microbial infection has been proposed.
We have recently reported that CA from brain tissue of AD, ALS and patients with Parkinson’s disease contain fungal components, as revealed by immunoreactivity against antifungal antibodies32. However, the exact proteins present in CA were not identified. We have proposed that CA represent a response to the microbial infection present in these patients. Thus, this proposal reconciles the idea that CA are built up by cellular breakdown products, which originate from the microbial infection that exists in the CNS from patients with these neurodegenerative diseases. Indeed, there is strong evidence for the presence of fungal proteins and DNA in CNS from AD patients33–35, and also in patients diagnosed with ALS36,37. In the current study, we have characterized in detail the fungal and bacterial proteins that can be found in purified CA from two AD patients. In addition, the human proteins present in CA reveal a great variety of cellular polypeptides, consistent with the concept that CA represent breakdown products of neural cells.
Results
Purification of CA from brain tissue
Our initial goal was to develop a protocol to purify CA from brain tissue. We analyzed tissue from two patients (AD1 and AD2) diagnosed with AD; patient AD2 was also diagnosed with dementia with Lewy bodies. Prior results from our group have demonstrated the existence of fungal proteins in CA from brain sections of AD patients32. Accordingly, the presence of CA was first examined by periodic acid-Schiff (PAS) staining and by immunohistochemistry using antibodies against human α-tubulin, C. albicans and also bacterial peptidoglycan. Abundant CA were observed in insular cortex (INCO) tissue sections in patient AD1 by PAS staining and by immunohistochemistry (Fig. 1). For this reason, brain tissue from this INCO region was selected. Interestingly, CA immunoreacted with both antifungal and antibacterial antibodies, suggesting the presence of microbial components in CA. Similar results were found in patient AD2, although CA were less abundant in the sections examined from this patient (Supplementary Fig. 1A).
Figure 1.
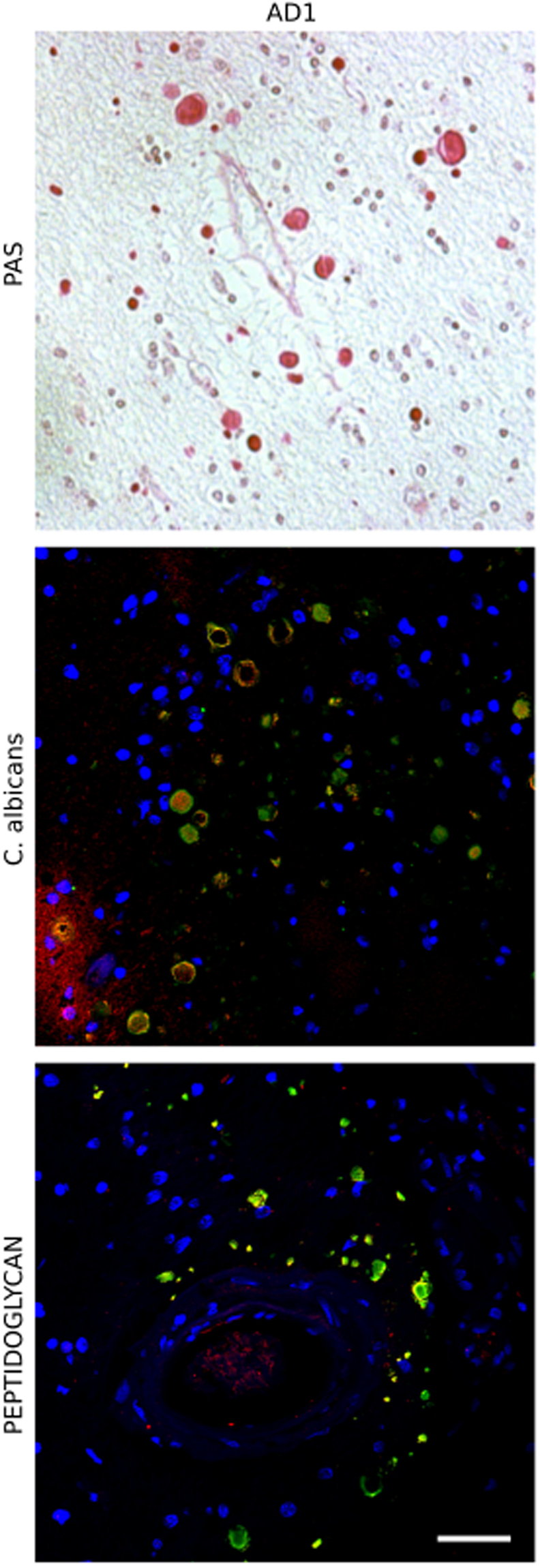
Histochemistry analysis of brain sections from patient AD1. Evidence of CA in the brain cortex containing microbial proteins. Insula cortex (INCO) sections from patient AD1 stained with Periodic acid–Schiff reagent (upper panel); immunostained with a rabbit polyclonal anti-C. albicans antibody (green) used at 1:100 dilution, and a mouse monoclonal anti-human α-tubulin antibody (red) used at 1:50 dilution (middle panel); immunostained with a mouse monoclonal anti-peptidoglycan antibody (green) used at 1:20 dilution, and a rabbit polyclonal anti-C. albicans antibody (red) used at 1:100 dilution (lower panel). DAPI staining of nuclei appears in blue. Scale bar: 50 μm.
To obtain purified CA, we used the protocol described in Materials and Methods. Homogenized tissue was pelleted through a series of centrifugation steps on sucrose cushions to obtain a final pellet named P7. The proteins of the first homogenate and the P7 fraction were analyzed by SDS-PAGE under reducing conditions and visualized by Coomassie blue staining. Analysis showed evident differences in protein content between the first homogenate and the P7 pellet (Supplementary Fig. 1B). In addition, the P7 pellet was immunostained with an anti-C. albicans antibody to visualize the purified CA in this fraction. Notably, CA was evidenced in P7 from both patients after immunostaining with the fungal antibody (Supplementary Fig. 1C). These findings clearly indicate that P7 contains CA bodies, but we cannot rule out the possibility that other human proteins present in organelles also form part of this fraction. It is unlikely that soluble cellular proteins obtained during the homogenization procedure remain in P7, since the components of this fraction were sedimented through 25–45% sucrose and this step was repeated several times. However, it should be possible that soluble cellular proteins interact with CA during their formation. To test this possibility directly, we subjected fractions H and P7 to western blotting using an antibody against the translation initiation factor eIF4GI. This factor was clearly detected in fraction H, but not in P7 (Supplementary Fig. 2A, see also Supplementary info file). Furthermore, whereas the translation elongation factor eEF2 was detected only sparingly in fraction H, CA clearly accumulated this protein. Also, human α-tubulin was found both in fractions H and P7. Interestingly, 18S rRNA was detected by RT-PCR in the P7 fraction from both patients. By contrast, mtDNA was only found in the H and P2 fractions, but not in P7, reflecting that mitochondria do not co-purify with CA and that these organelles are not recruited by CA (Supplementary Fig. 2B, see also Supplementary info file).
As regards to CA from control subjects, their amount was very low (Supplementary Fig. 3). Only some CA can be revealed in the entorhinal cortex/hippocampus (ERH) using antibodies against human α-tubulin. Therefore, purification of these bodies by our present protocol was precluded. The best characterization of CA from control subjects can be accomplished by immunohistochemistry. However, immunolabeling with anti-C. albicans antibodies was not found (Supplementary Fig. 3). Besides, no signal was observed with anti-peptidoglycan antibodies. These findings are in good agreement with previous results38.
Human proteins detected in purified CA
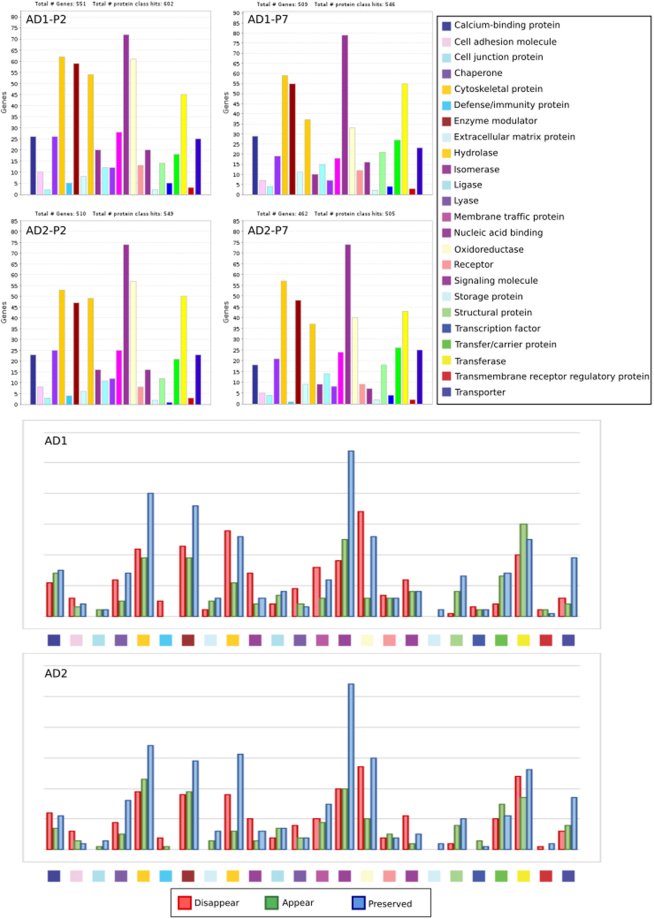
After determining that P7 contains CA, we next sought to characterize this fraction using proteomic analysis. Initially, we sought to identify the human proteins that constitute CA. To do this, CA samples were digested with trypsin and analyzed by MS. Supplementary Table I and Table II list the human proteins that are found in P2 and P7 fractions from both patient AD1 and AD2, with at least two peptides. The human proteins detected were classified according to their function (Fig. 2). Most of these peptides belong to nucleic acid binding proteins, cytoskeleton and enzymatic proteins. Interestingly, a number of these proteins were detected in P2, but not in P7 and vice versa, while others were common to both fractions (Supplementary Tables III and IV). The major finding of this proteomic analysis was the great variety of human proteins detected in P7, presumably forming part of CA. Evidently, some of these proteins could be minor components of CA and/or could be present in a small fraction of these bodies due to their heterogeneity.
Figure 2.
Proteomic analysis of P2 and P7 fractions from patients AD1 and AD2. Identification of the human proteins present in these fractions. Analysis of the human proteins in CA (fraction P7) by proteomics. Classification of the proteins found in P2 and P7 according to protein class function was carried out using Panther online software. Upper panels: P2 and P7 from AD1, and P2 and P7 from AD2. Lower panels: comparison of the proteins that appear in P2 and P7 from AD1 and AD2. Red: proteins absent in P7 and present in P2. Green: proteins present in P7 and absent in P2. Blue: proteins common to both P2 and P7.
Fungal proteins in CA
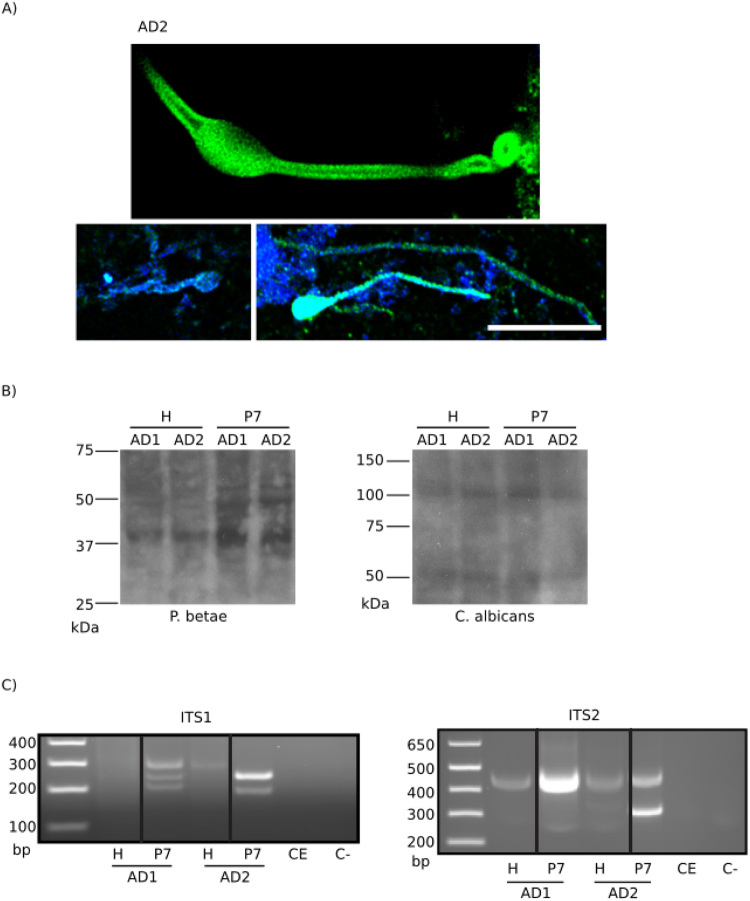
As stated above, fungal infection can be evidenced in AD brains by immunohistochemistry34,39, which is consistent with the detection of fungal proteins in CA from AD1 and AD2 (Fig. 1 and Supplementary Figure 1). We therefore investigated the presence of mycotic structures in the different fractions of CA. We found that the P2 fraction from both patients contained a significant proportion of yeast-like and hyphal structures that could be detected by immunostaining with a specific anti-C. albicans antibody (Fig. 3A). We also analyzed the fractions by western blotting using different antifungal antibodies. Figure 3B (see also Supplementary info file) shows a number of different protein bands that immunoreacted with the anti-Phoma betae and anti- C. albicans antibodies. To identify the fungal species present in the two patients, we performed nested PCR of the fungal ITS-1 and ITS-2 regions (Fig. 3C, see also Supplementary info file). Fungal DNA fragments were successfully amplified in fractions H and P7, which after sequencing, revealed a variety of fungal species present in both patients (Supplementary Table V). The fungal genera identified included Cladosporium, Malassezia or Rhodotorula. These fungal genera correspond with those previously described by our group34,40.
Figure 3.
Fungal proteins and DNA in the homogenate and P7 fractions from patients AD1 and AD2. Identification of fungal components in CA. (A) Pellet (P2) of AD2 immunostained with a rabbit polyclonal anti-C. albicans antibody (green) used at 1:100 dilution. DAPI staining of nuclei appears in blue. Scale bar: 10 μm. (B) Western blot of proteins from homogenate (H) and P7 fractions from AD1 and AD2. Left panel: Rabbit polyclonal anti-P. betae antibody used at 1:500 dilution; right panel: rabbit polyclonal anti-C. albicans antibody used at 1:100 dilution. (C) Nested PCR analysis of DNA extracted from patients AD1 and AD2. Agarose gel electrophoresis of the DNA fragments amplified by nested PCR. Left panel: PCR analysis of the homogenate (H) and P7 fractions from patients AD1 and AD2 using primers ITS-5 and ITS-3 to amplify the ITS-1 region. Right panel: PCR analysis of the H and P7 fractions from patients AD1 and AD2 using primers ITS-3 and ITS-4 to amplify the ITS-2 region. (C) Control of PCR without DNA. CE: Control of DNA extraction without DNA. H: Homogenate. P7: fraction 7. See also Supplementary info file.
The presence of fungal proteins in CA as revealed by immunohistochemistry using different antibodies does not identify the precise proteins in these bodies32 (Fig. 1). Thus, to demonstrate that fungal proteins form part of CA and to identify them with precision, the peptides obtained after tryptic digestion of P2 and P7 were analyzed against the fungi database using the Proteome Discoverer 1.4 tool. Obviously, the vast majority of peptides obtained after tryptic digestion were of human origin and we only considered bona fide peptides belonging to fungi. Through this analysis, several fungal peptides from P2 and P7 could be unambiguously ascribed in the most part to cytoskeleton proteins (Table 1). It seems logical that the most abundant proteins in fungal cells, i.e., cytoskeletal proteins, appear in CA.
Table 1.
Fungal peptides in P2 and P7 fractions from AD1 and AD2.
| PEPTIDE | PROTEIN | SPECIES | AD1-P2 | AD1-P7 | AD2-P2 | AD2-P7 |
|---|---|---|---|---|---|---|
| GHYTEGAELIDSVLDVVR | Beta tubulin | Several species | — | YES | YES | YES |
| GHYTEGAELVEAVLDVVR | Beta tubulin | Several species | YES | — | — | — |
| MAATFIGNSTAQQELFK | Beta tubulin | Several species | — | YES | — | YES |
| MSATFIGNSTSIQELFK | Beta tubulin | Several species | — | — | YES | — |
| MSVTFIGNSTAIQELFK | Beta tubulin | Several species | — | — | YES | YES |
| MSVTFLGNSTAIQELFK | Beta tubulin | Several species | — | YES | — | — |
| MSVTLIGNSTAIQELFK | Beta tubulin | Rhizophlyctis rosea | YES | — | — | — |
| NSSYYVEWIPNNVK | Beta tubulin | Blastocladiella emersonii | — | YES | — | — |
| SLGGGTGAGMGTLLISK | Beta tubulin | Several species | YES | — | — | — |
| AICMLSNTTAIAEAWAR | Alpha tubulin | Lichtheimia sp | — | — | YES | — |
| ALCMLSNTTAIAEAWAR | Alpha tubulin | Several species | YES | YES | — | YES |
| AVCMLSNTTAIAEAWSR | Alpha tubulin | Several species | — | — | — | YES |
| AVCMLSNTTAISEAWSR | Alpha tubulin | Geotrichum candidum | — | YES | — | YES |
| IHFPLATYAPIISAEK | Alpha tubulin | Several species | — | YES | YES | — |
| IHFPLATYAPLISADK | Alpha tubulin | Several species | — | YES | — | — |
| IHFPLATYAPLLSAEK | Alpha tubulin | Several species | YES | — | — | YES |
| LIAQVVSSITASLR | Alpha tubulin | Several species | YES | — | YES | — |
| DSYVGDEAESK | Actin | Fusarium oxysporum | — | — | YES | — |
| GEEEVAALVIDNGSGMCK | Actin | Cryptococcus depauperatus | YES | YES | YES | YES |
| SYELPDGQNITIGNER | Actin | Nematocida sp | YES | YES | YES | — |
| TYELPDGQVITIGNER | Actin | Several species | — | — | — | YES |
| LILEVAQHLGESTVR | ATP synthase subunit beta | Ophiostoma piceae | YES | — | YES | — |
| VALTGLTIAEYFR | ATP synthase subunit beta | Several species | YES | — | YES | — |
| VSLVFGQMNEPPGAR | ATP synthase subunit beta | Several species | YES | — | — | — |
| IEIIANDQGNR | Heat shock protein | Several species | YES | — | YES | — |
| SLTNDWEEHLAVK | Heat shock protein | Several species | YES | — | — | — |
| FFTPEEISSMVLTK | Unplaced genomic scaffold | Several species | — | — | YES | — |
Bacterial proteins detected in purified CA
To complement our studies on human and fungal proteins in CA, we extended our analysis to the possibility that viral or bacterial proteins are also present in these samples. Indeed, we and others have recently reported that a variety of bacterial species are detected in brain tissue from AD patients39,41. Moreover, prokaryotic-like cells immunopositive for peptidoglycan could be detected in INCO tissue sections (Fig. 1). To test the existence of bacterial proteins, the peptides obtained in P7 from both AD1 and AD2 patients were tested against bacterial databases belonging to different phyla. Only those peptides that unequivocally belong to bacteria were considered. Interestingly, the high sensitivity of the proteomic analyses identified a few peptides that could be ascribed to bacteria. The bacterial peptides, the corresponding proteins and the phyla are listed in Table 2. Only three prokaryotic peptides were found in AD1 and one in AD2. Of note, the number of bacterial peptides detected was lower than those of fungal origin, possibly reflecting the lower burden of bacterial infection as compared with mycotic infection. Finally, the peptides obtained after tryptic digestion of P2 and P7 were also analyzed against the database of DNA animal viruses to identify potential proteins from these viruses. No peptides corresponding to herpesviruses or any other DNA virus were found in P2 or P7 from both AD patients, which is consistent with our recent report that HSV-1 proteins are not detected in brain tissue from AD patients39.
Table 2.
Bacterial peptides in P2 and P7 fractions from AD1 and AD2.
| PEPTIDE | PROTEIN | AD1-P7 | AD2-P7 | PHYLUM |
|---|---|---|---|---|
| LINEPTAAALAYGLSR | Chaperone protein DnaK | YES | — | proteobacteria |
| LLNEPTAAALAYGVEK | Chaperone protein HscA | — | YES | proteobacteria |
| FTQAGSEISALLGR | ATP synthase subunit beta | YES | — | proteobacteria tenericutes |
| NETDDQVTIDAAEATKK | Isocitrate dehydrogenase [NADP] | YES | — | firmicutes |
In-depth analysis of fungal proteins
Our finding that fungal proteins are present in a lower proportion than human proteins prompted us to analyze in more detail these proteins in the P7 fraction from both patients. To do this, proteins were fractionated by a high-pH, reversed-phase fractionation spin column. Nine fractions were obtained and were subsequently pooled into three fractions F1, F2 and F3, as described in Materials and Methods. After MS analyses of F1, F2 and F3, the number of fungal peptides detected increased considerably (Table 3). Thus, the number of peptides unambiguously identified as fungal were 49 in P7 from AD1 and 70 in P7 from AD2, from those 23 were common to both patients. These peptides corresponded to a number of fungal proteins mostly belonging to tubulins and actins, in agreement with the fact that these proteins are very abundant. These findings reinforce our previous observations that fungal infection exists in AD patients.
Table 3.
Fungal proteins after column fractionation in P7 fractions.
| PEPTIDE | PROTEIN | SPECIES | AD1-P7 | Fract. AD1-P7 | AD2-P7 | Fract. AD2-P7 |
|---|---|---|---|---|---|---|
| MSGTFIGDSTAIQELFK | Beta tubulin | Absidia spinosa | — | — | YES | F1 |
| MSGTFIGNSTAIQELFK | Beta tubulin | Absidia spinosa | — | — | YES | F1 |
| AILVDLEVATMDAVR | Beta tubulin | Agaricus bisporus var. Burnettii | — | — | YES | F3 |
| AVLVDLEPGTMDTTR | Beta tubulin | Basidiobolus microsporus | YES | F1 | YES | F3-F3 |
| MAVTFVGNSTAIQELFK | Beta tubulin | Basidiobolus microsporus | YES | F1 | — | — |
| NSSYYVEWIPNNVK | Beta tubulin | Blastocladiella emersonii | YES | F3 | — | — |
| LGICDEPPTVVPGGDLAK | Alpha tubulin | Calocera sp. | YES | F1 | — | — |
| EVEDQMLSVQTK | Beta tubulin | Claviceps purpurea | YES | F3 | — | — |
| IAEQFTAMFR | Beta tubulin | Conidiobolus coronatus | YES | F1 | — | — |
| MCSTFIGNSTAIQELFK | Beta tubulin | Conidiobolus coronatus | YES | F1 | YES | F3 |
| CVSMLSNTTAIAEAWAR | Alpha tubulin | Dactylellina haptotyla | YES | F1 | YES | F1-F2 |
| LEAEAAAAAAAAAK | Uncharacterized protein | Daedalea quercina | — | — | YES | F1 |
| EVEEQMLAVQTK | Beta tubulin | Dentiscutata heterogama | YES | F1 | YES | F3 |
| MSATFIGNTTSIQEPFK | Beta tubulin | Geranomyces variabilis | — | — | YES | F2 |
| YLVVNADEGEPQTCK | NADH dehydrogenase | Gonapodya prolifera | YES | F2 | YES | F3 |
| AICMLSNTTAIAEAWAR | Alpha tubulin | Lichtheimia sp | YES | F3 | YES | F3 |
| SLFHPEQLITGK | Alpha tubulin | Lichtheimia sp. | YES | F3 | — | — |
| AVLVDLEPGTMDNVR | Beta tubulin | Microsporidia sp. | YES | F3 | — | — |
| AVSIPELTQQMFDAK | Beta tubulin | Mycena chlorophos | — | — | YES | F3 |
| ISVYYNEAGASK | Beta tubulin | Mycena chlorophos | YES | F2 | — | — |
| EVDVQMLNVQNK | Beta tubulin | Nowakowskiella elegans | YES | F1 | — | — |
| ALTVPELAQQMFDAK | Beta tubulin | Nowakowskiella hemisphaerospora | YES | F1 | YES | F1 |
| LILEVAQHLGESTVR | ATP synthase subunit beta | Ophiostoma piceae | — | — | YES | F2 |
| KSIQFVDWCPTGFK | Alpha tubulin | Parasitella parasitica | — | — | YES | F1 |
| IESLEEEIRK | Uncharacterized protein | Parasitella parasitica | — | — | YES | F1 |
| EIAESFLGK | Heat shock protein 70 | Penicillium sp. | — | — | YES | F3 |
| EVDEQMLNVHNK | Beta tubulin | Phlyctochytrium californicum | — | — | YES | F2 |
| AVCMLGNTTAIAEAWAR | Alpha tubulin | Phycomyces blakesleeanus | YES | F1 | — | — |
| LVVIGDSGVGK | Uncharacterized protein | Piloderma croceum | YES | F2 | — | — |
| MDQGFSTFFSETGAGK | Alpha tubulin | Pisolithus tinctorius | — | — | YES | F3 |
| GDDGFSTFFSETGAGK | Alpha tubulin | Puccinia | YES | F1 | YES | F1-F2 |
| AILIDLEPGTMDSVR | Beta tubulin | Puccinia sorghi | — | — | YES | F3 |
| ALCMLSNTTAIAEAWSR | Alpha tubulin | Rhizopus delemar | — | — | YES | F2 |
| EVDEQMLQVQNK | Beta tubulin | Rhopalomyces elegans | — | — | YES | F2 |
| VGEAMEEGEFSEAR | Alpha tubulin | Schizophyllum commune | YES | F1 | — | — |
| HQGVMVGMSNK | Actin | Schizopora paradoxa | — | — | YES | F2 |
| SMNLTRGINLLSIAEK | 19 S proteasome regulatory | Schizosaccharomyces octosporus | — | — | YES | F2 |
| AILVDLEPGTMDAVR | Beta tubulin | Several species | — | — | YES | F1 |
| AVLIDLEPGTMDSVR | Beta tubulin | Several species | — | — | YES | F3 |
| AVLVDLEPGTMDAIK | Beta tubulin | Several species | — | — | YES | F1 |
| AVLVDLEPGTMDAVR | Beta tubulin | Several species | YES | F3 | YES | F1 |
| GGGTGAGMGTLLISK | Beta tubulin | Several species | YES | F3 | — | — |
| GHYTEGAELIDSVLDVVR | Beta tubulin | Several species | — | — | YES | F1 |
| GTGAGMGTLLISK | Beta tubulin | Several species | YES | F1 | YES | F3 |
| INVYYNEASGGKYVPR | Beta tubulin | Several species | YES | F1 | — | — |
| ISVYYNEASGGKYVPR | Beta tubulin | Several species | — | — | YES | F1 |
| LAVNTVPFPR | Beta tubulin | Several species | YES | F1 | — | — |
| LGVNMVPFPR | Beta tubulin | Several species | — | — | YES | F1 |
| MAATFIGNSTAQQELFK | Beta tubulin | Several species | YES | F2 | YES | F2 |
| MSVTFIGNSTAIQELFK | Beta tubulin | Several species | YES | F1-F1 | YES | F2 |
| MSVTFLGNSTAIQELFK | Beta tubulin | Several species | YES | F1 | — | — |
| NSAYFVEWIPNNVK | Beta tubulin | Several species | — | — | YES | F1 |
| SGPFGKLFRPDNFVFGQ | Beta tubulin | Several species | — | — | YES | F2 |
| TAICDIPPR | Beta tubulin | Several species | — | — | YES | F3 |
| TGAGMGTLLISK | Beta tubulin | Several species | YES | F3 | — | — |
| VLVDLEPGTMDAVR | Beta tubulin | Several species | — | — | YES | F1 |
| VNDQFTAMFR | Beta tubulin | Several species | — | — | YES | F2 |
| ALCMLSNTTAIAEAWAR | Alpha tubulin | Several species | — | — | YES | F2 |
| AVCALSNTTAIAEAWSR | Alpha tubulin | Several species | — | — | YES | F2 |
| AVCMLSNTTAIAEAWKR | Alpha tubulin | Several species | — | — | YES | F1 |
| AVCMLSNTTAIAEAWNR | Alpha tubulin | Several species | — | — | YES | F1 |
| AVCMLSNTTAIAEAWSR | Alpha tubulin | Several species | — | — | YES | F2 |
| AVCMLSNTTAISEAWAR | Alpha tubulin | Several species | YES | F1 | YES | F2 |
| CVSMLSNTTAIAEAWSR | Alpha tubulin | Several species | — | — | YES | F2 |
| DVHASVATLK | Alpha tubulin | Several species | YES | F1 | — | — |
| IIAQVVSSITASLR | Alpha tubulin | Several species | — | — | YES | F2 |
| LIAQIVSSITASLR | Alpha tubulin | Several species | — | — | YES | F2 |
| RTVQFVDWCPTGFK | Alpha tubulin | Several species | YES | F3 | YES | F2 |
| SLCMLSNTTAIATAWSR | Alpha tubulin | Several species | YES | F1 | YES | F1 |
| SVTMLSNTTAIAEAWSR | Alpha tubulin | Several species | — | — | YES | F2 |
| TIQFVEWCPTGFK | Alpha tubulin | Several species | YES | F1 | YES | F1 |
| TVQFVDWCPTGFK | Alpha tubulin | Several species | YES | F1 | — | — |
| TVQMVDWCPTGFK | Alpha tubulin | Several species | — | — | YES | F3 |
| VGEGMEEGEFTEAR | Alpha tubulin | Several species | YES | F2 | YES | F2 |
| DLTDYLMR | Actin | Several species | — | — | YES | F3 |
| EEEVAALVIDNGSGMCK | Actin | Several species | YES | F3 | — | — |
| EEEVAALVVDNGSGMCK | Actin | Several species | YES | F2 | — | — |
| NYELPDGQVITIGNER | Actin | Several species | YES | F3 | YES | F1 |
| VAPEEHPVLLTEAPLNPR | Actin | Several species | YES | F1 | — | — |
| IEIIANDQGNR | Heat shock protein | Several species | — | — | YES | F1 |
| AMSIMNSFVNDIFER | Histone H2B | Several species | — | — | YES | F3 |
| HAVSEGTRAVTK | Histone H2B | Several species | YES | F2 | — | — |
| QDLPNAMQAAEITDK | ADP-ribosylation factor | Several species | YES | F1 | YES | F1 |
| SVCTEAGMYAIR | 26 s protease regulatory | Several species | — | — | YES | F2 |
| TFTTQETITNAESAR | Glucose-6-phosphate isomerase | Several species | YES | F1 | — | — |
| DAGTISGLNVLR | Several proteins | Several species | — | — | YES | F1 |
| IVLIGDSGVGK | Several proteins | Several species | — | — | YES | F2 |
| IVNEPTAAAIAYGLDK | Several proteins | Several species | YES | F2 | YES | F2 |
| VIVLGDSGVGK | Several proteins | Several species | — | — | YES | F2 |
| DVNAALPPSR | Alpha tubulin | Spizellomyces punctatus | YES | F3 | — | — |
| NPDDITNEEYAAFYK | Hsp82-like protein | Spizellomyces punctatus | — | — | YES | F2 |
| NPGYFVEWIPNNVK | Beta tubulin | Spraguea lophii | YES | F1 | YES | F1 |
| NLTERGYSFTTTAER | Actin | Suillus bovinus | — | — | YES | F1 |
| TIQFVDWCTTGFK | Alpha tubulin | Syncephalis depressa | YES | F1 | YES | F1 |
| CVPAAVLGSGAANGAR | Putative AMP-binding enzyme | Taphrina deformans | YES | F1 | YES | F1 |
| SSENAPAIVIDNGSGMCK | Actin | Trachipleistophora hominis | YES | F3 | — | — |
Discussion
Knowledge on the precise protein composition of CA from the CNS may shed light on the origin of these bodies. In this regard, our current findings provide evidence for the complexity of the proteins that form part of CA, consistent with the notion that these proteins could be debris of dead cells, or could appear in the intercellular space, or are extravasated polypeptides12,25. Our results show that P7 contains CA and they are likely devoid of contamination by soluble cellular proteins and mitochondria. The finding of ribosomal components in P7 suggests that ribosomes are recruited to CA when they formed. Alternatively, it should be possible that ribosomes cosediment with CA, although this possibility is unlikely due to the purification protocol employed. However, not all cellular proteins are represented in the same proportion in CA and in the CNS tissue since some of them, such as eIF4GI, appeared in the tissue homogenate but not in CA. Comparison of proteins from P2 and P7 fractions indicated that whereas some of them are common to both fractions, each fraction contains several unique protein species. Perhaps the stability of the proteins, or their propensity to interact with polyglucans or with other proteins, dictate their fate to form part of CA. Aside from this complexity, another important concept in CA is their heterogeneity. For example, immunohistochemistry analyses revealed that not all CA stained equally with a given antibody. Some of them contained a given human protein while others in the same preparation contained less or were devoid of that protein. Heterogeneity can also exist between CA from different brain regions, and more important, between those found in AD and in elderly subjects. This heterogeneity also raises questions about the concept that CA simply agglutinate proteins in a random fashion. If so, CA could be envisaged as scavengers of cellular debris products that would be generated after cell injury. In agreement with this idea, myelin injury in patients with AD leads to the degradation of myelin basic protein, which appears in CA42.
Our proposal that CA contain fungal proteins based on the observation that they immunoreact with antifungal antibodies32 is now substantiated by proteomic analysis. Moreover, we can conclude that both fungal and bacterial proteins can be found in purified fractions of CA from AD patients. We recently reported that prokaryotic-like cells and bacterial DNA can be detected in brain tissue from AD39, expanding our previous results of fungal infection of the CNS in these patients32,33. Consistent with our findings, bacterial DNA has also been recently detected by next generation sequencing in Alzheimer’s brains41. Moreover, in support of the idea that polymicrobial infections are present in AD CNS is the observation that the amyloid peptide exhibits antifungal and antibacterial activity43. In this regard, the production of amyloid has been considered as part of the innate immune system and is synthesized in brain tissue as a response to microbial infections44. We believe that these findings are important to understand the origin of CA and their potential function. Accordingly, we posited that CA originate from fungal infection32. We now extend this hypothesis to consider both fungal and bacterial infections as the trigger for CA formation. In this regard, CA may represent a response to agglutinate cellular debris provoked by cell damage that in turn is the result of microbial infection. In addition, fungal and bacterial proteins will also be agglutinated through the adhesive properties of polyglucans. The local decrease in pH as a result of these infections will also induce the precipitation of calcium salts, which will also be trapped in CA. According to our hypothesis CA in AD patients originate because there is a microbial infection in the CNS, and their function would be to remove breakdown components from human and microbial cells. However, other possibilities to explain the origin of CA could be envisaged. For instance, the modification of the blood-brain barrier by the previous formation of CA could facilitate microbial colonization of the CNS. Then, glial activation could proceed, leading to microbial destruction and the consequent accumulation of fungal proteins in CA.
The existence of CA in other tissues of the human body as well as in the CNS of elderly people without neurodegenerative diseases could also be a consequence of microbial cells that may exist in much lower amounts than those found in AD patients. Indeed, bacteria have been detected in the CNS of control human subjects39,41,45. These microbial cells may have a tendency to locate in close proximity to blood vessels, where oxygen and nutrients are more abundant. This is in accord with the finding that CA are distributed in perivascular areas10–12. Remarkably, bacterial cells have also been found in human atherosclerosis and in aortic aneurysms46–48. In addition, bacteria can also be detected in internal tissues, that otherwise should be “sterile”49. The possibility that fungi coexist with these bacteria in some human organs or tissues remains to be explored. Therefore, the presence of CA in other tissues may be a consequence of low burdens of microbial cells that, with time, may increase in number and provoke clinical symptoms. Indeed, in the case of CA from prostate, the suggestion that they are a consequence of bacterial or viral infections has been proposed31,50.
Materials and Methods
Description of subjects
The study comprised two women: one aged 92, diagnosed with AD (hereafter described as AD1), and the other aged 76, diagnosed with AD and dementia with Lewy bodies (hereafter described as AD2). The two cases were diagnosed according to current neuropathological consensus guidelines51. Samples were supplied by Banco de Tejidos, Fundación CIEN (Centro de Investigación de Enfermedades Neurológicas, Madrid. Spain).Brain samples from two control subjects were also tested. These two controls did not suffer any neurodegenerative disease. Control 1 was a woman aged 48, while Control 2 was a man aged 78. The brain donors were anonymous to the investigators who participated in the study. All ethico-legal documents of the brain bank, including written informed consent, were approved by an ethics committee external to the bank. The study was approved by the ethics committee of Universidad Autónoma de Madrid. The transfer of samples was carried out according to national regulations concerning research on human biological samples. In addition to the informed consents, all experiments were performed in accordance with relevant guidelines and regulations.
Frozen brain samples and paraffin-fixed sections from insular cortex (INCO) were used for DNA purification and immunohistochemistry analyses, respectively. Brain and sample processing were carried out as described previously in detail32. Briefly, rapid neuropathological autopsy was performed upon call by the donor’s proxies (mean post-mortem interval, 4.5 h). Immediately after brain extraction, two symmetrical brain halves were obtained in fresh through a mid-sagital cut. The right half was frozen in −50 °C isopentane after serial cutting of the brain hemisphere, the cerebellar hemisphere and the brainstem. The left half was fixed in 4% phosphate-buffered formaldehyde during 3 weeks, and thereafter it was cut and sampled. A total 25 tissue blocks were obtained and embedded in paraffin. Samples from the frozen tissue were obtained with sterile instruments in a laminar flow hood, taking all measures to avoid contamination.
CA purification
INCO tissue (350 mg) was suspended in 1 ml phosphate buffered saline with calcium (PBS-Ca). The tissue was homogenized gently using a Miccra D-1 homogenizer (Miccra, Müllheim; Germany). A 30 μl aliquot of this homogenate was removed for later analysis. The remaining of the homogenate was centrifuged through a series of sucrose solutions at 20,000 × g. Accordingly, the pellet from the first centrifugation step (5 min) was resuspended in 300 μl PBS-Ca (P1) and loaded onto 1.5 ml of 25% sucrose in PBS-Ca. After centrifugation (10 min), the pellet was again resuspended in 300 μl PBS-Ca (P2). This pellet (P2) was treated with 1% sodium deoxycholate and the resulting suspension was loaded onto 1.5 ml 25% sucrose in PBS-Ca and centrifuged as before. This pellet was resuspended in 300 μl PBS-Ca (P3) and treated with 1,000 IU RNAse T1 (Thermoscientific, MA, USA) and 15 IU DNAse 1 (TAKARA Clontech, CA, USA) at 37 °C for 5 min. The resulting suspension was loaded onto 1.5 ml 25% sucrose in PBS-Ca and centrifuged as before. The pellet was again resuspended in 300 μl PBS-Ca (P4). This pellet (P4) was treated with 1.5% sodium deoxycholate and the resulting suspension was loaded onto 1.5 ml 35% sucrose in PBS-Ca and centrifuged as before. After this centrifugation step, this pellet was resuspended in 300 μl PBS-Ca (P5) and treated with 1,000 IU RNAse T1 and 15 IU DNAse 1 at 37 °C for 5 min. The resulting suspension was layered onto 1.5 ml of 35% sucrose in PBS-Ca and centrifuged and resuspended in 300 μl PBS-Ca to obtain pellet P6. Finally, the P6 pellet obtained was treated with 1,000 IU RNAse T1 and 15 IU DNAse 1 at 37 °C for 5 min and layered onto 1.5 ml 45% sucrose in PBS-Ca and centrifuged. It was then resuspended in 300 μl of PBS-Ca to obtain pellet P7.
Immunohistochemistry
Tissue sections from the CNS (5 μm) were fixed in 10% buffered formalin for 24 h and embedded in paraffin following standard protocols. Methods for immunohistochemical analysis have been previously described in detail34.
Western blotting
CA proteins were separated by SDS-PAGE (4–12% polyacrylamide), transferred to nitrocellulose membranes by wet immunotransfer and processed for western blotting as described38 after blocking the membranes with 1% bovine serum albumin. Rabbit polyclonal antibodies against eIF4GI52, Candida albicans and Phoma betae34 have been described previously. Mouse monoclonal antibodies against peptidoglycan and human α-tubulin were purchased from Sigma and Thermo Scientific, respectively. The goat polyclonal antibody against elongation factor eEF2 was purchased from Santa Cruz Biotechnology. Stripping of the nitrocellulose membrane was accomplished in some instances to test different antibodies.
Mass Spectrometry Analysis
The methodology for mass spectrometry (MS) analysis has been described in detail elsewhere36. Briefly, proteins from INCO samples were separated by PAGE under reducing conditions using a 12.5% separating gel and a 5% stacking gel. Protein staining was carried out with GelCode Blue Stain Reagent (Thermo Scientific). Proteins were digested in situ with sequencing grade trypsin (Promega, Madison, WI). Digestion was stopped by the addition of 1% trifluroacetic acid. The desalted protein digest was dried, resuspended in 10 µl of 0.1% formic acid and analyzed by RP-LC-MS/MS in an Easy-nLC II system coupled to a LTQ-Orbitrap Velos Pro hybrid mass spectrometer (Thermo Scientific). ESI ionization was performed using a stainless steel nano-bore emitter (Proxeon, ID 30 μm). The Orbitrap mass resolution was set at 30,000.
To perform a more in-depth analysis of the peptides obtained after trypsin digestión, whole supernatants were dried down, reconstituted in 0.1% TFA and then loaded onto a high-pH, reversed-phase fractionation spin column (Pierce). A step gradient of increasing acetonitrile concentrations in a volatile high-pH solution was applied to elute bound peptides into nine different fractions. After collecting together alternate fractions into three groups (F1 = 1 + 4 + 7, F2 = 2 + 5 + 8, F3 = 3 + 6 + 9), samples were dried and stored for MS analysis. To do this, each pool of fractions was resuspended in 10 μl of 0.1% formic acid and analyzed as described above.
Processing of the MS data was carried out as previously described36. Database searching was performed against uniprot-homo fasta and uniprot-fungi fasta.
Nested PCR assay
DNA was extracted from frozen tissue using the QIAmp Genomic DNA Isolation Kit (Qiagen, Hilden, Germany) as previously described33. To amplify the intergenic sequences 1 and 2 (ITS-1 and ITS-2) from fungal DNA, we performed nested PCR using the oligonucleotide primers and conditions described36. In addition, human mitochondrial DNA (mtDNA, D-loop region) was amplifed following the protocol described by Ghatak et al.53. RT-PCR analysis of the human 18S rRNA was performed using the primers 18SF 5′GTAACCCGTTGAACCCCATT 3′and 18SR 5′ CCATCCAATCGGTAGTAGCG 3′.
Electronic supplementary material
Acknowledgements
The financial support of Pharma Mar, S.A. is acknowledged. We also acknowledge an institutional grant to Centro de Biología Molecular “Severo Ochoa” from the Fundación Ramón Areces.
Author Contributions
D.P., R.A. carried out the immunofluorescence and PCR experiments A.I.M. carried out the proteomic study A.R. prepared the samples from frozen CNS material and obtained the paraffin sections D.P., R.A. and L.C. designed the experiments L.C. designed the study and wrote the paper. D.P., R.A. prepared the figures. All authors discussed the data obtained. All authors reviewed and provided comments upon preparation of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28231-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Leel-Ossy L. Corpora amylacea in hippocampal sclerosis. J Neurol Neurosurg Psychiatry. 1998;65:614. doi: 10.1136/jnnp.65.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selmaj K, et al. Corpora amylacea from multiple sclerosis brain tissue consists of aggregated neuronal cells. Acta Biochim Pol. 2008;55:43–49. [PubMed] [Google Scholar]
- 3.Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Brain Res Rev. 1999;29:265–295. doi: 10.1016/S0165-0173(99)00003-X. [DOI] [PubMed] [Google Scholar]
- 4.Delgado-Escueta AV. Advances in lafora progressive myoclonus epilepsy. Curr Neurol Neurosci Rep. 2007;7:428–433. doi: 10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 5.Robitaille Y, Carpenter S, Karpati G, DiMauro SD. A distinct form of adult polyglucosan body disease with massive involvement of central and peripheral neuronal processes and astrocytes: a report of four cases and a review of the occurrence of polyglucosan bodies in other conditions such as Lafora’s disease and normal ageing. Brain. 1980;103:315–336. doi: 10.1093/brain/103.2.315. [DOI] [PubMed] [Google Scholar]
- 6.Keller JN. Age-related neuropathology, cognitive decline, and Alzheimer’s disease. Ageing Res Rev. 2006;5:1–13. doi: 10.1016/j.arr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Mrak RE, Griffin ST, Graham DI. Aging-associated changes in human brain. J Neuropathol Exp Neurol. 1997;56:1269–1275. doi: 10.1097/00005072-199712000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Song W, et al. Astroglial heme oxygenase-1 and the origin of corpora amylacea in aging and degenerating neural tissues. Exp Neurol. 2014;254:78–89. doi: 10.1016/j.expneurol.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacs GG, Risser D. Clinical Neuropathology image 6-2014: Corpora amylacea replacing cornu ammonis (CACA) Clin Neuropathol. 2014;33:378–379. doi: 10.5414/NP300831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishio S, et al. Corpora amylacea replace the hippocampal pyramidal cell layer in a patient with temporal lobe epilepsy. Epilepsia. 2001;42:960–962. doi: 10.1046/j.1528-1157.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 11.Pirici I, et al. Corpora amylacea in the brain form highly branched three-dimensional lattices. Rom J Morphol Embryol. 2014;55:1071–1077. [PubMed] [Google Scholar]
- 12.Rohn, T. T. Corpora Amylacea in Neurodegenerative Diseases: Cause or Effect? Int J Neurol Neurother2 (2015). [DOI] [PMC free article] [PubMed]
- 13.Badea P, Petrescu A, Moldovan L, Zarnescu O. Structural heterogenity of intraluminal content of the prostate: a histochemical and ultrastructural study. Microsc Microanal. 2015;21:368–376. doi: 10.1017/S1431927615000197. [DOI] [PubMed] [Google Scholar]
- 14.Christian JD, Lamm TC, Morrow JF, Bostwick DG. Corpora amylacea in adenocarcinoma of the prostate: incidence and histology within needle core biopsies. Mod Pathol. 2005;18:36–39. doi: 10.1038/modpathol.3800250. [DOI] [PubMed] [Google Scholar]
- 15.Hechtman JF, Gordon RE, Harpaz N. Intramuscular corpora amylacea adjacent to ileal low-grade neuroendocrine tumours (typical carcinoids): a light microscopic, immunohistochemical and ultrastructural study. J Clin Pathol. 2013;66:569–572. doi: 10.1136/jclinpath-2012-201415. [DOI] [PubMed] [Google Scholar]
- 16.Morales E, et al. Characterization of corpora amylacea glycoconjugates in normal and hyperplastic glands of human prostate. J Mol Histol. 2005;36:235–242. doi: 10.1007/s10735-005-5784-z. [DOI] [PubMed] [Google Scholar]
- 17.Goebel HH, et al. Adult polyglucosan body myopathy. J Neuropathol Exp Neurol. 1992;51:24–35. doi: 10.1097/00005072-199201000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Rocken C, Linke RP, Saeger W. Corpora amylacea in the lung, prostate and uterus. A comparative and immunohistochemical study. Pathol Res Pract. 1996;192:998–1006. doi: 10.1016/S0344-0338(96)80041-0. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura A, et al. The carbohydrate deposits detected by histochemical methods in the molecular layer of the dentate gyrus in the hippocampal formation of patients with schizophrenia, Down’s syndrome and dementia, and aged person. Glycoconj J. 2000;17:815–822. doi: 10.1023/A:1010996911581. [DOI] [PubMed] [Google Scholar]
- 20.Kodaka T, et al. Fine structure and mineral components of primary calculi in some human prostates. J Electron Microsc (Tokyo) 2008;57:133–141. doi: 10.1093/jmicro/dfn013. [DOI] [PubMed] [Google Scholar]
- 21.Magura, C. E. & Spector, M. Scanning electron microscopy of human prostatic corpora amylacea and corpora calculi, and prostatic calculi. Scan Electron Microsc, 713–720 (1979). [PubMed]
- 22.Nakamura KT, Nakahara H, Nakamura M, Tokioka T, Kiyomura H. Ultrastructure and x-ray microanalytical study of human pineal concretions. Ann Anat. 1995;177:413–419. doi: 10.1016/S0940-9602(11)80146-9. [DOI] [PubMed] [Google Scholar]
- 23.Singhrao SK, Neal JW, Piddlesden SJ, Newman GR. New immunocytochemical evidence for a neuronal/oligodendroglial origin for corpora amylacea. Neuropathol Appl Neurobiol. 1994;20:66–73. doi: 10.1111/j.1365-2990.1994.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 24.Day RJ, Mason MJ, Thomas C, Poon WW, Rohn TT. Caspase-Cleaved Tau Co-Localizes with Early Tangle Markers in the Human Vascular Dementia Brain. Plos One. 2015;10:e0132637. doi: 10.1371/journal.pone.0132637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng H, Zhang X, Blaivas M, Wang MM. Localization of blood proteins thrombospondin1 and ADAMTS13 to cerebral corpora amylacea. Neuropathology. 2009;29:664–671. doi: 10.1111/j.1440-1789.2009.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singhrao SK, Morgan BP, Neal JW, Newman GR. A functional role for corpora amylacea based on evidence from complement studies. Neurodegeneration. 1995;4:335–345. doi: 10.1016/1055-8330(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 27.Cisse S, Perry G, Lacoste-Royal G, Cabana T, Gauvreau D. Immunochemical identification of ubiquitin and heat-shock proteins in corpora amylacea from normal aged and Alzheimer’s disease brains. Acta Neuropathol. 1993;85:233–240. doi: 10.1007/BF00227716. [DOI] [PubMed] [Google Scholar]
- 28.Botez G, Rami A. Immunoreactivity for Bcl-2 and C-Jun/AP1 in hippocampal corpora amylacea after ischaemia in humans. Neuropathol Appl Neurobiol. 2001;27:474–480. doi: 10.1046/j.1365-2990.2001.00362.x. [DOI] [PubMed] [Google Scholar]
- 29.Sfanos KS, Wilson BA, De Marzo AM, Isaacs WB. Acute inflammatory proteins constitute the organic matrix of prostatic corpora amylacea and calculi in men with prostate cancer. Proc Natl Acad Sci USA. 2009;106:3443–3448. doi: 10.1073/pnas.0810473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyaux D, et al. S100 proteins in Corpora amylacea from normal human brain. Brain Res. 2000;867:280–288. doi: 10.1016/S0006-8993(00)02393-3. [DOI] [PubMed] [Google Scholar]
- 31.Sfanos KS, Hempel HA, De Marzo AM. The role of inflammation in prostate cancer. Adv Exp Med Biol. 2014;816:153–181. doi: 10.1007/978-3-0348-0837-8_7. [DOI] [PubMed] [Google Scholar]
- 32.Pisa D, Alonso R, Rabano A, Carrasco L. Corpora Amylacea of Brain Tissue from Neurodegenerative Diseases Are Stained with Specific Antifungal Antibodies. Front Neurosci. 2016;10:86. doi: 10.3389/fnins.2016.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso R, et al. Fungal infection in patients with Alzheimer’s disease. J Alzheimers Dis. 2014;41:301–311. doi: 10.3233/JAD-132681. [DOI] [PubMed] [Google Scholar]
- 34.Pisa D, Alonso R, Rabano A, Rodal I, Carrasco L. Different Brain Regions are Infected with Fungi in Alzheimer’s Disease. Sci Rep. 2015;5:15015. doi: 10.1038/srep15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pisa D, Alonso R, Juarranz A, Rabano A, Carrasco L. Direct visualization of fungal infection in brains from patients with Alzheimer’s disease. J Alzheimers Dis. 2015;43:613–624. doi: 10.3233/JAD-141386. [DOI] [PubMed] [Google Scholar]
- 36.Alonso R, et al. Evidence for fungal infection in cerebrospinal fluid and brain tissue from patients with amyotrophic lateral sclerosis. Int J Biol Sci. 2015;11:546–558. doi: 10.7150/ijbs.11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso, R., Pisa, D., Fernández-Fernández, A., Rabano, A. & Carrasco, L. Fungal Infection In Neural Tissue Of Patients With Amyotrophic Lateral Sclerosis. Neurobiology of disease (2017). [DOI] [PubMed]
- 38.Pisa D, Alonso R, Rabano A, Horst MN, Carrasco L. Fungal Enolase, beta-Tubulin, and Chitin Are Detected in Brain Tissue from Alzheimer’s Disease Patients. Front Microbiol. 2016;7:1772. doi: 10.3389/fmicb.2016.01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pisa D, Alonso R, Fernandez-Fernandez AM, Rabano A, Carrasco L. Polymicrobial Infections In Brain Tissue From Alzheimer’s Disease Patients. Sci Rep. 2017;7:5559. doi: 10.1038/s41598-017-05903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso R, Pisa D, Aguado B, Carrasco L. Identification of Fungal Species in Brain Tissue from Alzheimer’s Disease by Next-Generation Sequencing. J Alzheimers Dis. 2017;58:55–67. doi: 10.3233/JAD-170058. [DOI] [PubMed] [Google Scholar]
- 41.Emery DC, et al. 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer’s Post-Mortem Brain. Front Aging Neurosci. 2017;9:195. doi: 10.3389/fnagi.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan X, et al. Myelin injury and degraded myelin vesicles in Alzheimer’s disease. Curr Alzheimer Res. 2014;11:232–238. doi: 10.2174/1567205011666140131120922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soscia SJ, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. Plos One. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar DK, et al. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci Transl Med. 2016;8:340ra372. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Branton WG, et al. Brain microbial populations in HIV/AIDS: alpha-proteobacteria predominate independent of host immune status. Plos One. 2013;8:e54673. doi: 10.1371/journal.pone.0054673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marques da Silva R, et al. Bacterial diversity in aortic aneurysms determined by 16S ribosomal RNA gene analysis. J Vasc Surg. 2006;44:1055–1060. doi: 10.1016/j.jvs.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Ott SJ, et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113:929–937. doi: 10.1161/CIRCULATIONAHA.105.579979. [DOI] [PubMed] [Google Scholar]
- 49.Padgett KA, et al. Phylogenetic and immunological definition of four lipoylated proteins from Novosphingobium aromaticivorans, implications for primary biliary cirrhosis. J Autoimmun. 2005;24:209–219. doi: 10.1016/j.jaut.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Yanamandra K, et al. Amyloid formation by the pro-inflammatory S100A8/A9 proteins in the ageing prostate. Plos One. 2009;4:e5562. doi: 10.1371/journal.pone.0005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montine TJ, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123:1–11. doi: 10.1007/s00401-011-0910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novoa I, Feduchi E, Carrasco L. Hybrid proteins between Pseudomonas exotoxin A and poliovirus protease 2Apro. FEBS Lett. 1994;355:45–48. doi: 10.1016/0014-5793(94)01157-5. [DOI] [PubMed] [Google Scholar]
- 53.Ghatak S, Muthukumaran RB, Nachimuthu SK. A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. J Biomol Tech. 2013;24:224–231. doi: 10.7171/jbt.13-2404-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.