Abstract
Using publicly available genomic data, combined with RT-PCR validation, we explore structural genomic variation for two major ion channels across insect classes. We have manually curated ryanodine receptor (RyR) and inositol 1,4,5-trisphosphate receptor (IP3R) ORFs and their corresponding genomic structures from 26 different insects covering major insect orders. We found that, despite high protein identity for both RyRs (>75%) and IP3Rs (~67%), the overall complexity of the gene structure varies greatly between different insect orders with the simplest genes (fewest introns) found in Diptera and the most complex in Lepidoptera. Analysis of intron conservation patterns indicated that the majority of conserved introns are found close to the 5′ end of the channels and in RyR around the highly conserved mutually exclusive splice site. Of the two channels the IP3Rs appear to have a less well conserved organisation with a greater overall number of unique introns seen between insect orders. We experimentally validated two of the manually curated ORFs for IP3Rs and confirmed an atypical (3799aa) IP3R receptor in Myzus persicae, which is approximately 1000 amino acids larger than previously reported for IP3Rs.
Abbreviations: ORF, open reading frame; RyR, ryanodine receptor; IP3R, inositol 1,4,5-trisphosphate receptor; RT-PCR, reverse transcription polymerase chain reaction; TM, trans-membrane
Keywords: Ryanodine receptor; Inositol 1,4,5-trisphosphate receptor; Insect
Highlights
-
•
Mapped the genomic structures of RyR and IP3R from 26 different insect species
-
•
Overall complexity of RyR and IP3R gene structure varies greatly between different insect orders.
-
•
Identified and experimentally validated an atypically large IP3R receptor in aphid species
1. Introduction
Recent advances in sequencing technologies have led to a rapid increase in the number of publicly available genomes, including those of insects. The insect genomes have been found to be incredibly diverse in size; the smallest genome, that of Belgica antarctica, comprising only 99 megabases (Kelley et al., 2014) whilst the ~6.5 gigabase genome of Locusta migratoria is the largest genome within the animal kingdom (Wang et al., 2014). This diversity in genome size is also reflected in gene architecture complexity; in other words, despite a high degree of similarity at the protein level, the genomic architecture of genes can vary greatly between insect orders. Recent genome annotation efforts suggest that insect ryanodine receptors (RyRs) are a classic example of this type of complexity e.g. the number of exons present within this gene can vary from 26 in the fruit fly D. melanogaster (Takeshima et al., 1994) to 55 in the red flour beetle T. castaneum (Liu et al., 2014) and 98 in aphids (e.g. Myzus persicae and Acyrthosiphon pisum) (Troczka et al., 2015a; Dale et al., 2010).
Ryanodine receptors (RyRs) and inositol 1,4,5-trisphosphate receptors (IP3Rs) are large and complex calcium release channels; whilst RyRs are primarily located in the endo (sarco) plasmic reticulum of muscle cells, IP3Rs and RyRs are also found in many other cell types (Foskett et al., 2007; Hamilton and Serysheva, 2009). Both types of channels are inherently involved in the release of Ca2+ from internal stores - however, each channel has a distinct biological function; RyRs are involved in the regulation of calcium release during excitation-contraction coupling in muscle tissues and are primarily activated either by free Ca2+ or direct interaction with Cav1.2 located in the plasma membrane (depending on channel isoform) (Fill and Copello, 2002), whereas IP3Rs are involved in complex spatio-temporal Ca2+ dynamics that have been implicated in a wide variety of biological functions from gene expression and apoptosis to learning and memory, and are primarily activated by the secondary messenger inositol 1,4,5-trisphosphate (IP3) (Foskett et al., 2007).
Partly due to the effectiveness and consequent commercial success of diamide insecticides, namely flubendiamide and chlorantraniliprole, which act as selective activators of insect RyRs (Cordova et al., 2006; Ebbinghaus-Kintscher et al., 2007), these channels have recently been receiving a considerable amount of attention from the scientific community, especially from researchers interested in understanding the mechanisms underlying the development of diamide resistance. Consequently, the number of RyR cDNAs being sequenced and cloned from a variety of agriculturally important pest species has rapidly increased (Nauen and Steinbach, 2016; Troczka et al., 2017). In comparison, IP3Rs remain significantly understudied in invertebrates. To date, only two insect IP3Rs have been cloned and experimentally tested; one from the Drosophila melanogaster (Yoshikawa et al., 1992) and the other from Tribolium castaneum (Liu et al., 2014). Functional expression of the D. melanogaster IP3R showed that its physiological properties are highly conserved in relation to its mammalian counterparts (Srikanth et al., 2004). The genomic organisation of IP3Rs however remains largely uncharacterised in insects - to date only T. castaneum IP3R has had its intron/exon organisation reported (Liu et al., 2014).
Although automated gene prediction tools are indispensable for genome annotations, these can occasionally generate incorrect gene structures (Yandell and Ence, 2012). The aim of this paper has therefore been to catalogue and validate a number of RyR and IP3R protein sequences, together with their genomic organisations, in 26 representative insect species, to help with future annotation of calcium release channels in genomic datasets and to further our understanding of the receptors' diversity in insects. Understanding the genomic structures of the insect channels could also contribute to a deeper appreciation of the evolution and regulation of these genes. IP3Rs are also candidate targets for the development of new classes of anti-insect molecules.
2. Materials and methods
2.1. In silico annotation and data mining
26 insect species from 5 insect orders with publicly available genomes (Diptera, Coleoptera, Hemiptera, Hymenoptera, Lepidoptera), 3 representatives of other insect orders (Blattellidae, Locusta, Phthiraptera) and one outlying acarine species (Tetranychus urticae) of the order Trombidiformes were included in the study (for a list of genome projects used for data generation see Appendix A). For each of the 5 main insect orders studied, a ‘reference species’ sequence was chosen to BLAST against the other available genomes within that particular order. The choice of ‘reference species’ sequence was based on a previous cloning and annotation of the sequence and the overall quality of the available genome. In the case of RyR the 5 reference species were: Dipteran = D. melanogaster, Hemipteran = Myzus persicae, Coleopteran = T. castaneum, Hymenopteran = Apis mellifera, Lepidopteran = Bombyx mori. For the IP3R, D. melanogaster and T. castaneum IP3Rs were the only sequences available to be used as a reference. PCR validation of Hymenopteran and Hemipteran IP3R genes was carried out to confirm the automated annotations. Three additional species, Blattella germanica, Locusta migratoria and Pediculus humanus corporis, were chosen for analysis based on the ‘completeness’ of their RyR genomic region, low contig fragmentation and few intra-contig gaps, determined by tblastn results against the WGS contig database with a reference RyR sequence.
Relevant contigs were downloaded from the NCBI database and manually curated using Geneious software v. 8.1.3 (Biomatters, Ltd., New Zealand). Translated exon sequences from the ‘reference species’ were blasted against databases created in Geneious using the tblastn algorithm. Megablast was used when DNA sequences were compared. Multiple sequence alignments were done using the MAFFT plugin in Geneious.
Splign (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi) (Kapustin et al., 2008) and manual curation was used to map intron-exon junctions, using as a curation guide the sequences of the most closely related reference species. Predicted RyR ORFs were extracted from the genomic sequence and validated with multiple alignments against a database of 25 manually curated insect and mammalian RyR sequences obtained from the NCBI database (see Appendix B). Gene structure graphs were generated using WebScipio (http://www.webscipio.org/) (Hatje et al., 2011). A web version of GenePainter (http://www.motorprotein.de/genepainter/genepainter) (Muhlhausen et al., 2015; Hammesfahr et al., 2013) was then used to generate intron phylogeny analysis based on the structure of the genes.
Structural features of analysed RyRs were determined based on multiple alignments of protein sequences with the previously annotated P. xylostella RyR (NCBI accession AET09964) (Troczka et al., 2017) and Pfam annotations (http://pfam.xfam.org/).
2.2. PCR validation of IP3R receptors
Total RNA from M. persicae and B. terrestris were extracted using Trizol reagent (Life technologies, CA, USA) or ISOLATE II RNA mini kit (Bioline, UK) following the manufacturer's guide. 4 μg of total RNA was used to synthesise cDNA in 20 μl reactions containing SuperScript® III Reverse Transcriptase (Thermo Fisher Scientific, USA) and Oligo dT (15) primers (Promega, USA), following the manufacturer's instructions. PCR primers were designed based on the in silico predicted sequences using Geneious software v. 8.1.3 (Biomatters, Ltd., New Zealand). PCR reactions were performed in 25 μl volumes using 2× Dreamtaq mastermix (ThermoFisher Scinetific, USA) with 10pmols of each primer and 1 μl of cDNA. Details of the PCR primers used can be found in Supplementary Tables 2 and 3. All PCR reactions were analysed on 1% (w/v) TAE agarose gels and visualised using ethidium bromide staining and UV light. PCR products were purified from the agarose gel using QIAquick gel extraction kit (Qiagen, Germany), following the manufacturer's recommended protocol, and directly sequenced using Eurofins Genomics Value Read service.
3. Results & discussion
3.1. Structural organisation of insect RyR's
RyR cDNA sequences and their corresponding genomic organisation have been previously confirmed experimentally in D. melanogaster, M. persicae, T. castaneum and B. mori (Takeshima et al., 1994; Liu et al., 2014; Troczka et al., 2015a; Xu et al., 2000; Kato et al., 2009). Here we have manually annotated the RyRs of a further 23 insect species. A summary of the predicted channels and intron/exon arrangements of all 26 insect species can be found in Table 1 and Fig. 1. Additional information can be found in Supplementary excel file (SUP Table 1).
Table 1.
Summary of annotated sequences for ryanodine receptors.
| Order | Species | Exon | Accession no. | Protein (AA) | Gene (bp) | Genome (Mb) |
|---|---|---|---|---|---|---|
| Diptera | Anopheles gambiae (African malaria mosquito) | 29 | XP_318561 | 5109 | 29,159 | 265.027 |
| Ceratitis capitata (Mediterranean fruit fly) | 31 | XP_012158404 | 5140 | 38,927 | 479.048 | |
| Drosophila melanogaster (fruit fly) | 26 | NP_001246211 | 5134 | 24,856 | 143.726 | |
| Musca domestica (house fly) | 30 | XP_011296550 | 5127 | 71,276a | 750.404 | |
| Belgica antarctica (Antartic midge) | 28 | 5088 | 21,215 | 89.5837 | ||
| Hymenoptera | Apis mellifera (honey bee) | 54 | 5107 | 30,266 | 250.287 | |
| Bombus terrestris (buff-tailed bumblebee) | 54 | XP_012175586 | 5108 | 37,861 | 248.654 | |
| Nasonia vitripennis (jewel wasp) | 56 | XP_003425568 | 5099 | 30,168 | 295.781 | |
| Megachile rotundata (alfalfa leafcutting bee) | 55 | 5109 | 30,560 | 272.661 | ||
| Harpegnathos saltator (Jerdon's jumping ant) | 54 | 5101 | 41,561 | 294.466 | ||
| Coleoptera | Tribolium castaneum (red flour beetle) | 54 | NP_001308588 | 5094 | 97,420 | 165.944 |
| Dendroctonus ponderosae (mountain pine beetle) | 70 | 5137 | 50,615 | 252.848 | ||
| Anoplophora glabripennis (Asian longhorned beetle) | 63 | 5091 | 30,477 | 707.712 | ||
| Hypothenemus hampei (coffee berry borer) | 66 | 5107 | 21,874 | 151.272 | ||
| Hemiptera | Myzus persicae (green peach aphid) | 98 | AJA41114 | 5101 | 59,601 | 347.313 |
| Cimex lectularius (bed bug) | 91 | XP_014249567 | 5093 | 62,674 | 650.478 | |
| Rhodnius prolixus (Assassin bug) | 103 | 5103 | 117,911 | 706.824 | ||
| Ferrisia virgata (striped mealybug) | 100 | 4982 | 47,978 | 304.571 | ||
| Lepidoptera | Bombyx mori (domestic silkworm) | 110 | 5121 | 137,409 | 481.819 | |
| Papilio xuthus (Asian swallowtail butterfly) | 109 | 5124 | 92,730 | 243.89 | ||
| Manduca sexta (tobacco hornworm) | 110 | 5127 | 114,469 | 419.424 | ||
| Phoebis sennaea (cloudless sulphur butterfly) | 108a | 5109 | 97,628 | 287.49 | ||
| Spodoptera frugiperda (fall armyworm) | 109 | 5127 | 131,979 | 358.048 | ||
| Acari | Tetranychus urticae (two-spotted spider mite) | 12 | XP_015783312 | 5200 | 18,117 | 90.8286 |
| Other orders | Blattella germanica (German cockroach) | 103 | 5120 | 159,200 | 2037.2 | |
| Pediculus humanus corporis (human body louse) | 78 | XP_002424547 | 5058 | 23,243 | 110.781 | |
| Locusta migratoriab (migratory locust) | 103b | 5130 | 371,221 | 5759.8 |
The ORF in Pheboes sennae, although located on a single scaffold, remains incomplete as exons 48, 102, 103 and part of exon 54 are missing. This is likely due to gaps in the published scaffold sequence.
The ORF in Locusta migratoria also remains incomplete with predicted exons 44, 68 and 78 being missing and sequencing gaps are present in exons 52, 58, 80 and 88. Due to missing sequence information the number of predicted exons for these two genes differs on manual curation compared with the webscipio/gene painter analysis.
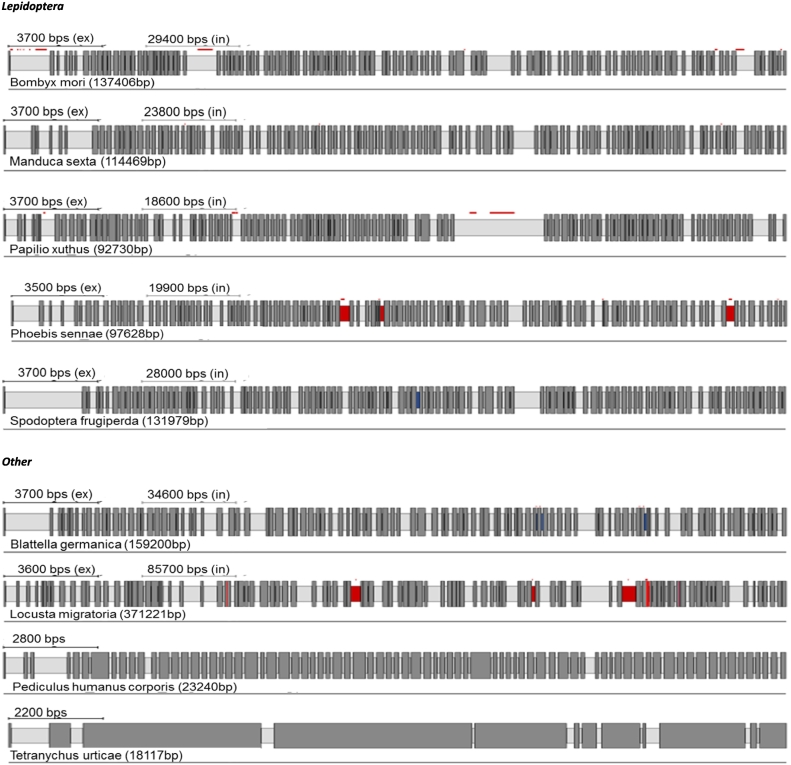
Fig. 1.
RyR gene structures generated by Webscipio. Dark grey regions correspond to exons. Red dashes indicate gaps in the sequencing, blue dashes indicate some uncertainty in intron assignment (non-canonical intron boundaries). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Protein alignments and Pfam database searches were performed to compare RyRs from different insect species. As expected, all the insect RyRs presented the classical cytosolic motifs and domains previously annotated in P. xylostella (Troczka et al., 2017) including one MIR (220–400), two SPRY domains, (726–862 and 1702–1841), four RYR domains (917–1007, 1031–1120, 3075–3166 and 3220–3303), three RIH domains (503–702, 2433–2682, 4307–4423) and EF calcium binding motifs (4531–4576). The numbering is based on a consensus sequence of a MAFFT alignment of all 27 RyR sequences with the annotated P. xylostella RyR (Fig. 2). The transmembrane (TM) region of the channel, which according to three-dimensional reconstructions using Cryo EM structures consist of six α-helixes (Yan et al., 2015; Zalk et al., 2015), was the most highly conserved region of the protein across all examined insect species.
Fig. 2.
MAFFT alignment and Pfam domain mapping of annotated RyR sequences vs P. xylostella RyR. Red arrows indicate individual transmembrane helixes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The TM region of RyR is thought to be the binding site of diamide insecticides (Kato et al., 2009). In support of that hypothesis a glycine to glutamic acid substitution at position 4946 of the transmembrane domain of Plutella xylostella RyR has been shown to be a major cause of diamide resistance in this pest insect (Troczka et al., 2015b; Troczka et al., 2012; Steinbach et al., 2015). Recently, a novel glycine to valine substitution at this position has been identified in diamide resistant populations of Tuta absoluta (Roditakis et al., 2017). Additionally, three other mutations (E1338D, Q4594L and I4790M) have been associated with diamide resistant strains of P. xylostella (Guo et al., 2014). The equivalent of I4790M, located in the second TM helix and coming to close proximity to G4946 in a 3D model of insect RyR, was also found in some resistant populations of T. absoluta (Roditakis et al., 2017). Interestingly, the glycine residue at position 4946 appears to be conserved in most insect species, apart from B. antarctica and T. utricae which have an alanine in this position and F. virgata which has an arginine. It remains to be tested, however, whether these changes have any implications on diamide binding. Additionally, the neighboring amino acids in these later three species have a lower level of conservation in comparison to other insects (SUP Fig. 1). This amino acid residue “flexibility” within a highly conserved region could be one of the reasons diamide resistance mutations have emerged relatively quickly and without any apparent fitness costs (Ribeiro et al., 2014). In contrast to G4946 the isoleucine at position 4790 appears to be unique to lepidopterans, with all other insect species analysed in this study having a methionine or valine (found only in F. virgata). Therefore, it is hypothesised that an isoleucine at position 4790 may confer selectivity for some diamides towards lepidopteran insects. Q4594 is located in divergent region one with several different amino acids being found at this position, the most common being lysine (present in Coleoptera, Hymentoptera and some Diptera), glutamine (Lepidoptera), histidine (some Hemiptera), arginine (A. gambiae) and asparagine (M. persicae). E1338 is located between the first SPRY domain and divergent region two and like Q45954 shows little amino acid conservation with various amino acids (valine, glycine, serine, glutamine, aspartic acid, glutamine) found in different species.
3.2. Structural organisation of insect IP3R's
IP3R cDNA sequences and their corresponding genomic organisation have been previously confirmed experimentally only in D. melanogaster and T. castaneum (Liu et al., 2014; Yoshikawa et al., 1992). Here we have manually annotated the IP3Rs of a further 24 insect species. A summary of the predicted channels and intron/exon arrangements of all 26 insect species can be found in Table 2 and Fig. 3.
Table 2.
Summary of annotated sequences for inositol 1,4,5-triphosphate receptors.
| Order | Species | Exon | Accession no. | Protein (AA) | Gene (bp) | Genome (Mb) |
|---|---|---|---|---|---|---|
| Diptera | Anopheles gambiae (African malaria mosquito) | 16 | XP_557157 | 2830 | 14,149 | 265.027 |
| Ceratitis capitata (Mediterranean fruit fly) | 22 | XP_012156517 | 2886 | 18,431 | 479.048 | |
| Drosophila melanogaster (fruit fly) | 19 | P29993 | 2838 | 10,830 | 143.726 | |
| Musca domestica (house fly) | 22 | 2800 | 32,699 | 750.404 | ||
| Belgica antarctica (Antartic midge) | 8 | 2694 | 8468 | 89.5837 | ||
| Hymenoptera | Apis mellifera (honey bee) | 43 | 2727 | 27,950 | 250.287 | |
| Bombus terrestris (buff-tailed bumblebee) | 43 | XP_003394052 | 2727 | 34,077 | 248.654 | |
| Nasonia vitripennis (jewel wasp) | 38 | XP_016839078 | 2745 | 26,320 | 295.781 | |
| Megachile rotundata (alfalfa leafcutting bee) | 42 | 2724 | 39,393 | 272.661 | ||
| Harpegnathos saltator (Jerdon's jumping ant) | 44 | XP_011151642 | 2741 | 37,606 | 294.466 | |
| Coleoptera | Tribolium castaneum (red flour beetle) | 27 | NP_001308600 | 2724 | 11,552 | 165.944 |
| Dendroctonus ponderosae (mountain pine beetle) | 27 | 2700 | 15,589 | 252.848 | ||
| Anoplophora glabripennis (Asian longhorned beetle) | 26 | 2706 | 12,268 | 707.712 | ||
| Hypothenemus hampei (coffee berry borer) | 26 | 2692 | 10,408 | 151.272 | ||
| Hemiptera | Myzus persicae (green peach aphid) | 49 | 3790 | 24,399 | 347.313 | |
| Cimex lectularius (bed bug) | 49 | XP_014243514 | 2735 | 26,666 | 650.478 | |
| Rhodnius prolixus (assassin bug) | 43 | 2634 | 40,691 | 706.824 | ||
| Ferrisia virgata (striped mealybug) | 45 | 2645 | 25,926 | 304.571 | ||
| Lepidoptera | Bombyx mori (domestic silkworm) | 58 | 2713 | 74,323 | 481.819 | |
| Papilio xuthus (Asian swallowtail butterfly) | 58 | 2722 | 32,751 | 243.89 | ||
| Manduca sexta (tobacco hornworm) | 58 | 2717 | 51,935 | 419.424 | ||
| Spodoptera frugiperda (fall armyworm) | 58 | 2707 | 58,102 | 358.048 | ||
| Acari | Tetranychus urticae (two-spotted spider mite) | 6 | XP_015786315 | 2754 | 9348 | 90.8286 |
| Other orders | Blattella germanica (German cockroach) | 46 | 2661 | 219,647 | 2037.2 | |
| Pediculus humanus corporis (human body louse) | 2 | XP_002425693 | 2680 | 8124 | 110.781 | |
| Locusta migratoria (migratory locust) | 46 | 2680 | 277,815 | 5759.8 |
Fig. 3.
IP3R gene structures generated by Webscipio. Dark grey regions correspond to exons. Red dashes indicate sequence gaps, blue indicate some uncertainty in intron assignment (non-canonical intron boundaries). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A Pfam search of conserved domains for the IP3Rs identified 6 such domains across all 26 annotated channels. They include the IP3 binding region (44–398), MIR domain (402–703), two RIH domains (747–987, 1844–2031), RIH associated domain (3070–3177) and the transmembrane ion transport region (3508–3940). All numbering is based on the consensus sequence of the MAFFT alignment of the 26 annotated IP3Rs (Fig. 4). The transmembrane region of IP3R, as for RyRs, is thought to consist of 6 transmembrane helixes, which has been confirmed in a 3D structure of mammalian IP3R1 elucidated by Cryo-EM (Ludtke et al., 2011). Insect IP3Rs appear to have overall higher homology than RyRs to their mammalian counterparts with approximately 60% amino acid sequence identity (Srikanth et al., 2004).
Fig. 4.
MAFFT alignment and Pfam domain map of annotated insect IP3R sequences.
Due to very little experimental information being available in the literature on cloning and sequencing of IP3R a decision was taken to PCR and sequence two of the insect IP3Rs channels, from M. persicae and Bombus terrestris, to validate our manual annotation predictions.
In silico annotation of M. persicae IP3R predicted a relatively larger channel compared to other insects. With a predicted ORF of 11,373 bp, M. persicae IP3R was projected to encode a 3790 aa protein, making it approximately 1000 aa larger than any other insect IP3R reported in this study, including those insects in the order hemiptera (e.g. Bemisia tabaci (Guo et al., 2017)). Protein alignment of the M. persicae channel with D. melanogaster IP3R indicated that the additional amino acids are scattered across the entire length of the protein (Fig. 5). Some of these insertions are present in functionally important domains, which could have a significant impact on the channels function. Interestingly the majority of insertions appear to be within the middle of the exons as opposed to 5′ or 3′ ends. Protein blast analysis of this predicted protein provided a good match up (over 90% identity) with other computationally predicted aphid IP3Rs, from pea aphid (Acyrthosiphon pisum, NCBI Acc. XP_001947344.2, encoding a 3831aa protein) and two predicted Russian wheat aphid alternatively spliced forms (Diuraphis noxia, NCBI Acc. XP_015373414.1 and XP_015373416.1, encoding proteins of 3816 and 3788 aa respectively). RT-PCR validation of the in-silico prediction for M. persicae IP3R confirmed that the receptor is much larger in this species in comparison to other insects, and that the predicted extra amino acids are present in the cDNA and are not intronic sequences. These larger IP3R channels appear to be unique to aphids. Pfam analysis of the protein sequence matches it with IP3R receptors, with conserved domains: Ins145_P3_rec (45–313), two MIR domains (345–476, 530–583), RIH (627–860) and the ion transport domain (3404–3624). The reason for a much larger IP3R in aphids is not apparent and we have found no evidence of similarly enhanced IP3Rs in other insect orders, so there is no clear evolutionary lineage. IP3R is the more ancient of the two channels studied (Alzayady et al., 2015). Evidence for larger IP3Rs-like channels (over 3000 amino acids) was reported in protozoan species such as Paramecium (Ladenburger and Plattner, 2011) and the filasterean Capsaspora owczarzaki (Alzayady et al., 2015); however these channels show very little conservation to the aphid protein. A significantly increased receptor size clearly has the potential to have a substantial impact on the channel's physiology and regulation. It has also been previously reported that aphids have an unusual heterodimeric voltage-gated cation channel, with close sequence homology with the voltage-gated sodium channel in other insects, albeit with an altered selectivity filter and being encoded by two unique heteromers (Amey et al., 2015; Zuo et al., 2016; Jiang et al., 2017). It is worth speculating that significant structural changes to at least two important ion channels could be indicative of a unique ion physiology in aphids in comparison to other insects.
Fig. 5.
Alignment of D. melanogaster IP3R with M. persicae IP3R. Black arrows indicate individual exons. The approximate location of additional amino acids found in the aphid channel are indicated in purple. There are 36 individual insertions in the aphid channel with the largest being 63 amino acids in length. IP3R functional domains are mapped on the D. melanogaster sequence. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Initial computational prediction of B. terrestris IP3R projected an ORF of 8370 bp (NCBI Acc. XP_012175773.1). However, RT-PCR validation followed by cDNA sequencing gave an ORF of 8184 bp (encoding a 2727 aa protein), which is a perfect match to another automatically predicted isoform of B. terrestris IP3Rs (NCBI Acc. XP_003394052). In comparison to the initial in silico prediction, the PCR validated sequence is missing two predicted exons (exons 23 and 28). We therefore assume that these missing exons are not part of the canonical channel, as we did not detect them in our sequenced PCR fragments. They might, however, represent rare transcripts specific to B. terrestris and other Hymenopterans, as BLAST results for both exons generate hits exclusively to species in this order.
The overall protein sequence similarity between different insect orders for the RyRs and IP3Rs was 80% and 70% respectively; the Hymenoptera was the least diverse order with over 90% identity among analysed species for both channels (93.5% for RyR and 92.29% for IP3Rs).
3.3. History of intron gain and loss within and across insect orders
There is a considerable variation in the number of introns present in RyR and IP3R genes across insect orders and other arthropods, reflected in the number of exons recorded for each species (Table 1, Table 2). However, there is generally only a small variation in the number of introns within each order, irrespectively of the genome size. For example, in Diptera, Musca domestica RyR has only two additional introns in comparison to B. antarctica despite its genome being 8.4 times larger (750 Mb and 89 Mb respectively). Substantial variation in intron number within a single order was only observed for Dipteran IP3R genes, with the flightless midge, B. antarctica, having only 7 introns compared with 21 introns for M. domestica and C. capitata. Species with the most complex IP3R belong to Lepidopterans (58 exons), followed by Hemipterans and Hymenopterans (over 40 exons) making them almost as complex as human IP3R genes with 63 exons for human type 1 and 2 isoforms (NCBI gene ID: 3708 and 3709). The RyRs of the representative species of the orders Diptera, Hymenoptera and Coleoptera showed significant changes in intron number in comparison to mammalian orthologues. For Lepidoptera, Hemiptera and Orthopteroida the total intron numbers recorded are close to the human isoforms (~105 introns) (Phillips et al., 1996).
A summary of loss and gain of introns across species is presented in the Genepainter phylogenetic ‘intron distribution’ trees for RyR and IP3Rs (Fig. 6, Fig. 7). Notably within the Paraneoptera, the entire IP3R gene of P. humanus corporis is made up of only 2 large exons. This dramatic reduction in intron number could be related to the ectoparasitic lifestyle of the species and small genome size (Sundberg and Pulkkinen, 2015). Within the Endopterygota, a significant intron loss has occurred within RyR genes in 3 of the 4 main orders (Diptera, Coleoptera, Hymenoptera, Lepidoptera), with almost 75% of ancestral introns being lost in some Dipteran species (e.g. D. melanogaster with 25 introns). Only Lepidoptera (Obtectomera) have maintained a relatively high ancestral intron number whilst simultaneously showing evidence of intron gain, making their RyR genes the most complex in all of the targeted insect species included in this study. They contain up to 110 exons on average, comparable in complexity to their mammalian counterparts (Phillips et al., 1996). The same pattern is observed for the IP3Rs genes, with the most complex architecture being found in Lepidoptera (58 exons), whilst the Diptera display the greatest overall reduction in intron number (25 introns) of all the insect orders studied (with the notable exception of P. humanus corporis as discussed above).
Fig. 6.
Phylogenetic distribution of intron positions for insect RyRs generated by GenePainter. The phylogenetic tree indicates those taxon branches at which introns were lost (coloured red) or gained (coloured green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Phylogenetic distribution of intron positions for insect IP3R's generated by GenePainter. The phylogenetic tree indicates those taxon branches at which introns were lost (coloured red) or gained (coloured green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite an overall high level of conservation at the protein (amino acid) level for both RyR and IP3R channels in insects, an extensive remodelling of the genomic structure has resulted in a low conservation of common introns across insect orders. GenePainter analysis showed only 8 common introns conserved within RyR across the insect species looked at in this study. This number falls to only 3 when T. urticae (Acari) is included in the analysis. Interestingly, 4 of the common (conserved) introns are located within the first 10 introns, with a further 2 encompassing a highly-conserved exon (exon 20 in D. melanogaster, 39 in A. mellifera, and B. mori, 71 in M. persicae). Looking at the overall distribution of intron positions, it appears that the greatest number of intron losses occur towards the 3′ end of the ORF (see SUP Table 1), possibly indicating the involvement of reverse transcriptase in this evolutionary process (Cohen et al., 2012). The conservation of intron-exon organisation does not appear to be linked with any particular structural features. All of the conserved intron-exon junctions occur within the cytosolic portion of the protein. The highest level of conservation is found in the introns surrounding a mutually exclusive splice site in the second SPRY domain, with M. persicae being the outlier. The second highest conserved pair of introns is found around the predicted calmodulin/Apo-calmodulin interaction site, which also appears to be alternatively spliced in some species. In contrast, there are 34 unique introns detected across all species (Fig. 8). There are no conserved introns for IP3Rs and only 2 conserved introns if T. urticae and P. humanus corporis are excluded. However, there are 56 unique introns found across the 26-species studied (Fig. 9). In comparison to the RyRs, the IP3R gene structure is less well conserved with a much higher proportion of unique introns in relation to the total intron number.
Fig. 8.
Gene structure alignment for insect RyRs obtained from Genepainter. Each dash corresponds to an intron overlaid on a protein alignment. To improve clarity the size of each exon is not representative of the actual exon size. Red dashes indicate unique introns and their number in individual species. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Gene structure alignment for IP3Rs obtained from Genepainter. Each dash corresponds to an intron. To improve clarity the size of each exon is not representative of the actual exon size. Red dashes indicate unique introns and their number in individual species. In total there are 56 unique introns mapped on the alignment. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Alternative splicing of RyR and IP3R
Fully annotated gene structures can elucidate further useful information such as the regulatory mechanisms governing gene expression and the probability of alternative splicing (Chorev and Carmel, 2012). Understanding splicing regulation is a difficult challenge as the spliceosome is one of the most complicated molecular complexes, consisting of over 150 proteins (Wahl et al., 2009). Point mutations in the genomic structure may lead to modulation of the splicing machinery resulting in exon skipping (Cartegni et al., 2002). Such an event in the nicotinic acetylcholine receptor has already been linked with resistance to the insecticide spinosad in Tuta absoluta (Berger et al., 2016). Intron size and number have also been shown to directly correspond to splicing diversity (Chorev and Carmel, 2012; Fox-Walsh et al., 2005).
RyRs are known to possess an inherently complex genomic organisation and several alternative spliced isoforms have been reported in insects (Xu et al., 2000; X. Wang et al., 2012; Puente et al., 2000). A common (mutually exclusive) alternative splice site has been found in 21 of the 26 studied species, the exceptions being P. humanus, T. urticae, B. antarctia, C. lectularius and the previously reported M. persicae (Troczka et al., 2015a). This site is located within the second SPRY domain in the N-terminal part of the channel (Fig. 10). SPRY domains are found in many mammalian proteins and are thought to be linked with immune responses (D'Cruz et al., 2013). In mammalian RyR1, the second SPRY domain is a site of interaction with the II-III loops of Cav1.1 and also with scorpion toxin A (Tae et al., 2009). Although insect RyRs are thought not to be directly linked with Cav1.1 channels (Takekura and Franzini-Armstrong, 2002), alternative exons in this region might play important roles in modulating the interaction of insect RyRs with other regulatory proteins. To date alternative exons have been best described in Lepidopteran species (Wang et al., 2013; J. Wang et al., 2012; Sun et al., 2015; Wu et al., 2013; Cui et al., 2013). As highlighted in Fig. 8, exon version A is present in all species lacking an alternative exon. Thus, version A is likely to be the most common splice form. In M. persicae this exon is fused with a neighboring exon indicating a clear intron loss event (Troczka et al., 2015a).
Fig. 10.
Multiple alignment of a mutually exclusive splice site found in 21 of the analysed insect RyR sequences.
Apart from mutually exclusive exons, there is growing evidence, especially in Lepidoptera, of a number of deletions and insertions which result in a greater diversity of detectable splice forms in comparison to other insect species, as shown by the extensive splice forms detected in P. xylostella (X. Wang et al., 2012).
We did not attempt to map mutually exclusive exons in insect IP3Rs due to the scarcity of experimentally obtained mRNA sequences. However, due to this paucity of validated cDNA's the existence of such exons cannot be dismissed. Alternative splicing of IP3Rs is well documented in mammals (Foskett et al., 2007) and at least one of these splice sites appears to be conserved in D. melanogaster (Sorrentino et al., 2000).
4. Conclusions
Despite an ever-growing number of insect genomes becoming publicly available the quality of many of them remains problematic. Short contig lengths and a high degree of fragmentation make it challenging to fully annotate large genes such as RyRs and IP3Rs (Mackrill, 2012). Additionally, automatic annotations can omit certain sections of the gene (usually the 1st exon) if it is positioned a long distance from the rest of the gene and no reference transcriptome data is available. Manual curation of important genes and gene families is required to verify and improve genomic data. In the case of insects, validation of automatically annotated genes has not been done for many species. Certain insect orders remain over represented in the wet biology due to their importance to agriculture or disease control. Our study shows that despite a relatively high protein homology, the gene architecture of proteins can differ substantially among different insect orders. We have shown that there is a substantial variation in exon number and overall gene size for both RyR and IP3R channels and very little intron conservation across different species. Structural variation of genes coding for highly conserved proteins is likely to contribute to a great diversity in potential mRNAs among insects allowing for the existence of species and order-specific splice variants.
The following are the supplementary data related to this article.
Summary of exon assignments.
Supplementary Tables 1,2,3, Supplementary Figure 1
Conflicts of interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Acknowledgments
Dr. T.G. Emyr Davies and Dr. Rafael A. Homem receive grant-aided support from the Biotechnology and Biological Sciences Research Council's Industrial Strategy Challenge Fund (BBS/OS/CP/000001). Ewan Richardson is in receipt of an Industrial studentship jointly funded by the BBSRC and Bayer CropScience (BB/N504075/1).
Contributor Information
Bartlomiej J. Troczka, Email: bartek.troczka@rothamsted.ac.uk.
Ewan Richardson, Email: ewan.richardson@rothamsted.ac.uk.
Rafael A. Homem, Email: rafael.homem@rothamsted.ac.uk.
T.G. Emyr Davies, Email: emyr.davies@rothamsted.ac.uk.
Appendix A. List of genome projects and Accessions used for data generation with associated peer reviewed publications (where appropriate)
Tetranychus urticae (BioProjects: PRJNA315122, PRJEA71041) (Grbic et al., 2011), Drosophila melanogaster (chromosome 2R ref. seq: NT_033778.4) (Hoskins et al., 2007), Anopheles gambiae (chromosome 3L ref. seq: NT_078267.5) (Holt et al., 2002), Ceratitis capitata (BioProjects: PRJNA201381, PRJNA168120) (Papanicolaou et al., 2016), Belgica antarctica (BioProject: PRJNA172148) (Kelley et al., 2014), Musca domestica (BioProject: PRJNA210139) (Scott et al., 2014), Megachile rotundata (BioProjects: PRJNA87021, PRJNA66515), Apis mellifera (Linkage group: NC_007071.3, BioProject: PRJNA10625) (Honeybee Genome Sequencing Consortium, 2006), Bombus terrestris (BioProjects: PRJNA68545, PRJNA45869) (Sadd et al., 2015), Nasonia vitripennis (BioProjects: PRJNA20073, PRJNA13660) (Werren et al., 2010), Harpegnathos saltator (BioProjects: PRJNA273397, PRJNA50203) (Bonasio et al., 2010), Tribolium castaneum (BioProjects: PRJNA15718, PRJNA12540) (Tribolium Genome Sequencing Consortium, 2008), Dendroctonus ponderosae (BioProject: PRJNA162621) (Keeling et al., 2013), Anoplophora glabripennis (BioProjects: PRJNA348318, PRJNA167479), Hypothenemus hampei (BioProject: PRJNA279497) (Vega et al., 2015), Myzus persicae (BioProjects: PRJNA397782, PRJNA319804), Cimex lectularius (BioProject: PRJNA167477) (Rosenfeld et al., 2016), Rhodnius prolixus (BioProject: PRJNA13648) (Mesquita et al., 2015), Ferrisia virgata (BioProject: PRJEB12067), Blattella germanica (BioProject: PRJNA203136), Locusta migratoria (BioProject: PRJNA185471) (Wang et al., 2014), Pediculus humanus corporis (BioProjects: PRJNA19807, PRJNA16223) (Kirkness et al., 2010), Bombyx mori (BioProject: PRJDA20217) (International Silkworm Genome Consortium, 2008), Papilio xuthus (BioProject: PRJDB2956), Manduca sexta (BioProject: PRJNA81037) (Kanost et al., 2016), Spodoptera frugiperda (BioProject: PRJNA257248) (Kakumani et al., 2014), Phoebis sennae (BioProject: PRJNA308118) (Cong et al., 2016).
Appendix B. Ryanodine receptor sequences (and associated NCBI Accession numbers) used for the validation alignments
Aphis citricidus (AKM95171.1), Atta colombica (KYM79740.1), Bemisia tabaci (AFK84957.1), Cnaphalocrocis medinalis (AFI80904.1), Cerapachys biroi (EZA52107.1), Carposina sasakii (AHN16453.1), Chilo suppressalis (AFN70719.1), Daphnia pulex (EFX89429.1), Dialeurodes citri (AKM95170.1), Helicoverpa armigera (AHB33498.1), Homo sapiens RyR1 (AAC51191.1), Homo sapiens RyR2 (CAA66975.1), Homo sapiens RyR3 (CAA04798.1), Laodelphax striatella (AFK84959.1), Leptinotarsa decemlineata (AHW99830), Lucilia cuprina (KNC23059.1), Melipona quadrifasciata (KOX73585.1), Nilaparvata lugens (KF306296.1), Ostrinia furnacalis (AGH68757.1), Pieris rapae (AGI62938.1), Plutella xylostella (AET09964.1), Sogatella furcifera (AIA23859), Spodoptera exigua (AFC36359.1), Trachymyrmex cornetzi (KYN11971.1), Tuta absoluta (APC65631.1).
References
- Alzayady K.J. Tracing the evolutionary history of inositol, 1, 4, 5-trisphosphate receptor: insights from analyses of Capsaspora owczarzaki Ca2+ release channel orthologs. Mol. Biol. Evol. 2015;32(9):2236–2253. doi: 10.1093/molbev/msv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amey J.S. An evolutionarily-unique heterodimeric voltage-gated cation channel found in aphids. FEBS Lett. 2015;589(5):598–607. doi: 10.1016/j.febslet.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. Insecticide resistance mediated by an exon skipping event. Mol. Ecol. 2016;25(22):5692–5704. doi: 10.1111/mec.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science. 2010;329(5995):1068–1071. doi: 10.1126/science.1192428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3(4):285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chorev M., Carmel L. The function of introns. Front. Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N.E., Shen R., Carmel L. The role of reverse transcriptase in intron gain and loss mechanisms. Mol. Biol. Evol. 2012;29(1):179–186. doi: 10.1093/molbev/msr192. [DOI] [PubMed] [Google Scholar]
- Cong Q. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol. Evol. 2016;8(3):915–931. doi: 10.1093/gbe/evw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova D. Anthranilic diamides: a new class of insecticides with a novel mode of action, ryanodine receptor activation. Pestic. Biochem. Physiol. 2006;84(3):196–214. [Google Scholar]
- Cui L. Molecular cloning, characterization and expression profiling of a ryanodine receptor gene in Asian corn borer, Ostrinia furnacalis (Guenee) PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0075825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R.P. Identification of ion channel genes in the Acyrthosiphon pisum genome. Insect Mol. Biol. 2010;19:141–153. doi: 10.1111/j.1365-2583.2009.00975.x. [DOI] [PubMed] [Google Scholar]
- D'Cruz A.A. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 2013;22(1):1–10. doi: 10.1002/pro.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbinghaus-Kintscher U. Flubendiamide, the first insecticide with a novel mode of action on insect ryanodine receptors. Pflanzenschutz-Nachrichten Bayer. 2007;60(2):117–140. [Google Scholar]
- Fill M., Copello J.A. Ryanodine receptor calcium release channels. Physiol. Rev. 2002;82(4):893–922. doi: 10.1152/physrev.00013.2002. [DOI] [PubMed] [Google Scholar]
- Foskett J.K. Inositol trisphosphate receptor Ca2+ release channels. Physiol. Rev. 2007;87(2):593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh K.L. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. U. S. A. 2005;102(45):16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic M. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479(7374):487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. Novel mutations and mutation combinations of ryanodine receptor in a chlorantraniliprole resistant population of Plutella xylostella (L.) Sci. Rep. 2014;4:6924. doi: 10.1038/srep06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. Silence of inositol 1,4,5-trisphosphate receptor expression decreases cyantraniliprole susceptibility in Bemisia tabaci. Pestic. Biochem. Physiol. 2017;142:162–169. doi: 10.1016/j.pestbp.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Hamilton S.L., Serysheva I.I. Ryanodine receptor structure: progress and challenges. J. Biol. Chem. 2009;284(7):4047–4051. doi: 10.1074/jbc.R800054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammesfahr B. GenePainter: a fast tool for aligning gene structures of eukaryotic protein families, visualizing the alignments and mapping gene structures onto protein structures. BMC Bioinf. 2013;14:77. doi: 10.1186/1471-2105-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatje K. Cross-species protein sequence and gene structure prediction with fine-tuned Webscipio 2.0 and Scipio. BMC. Res. Notes. 2011;4:265. doi: 10.1186/1756-0500-4-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R.A. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis melliferaNature. 2006;443(7114):931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R.A. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316(5831):1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008;38(12):1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Jiang X.Z. Characterization of an insect heterodimeric voltage-gated sodium channel with unique alternative splicing mode. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017;203:149–158. doi: 10.1016/j.cbpb.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Kakumani P.K. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics. 2014;104(2):134–143. doi: 10.1016/j.ygeno.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Kanost M.R. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem. Mol. Biol. 2016;76:118–147. doi: 10.1016/j.ibmb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustin Y. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol. Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K. Molecular characterization of flubendiamide sensitivity in the Lepidopterous ryanodine receptor Ca2+ release channel. Biochemistry. 2009;48(43):10342–10352. doi: 10.1021/bi900866s. [DOI] [PubMed] [Google Scholar]
- Keeling C.I. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14(3):R27. doi: 10.1186/gb-2013-14-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J.L. Compact genome of the Antarctic midge is likely an adaptation to an extreme environment. Nat. Commun. 2014;5:4611. doi: 10.1038/ncomms5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness E.F. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. U. S. A. 2010;107(27):12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladenburger E.M., Plattner H. Calcium-release channels in paramecium. Genomic expansion, differential positioning and partial transcriptional elimination. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.P. Comparative characterization of two intracellular Ca2+-release channels from the red flour beetle, Tribolium castaneum. Sci. Rep. 2014;4 doi: 10.1038/srep06702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J. Flexible architecture of IP3R1 by Cryo-EM. Structure. 2011;19(8):1192–1199. doi: 10.1016/j.str.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackrill J.J. Ryanodine receptor calcium release channels: an evolutionary perspective. Adv. Exp. Med. Biol. 2012;740:159–182. doi: 10.1007/978-94-007-2888-2_7. [DOI] [PubMed] [Google Scholar]
- Mesquita R.D. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl. Acad. Sci. U. S. A. 2015;112(48):14936–14941. doi: 10.1073/pnas.1506226112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlhausen S., Hellkamp M., Kollmar M. GenePainter v. 2.0 resolves the taxonomic distribution of intron positions. Bioinformatics. 2015;31(8):1302–1304. doi: 10.1093/bioinformatics/btu798. [DOI] [PubMed] [Google Scholar]
- Nauen R., Steinbach D. In: Resistance to diamide insecticides in Lepidopteran pests, in Advances in Insect Control and Resistance Management. Horowitz A.R., Ishaaya I., editors. Springer International Publishing; Cham: 2016. pp. 219–240. [Google Scholar]
- Papanicolaou A. The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species. Genome Biol. 2016;17(1):192. doi: 10.1186/s13059-016-1049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.S. The structural organization of the human skeletal muscle ryanodine receptor (RYR1) gene. Genomics. 1996;34(1):24–41. doi: 10.1006/geno.1996.0238. [DOI] [PubMed] [Google Scholar]
- Puente E. Identification of a polymorphic ryanodine receptor gene from Heliothis virescens (Lepidoptera: Noctuidae) Insect Biochem. Mol. Biol. 2000;30(4):335–347. doi: 10.1016/s0965-1748(00)00009-6. [DOI] [PubMed] [Google Scholar]
- Ribeiro L.M.S. Fitness costs associated with field-evolved resistance to chlorantraniliprole in Plutella xylostella (Lepidoptera: Plutellidae) Bull. Entomol. Res. 2014;104(1):88–96. doi: 10.1017/S0007485313000576. [DOI] [PubMed] [Google Scholar]
- Roditakis E. Ryanodine receptor point mutations confer diamide insecticide resistance in tomato leafminer, Tuta absoluta (Lepidoptera: Gelechiidae) Insect Biochem. Mol. Biol. 2017;80:11–20. doi: 10.1016/j.ibmb.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J.A. Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius. Nat. Commun. 2016;7 doi: 10.1038/ncomms10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd B.M. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.G. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014;15(10):466. doi: 10.1186/s13059-014-0466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V., Barone V., Rossi D. Intracellular Ca(2+) release channels in evolution. Curr. Opin. Genet. Dev. 2000;10(6):662–667. doi: 10.1016/s0959-437x(00)00139-8. [DOI] [PubMed] [Google Scholar]
- Srikanth S. Functional properties of the Drosophila melanogaster inositol 1,4,5-trisphosphate receptor mutants. Biophys. J. 2004;86(6):3634–3646. doi: 10.1529/biophysj.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach D. Geographic spread, genetics and functional characteristics of ryanodine receptor based target-site resistance to diamide insecticides in diamondback moth, Plutella xylostella. Insect Biochem. Mol. Biol. 2015;63:14–22. doi: 10.1016/j.ibmb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Sun L.N. Molecular characterization of a ryanodine receptor gene from Spodoptera exigua and its upregulation by chlorantraniliprole. Pestic. Biochem. Physiol. 2015;123:56–63. doi: 10.1016/j.pestbp.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Sundberg L.R., Pulkkinen K. Genome size evolution in macroparasites. Int. J. Parasitol. 2015;45(5):285–288. doi: 10.1016/j.ijpara.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Tae H., Casarotto M.G., Dulhunty A.F. Ubiquitous SPRY domains and their role in the skeletal type ryanodine receptor. Eur. Biophys. J. 2009;39(1):51–59. doi: 10.1007/s00249-009-0455-8. [DOI] [PubMed] [Google Scholar]
- Takekura H., Franzini-Armstrong C. The structure of Ca2+ release units in arthropod body muscle indicates an indirect mechanism for excitation-contraction coupling. Biophys. J. 2002;83(5):2742–2753. doi: 10.1016/S0006-3495(02)75284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H. Isolation and characterization of a gene for a ryanodine receptor/calcium release channel in Drosophila melanogaster. FEBS Lett. 1994;337(1):81–87. doi: 10.1016/0014-5793(94)80634-9. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Troczka B. Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect Biochem. Mol. Biol. 2012;42(11):873–880. doi: 10.1016/j.ibmb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Troczka B.J. Molecular cloning, characterisation and mRNA expression of the ryanodine receptor from the peach-potato aphid, Myzus persicae. Gene. 2015;556(2):106–112. doi: 10.1016/j.gene.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troczka B.J. Stable expression and functional characterisation of the diamondback moth ryanodine receptor G4946E variant conferring resistance to diamide insecticides. Sci. Rep. 2015;5 doi: 10.1038/srep14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troczka B.J. Rapid selection for resistance to diamide insecticides in Plutella xylostella via specific amino acid polymorphisms in the ryanodine receptor. Neurotoxicology. 2017;60:224–233. doi: 10.1016/j.neuro.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F.E. Draft genome of the most devastating insect pest of coffee worldwide: the coffee berry borer, Hypothenemus hampei. Sci. Rep. 2015;5 doi: 10.1038/srep12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang X. Molecular cloning, characterization and mRNA expression of a ryanodine receptor gene from diamondback moth, Plutella xylostella. Pestic. Biochem. Physiol. 2012;102(3):204–212. [Google Scholar]
- Wang J. Molecular characterization of a ryanodine receptor gene in the rice leaffolder, Cnaphalocrocis medinalis (Guenee) PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Molecular cloning and mRNA expression of a ryanodine receptor gene in the cotton bollworm, Helicoverpa armigera. Pestic. Biochem. Physiol. 2013;107:327–333. doi: 10.1016/j.pestbp.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Wang X.H. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014;5:1–9. doi: 10.1038/ncomms3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J.H. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327(5963):343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Molecular and cellular analyses of a ryanodine receptor from hemocytes of Pieris rapae. Dev. Comp. Immunol. 2013;41(1) doi: 10.1016/j.dci.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Xu X.H. Molecular cloning of cDNA encoding a Drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium release channel. Biophys. J. 2000;78(3):1270–1281. doi: 10.1016/S0006-3495(00)76683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517(7532):50. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M., Ence D. A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012;13(5):329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S. Molecular cloning and characterization of the inositol 1,4,5-trisphosphate receptor in Drosophila melanogaster. J. Biol. Chem. 1992;267(23):16613–16619. [PubMed] [Google Scholar]
- Zalk R. Structure of a mammalian ryanodine receptor. Nature. 2015;517(7532):44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y. Expression patterns, mutation detection and RNA interference of Rhopalosiphum padi voltage-gated sodium channel genes. Sci. Rep. 2016;6:30166. doi: 10.1038/srep30166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of exon assignments.
Supplementary Tables 1,2,3, Supplementary Figure 1