Abstract
Background
Emphysema has considerable variability in its regional distribution. Craniocaudal emphysema distribution is an important predictor of the response to lung volume reduction. However, there is little consensus regarding how to define upper lobe-predominant and lower lobe-predominant emphysema subtypes. Consequently, the clinical and genetic associations with these subtypes are poorly characterized.
Methods
We sought to identify subgroups characterized by upper-lobe or lower-lobe emphysema predominance and comparable amounts of total emphysema by analyzing data from 9,210 smokers without alpha-1-antitrypsin deficiency in the Genetic Epidemiology of COPD (COPDGene) cohort. CT densitometric emphysema was measured in each lung lobe. Random forest clustering was applied to lobar emphysema variables after regressing out the effects of total emphysema. Clusters were tested for association with clinical and imaging outcomes at baseline and at 5-year follow-up. Their associations with genetic variants were also compared.
Results
Three clusters were identified: minimal emphysema (n = 1,312), upper lobe-predominant emphysema (n = 905), and lower lobe-predominant emphysema (n = 796). Despite a similar amount of total emphysema, the lower-lobe group had more severe airflow obstruction at baseline and higher rates of metabolic syndrome compared with subjects with upper-lobe predominance. The group with upper-lobe predominance had greater 5-year progression of emphysema, gas trapping, and dyspnea. Differential associations with known COPD genetic risk variants were noted.
Conclusions
Subgroups of smokers defined by upper-lobe or lower-lobe emphysema predominance exhibit different functional and radiological disease progression rates, and the upper-lobe predominant subtype shows evidence of association with known COPD genetic risk variants. These subgroups may be useful in the development of personalized treatments for COPD.
Key Words: clustering, COPD, COPD disease progression, emphysema distribution, machine learning
Abbreviations: COPDGene, Genetic Epidemiology of COPD; GOLD, Global Initiative for Chronic Obstructive Lung Disease; GWAS, genome-wide association studies; HU, Hounsfield units; MMRC, Modified Medical Research Council; PCA, principal components analysis; % LAA-950, percent of CT low attenuation area < –950 HU at end-inspiration; Pi10, square root of the wall area of a hypothetical airway of 10-mm internal perimeter; PRM, Parametric Response Mapping; PRM-fSAD, functional small airways disease; SGRQ, St. George’s Respiratory Questionnaire
Emphysema exhibits considerable heterogeneity in its extent and spatial distribution.1 Although total lung emphysema is a strong predictor of many COPD-related outcomes, the craniocaudal distribution of emphysema is an independent predictor of mortality and the response to lung volume reduction.2, 3, 4, 5, 6 Emphysema distribution is partially genetically determined, with an estimated heritability of 20%, and genome-wide association studies (GWAS) of this trait as a continuous measure have identified multiple genome-wide significant associations.7, 8 Thus, emphysema distribution is a COPD phenotype with both clinical and biological relevance.
Most previous studies have evaluated emphysema distribution as a continuous trait, but for treatment decisions and clinical trials, it is often useful to define groups of subjects based on specific disease characteristics. However, although there is compelling evidence that emphysema distribution is an important facet of COPD clinical heterogeneity, approaches to define groups of subjects according to upper-lobe or lower-lobe emphysema predominance independent of other disease markers are needed to enable better characterization of these subtypes. In addition, defining emphysema distribution subgroups through the use of thresholds for an emphysema ratio often results in an imbalance in the amount of total emphysema between groups.
In the present study, we sought to identify subgroups of smokers characterized by upper-lobe or lower-lobe emphysema predominance and comparable amounts of total emphysema. We hypothesized that these emphysema distribution subtypes would have different clinical characteristics and genetic associations. To test this hypothesis, we used a data-driven clustering approach to identify upper lobe-predominant and lower lobe-predominant emphysema subgroups with comparable amounts of total emphysema, and we characterized these subgroups using extensive phenotypic and genotypic data from the COPDGene study at baseline and at 5-year follow-up.
Methods
Study Population
The COPDGene study is an ongoing large multicenter longitudinal study designed to investigate the genetic and epidemiologic characteristics of COPD. The protocols for subject recruitment and data collection for the COPDGene study have been previously described.9 During phase I, the COPDGene study enrolled 10,192 non-Hispanic white and African American subjects across the full spectrum of disease severity as defined by the Global Initiative for Chronic Obstructive Lung Disease (GOLD) spirometric grading system. Eight individuals had Pi ZZ or ZNull α1-antitrypsin deficiency and were excluded in this present study. During phase II, 5,000 subjects had completed their 5-year follow-up visit at the time of this analysis (COPDGene phase II data set, September 24, 2016). Nineteen of these 5,000 subjects underwent lung volume reduction surgery or lung transplantation between the two visits and were excluded from this analysis. Written informed consent was obtained from all participants. The study and consent forms were approved by the Brigham and Women's Hospital Institutional Review Board (No. 2007P000554/BWH).
CT Measurements
Using 3D SLICER analysis (www.chestimagingplatform.org) at visit 1 and Thirona software (www.thirona.eu) at visit 2, emphysema was quantified as the percentage of lung voxels with attenuation lower than –950 HU at maximal inspiration (% LAA-950).10 The lungs were automatically segmented into anatomically defined lung lobes, as previously described,11 and % LAA-950 was quantified in each lobe. The lingula was included in the left upper lobe measurements. The ratio of % LAA-950 in both upper lobes to that in both lower lobes was used to evaluate the lobar distribution of emphysema (U/L ratio).
Airway disease was assessed using VIDA software (VIDA Diagnostics, Inc.; www.vidadiagnostics.com) as gas trapping (percentage of low attenuation units < –856 Hounsfield units [HU] at end-expiration), airway wall thickness (obtained along the center line of the lumen, in the middle third of the airway segment, for one segmental airway of each lung lobe; the mean value across all lobes was used for analysis), and Pi10 (the square root of the wall area of a hypothetical airway of 10-mm internal perimeter).
Although small airway disease can be assessed by gas trapping, a significant limitation of this approach is that many lung regions that trapped gas on exhalation will also show emphysematous destruction when fully inflated to total lung capacity.12 We therefore also analyzed data from a recently developed CT analytic method, Parametric Response Mapping (PRM) by Imbio LLC, using Lung Density Analysis software (Imbio LLC).13 By co-registering inspiratory and expiratory CT images, the PRM method discriminates emphysema (PRM-emphysema) from nonemphysematous air trapping, termed functional small airways disease (PRM-fSAD).14 These PRM measurements were available for 3,073 subjects at visit 1 and 2.
Clustering and Statistical Analyses
Emphysema variables from visit 1 were log(x+1) transformed to reduce the impact of deviations from normality. To account for emphysema severity and generate lobe-specific measures, each lobar emphysema variable was regressed on total emphysema. Unsupervised random forest clustering was then performed on the residuals, using the approach described by Shi and Horvath.15
The identified clusters were tested for associations with visit 1 and visit 2 characteristics. The definitions of the various COPD characteristics and comorbidities are shown in e-Table 1. The longitudinal outcomes were calculated by subtracting the visit 1 value from the visit 2 value. Negative values represent a lower value of the outcome at visit 2.
Using PLINK, version 1.9, a GWAS was conducted comparing upper-lobe vs lower-lobe clusters adjusting for pack-years of smoking, current smoking, sex, and principal components of genetic ancestry.16 Separate results in non-Hispanic white and African American subjects were combined using fixed-effects meta-analysis implemented in METAL software (August 8, 2010).17 We also evaluated associations with 25 genetic variants previously associated with COPD susceptibility, emphysema, or emphysema distribution (“candidate single nucleotide polymorphism [SNP] analysis”).7, 8, 18, 19, 20, 21, 22, 23, 24 Additional methods are available in e-Appendix.
Results
Characteristics of Study Participants Included in the Analyses
We included in the analysis 9,210 smokers (6,195 non-Hispanic whites and 3,015 African Americans) with complete lobar imaging and phenotypic data from visit 1. The characteristics of these subjects are shown in e-Table 2. Compared with non-Hispanic whites, African American subjects were slightly younger and had a lower median exposure to smoking, a higher FEV1 % predicted, less severe total emphysema, and less advanced COPD.
Cluster Analysis
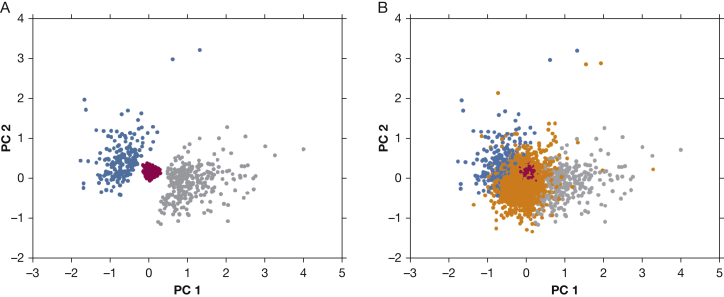
When the clustering variables were visualized using principal components analysis (PCA), it was apparent that subjects are spread along a continuum of emphysema distribution, and standard approaches that classify all subjects would not be guaranteed to identify a core of tightly clustered subjects (Fig 1). Based on this observation, we applied an unsupervised random forest clustering method designed to identify tightly clustered subjects and remove poorly clustered observations. This algorithm identified three clusters consisting of 3,013 subjects. Clustering criteria were not met in 6,197 subjects. The characteristics of these subjects, shown in e-Table 3, demonstrate that this “poorly clustered” group encompasses subjects across the range of COPD severity (GOLD 0-GOLD 4) with less total emphysema than either the upper lobe- predominant or lower lobe-predominant groups. Most importantly, the members of this group have neither marked upper-lobe or lower-lobe emphysema predominance (U/L ratio, 1.31 [IQR, 0.94-1.86]).
Figure 1.
Principal components analysis (PCA) plot of the residualized lobar emphysema variables. A, The three identified clusters: the minimal emphysema cluster is in red, the upper lobe-predominant emphysema cluster is in gray, and the lower lobe-predominant emphysema cluster is in blue. B, The unassigned group is in orange. To summarize the relationships between the five residualized lobar emphysema variables, we performed PCA on these data. The top two principal components (PC) explained almost 80% of the variance in these data (50.7% for PC1 and 26.2% for PC2). PC1 represents an “upper lobe emphysema axis,” and PC2 is a “lower-lobe emphysema” axis, based on their positive loadings for each kind of emphysema, respectively.
Visit 1 Imaging Characteristics
To determine whether these clusters did represent upper lobe-predominant and lower-lobe predominant emphysema subtypes, we evaluated the lobar emphysema values within each cluster and confirmed that the clusters were characterized by minimal emphysema (n = 1,312), upper lobe-predominant emphysema (upper-lobe predominant cluster; n = 905; U/L ratio: 4.60 [interquantile range [IQR], 3.31-7.08]), and lower lobe-predominant emphysema (lower-lobe predominant cluster [n = 796]; U/L ratio, 0.50 [IQR, 0.39-0.62]) (Table 1; e-Fig 1). Despite the pronounced difference in emphysema distribution, the upper lobe-predominant and lower lobe-predominant clusters had a similar extent of total emphysema (5.4 [IQR, 2.4-13.8] vs 5.5 [IQR, 2.2-16.7]; P = .85) (e-Fig 1). No difference in airway wall thickening or Pi10 was noted among the three clusters, but both upper lobe-predominant and lower lobe-predominant subjects had more gas trapping compared with subjects with minimal emphysema. e-Table 4 includes detailed characteristics of these subgroups.
Table 1.
Demographic, Imaging, and Clinical Characteristics of the Subjects in Each of the Emphysema Distribution Clusters at Visit 1
| Characteristic | Minimal Emphysema Cluster (n = 1,312) | Upper Lobe-Predominant Cluster (n = 905) | Lower Lobe-Predominant Cluster (n = 796) | P Value (All Groups Omnibus Test) | P Value (Upper-Lobe Predominant vs Lower-Lobe Predominant) |
|---|---|---|---|---|---|
| Demographics | |||||
| Age, y | 53.20 (49.18-59.26) | 59.30 (52.50-66.30) | 64.00 (57.40-69.83) | < .001 | < .001 |
| Male sex, % | 36.36 | 53.37 | 58.92 | < .001 | .07 |
| Non-Hispanic whites, % | 48.63 | 61.44 | 83.67 | < .001 | < .001 |
| BMI | 29.69 (25.91-33.71) | 26.64 (23.17-30.33) | 28.03 (24.21-32.55) | < .001 | < .001 |
| Smoking history (pack-years) | 35.80 (24.08-47.10) | 43.90 (33.00-61.30) | 45.00 (32.68-63.00) | < .001 | 1.00 |
| Current smoking, % | 75.15 | 57.79 | 40.83 | < .001 | < .001 |
| CT measurements | |||||
| Total emphysema, % LAA- 950) | 0.24 (0.14-0.36) | 5.44 (2.43-13.76) | 5.52 (2.21-16.67) | < .001 | .85 |
| U/L ratio | 1.08 (0.78-1.51) | 4.60 (3.31-7.08) | 0.50 (0.39-0.62) | < .001 | < .001 |
| Airway wall thickening | 61.82 (59.73-64.20) | 61.76 (59.49-63.86) | 61.48 (59.46-63.91) | .14 | .75 |
| Pi10 | 3.72 (3.66-3.81) | 3.68 (3.61-3.76) | 3.68 (3.59-3.76) | < .001 | .41 |
| Percent gas trapping | 4.05 (1.97-8.10) | 24.58 (13.80-41.81) | 31.30 (12.37-55.29) | < .001 | .23 |
| Spirometry | |||||
| FEV1, % predicted | 86.45 (74.80-97.80) | 71.70 (52.90-87.00) | 65.80 (39.40-86.20) | < .001 | < .001 |
| FEV1/FVC | 0.79 (0.74-0.83) | 0.64 (0.51-0.72) | 0.59 (0.41-0.74) | < .001 | .73 |
| Bronchodilator, % responsiveness (%) | 16.86 | 24.81 | 27.33 | < .001 | .78 |
| Symptoms and exacerbations | |||||
| MMRC dyspnea score | 1.24 ± 1.41 | 1.67 ± 1.45 | 1.63 ± 1.45 | < .001 | 1.00 |
| SGRQ score | 30.55 ± 25.86 | 38.65 ± 26.15 | 36.72 ± 26.80 | < .001 | .39 |
| Chronic bronchitis, % | 17.84 | 22.65 | 22.36 | .007 | 1.00 |
| Exacerbation frequency, % | 14.94 | 28.18 | 28.89 | < .001 | 1.00 |
| Comorbidities, % | |||||
| Coronary artery disease | 4.50 | 6.63 | 10.68 | < .001 | .01 |
| Diabetes | 16.08 | 10.06 | 14.82 | < .001 | .01 |
| Dyslipidemia | 35.37 | 39.34 | 46.23 | < .001 | .01 |
| Metabolic syndrome | 19.28 | 13.04 | 18.84 | < .001 | .004 |
| Obesity | 47.64 | 27.62 | 35.80 | < .001 | .001 |
| Sleep apnea | 15.10 | 14.07 | 22.48 | < .001 | < .001 |
Variables are expressed as mean and SD for continuous normally distributed variables, median and interquartile range (25th to 75th percentile) for continuous nonnormally distributed variables, and percentages for categorical variables. Omnibus P values are obtained using analysis of variance for the continuous normally distributed variables, the Kruskal-Wallis test for the continuous nonnormally distributed variables, and the χ2 test for proportions. P values of upper-lobe predominance vs lower-lobe predominance are from pairwise t tests, Nemenyi tests, and χ2 post hoc tests for pairwise comparisons between upper lobe- predominant and lower lobe-predominant clusters for the continuous normally distributed variables, nonnormally distributed variables, and categorical variables, respectively. P values < .05 are in bold.
Emphysema is defined as percent of CT low-attenuation area < –950 Hounsfield units at end-inspiration using SLICER software. U/L ratio: ratio of % LAA-950 in both upper lobes to that in both lower lobes. Airway wall area percent is the percentage of the wall area compared with the total bronchial area for segmental airways. Exacerbation frequency is the percentage of subjects reporting at least 1 COPD exacerbation in the previous year. Metabolic syndrome = 3 of the following 4: BMI ≥ 30 (measured), diabetes mellitus, hypertension, and high cholesterol (all self-reported). Obesity = BMI ≥ 30.
MMRC = Modified Medical Research Council; % LAA-950 = percent of CT low attenuation area < –950 HU at end-inspiration; Pi10 = square root of the wall area of a hypothetical airway of 10-mm internal perimeter; SGRQ = St. George’s Respiratory Questionnaire; U/L ratio = ratio of % LAA-950 in both upper lobes to that in both lower lobes.
Visit 1 Demographic, Physiological, and Clinical Characteristics
Relative to subjects with upper-lobe predominance, those in the lower-lobe cluster were slightly older, more likely to be non-Hispanic white, had more severe airway obstruction, and had a higher prevalence of metabolic syndrome. There was no significant difference between the upper-lobe and lower-lobe clusters regarding smoking exposure and exacerbation frequency (Table 1). The “minimal emphysema” cluster contained predominantly female subjects with fewer pack-years of smoking, minimal airway obstruction, more obesity, low comorbidity burden, and mildly impaired health-related quality of life.
Five-Year Prospective Change in Imaging
The 5-year changes in imaging measures are shown in Table 2. Over the 5-year follow-up, a more rapid progression of % LAA-950 was observed in subjects with upper-lobe predominance compared with those with lower-lobe predominance (absolute increase of 1.05% vs –0.01%; P < .001). The changes in emphysema as a percentage of emphysema at baseline are shown in e-Table 5. Although the absolute increase in emphysema in the group with upper-lobe predominance was modest, the percentage increase in emphysema was notable (emphysema change relative to visit 1, 23% vs –0.1%; P < .001), reflecting the fact that roughly half of the group with upper-lobe predominance had < 5% emphysema at baseline. These differences in radiological changes remain significant with a consistent direction of effect after accounting for age, sex, race, BMI, pack-years of smoking, change in smoking status between the two visits, and the baseline level of each respective outcome (Table 3). The emphysema distribution pattern remained similar between visit 1 and visit 2 with minimal change noted in the U/L ratio between the two visits for the upper lobe- predominant and lower lobe-predominant clusters (Table 2). Since CT emphysema measures were calculated with implementation of different software at visit 1 and visit 2, Thirona and SLICER % LAA-950 values were compared in 5,000 visit-1 scans processed by both methods, and the correlation was 0.99. A more rapid progression of PRM-emphysema and PRM-fSAD was observed in subjects with upper lobe predominance compared with those with lower lobe predominance (absolute change, 0.92% vs 0.04%; P < .001 for PRM-emphysema and 1.56% vs 0.11%, P = .01 for PRM-fSAD) (e-Table 6).
Table 2.
Five-Year Changes in Imaging Characteristics in Upper Lobe-Predominant vs Lower Lobe-Predominant Emphysema Clusters
| Variable | Upper Lobe-Predominant Cluster | Lower Lobe-Predominant Cluster | P Value (Upper-Lobe Predominance vs Lower-Lobe Predominance) |
|---|---|---|---|
| Change of total emphysema (% LAA-950) between visit 1 and visit 2 | |||
| All | 1.05 (–0.50 to 3.90) | –0.01 (–2.19 to 2.99) | < .001 |
| Current smokers at visit 1 and visit 2 | 0.95 (–0.25 to 3.18) | –0.17 (–1.40 to 3.20) | < .001 |
| Former smokers at visit 1 and visit 2 | 0.95 (–0.60 to 3.89) | –0.10 (–3.02 to 2.56) | .003 |
| U/L ratio | |||
| All | |||
| Visit 1 | 4.09 (2.98 to 7.17) | .65 (.49 to .81) | < .001 |
| Visit 2 | 3.80 (2.35 to 6.78) | .66 (.46 to .90) | < .001 |
| Current smokers at visit 1 and visit 2 | |||
| Visit 1 | 4.90 (3.38 to 8.09) | .55 (.43 to .69) | < .001 |
| Visit 2 | 4.40 (2.69 to 8.15) | .51 (.35 to .81) | < .001 |
| Former smokers at visit 1 and visit 2 | |||
| Visit 1 | 3.85 (2.89 to 5.91) | .72 (.55 to .84) | < .001 |
| Visit 2 | 3.59 (2.21 to 5.85) | .71 (.53 to .91) | < .001 |
| Change of % gas trapping between visit 1 and visit 2 | |||
| All | 3.66 (–2.79 to 10.50) | 1.04 (–4.10 to 6.46) | .001 |
| Current smokers at visit 1 and visit 2 | 5.97 (0.02 to 12.20) | 3.23 (–2.63 to 9.44) | .17 |
| Former smokers at visit 1 and visit 2 | 2.24 (–4.91 to 7.22) | –0.40 (–4.94 to 3.74) | .08 |
| Change of total lung capacity on CT between visit 1 and visit 2 | |||
| All | 0.06 (–0.26 to 0.31) | –0.08 (–0.31 to 0.15) | .001 |
| Current smokers at visit 1 and visit 2 | 0.12 (–0.24 to 0.36) | –0.03 (–0.27 to 0.25) | .16 |
| Former smokers at visit 1 and visit 2 | –0.01 (–0.26 to 0.25) | –0.10 (–0.31 to 0.08) | .02 |
Thirona software was used for % LAA-950. Percent gas trapping was measured at end-exhalation and defined as the percentage of lung voxels with density less than –856 Hounsfield units.
Variables are expressed as median and interquartile range (25th to 75th percentile). Change between visit 1 and visit 2 variables is defined as (value at visit 2 – value at visit 1). Negative values indicate decrease of the analyzed variable at visit 2. P values of upper-lobe predominance vs lower lobe-predominance are from Nemenyi tests for pairwise comparisons between upper lobe-predominant and lower lobe-predominant clusters following omnibus Kruskal-Wallis tests.
Sample sizes in each subgroup: All (n = 437 [upper-lobe predominance]; n = 394 [lower-lobe predominance]); current smokers at visit 1 and visit 2 (n = 185 [upper-lobe predominance]; n = 102 [lower-lobe predominance]); former smokers at visit 1 and visit 2 (n = 192 [upper-lobe predominance]; n = 222 [lower-lobe predominance]). P values < .05 are in bold.
See Table 1 legend for expansion of abbreviations.
Table 3.
Multivariate Models of 5-Year Changes in Spirometry, Functional Measures, and Imaging Characteristics in Upper Lobe-Predominant vs Lower Lobe-Predominant Emphysema Clusters
| Upper Lobe-Predominant Cluster vs Lower Lobe-Predominant Cluster | Univariate Model |
Multivariate Model 1a |
Multivariate Model 2b |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P Value | Beta | SE | P Value | Beta | SE | P Value | |
| Change of total emphysema between visit 1 and visit 2, % LAA-950/y | 0.19 | 0.05 | < .001 | 0.14 | 0.05 | 0.002 | 0.15 | 0.04 | < .001 |
| Change of % gas trapping between visit 1 and visit 2, %/y | 0.48 | 0.19 | .01 | 0.47 | 0.18 | 0.007 | 0.50 | 0.16 | .002 |
| Change of FEV1 between visit 1 and visit 2, mL/y | –9.00 | 4.62 | .05 | –3.51 | 3.78 | 0.35 | –2.57 | 4.24 | .59 |
| Change of FEV1 between visit 1 and visit 2, % predicted/y | –0.16 | 0.15 | .29 | –0.19 | 0.15 | 0.23 | –0.11 | 0.14 | .42 |
| Change of 6-min walk distance between visit 1 and visit 2, ft | 56.00 | 22.84 | .01 | 22.89 | 25.73 | 0.37 | –2.03 | 29.21 | .95 |
| Change of SGRQ score between visit 1 and visit 2 | –0.42 | 0.79 | .59 | 0.35 | 0.97 | 0.72 | 0.77 | 0.95 | .42 |
| Change of MMRC dyspnea score between visit 1 and visit 2 | 0.21 | 0.13 | .10 | 0.36 | 0.13 | 0.008 | 0.42 | 0.14 | .002 |
Total emphysema: Percent of CT low attenuation area < -950 Hounsfield units at end-inspiration using Thirona software. Percent gas trapping is measured at end-exhalation and defined as the percentage of lung voxels with density < –856 Hounsfield units.
The longitudinal outcomes of change per year for each individual are defined as (value at visit 2 – value at visit 1)/(time between visit 1 and visit 2). Median quantile regression models were used for all the variables except MMRC, in which an ordinal logistic regression model was used. The lower lobe-predominant cluster is the reference cluster. P values < .05 are in bold.
MMRC = Modified Medical Research Council; SGRQ = St. George’s Respiratory Questionnaire. See Table 1 legend for expansion of other abbreviations.
Model 1 includes the following covariates: visit 1 value of the outcome of interest and change in smoking status between visit 1 and visit 2.
Model 2 includes the following covariates: visit 1 value of the outcome of interest, change in smoking status between visit 1 and visit 2, age, sex, race, BMI, and pack-years of smoking. Change in smoking status between visit 1 and visit 2 was defined as: “Current to former smoker”, “former to current smoker”, “current; unchanged”, or “former, unchanged”.
Using stratified analyses, we explored how the relationship between emphysema distribution and emphysema progression is modified by GOLD grade, overall severity of emphysema, BMI, and current smoking. Interestingly, in the lower-lobe cluster, radiological decline was inversely associated with BMI, although the differential associations of upper-lobe clusters vs lower-lobe clusters with total emphysema and emphysema distribution were consistent across BMI strata (e-Table 7). Although more severe in the subjects with total emphysema > 5% (e-Table 8), radiological progression was similar at all GOLD grades (e-Table 9). As noted in Table 2, the differences in the % change of gas trapping between visit 1 and visit 2 in the upper-lobe vs lower-lobe clusters had a P value of .17 in current smokers and .08 in former smokers. The differences in the % change of CT-total lung capacity had a P value of .16 in current smokers and .02 in former smokers. Conversely, the differences in the % change of total emphysema were statistically significant in both current and former smokers.
Five-Year Prospective Change in Spirometry and Functional Measures
Although suggestive but not nominally significant in univariate analysis (P = .10), the association between the group with upper-lobe predominance and an increase in the Modified Medical Research Council (MMRC) score relative to the group with lower-lobe predominance was strongly significant after adjustment for the baseline MMRC score (P = .006). No significant differences were detected in multivariate analyses of longitudinal changes of FEV1 % predicted, 6-min walk distance, or St. George's Respiratory Questionnaire (SGRQ) (Table 3). The statistically significant difference seen in the univariate analysis of FEV1 (mL/y) was attenuated after adjustment for potential confounders. Stratified analyses suggest that the difference in FEV1 decline between the group with upper-lobe predominance and the group with lower-lobe predominance is greatest in early GOLD grades (e-Table 8).
Genome-Wide Association Studies
The quantile-quantile plot of the P value distributions and genomic control values demonstrated absence of a systematic inflation (e-Fig 2). No SNPs reached genome-wide significance. However, regions of interest were identified near genome-wide significance, with SNPs at meta-analysis P values < 5 × 10–6 (e-Table 10). Nominally significant associations were observed near the HHIP, AGPHD1, PPT2/AGER, TRAPPC9, MYO1D, KIAA1462, and IREB2/CHRNA3/5 genes (Table 4). Interestingly, for four of the five loci with nominally significant associations with both emphysema distribution subtypes and COPD, the COPD risk allele is associated with upper-lobe predominance.
Table 4.
Associations With Known COPD Susceptibility or Emphysema Distribution Genetic Variants, or Both, Comparing the Upper Lobe-Predominant Cluster and the Lower Lobe-Predominant Cluster
| Candidate SNP | Locus | Nearest Gene | Effect Allele | Effect Allele Frequency in Non-Hispanic Whites | Effect Allele Frequency in African Americans | Meta-analysis GWAS: Upper Lobe vs Lower-Lobe Clustersa |
GWAS: COPD Cases vs Control Subjectsb |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | SE | P Value | Case-Control OR | Case-Control SE | Case-control P Value | ||||||
| rs13141641 | 4q31 | HHIP | T | 0.61 | 0.91 | 1.45 | 1.09 | 1.20 × 10–5 | 1.27 | 1.03 | 4.6 × 10–13 |
| rs9788721 | 15q25 | AGPHD1 | T | 0.60 | 0.61 | 0.78 | 1.08 | .0009 | 0.79 | 1.03 | 5.2 × 10–13 |
| rs12914385 | 15q25 | IREB2/CHRNA3/5 | T | 0.41 | 0.19 | 1.30 | 1.08 | .0009 | 1.29 | 1.03 | 2.1 × 10–15 |
| rs75755010 | 8q24 | TRAPPC9 | A | 0.95 | 0.88 | 0.66 | 1.18 | .009 | 1.01 | 1.07 | .86 |
| rs2070600 | 6p22 | PPT2/AGER | T | 0.04 | NA | 1.77 | 1.25 | .01 | 0.71 | 0.08 | 2.9 × 10–5 |
| rs35374984 | 10p12 | KIAA1462 | A | 0.88 | 0.47 | 1.29 | 1.11 | .02 | 1.11 | 1.05 | .02 |
| rs379123 | 17q11 | MYOID | T | 0.41 | 0.34 | 0.85 | 1.08 | .03 | 0.94 | 1.03 | .05 |
| rs2645694 | 4q13 | SOWAHB | T | 0.74 | 0.58 | 1.17 | 1.09 | .07 | 1.00 | 1.04 | .9 |
| rs7957346 | 12q22 | SNRPF | A | 0.56 | 0.60 | 0.88 | 1.08 | .08 | 1.08 | 1.03 | .02 |
| rs7733088 | 5q32 | HTR4 | A | 0.40 | 0.32 | 0.88 | 1.07 | .08 | 0.91 | 1.03 | .004 |
| rs754388 | 14q32 | RIN3 | C | 0.82 | 0.86 | 1.19 | 1.11 | .09 | 1.29 | 1.04 | 1.5 × 10–9 |
| rs75200691 | 8p22 | DLC1 | T | 0.88 | 0.92 | 0.82 | 1.13 | .1 | 1.05 | 1.05 | .34 |
| rs7698250 | 4p15 | DHX15 | T | NA | 0.94 | 0.66 | 1.40 | .21 | 0.94 | 0.15 | .67 |
| rs56113850 | 19q13 | CYP2A6/ADCK4 | T | 0.61 | 0.55 | 0.88 | 1.11 | .23 | 0.8 | 1.04 | 1.1 × 10–7 |
| rs7221059 | 17q25 | MGAT5B | A | 0.81 | 0.69 | 0.90 | 1.10 | .27 | 1.02 | 1.04 | .64 |
| rs9590614 | 13q14 | VWA8 | C | 0.60 | 0.85 | 0.95 | 1.08 | .49 | 1.02 | 1.03 | .5 |
| rs2234922 | 1q41 | EPHX1 | A | 0.19 | 0.33 | 1.05 | 1.09 | .56 | 0.96 | 1.04 | .26 |
| rs626750 | 11q22 | MMP12 | A | 0.82 | 0.73 | 1.05 | 1.10 | .61 | 0.83 | 1.04 | 2.9 × 10–6 |
| rs10411619 | 19p13 | MAN2B | T | 0.01 | 0.21 | 1.08 | 1.18 | .64 | 0.96 | 1.08 | .58 |
| rs10844154 | 12p11 | BICD1 | A | 0.44 | 0.43 | 1.03 | 1.08 | .65 | 1.02 | 1.03 | .5 |
| rs2250889 | 20q13 | MMP9 | C | 0.04 | 0.16 | 1.06 | 1.16 | .68 | 1.00 | 1.06 | .97 |
| rs4846480 | 1q41 | TGFB2 | A | 0.75 | 0.67 | 1.03 | 1.08 | .68 | 1.17 | 1.03 | 5.6 × 10–6 |
| rs4416442 | 4q22 | FAM13A | T | 0.58 | 0.46 | 0.98 | 1.08 | .76 | 0.79 | 1.03 | 3.5 × 10–14 |
| rs45505795 | 14q32 | SERPINA10 | C | 0.96 | 0.99 | 1.05 | 1.25 | .84 | 1.52 | 1.11 | 3.4 × 10–5 |
| rs9478940 | 6q25 | GSTP1 | A | 0.83 | 0.41 | 1.00 | 1.10 | .96 | 1.04 | 1.04 | .28 |
A total of 25 SNPs were considered in this analysis. They were selected on the basis of previously published genome-wide significant associations with COPD case-control status, qualitative or quantitative CT measurements of emphysema, and emphysema distribution. The analyses were adjusted for pack-years of smoking, current smoking, genetic ancestry, and sex.
OR was calculated with the upper-lobe predominant cluster as the reference group in the upper-lobe vs lower-lobe GWAS, and COPD cases were the reference group in the case-control GWAS. P values < .05 are bold.
GWAS = genome-side association study; NA = not available; SNP = single nucleotide polymorphism.
The present study.
The published GWAS of COPD case-control status.18
Discussion
In this large cohort of smokers without alpha-1-antitrypsin deficiency, random forest clustering identified tightly clustered groups of subjects with distinct craniocaudal emphysema distribution patterns but similar degrees of total emphysema, measurable airway disease, and cigarette smoking. Compared with lower lobe-predominant emphysema, subjects with mostly upper-lobe emphysema had less severe lung function impairment and lower rates of obesity and metabolic syndrome at baseline but greater progression of radiological emphysema, gas trapping, and dyspnea after 5 years of follow-up. These two clusters also demonstrated different patterns of association with established COPD genetic risk variants.
Data mining methods such as cluster analysis enable the identification of discrete groups of patients with similar disease characteristics. These techniques have been previously used in COPD, but no investigation to date has focused on emphysema distribution.25, 26, 27 In a recent study, Castaldi et al28 directly assessed the reproducibility of different clustering methods applied commonly to COPD clinical variables in 10 independent cohorts of patients with COPD. This analysis revealed that there was higher reproducibility for clustering methods that excluded individuals who did not clearly belong to any cluster. Based on these novel findings, we selected the random forests clustering approach used for this study precisely because of its ability to discriminate well-clustered from poorly clustered subjects. Thus, our primary goal for this project was not to cluster all subjects with COPD but rather to identify relatively pure subgroups of subjects characterized by upper or lower emphysema predominance. The clustering approach used here was able to identify clearly differentiated upper lobe-predominant and lower lobe-predominant emphysema subgroups that had a similar extent of total emphysema, airway disease, and cigarette smoking, enabling the further characterization of these specific subgroups. These subgroups would not have been identified with the more traditional approach of thresholding based on differences or ratios in the percentages of emphysema in upper vs lower lobes.
Our findings are consistent with prior reports of more severe airway obstruction in lower zone-predominant emphysema and are reflective of the dominant contribution of the lower lobes to the forced expired maneuver.29, 30, 31, 32 Since most of these studies included more advanced stages of COPD, our observations extend those findings to subjects with mild to moderate COPD. Investigating 115 smokers with mild to moderate COPD (58 of whom had emphysema), de Torres et al33 did not demonstrate any significant association between craniocaudal emphysema distribution and lung function parameters, likely because of the smaller sample size and the differences in the methods of assessing emphysema distribution compared with our study.
To our knowledge, this is the first report of an association between an upper-lobe emphysema subtype and faster rates of overall emphysema progression. In a cohort of 587 male smokers followed for 3 years, a faster spirometric decline was observed in upper-lobe emphysema compared with lower-lobe emphysema, but emphysema progression was not assessed.34 Our study also demonstrated an association between upper-lobe emphysema and more rapid disease progression, but our significant findings were primarily limited to emphysema progression. Although we did observe a significant univariate association demonstrating more rapid decline in FEV1 (mL/y) in the group with upper-lobe predominance, this association was no longer significant after adjustment for baseline FEV1. Bhatt et al14 showed that decline in FEV1 in mild to moderate disease is associated with airway disease and emphysema, and in our study, the upper-lobe and lower-lobe clusters had similar degrees of measurable airway disease and overall emphysema, potentially explaining the lack of significant difference in FEV1 decline between the two clusters.
Diabetes and the metabolic syndrome are frequent comorbidities in patients with COPD. These comorbidities have been shown to be more frequent in airway-predominant COPD,35, 36 and this study extends those results to subjects with lower lobe-predominant emphysema. These comorbidities are all related to systemic inflammation, which may explain the co-occurrence. It is also possible that this is a reflection of worse lower-lobe mechanics leading to more immobility, obesity, and associated cardiovascular disease.
In our prior GWAS of emphysema distribution,8 we identified five loci associated with continuous emphysema distribution measures at genome-wide significance. In this current study, we sought to supplement these findings by determining the genetic associations in distinct groups of subjects with upper lobe-predominant vs lower lobe-predominant emphysema, but we did not identify any novel genome-wide significant associations, likely because this analysis was limited to a subset of subjects from the overall cohort and therefore had less power to detect genetic associations. In addition to the top GWAS signals, we chose to also report the associations with SNPs already known to be associated with COPD or emphysema, since these SNPs are already of interest to the study of COPD and its related phenotypes. Although the number of nominally significant associations in this set of known COPD/emphysema variants suggests that genome-wide significant associations may be identified with a larger sample size, these non-genome-wide significant associations should be interpreted cautiously.
One of the interesting findings from this analysis is the observed higher prevalence of the COPD risk allele in subjects with upper-lobe predominance that suggests a potential link between upper-lobe predominance and common risk variants for COPD. As suggested by the pathway analyses that we reported in our previous publication,8 emphysema distribution shares some of the processes that occur in general COPD pathogenesis and progression, but other pathways may be specific to the pathogenesis of emphysema distribution. The precise reasons for the upper lobe predominance in smokers without alpha-1-antitrypsin deficiency and emphysema are unclear, but it has been attributed to regional differences in perfusion, transit time of leukocytes, clearance of deposited dust, mechanical stress, and pleural pressure.20, 24, 37, 38 It is also noteworthy that in this current manuscript, we analyzed common genetic variants only (ie, variants with major allele frequency > 1%) and had limited power to detect small effect sizes. We therefore cannot exclude the possibility that rare genetic variants or common variants with smaller effect sizes, or both, are involved and have differential associations with COPD susceptibility and emphysema distribution.
Our study has a number of strengths. It is the first study designed to identify emphysema distribution subtypes using data-driven methods, and it is the largest analysis to date of emphysema distribution in well-characterized smokers with a wide range of COPD disease severity and longitudinal follow-up data. In addition, the lobar segmentation approach used to generate our emphysema measures was shown to provide improved characterization of emphysema distribution compared with nonanatomic approaches.39
This study has several limitations. First, we focused exclusively on craniocaudal emphysema distribution variables. However, other measures of emphysema distribution have been described in the literature, such as core-rind and centrilobular/panlobular/paraseptal distributions40 and warrant future explorations. Second, unsupervised clustering is an exploratory analysis method, and the generalizability of these results to other cohorts cannot be assumed. Third, BMI has a strong inverse association with total emphysema14, 30, 40 and may also confound emphysema measures due to technical artifact induced by scatter effects.41, 42 However, apicobasal emphysema is effectively standardized within the same subject, making this measure more likely to be robust to technical artifacts than is total emphysema.41 In addition, most of our observed associations remained significant after adjusting for BMI.
In conclusion, this cluster analysis using lobar emphysema identifies subgroups of smokers that differ by craniocaudal emphysema distribution patterns, disease progression, and genetic associations. These findings may be of importance in optimizing participant selection in future clinical trials to ultimately improve and personalize therapy.
Acknowledgments
Author contributions: P. J. C. had full access to all the data in the study, takes responsibility for the integrity of the data and the accuracy of the data analysis, and had authority over manuscript preparation and the decision to submit the manuscript for publication.: A. B. and P. J. C. contributed to the study concept and design. All authors contributed to acquisition, analysis, or interpretation of the data. A. B. and P. J. C. contributed to drafting the manuscript. All authors contributed to the critical revision of the manuscript for important intellectual content. A. B. and P. J. C. contributed to statistical analysis. P. J. C., J. D. C., and E. K. S. obtained funding. All authors contributed to study supervision, gave final approval of the version to be published, and have agreed to be accountable for all aspects of the work.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: M. H. C. reports grants from GSK and National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute during the conduct of the study. G. R. W. reports grants from Beohringer Ingelheim, Genentech, GlaxoSmithKline, PulmonX, and Janssen Pharmaceuticals outside the submitted work. D. L. D. reports grants from NIH and personal fees from Novartis outside the submitted work. E. K. S. reports grants from NIH during the conduct of the study, personal fees from Novartis, and grant and travel support from GlaxoSmithKline outside the submitted work. P. J. C. reports grants and advisory board membership from GSK outside the submitted work. None declared (A. B., Y. C., R. S. J. E., R. P. B., J. D. D., J. G. D.).
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Collaborators: COPDGene Investigators: Core Units:Administrative Core: James Crapo, MD (principal investigator [PI]), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth Regan, MD, PhD; Genetic Analysis Core: Terri Beaty, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael H. Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Peter Castaldi, MD, Merry-Lynn McDonald, PhD, Emily Wan, MD, Megan Hardin, MD, Jacqueline Hetmanski, MS, Margaret Parker, MS, Marilyn Foreman, MD, Brian Hobbs, MD, Robert Busch, MD, Adel Boueiz, MD, Peter Castaldi, MD, Megan Hardin, MD, Dandi Qiao, PhD, Elizabeth Regan, MD, Eitan Halper-Stromberg, Ferdouse Begum, Sungho Won, and Sharon Lutz, PhD; Imaging Core: David A. Lynch, MB, Harvey O. Coxson, PhD, MeiLan K. Han, MD, Eric A. Hoffman, PhD, Stephen Humphries, MS, Francine L. Jacobson, MD, Philip F. Judy, PhD, Ella A. Kazerooni, MD, John D. Newell, Jr, MD, Elizabeth Regan, MD, James C. Ross, PhD, Raul José Estépar, PhD, Berend C. Stoel, PhD, Juerg Tschirren, PhD, Eva van Rikxoort, PhD, Bram van Ginneken, PhD, George R, Washko, MD, Carla G. Wilson, MS, Mustafa Al Qaisi, MD, Teresa Gray, Alex Kluiber, Tanya Mann, Jered Sieren, Douglas Stinson, Joyce Schroeder, MD, and Edwin Van Beek, MD, PhD; PFT QA Core, Salt Lake City, UT: Robert Jensen, PhD; Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, PhD, Anna Faino, MS, Matt Strand, PhD, and Carla Wilson, MS; Epidemiology Core, University of Colorado Anschutz Medical Campus, Aurora, CO: John E. Hokanson, MPH, PhD, Gregory Kinney, MPH, PhD, Sharon Lutz, PhD, Kendra Young, PhD, Katherine Pratte, MSPH, and Lindsey Duca, MS; COPDGene Investigators: Clinical Centers:Ann Arbor VA: Jeffrey L. Curtis, MD, Carlos H. Martinez, MD, MPH, Perry G. Pernicano, MD; Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, Philip Alapat, MD, Venkata Bandi, MD, Mustafa Atik, MD, Aladin Boriek, PhD, Kalpatha Guntupalli, MD, Elizabeth Guy, MD, Amit Parulekar, MD, and Arun Nachiappan, MD; Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, MD, MPH, Craig Hersh, MD, MPH, George R. Washko, MD, Francine Jacobson, MD, MPH; Columbia University, New York, NY: R. Graham Barr, MD, DrPH, Byron Thomashow, MD, John Austin, MD, Belinda D’Souza, MD, Gregory D.N. Pearson, MD, and Anna Rozenshtein, MD, MPH; Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr, MD, Lacey Washington, MD, and H. Page McAdams, MD; Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH and Joseph Tashjian, MD; Johns Hopkins University, Baltimore, MD: Robert Wise, MD, Nadia Hansel, MD, MPH, Robert Brown, MD, Karen Horton, MD, and Nirupama Putcha, MD, MHS; Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Torrance, CA: Richard Casaburi, PhD, MD, Alessandra Adami, PhD, Janos Porszasz, MD, PhD, Hans Fischer, MD, PhD, Matthew Budoff, MD, and Harry Rossiter, PhD; Michael E. DeBakey VA Medical Center, Houston, TX: Amir Sharafkhaneh, MD, PhD and Charlie Lan, DO; Minneapolis VA: Christine Wendt, MD and Brian Bell, MD; Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS, Gloria Westney, MD, MS, and Eugene Berkowitz, MD, PhD; National Jewish Health, Denver, CO: Russell Bowler, MD, PhD and David Lynch, MD; Reliant Medical Group, Worcester, MA: Richard Rosiello, MD and David Pace, MD; Temple University, Philadelphia, PA: Gerard Criner, MD, David Ciccolella, MD, Francis Cordova, MD, Chandra Dass, MD, Gilbert D’Alonzo, DO, Parag Desai, MD, Michael Jacobs, PharmD, Steven Kelsen, MD, PhD, Victor Kim, MD, A. James Mamary, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, Kartik Shenoy, MD, Robert M. Steiner, MD, Alex Swift, MD, Irene Swift, MD, and Maria Elena Vega-Sanchez, MD; University of Alabama, Birmingham, AL: Mark Dransfield, MD, William Bailey, MD, J. Michael Wells, MD, Surya Bhatt, MD, and Hrudaya Nath, MD; University of California, San Diego, CA: Joe Ramsdell, MD, Paul Friedman, MD, Xavier Soler, MD, PhD, and Andrew Yen, MD; University of Iowa, Iowa City, IA: Alejandro Cornellas, MD, John Newell, Jr, MD, and Brad Thompson, MD; University of Michigan, Ann Arbor, MI: MeiLan Han, MD, Ella Kazerooni, MD, and Carlos Martinez, MD; University of Minnesota, Minneapolis, MN: Joanne Billings, MD and Tadashi Allen, MD; University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD, Divay Chandra, MD, Joel Weissfeld, MD, MPH, Carl Fuhrman, MD, and Jessica Bon, MD; University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD, Sandra Adams, MD, Diego Maselli-Caceres, MD, and Mario E. Ruiz, MD.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: This work was supported by NHLBI [Grants R01HL089897, R01HL089856, R01HL124233, R01HL126596, R01HL113264, P01105339, P01HL114501, and 2T32HL07427-32]. The COPDGene study [Grant NCT00608764] is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, GlaxoSmithKline, Siemens, and Sunovion. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Contributor Information
Peter J. Castaldi, Email: repjc@channing.harvard.edu.
COPDGene Investigators:
James Crapo, Edwin Silverman, Barry Make, Elizabeth Regan, Terri Beaty, Nan Laird, Christoph Lange, Michael H. Cho, Stephanie Santorico, John Hokanson, Dawn DeMeo, Nadia Hansel, Craig Hersh, Peter Castaldi, Merry-Lynn McDonald, Emily Wan, Megan Hardin, Jacqueline Hetmanski, Margaret Parker, Marilyn Foreman, Brian Hobbs, Robert Busch, Adel Boueiz, Peter Castaldi, Megan Hardin, Dandi Qiao, Elizabeth Regan, Eitan Halper-Stromberg, Ferdouse Begum, Sungho Won, Sharon Lutz, David A. Lynch, Harvey O. Coxson, MeiLan K. Han, Eric A. Hoffman, Stephen Humphries, Francine L. Jacobson, Philip F. Judy, Ella A. Kazerooni, John D. Newell, Jr., Elizabeth Regan, James C. Ross, Raul José Estépar, Berend C. Stoel, Juerg Tschirren, Eva van Rikxoort, Bram van Ginneken, George R. Washko, Carla G. Wilson, Mustafa Al Qaisi, Teresa Gray, Alex Kluiber, Tanya Mann, Jered Sieren, Douglas Stinson, Joyce Schroeder, Edwin Van Beek, Robert Jensen, Douglas Everett, Anna Faino, Matt Strand, Carla Wilson, John E. Hokanson, Gregory Kinney, Sharon Lutz, Kendra Young, Katherine Pratte, Lindsey Duca, Jeffrey L. Curtis, Carlos H. Martinez, Perry G. Pernicano, Nicola Hanania, Philip Alapat, Venkata Bandi, Mustafa Atik, Aladin Boriek, Kalpatha Guntupalli, Elizabeth Guy, Amit Parulekar, Arun Nachiappan, Dawn DeMeo, Craig Hersh, George R. Washko, Francine Jacobson, R. Graham Barr, Byron Thomashow, John Austin, Belinda D’Souza, Gregory D.N. Pearson, Anna Rozenshtein, Neil MacIntyre, Jr., Lacey Washington, H. Page McAdams, Charlene McEvoy, Joseph Tashjian, Robert Wise, Nadia Hansel, Robert Brown, Karen Horton, Nirupama Putcha, Richard Casaburi, Alessandra Adami, Janos Porszasz, Hans Fischer, Matthew Budoff, Harry Rossiter, Amir Sharafkhaneh, Charlie Lan, Christine Wendt, Brian Bell, Marilyn Foreman, Gloria Westney, Eugene Berkowitz, Russell Bowler, David Lynch, Richard Rosiello, David Pace, Gerard Criner, David Ciccolella, Francis Cordova, Chandra Dass, Gilbert D’Alonzo, Parag Desai, Michael Jacobs, Steven Kelsen, Victor Kim, A. James Mamary, Nathaniel Marchetti, Aditi Satti, Kartik Shenoy, Robert M. Steiner, Alex Swift, Irene Swift, Maria Elena Vega-Sanchez, Mark Dransfield, William Bailey, J. Michael Wells, Surya Bhatt, Hrudaya Nath, Joe Ramsdell, Paul Friedman, Xavier Soler, Andrew Yen, Alejandro Cornellas, John Newell, Jr., Brad Thompson, MeiLan Han, Ella Kazerooni, Carlos Martinez, Joanne Billings, Tadashi Allen, Frank Sciurba, Divay Chandra, Joel Weissfeld, Carl Fuhrman, Jessica Bon, Antonio Anzueto, Sandra Adams, Diego Maselli-Caceres, and Mario E. Ruiz
Supplementary Data
References
- 1.Han M.K., Bartholmai B., Liu L.X. Clinical significance of radiologic characterizations in COPD. COPD. 2009;6(6):459–467. doi: 10.3109/15412550903341513. [DOI] [PubMed] [Google Scholar]
- 2.Venuta F., Anile M., Diso D. Long-term follow-up after bronchoscopic lung volume reduction in patients with emphysema. Eur Respir J. 2012;39(5):1084–1089. doi: 10.1183/09031936.00071311. [DOI] [PubMed] [Google Scholar]
- 3.Deslee G., Mal H., Dutau H. Lung volume reduction coil treatment vs usual care in patients with severe emphysema: The REVOLENS randomized clinical trial. JAMA. 2016;315(2):175–184. doi: 10.1001/jama.2015.17821. [DOI] [PubMed] [Google Scholar]
- 4.Sciurba F.C., Chandra D., Bon J. Bronchoscopic lung volume reduction in COPD: lessons in implementing clinically based precision medicine. JAMA. 2016;315(2):139–141. doi: 10.1001/jama.2015.17714. [DOI] [PubMed] [Google Scholar]
- 5.Fishman A., Martinez F., Naunheim K. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348(21):2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 6.Martinez F.J., Foster G., Curtis J.L. Predictors of mortality in patients with emphysema and severe airflow obstruction. Am J Respir Crit Care Med. 2006;173(12):1326–1334. doi: 10.1164/rccm.200510-1677OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manichaikul A., Hoffman E.A., Smolonska J. Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189(4):408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boueiz A., Lutz S.M., Cho M.H. Genome-wide association study of the genetic determinants of emphysema distribution. Am J Respir Crit Care Med. 2017;195(6):757–771. doi: 10.1164/rccm.201605-0997OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regan E.A., Hokanson J.E., Murphy J.R. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coxson H.O., Rogers R.M., Whittall K.P. A quantification of the lung surface area in emphysema using computed tomography. Am J Respir Crit Care Med. 1999;159(3):851–856. doi: 10.1164/ajrccm.159.3.9805067. [DOI] [PubMed] [Google Scholar]
- 11.Ross J.C., Kindlmann G.L., Okajima Y. Pulmonary lobe segmentation based on ridge surface sampling and shape model fitting. Med Phys. 2013;40(12):121903. doi: 10.1118/1.4828782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han M.K. Clinical correlations of computed tomography imaging in chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2013;10(suppl):S131–S137. doi: 10.1513/AnnalsATS.201303-046AW. [DOI] [PubMed] [Google Scholar]
- 13.Galban C.J., Han M.K., Boes J.L. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med. 2012;18(11):1711–1715. doi: 10.1038/nm.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatt S.P., Soler X., Wang X. Association between functional small airway disease and FEV1 decline in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;194(2):178–184. doi: 10.1164/rccm.201511-2219OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi T., Horvath S. Unsupervised learning with random forest predictors. J Comput Graph Stat. 2006;15:118–138. [Google Scholar]
- 16.Purcell S., Neale B., Todd-Brown K. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho M.H., McDonald M.L., Zhou X. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2(3):214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho M.H., Castaldi P.J., Hersh C.P. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192(5):559–569. doi: 10.1164/rccm.201501-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeMeo D.L., Hersh C.P., Hoffman E.A. Genetic determinants of emphysema distribution in the national emphysema treatment trial. Am J Respir Crit Care Med. 2007;176(1):42–48. doi: 10.1164/rccm.200612-1797OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilk J.B., Shrine N.R., Loehr L.R. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186(7):622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong X., Cho M.H., Anderson W. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183(1):43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castaldi P.J., Cho M.H., San Jose Estepar R. Genome-wide association identifies regulatory loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190(4):399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito I., Nagai S., Handa T. Matrix metalloproteinase-9 promoter polymorphism associated with upper lung dominant emphysema. Am J Respir Crit Care Med. 2005;172(11):1378–1382. doi: 10.1164/rccm.200506-953OC. [DOI] [PubMed] [Google Scholar]
- 25.Rennard S.I., Locantore N., Delafont B. Identification of five chronic obstructive pulmonary disease subgroups with different prognoses in the ECLIPSE cohort using cluster analysis. Ann Am Thorac Soc. 2015;12(3):303–312. doi: 10.1513/AnnalsATS.201403-125OC. [DOI] [PubMed] [Google Scholar]
- 26.Castaldi P.J., Dy J., Ross J. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69(5):415–422. doi: 10.1136/thoraxjnl-2013-203601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho M.H., Washko G.R., Hoffmann T.J. Cluster analysis in severe emphysema subjects using phenotype and genotype data: an exploratory investigation. Respir Res. 2010;11:30. doi: 10.1186/1465-9921-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castaldi PJ, Benet M, Petersen H, et al. Do COPD subtypes really exist? COPD heterogeneity and clustering in 10 independent cohorts [published online ahead of print June 21, 2017]. Thorax.https://doi.org/10.1136/thoraxjnl-2016-209846. [DOI] [PMC free article] [PubMed]
- 29.Saitoh T., Koba H., Shijubo N., Tanaka H., Sugaya F. Lobar distribution of emphysema in computed tomographic densitometric analysis. Invest Radiol. 2000;35(4):235–243. doi: 10.1097/00004424-200004000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Mair G., Miller J.J., McAllister D. Computed tomographic emphysema distribution: relationship to clinical features in a cohort of smokers. Eur Respir J. 2009;33(3):536–542. doi: 10.1183/09031936.00111808. [DOI] [PubMed] [Google Scholar]
- 31.Gurney J.W., Jones K.K., Robbins R.A. Regional distribution of emphysema: correlation of high-resolution CT with pulmonary function tests in unselected smokers. Radiology. 1992;183(2):457–463. doi: 10.1148/radiology.183.2.1561350. [DOI] [PubMed] [Google Scholar]
- 32.Haraguchi M., Shimura S., Hida W., Shirato K. Pulmonary function and regional distribution of emphysema as determined by high-resolution computed tomography. Respiration. 1998;65(2):125–129. doi: 10.1159/000029243. [DOI] [PubMed] [Google Scholar]
- 33.de Torres J.P., Bastarrika G., Zagaceta J. Emphysema presence, severity, and distribution has little impact on the clinical presentation of a cohort of patients with mild to moderate COPD. Chest. 2011;139(1):36–42. doi: 10.1378/chest.10-0984. [DOI] [PubMed] [Google Scholar]
- 34.Mohamed Hoesein F.A., van Rikxoort E., van Ginneken B. Computed tomography-quantified emphysema distribution is associated with lung function decline. Eur Respir J. 2012;40(4):844–850. doi: 10.1183/09031936.00186311. [DOI] [PubMed] [Google Scholar]
- 35.Hersh C.P., Make B.J., Lynch D.A. Non-emphysematous chronic obstructive pulmonary disease is associated with diabetes mellitus. BMC Pulm Med. 2014;14:164. doi: 10.1186/1471-2466-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han M.K., Kazerooni E.A., Lynch D.A. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: associated radiologic phenotypes. Radiology. 2011;261(1):274–282. doi: 10.1148/radiol.11110173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurney J.W. Cross-sectional physiology of the lung. Radiology. 1991;178(1):1–10. doi: 10.1148/radiology.178.1.1984285. [DOI] [PubMed] [Google Scholar]
- 38.West J.B. Distribution of mechanical stress in the lung, a possible factor in localisation of pulmonary disease. Lancet. 1971;1(7704):839–841. doi: 10.1016/s0140-6736(71)91501-7. [DOI] [PubMed] [Google Scholar]
- 39.Owsijewitsch M., Ley-Zaporozhan J., Kuhnigk J.M. Quantitative emphysema distribution in anatomic and non-anatomic lung regions. COPD. 2015;12(3):257–266. doi: 10.3109/15412555.2014.933950. [DOI] [PubMed] [Google Scholar]
- 40.Stratelis G., Fransson S.G., Schmekel B., Jakobsson P., Molstad S. High prevalence of emphysema and its association with BMI: a study of smokers with normal spirometry. Scand J Prim Health Care. 2008;26(4):241–247. doi: 10.1080/02813430802452732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman E.A., Ahmed F.S., Baumhauer H. Variation in the percent of emphysema-like lung in a healthy, nonsmoking multiethnic sample. The MESA lung study. Ann Am Thorac Soc. 2014;11(6):898–907. doi: 10.1513/AnnalsATS.201310-364OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mobberley S.D., Fuld M.K., Sieren J.P., Primak A.N., Hoffman E.A. Scatter correction associated with dedicated dual-source CT hardware improves accuracy of lung air measures. Acad Radiol. 2013;20(11):1334–1343. doi: 10.1016/j.acra.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.