Abstract
Saccharomyces cerevisiae Cin8p belongs to the BimC family of kinesin-related motor proteins that are essential for spindle assembly. Cin8p levels were found to oscillate in the cell cycle due in part to a high rate of degradation imposed from the end of mitosis through the G1 phase. Cin8p degradation required the anaphase-promoting complex ubiquitin ligase and its late mitosis regulator Cdh1p but not the early mitosis regulator Cdc20p. Cin8p lacks a functional destruction box sequence that is found in the majority of anaphase-promoting complex substrates. We carried out an extensive mutagenesis study to define the cis-acting sequence required for Cin8p degradation in vivo. The C terminus of Cin8p contains two elements required for its degradation: 1) a bipartite destruction sequence composed of a KEN-box plus essential residues within the downstream 22 amino acids and 2) a nuclear localization signal. The bipartite destruction sequence appears in other BimC kinesins as well. Expression of nondegradable Cin8p showed very mild phenotypic effects, with an increase in the fraction of mitotic cells with broken spindles.
INTRODUCTION
Eukaryotic chromosome segregation is accomplished by the microtubule-based mitotic spindle, a dynamic structure. During mitosis, spindle microtubules undergo dramatic changes in structural organization. Microtubule-based motor proteins orchestrate many of these changes. A major question in the mitosis field concerns the mechanisms by which the activities of the numerous spindle motors are regulated.
The actions of five kinesin-related proteins and a dynein contribute to spindle structure and function in the budding yeast Saccharomyces cerevisiae (Hildebrandt and Hoyt, 2000). Two of the kinesins, Cin8p and Kip1p, belong to the conserved BimC subfamily and perform an overlapping and essential function (Hoyt et al., 1992; Roof et al., 1992; Saunders and Hoyt, 1992). Studies of numerous eukaryotic species have demonstrated that the BimC motors are required for spindle assembly (Enos and Morris, 1990; Hagan and Yanagida, 1990; Hoyt et al., 1992; Roof et al., 1992; Sawin et al., 1992; Heck et al., 1993; Blangy et al., 1995). They accomplish this by forming bipolar tetramers with two motor domains at opposite ends of an extended coiled-coil rod (Cole et al., 1994; Gordon and Roof, 1999). The ability of BimC motors to cross-link microtubules from each half spindle presumably is utilized to construct bipolar spindles (Kashina et al., 1996; Sharp et al., 1999). In addition to their role in spindle assembly, Cin8p and Kip1p produce the bulk of spindle elongating activity during anaphase (Saunders et al., 1995). This is probably accomplished by the plus end-directed sliding of cross-linked antiparallel microtubules. BimC motors in other species are believed to perform a similar spindle elongation function (Sharp et al., 2000). Phenotypic analysis has revealed that Cin8p is the most crucial of the S. cerevisiae mitotic motors. Indeed, under the appropriate conditions, Cin8p can uniquely support the viability of cells deficient for most of the activities of the five other spindle motors (Cottingham et al., 1999).
The conserved and essential nature of the spindle role of BimC motors has spurred interest in mechanisms that regulate their activity. Previous studies have implicated both transcriptional and posttranslational regulatory mechanisms operating on BimC motors. Phosphorylation of Eg5, the vertebrate BimC family member, by cdc2 (also known as cyclin-dependent kinase 1) and Aurora kinases has been observed (Blangy et al., 1995; Sawin and Mitchison, 1995; Giet et al., 1999). cdc2 phosphorylation is believed to alter Eg5 localization in a cell cycle-specific manner. Some BimC motor mRNA and protein levels have been reported to fluctuate in a cell cycle-dependent manner (Hagan and Yanagida, 1992; Giet et al., 1999; Uzbekov et al., 1999). The transcript levels of CIN8 and KIP1 fluctuate, peaking in S phase (Cho et al., 1998; Spellman et al., 1998). Our finding that elevated levels of Cin8p interfere with spindle assembly and function suggested the requirement for tight regulation of Cin8p activity (Saunders et al., 1997; Hildebrandt and Hoyt, unpublished observation). In this paper, we report that Cin8p protein levels are also regulated by a cell cycle-specific degradative mechanism.
Targeted degradation is a precise and rapid mechanism for promoting cell cycle transitions. Many mitotic proteins are eliminated by the covalent attachment of ubiquitin and subsequent degradation by the proteosome (King et al., 1996a; Zachariae and Nasmyth, 1999). The mitotic targets for proteolysis are selected by a specific ubiquitin ligase, the anaphase-promoting complex or cyclosome (here referred to as APC). Directed by its association with two homologous regulators, the APC acts at two temporally distinct steps in the cell cycle. In association with Cdc20p, the APC targets the degradation of securin proteins (i.e., S. cerevisiae Pds1p), whose elimination is required to initiate anaphase (Cohen-Fix et al., 1996). Exit from mitosis and passage into G1 of the next cycle is facilitated by the APC in association with Cdh1p (also known as Hct1p), a Cdc20p homologue. Known APCCdh1 targets include B-type mitotic cyclins and the spindle midzone protein Ase1p (Juang et al., 1997; Schwab et al., 1997; Visintin et al., 1997). Interestingly, the chromatin-associated kinesin Xkid is an APC substrate, and its destruction is critical for proper chromosome segregation in anaphase (Antonio et al., 2000; Funabiki and Murray, 2000). This suggests that ubiquitin-mediated proteolysis may be an important mechanism for regulating mitotic kinesins.
Substrates recognized by the APC contain a destruction signal sequence, frequently but not exclusively located near the N termini of target proteins. Two types of these destruction sequences have been found: the destruction box (D-box), with a consensus of RxxLxxxxN (where “x” is any amino acid, Glotzer et al., 1991; King et al., 1996b), and the KEN-box with a consensus of KENxxxN/D (Pfleger and Kirschner, 2000). D-box–containing substrates typically appear to be recognized by either APCCdc20 or APCCdh1. In the case of certain B-type cyclins and S. cerevisiae Hsl1p, it has been reported that the D-box is recognized by both APC forms (Schwab et al., 1997; Baumer et al., 2000a; Burton and Solomon, 2000; Pfleger and Kirschner, 2000; Yeong et al., 2000). The KEN-box is believed to be recognized exclusively by APCCdh1. The KEN-box was recently defined by in vitro ubiquitination and proteolysis studies (Pfleger and Kirschner, 2000). Its role in vivo has been demonstrated for human Cdc6, securin, and Drosophila cyclin A, where it partially contributes to the instability of these proteins (Petersen et al., 2000; Zur and Brandeis, 2001; Jacobs et al., 2001).
In this study, we demonstrate that Cin8p protein levels are regulated by the APC and its regulator Cdh1p. No functional D-box was found in Cin8p, but its degradation was dependent on two C-terminal sequence elements: a nuclear localization signal (NLS) and an APC-targeting sequence that contains a KEN-box plus essential amino acids located up to 22 positions downstream. Based on the phenotype of cells expressing a stable form of Cin8p, we propose that Cin8p destruction in late mitosis might be needed to prevent defects in spindle formation in the next cell cycle.
MATERIALS AND METHODS
Yeast Strains and Media
Rich (YPD) and synthetic (S) media were as described by Sherman et al. (1983). α-Factor (US Biologicals, Swampscott, MA) was added to 4 μg/ml in pH 4.0 medium except for bar1Δ strains for which 0.4 μg/ml α-factor was added and the medium pH was not adjusted. bar1Δ strains are hypersensitive to the actions of α-factor (Chan and Otte, 1982). Hydroxyurea (Sigma, St. Louis, MO) was added to 0.1 M in pH 5.8 medium. Nocodazole (Sigma) in dimethyl sulfoxide (DMSO) was added to 10 μg/ml. Strains and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study
| Genotypea | |
|---|---|
| Strains | |
| K1534* | MATa ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3 ssd1 bar1∷hisG (Irniger and Nasmyth, 1997) |
| K4438* | K1534 cdc16-123 (Irniger and Nasmyth, 1997) |
| MAY2063 | MATa ade2-101 lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cin8∷URA3 |
| MAY2422 | MATa lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cyh2R |
| MAY4751 | MATa ade2-101 his3-Δ200 leu2-3, 112 ura3-52 cin8∷CIN8-3HA cdc15-2 |
| MAY4822 | MATa lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cin8∷CIN8-3HA |
| MAY4823 | MATa ade2-101 lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cin8∷CIN8-3HA |
| MAY5017 | MATa lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cdc20-1 cin8∷CIN8-3HA |
| MAY5353* | K1534 cdh1∷KANR |
| MAY5402* | MATa ade2-1 trp1-1 leu2-3, 112 his3 ura3 bar1∷hisG cdc20-1 |
| MAY5466 | MAY4822 cdh1∷KANR |
| MAY5590 | MATa ade2-101 lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cyh2 cin8∷HIS3 kip1∷HIS3 pTK97 |
| MAY6752 | MATα lys2-801 his3-Δ200 leu2-3, 112 ura3-52 cyh2 cin8∷HIS3 dyn1-Δ3∷HIS3 pEH24 |
| MAY6810* | K1534 clb2∷URA3 |
| MAY6812* | K1534 clb2∷URA3 cdh1∷KANR |
| Plasmids | |
| pEH24 | CIN8 URA3 CYH2 CEN |
| pEH55 | PGAL1 > CIN8-3HA LEU2 CEN |
| pEH70 | PGAL1 > 6myc-CIN8 LEU2 CEN |
| pEH87 | PGAL1 > ΔN70-CIN8-3HA LEU2 CEN |
| pEH113 | CIN8 LEU2 CEN |
| pEH117 | 6myc-CIN8-1031 LEU2 2μ |
| pEH136 | PGALS > CIN8 LEU2 CEN |
| pEH157 | 6myc-SV40NLS-CIN8-1031 LEU2 2μ |
| pEH170 | PGAL1 > 6myc-SV40NLS-CIN8 LEU2 CEN |
| pEH287 | PGALS > CIN8 HIS3 CEN |
| pEH289 | PGAL1 > GAL4BD-cin8 LEU2 CEN |
| pEH290 | PGAL1 > GAL4BD-cin8-Δ996-1004 LEU2 CEN |
| pEH291 | PGAL1 > GAL4BD-cin8-alaKEN LEU2 CEN |
| pEH292 | PGAL1 > GAL4BD LEU2 CEN |
| pTK97 | KIP1 URA3 CEN |
| pTK138 | 6myc-CIN8 LEU2 2μ |
Strains marked with an asterisk are derivatives of W303. All other strains are derivatives of S288C.
Standard techniques were utilized for DNA manipulations (Sambrook et al., 1989). CLB2 and CDH1 disruptions were made by one-step gene replacement (Rothstein, 1983) and deletions were confirmed by polymerase chain reaction (PCR). The clb2::URA3 allele consists of a URA3-containing restriction fragment replacing nucleotides 361-1161 of the CLB2 open reading frame (ORF). The cdh1::kanR disruption consists of a kanR-containing restriction fragment replacing nucleotides −80 in the CDH1 promoter to 1326 in the CDH1 ORF. The dyn1-Δ3::HIS3 disruption consists of a restriction fragment containing HIS3 replacing nucleotides 912 to the end of the DYN1 ORF. The cin8::URA3, cin8::HIS3, and kip1::HIS3 disruptions were described previously (Hoyt et al., 1992; Roof et al., 1992; Saunders et al., 1995).
CIN8–3HA encodes three tandem copies of the hemagglutinin (HA) tag fused in-frame to Cin8p at the C terminus. CIN8–3HA was able to fully complement a number of cin8Δ mutant phenotypes. cin8::CIN8–3HA was created by replacing the genomic copy of CIN8 with CIN8–3HA by transplacement (Scherer and Davis, 1979). 6myc-CIN8 contains six tandem copies of the myc epitope tag inserted after the third CIN8 codon. 6myc-SV40NLS constructs had the SV40NLS inserted in-frame directly after the 6myc tag at amino acids 96–113.
The following CIN8 C-terminal truncation alleles were constructed by restriction digests with the indicated sites within CIN8 (and often a downstream site), followed by fill-in reactions and religation: CIN8–1031, XhoI(3090); CIN8–1013, SphI(3039); cin8–955, EcoRV(2863); cin8–871, EcoRI(2611). Other CIN8 alleles were generated with the use of mutagenic oligonucleotides and either the USE mutagenesis (Nicolas et al., 1997) or megaprimer mutagenesis (Sarkar and Sommer, 1990) protocols. Most mutagenic primers were engineered with diagnostic restriction site changes. All mutations were confirmed by sequencing. ΔN70-CIN8 was constructed by deleting between the MscI site and a ClaI site engineered at nucleotide 215 in CIN8. The resulting gene product is missing amino acids 4–73.
The CIN8 random and site-directed mutant alleles were subcloned into the base set of CIN8 plasmids as follows (also see Table 1): pEH113 was used for testing Cin8p function in vivo; pEH70 and pEH170 were used for pulse-chase degradation assays on C-terminal mutants; pEH287 was used for moderate overexpression studies.
GAL1 promoter (PGAL1)-driven GAL4BD-CIN8 fusions were generated in two steps. First, a 227-bp EcoRV to XhoI fragment from CIN8 was cloned into SmaI/SalI-cut pGBD-C1 (James et al., 1996). The resulting plasmid was cut with HindIII, and the fragment with GAL4BD-CIN8 was cloned into HindIII of p415-GAL1 (Mumberg et al., 1994).
Mutagenic PCR and Screen for Stable CIN8 Mutants
A 3′ region of the CIN8 ORF, between EcoRV and SphI (176 bp), was PCR amplified under mutagenic conditions (Cadwell and Joyce, 1995). The products were used to replace the wild-type EcoRV to SphI fragment of pEH287. The ligation reaction was transformed into Escherichia coli, 1150 colonies were pooled, and DNA was isolated. Yeast strain MAY2422 was transformed with the mutant DNA pool, and colonies were selected on SD-histidine medium. A total of 6300 individual colonies were replica plated to 3% glycerol/2% lactate-containing minimal medium, grown for 2 d, and then replica plated to 2% galactose-containing minimal medium. The 216 colonies that failed to grow on the galactose medium were identified; 40 of these mutants were retested after plasmid isolation and retransformation. These 40 mutants were sequenced.
In Vivo Studies
To collect small G1 cells from asynchronous cultures, we used Ficoll gradients (Sloat and Pringle, 1978; Sullivan and Huffaker, 1992). Briefly, cultures were grown in YPD medium to log phase. Cells were harvested, resuspended in 2% Ficoll, and layered onto the top of a gradient with steps of 4, 6, 8, and 10% Ficoll. Gradients were centrifuged for 25 min at 120 × g. Fractions were collected from the top of the gradient, and fractions with >90% unbudded cells as judged by microscopy were pooled.
Cin8p expression-pulse assay: To examine the stability of Cin8p in arrested G1 cells, cultures were grown to mid-log phase in S-medium with 2% raffinose at 30°C. α-Factor was added for 3 h followed by the addition of galactose to induce Cin8p expression. Induction times depended on the epitope tag and our ability to detect it by Western blot (i.e., 3HA- and 6myc-tagged Cin8p were induced for 30 min; 6myc-SV40NLS-Cin8p derivatives were induced for 90 min; Gal4BD-Cin8p fusions were induced for 360 min). Cycloheximide (1 mg/ml) and 2% glucose were then added (at t = 0 min.) to terminate gene expression, and samples were collected at times thereafter.
To test the in vivo function of CIN8 alleles, a plasmid derived from pEH113 but harboring the CIN8 mutant of interest was transformed into both cin8Δ kip1Δ and cin8Δ dyn1Δ strains (MAY5590 and MAY6752, respectively). These normally inviable genotypes are supported by a plasmid-borne copy of CIN8. The plasmid also carries URA3. Therefore, these strains can survive only on 5-fluoroorotic acid-containing media if transformed with another active allele of CIN8. CIN8 mutant allele transformants were tested on 5-fluoroorotic acid at 26 and 37°C. Alleles that supported viability at both temperatures were considered fully proficient for the essential functions of CIN8.
Yeast Extracts and Protein Analysis
Yeast protein extracts were prepared one of two ways. Most extracts of cells were made by mechanical lysis with glass beads as previously described (Farr and Hoyt, 1998) in 50 mM Tris-HCl, pH 8, 500 mM NaCl, 5 mM EDTA, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride. For Gal4BD-Cin8p fusions it was necessary to use alkaline-mediated hydrolysis and trichloroacetic acid precipitation (Fujimura-Kamada et al., 1997) to see the Gal4BD epitope by Western blot. Protein samples were separated by SDS-PAGE, and the proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA) with the use of standard techniques (Sambrook et al., 1989). Equal protein loading was confirmed by India ink staining of the membrane. The HA epitope was detected with 12CA-5 (Boehringer Mannheim, Indianapolis, IN), the myc epitope was detected with 9E10 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and Gal4BD-Cin8p fusions were detected with anti-Gal4 (Zymed, S. San Francisco, CA). Anti-Clb2 was a generous gift of D. Kellogg. Alkaline phosphatase-conjugated secondary antibodies were from Jackson ImmunoResearch Labs (West Grove, PA). Reagents for chemiluminescence were obtained from Tropix (Bedford, MA).
Microscopy
Indirect immunofluorescence microscopy of 6myc-tagged Cin8p was performed in MAY5590 transformed with the appropriate plasmid. Cells were arrested with preanaphase spindles by growth in minimal glucose medium with 0.1 M hydroxyurea. Cells were fixed for 7 min at 42°C and prepared for immunofluorescence as described before (Pringle et al., 1991; Cottingham and Hoyt, 1997). Myc was stained with 9E10 antibody and a Cy2-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Labs). Tubulin was stained with YOL 1/34 (Harlan Bioproducts for Science, Indianapolis, IN) and rhodamine-conjugated goat anti-rat (Jackson ImmunoResearch Labs) or Alexa Fluor 488-conjugated goat anti-rat (Molecular Probes, Eugene, OR).
RESULTS
Cin8p Levels Fluctuate in the Cell Cycle in Part due to G1-specific Instability
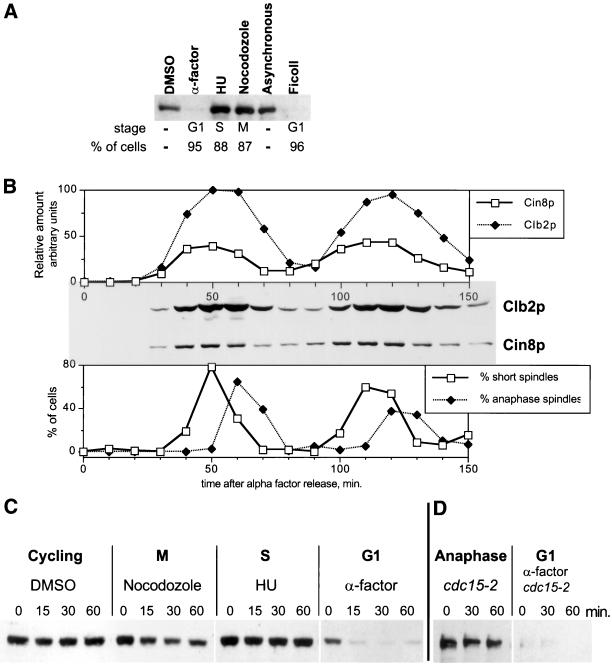
Because Cin8p has a major role in governing mitotic spindle dynamics, we first examined whether its levels fluctuate in the cell cycle. To assay Cin8p levels, we used a 3HA epitope-tagged version of CIN8 integrated at its normal genomic locus. Cell cycle inhibitors were used to synchronize cells at various cell cycle stages. Cin8p appeared equally abundant in cells arrested in S- and M-phase and by hydroxyurea and nocodazole treatment, respectively, but it was nearly undetectable in G1 cells arrested with α-factor (Figure 1A). To ensure that the disappearance of Cin8p in G1 cells was not a specific effect of α-factor treatment (i.e., specific for cells preparing to mate), we collected small unbudded G1 cells from an untreated asynchronous culture with the use of a Ficoll gradient (Sloat and Pringle, 1978; Sullivan and Huffaker, 1992). Whereas Cin8p was abundant in the total asynchronous culture, it was absent in the G1 cell fraction.
Figure 1.
Cin8p oscillates and is unstable in G1. (A) Cin8p is absent from G1 cells. Yeast cells (MAY4823) expressing genomic CIN8-3HA were treated with α-factor, hydroxyurea (HU), nocodazole, and DMSO only for 3.7 h. Asynchronous cells were applied to a Ficoll gradient for collection of small G1 cells. The percentage of cells with the morphology characteristic of the respective cell cycle arrest is shown below the blot. (B) Cin8p levels fluctuate in the cell cycle. Yeast cells expressing genomic CIN8-3HA (MAY4822) were synchronized with α-factor in G1. Cells were washed and released into medium without α-factor. Samples were collected every 10 min and analyzed for Cin8p and Clb2p levels by Western blot (middle) and for the presence of bipolar spindles by tubulin immunofluorescence (bottom). The top panel shows quantification of Cin8p and Clb2p levels. (C) Cin8p is unstable in G1. Strain K1534 carrying a PGAL1 > CIN8-3HA plasmid was grown in medium with 2% raffinose. Cells were treated with α-factor, hydroxyurea, nocodazole, or DMSO for 3 h. Cin8p was then induced by incubation with 2% galactose, followed by addition of cycloheximide and glucose (t = 0 minutes). Samples were collected at the indicated times (minutes), and protein extracts were prepared. Equal amounts of protein were separated by gel electrophoresis and analyzed for Cin8p levels by Western blotting. (D) Cin8p is stable in late anaphase. A cdc15-2 (MAY4751) strain carrying a PGAL1 > 6myc-SV40NLS-CIN8 plasmid was grown in synthetic medium plus raffinose at 22°C. Part of the culture was shifted to 37°C for 4.5 h to arrest in late anaphase. Another portion was treated with α-factor at 22°C for 4 h and then shifted to 37°C for 30 min to inactivate Cdc15-2p. Cin8p was induced with galactose for 90 min, followed by addition of cycloheximide and glucose (t = 0). In the α-factor–treated sample nearly all of the Cin8p was degraded by the time the first time sample was processed.
We also examined the cell cycle profile of Cin8p expression in a synchronized culture, beginning with release from α-factor arrest in G1 (Figure 1B). Cin8p levels peaked during spindle assembly. The level stayed high as cells entered anaphase and then declined as bipolar spindles disappeared. Cin8p reaccumulated in the next round of spindle assembly and anaphase. The cycling profile of Cin8p levels closely paralleled the profile for the B-type mitotic cyclin Clb2p.
Two types of regulation could be contributing to Cin8p's cell cycle oscillation: transcription and degradation. It has been demonstrated that CIN8 transcription is cell cycle regulated and peaks in S-phase (Cho et al., 1998; Spellman et al., 1998). We investigated whether Cin8p stability varies in a cell cycle-dependent manner by examining the fate of Cin8p heterologously expressed in cells arrested in various cell cycle stages (Figure 1C). A pulse of Cin8p was induced from the regulatable, high-level PGAL1 for 30 min in the presence of galactose. Expression was then terminated by the addition of glucose and cycloheximide (t = 0 min) to prevent further transcription and translation. Samples were collected at times thereafter, and Cin8p levels were determined by Western blot analysis. Cin8p was unstable in G1-arrested cells, disappearing within 15 min of the pulse. In contrast, Cin8p was stable in S-phase– and M-phase–arrested cells. We also found with the use of the temperature-sensitive cdc15-2 mutant that Cin8p was stable in cells arrested in late anaphase. However, we found Cin8p was very unstable in the same cdc15-2 cells if they were first arrested in G1 with α-factor and then shifted to 37°C (Figure 1D). This finding indicates that Cdc15p activity is not directly required for Cin8p degradation. The stabilization of Cin8p observed in cdc15-2–arrested cells, therefore, is due to the late mitosis block imposed by this mutant. We conclude that Cin8p becomes unstable during the period between late anaphase and S-phase of the next cell cycle.
Cin8p Instability in G1 Requires APCCdh1
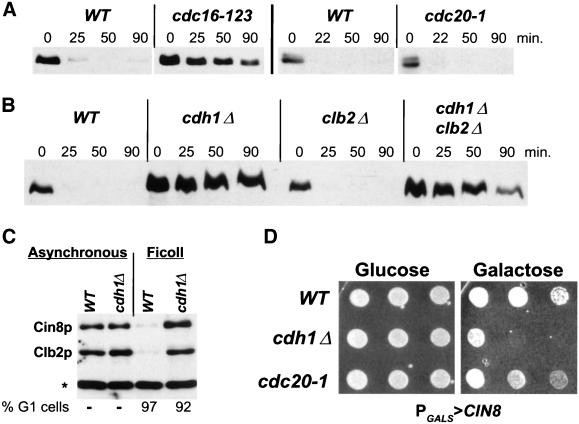
The instability of Cin8p in G1 cells is typical of proteins degraded via the APC. The requirement for the APC in Cin8p degradation was examined with the use of the cdc16-123 mutant that is temperature sensitive for function of an essential APC subunit (King et al., 1995; Tugendreich et al., 1995). Cells were arrested in G1 with α-factor and shifted to the nonpermissive temperature, and then the galactose-induced pulse of Cin8p was examined as described above. Cin8p was stable in the cdc16-123 strain at 37°C in contrast to the isogenic wild-type strain (Figure 2A) or a cdc16-123 strain carrying a CDC16 plasmid (Hildebrandt and Hoyt, unpublished results). In a similar type of experiment, we found that Cin8p is stable in α-factor–arrested cdh1Δ cells (Figure 2B). These findings indicate that Cin8p degradation requires the Cdc16p subunit of the APC and the APC regulator Cdh1p.
Figure 2.
Cin8p degradation requires the APC subunit Cdc16p and the regulator Cdh1p but not Cdc20p. (A) Cin8p's instability in G1 requires CDC16, not CDC20. Wild-type (K1534, WT), cdc16-123 (K4438), and cdc20-1 (MAY5402) strains with PGAL1 > CIN8-3HA were examined by the G1 expression-pulse protocol described in Figure 1C except that the temperature was increased to 37°C beginning 30 min before the Cin8p pulse. (B) Cin8p is stabilized in G1-arrested cdh1Δ cells. Wild-type (K1534), cdh1Δ (MAY5353), clb2Δ (MAY6810), and cdh1Δ clb2Δ (MAY6812) strains with PGAL1 > CIN8-3HA were examined by the G1 expression-pulse protocol described in Figure 1C. (C) Cin8p is absent from G1 cells isolated from an asynchronous culture. Unbudded wild-type (MAY4822) and cdh1Δ (MAY5466) cells with genomic CIN8-3HA were isolated from a Ficoll gradient and analyzed for Cin8p and Clb2p levels. The asterisk indicates a background band recognized by the Clb2 antibody. (D) Moderate overexpression of CIN8 kills cdh1Δ but not cdc20-1 cells. Wild-type (MAY4822), cdh1Δ (MAY5466), and cdc20-1 (MAY5017) with PGALS > CIN8 were spotted in serial dilution (left to right) onto solid synthetic medium containing either glucose (CIN8 expression off) or galactose (CIN8 expression on) and grown at 26°C.
It was reported previously that cdh1Δ cells cannot maintain an α-factor–mediated G1 arrest because of the accumulation of B-type cyclins that promote DNA synthesis (Amon et al., 1994; Irniger and Nasmyth, 1997; Schwab et al., 1997). In our experiments >90% of cdh1Δ cells treated with α-factor arrested with the unbudded morphology characteristic of G1 cells, but ∼50% of the cells accumulated a 2n DNA content as judged by flow cytometric analysis (Hildebrandt and Hoyt, unpublished results). Therefore, it was possible that the stability of Cin8p observed in cdh1Δ cells is due to their leakage past the α-factor arrest into S-phase. However, two observations demonstrate that Cin8p degradation is indeed Cdh1p dependent. First, small unbudded G1 cells collected from asynchronous cdh1Δ cultures showed very high levels of Cin8p and Clb2p compared with wild-type G1 cells, in which Cin8p is barely detectable (Figure 2C). Second, as previously reported, we found that the deletion of CLB2 from cdh1Δ cells reduced B-type cyclin levels sufficiently to allow α-factor to maintain a G1 arrest (Schwab et al., 1997; and confirmed by Hildebrandt and Hoyt, unpublished results). As observed for the cdh1Δ single mutant, Cin8p was stable in the α-factor–arrested cdh1Δ clb2Δ double mutant (Figure 2B). The deletion of CLB2 alone had no effect on Cin8p degradation.
We next investigated whether the APC regulator Cdc20p also affects Cin8p stability. Clb2p has been found to be partially degraded immediately after anaphase initiation via APCCdc20, whereas the remainder of its degradation occurs late in anaphase via APCCdh1 (Baumer et al., 2000a; Yeong et al., 2000). The Cin8p degradation assay was performed in a cdc20-1 temperature-sensitive strain arrested in α-factor. Cin8p was observed to be as unstable in the cdc20-1 strain as in the wild-type control (Figure 2A), suggesting that Cdc20p does not affect Cin8p stability.
Cdc20p is also subject to APCCdh1 degradation in G1 cells (Prinz et al., 1998; Shirayama et al., 1998; Goh et al., 2000; Pfleger and Kirschner, 2000). We were concerned that in our degradation assay with the use of G1 cells Cdc20p was not present to activate the APC. Therefore, this assay may not be a fair test for Cdc20p's possible role in Cin8p degradation. To further address this possibility we utilized a second in vivo assay that does not depend on G1 arrest. Cin8p overexpression (i.e., from PGAL1) is toxic and kills cells (Saunders et al., 1997). However, the amount expressed from a moderate promoter, such as PGALS or PMET25 (Mumberg et al., 1994), does not kill (Figure 2D). We speculated that moderate overexpression of Cin8p may kill cells if it cannot be degraded. Indeed, we found that expression of Cin8p from PGALS kills cdh1Δ cells when plated on galactose-containing medium on which the promoter is turned on but not on glucose medium on which the promoter is repressed (Figure 2D). In contrast, wild-type and cdc20-1 cells were able to tolerate moderate overexpression of Cin8p. Similar effects were found with the use of PMET25-expressed Cin8p (Hildebrandt and Hoyt, unpublished results). Therefore, both assays indicate that it is unlikely that Cin8p degradation is dependent on the APCCdc20 pathway (see DISCUSSION).
Cin8p Degradation Does Not Require a Canonical d-Box but Does Require Nuclear Localization and C-Terminal Sequences
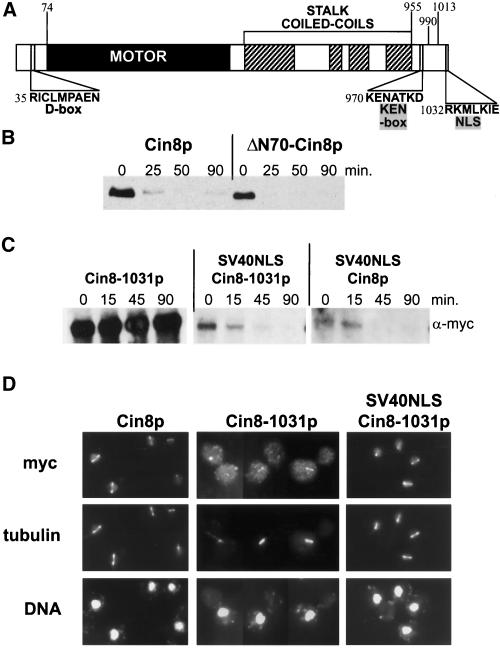
The APC targets the degradation of substrates containing at least one of two destruction signal sequences, the d-box and the KEN-box (see INTRODUCTION). Cin8p has sequences that resemble both elements: a potential d-box (RICLMPAEN), starting at position 35 in its N terminus, and a potential KEN-box (KENATKD), starting at position 970 in the C terminus (Figure 3A). The N-terminal 70 amino acids of Cin8p, upstream of the conserved motor domain, are not required for Cin8p function in vivo (Hildebrandt and Hoyt, unpublished observation). We examined the stability of Cin8p missing the first 70 amino acids (ΔN70-Cin8p). This truncated Cin8p was still degraded with kinetics similar to wild-type (Figure 3B). Although the sequence beginning at amino acid 35 is the only perfect match to the d-box consensus, related sequences can be found beginning at amino acids 196, 292, 621, 751, and 1032 (matching the most highly conserved part of the d-box, RxxL/F; King et al., 1996b). We altered each of these sites by either point mutation or deletion (example given below). In addition, a number of these were combined, creating double and triple mutants. None of these mutant Cin8p forms displayed altered stability (Hildebrandt and Hoyt, unpublished results). In contrast, single amino acid changes in a non–D-box sequence were able to greatly protect Cin8p from degradation (see below). Therefore, we conclude that the d-box-like sequences in Cin8p are not targeting it for degradation.
Figure 3.
Cin8p degradation is d-box independent but requires localization to the nucleus. (A) Schematic of Cin8p domains drawn to scale. The potential d-box and KEN-box sequences are indicated as well as the region required for Cin8p nuclear localization. (B) Deletion of the d-box does not stabilize Cin8p. Wild-type (K1534) cells with plasmids expressing full-length Cin8p (PGAL1 > CIN8-3HA) or Cin8p with the d-box deleted (PGAL1 > ΔN70-CIN8-3HA) were subjected to the G1 expression-pulse protocol described in Figure 1C. (C) Cin8p degradation requires a functional NLS. A wild-type strain (K1534) carrying plasmids expressing Cin8p derivatives with Cin8p's C-terminal NLS and/or an N-terminal SV40NLS (PGAL1 > 6myc-CIN8-1031; PGAL1 > 6myc-SV40NLS-CIN8-1031; PGAL1 > 6myc-SV40NLS-CIN8, from left to right) were subjected to the G1 expression-pulse protocol described in Figure 1C. (D) Demonstration that the SV40NLS restores nuclear localization to CIN8-1031. A cin8Δ kip1Δ strain (MAY5590) carrying 6myc-tagged versions of CIN8 on high-copy plasmids (6myc-CIN8; 6myc-CIN8-1031; 6myc-SV40NLS-CIN8-1031, from left to right) were arrested at the short spindle stage with hydroxyurea and stained for Cin8p (myc), microtubules (tubulin), and DNA. Although most of Cin8-1031p is cytoplasmic, a portion still binds spindles.
To assess the role of the C-terminal KEN-box (amino acids 970–976) we used an N-terminal 6myc-tagged Cin8p and made a series of C-terminal truncations. We found that deletion of as few as the seven C-terminal amino acids resulted in dramatic stabilization of Cin8p (Cin8-1031p, Figure 3C). In this case, the missing sequence (RKMLKIE) partially resembles the d-box consensus. However, mutation of the critical d-box residues, R1032 and L1035 to alanine, had no effect on degradation (Hildebrandt and Hoyt, unpublished results). CIN8-1031 was able to complement cin8Δ. Although its protein product partially localized to spindle microtubules, most of it was cytoplasmic (Figure 3D). This is in contrast to the primarily nuclear microtubule localization of the wild type. We could restore localization to the nucleus and mitotic spindle by fusing an SV40NLS to the N terminus of Cin8-1031p, immediately after the 6myc tag (SV40NLS-Cin8-1031p, Figure 3D). Therefore, we have identified a sequence in Cin8p that partially contributes to nuclear localization. This sequence was not identified as an NLS by motif-searching computer programs (Nakai and Horton, 1999). The addition of the 6myc-SV40NLS tag to Cin8p missing its own NLS also restored Cin8p degradation (Figure 3C). We conclude that Cin8p must be properly localized to the nucleus to be targeted for degradation by the APC.
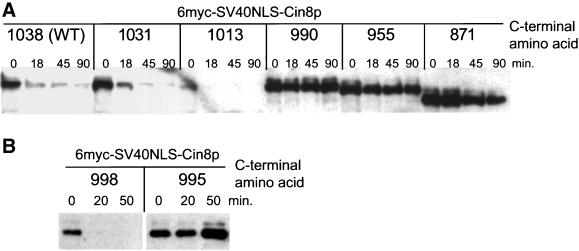
To examine the role of the KEN-box without disturbing Cin8p localization, we made a series of C-terminal truncations in a version of Cin8p expressed from PGAL1 and containing the 6myc-SV40NLS tag at the N terminus. Surprisingly, a truncation to amino acid 990, which leaves the KEN-box (970–976) intact, was highly stabilized. In marked contrast, a longer form of Cin8p, ending at amino acid 1013, was still degraded with wild-type kinetics (Figure 4A). More extensively truncated forms that removed the KEN-box (955, 871) were stabilized to a similar extent as Cin8-990p. Therefore, if the KEN-box is involved in Cin8p degradation, it is not sufficient to act as the targeting signal. Additional information required for degradation must lie between amino acids 990 and 1013. In a further dissection of this region, we found a truncation to amino acid 998 did not affect Cin8p degradation (Figures 4B and 6C). Therefore, there are no residues essential for Cin8p degradation after amino acid 998. In contrast, removal of three additional residues, to amino acid 995, completely stabilized Cin8p. This indicates that critical residues for Cin8p's degradation lie between amino acids 995 and 998.
Figure 4.
Cin8p's degradation signal is in the C terminus. Wild-type (K1534, WT) cells expressing versions of PGAL1 > 6myc-SV40NLS-CIN8, truncated at the indicated amino acid, were subjected to the G1 expression-pulse protocol described in Figure 1C.
Figure 6.
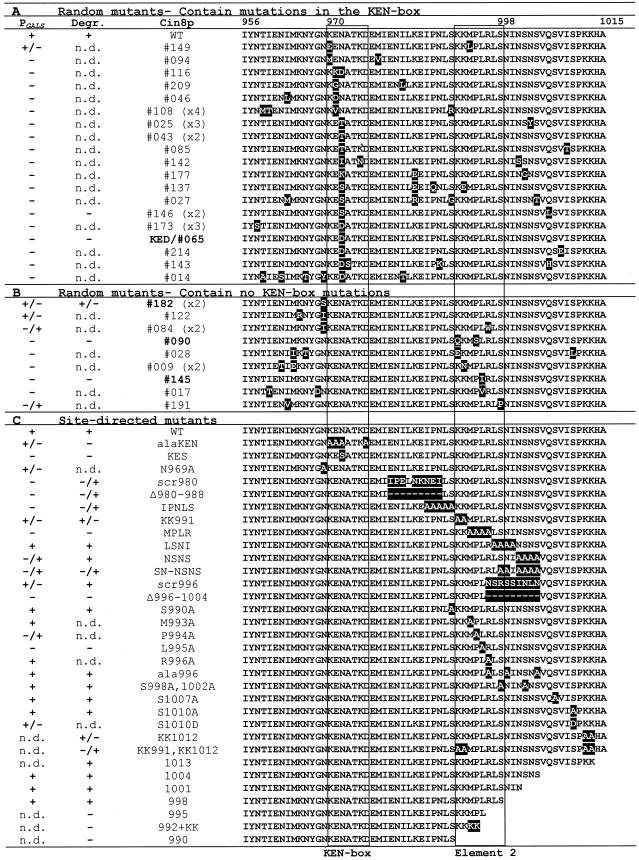
Summary of mutants in the region of KEN-box and element 2. Summary of findings from the two assays (PGALS and degradation assay) to test the stability of random and site-directed Cin8p mutants. A “+” under PGALS indicates wild-type levels of growth on galactose-containing media, and a “−” indicates death on galactose. A “+” in the degradation column (Degr.) indicates a wild-type (WT) level of instability, and a “−” indicates that the mutant was stable. “n.d.” means the mutant was not tested. Numbers given in parentheses (e.g., x2) in the Cin8p column indicate the number of times a particular random mutant was isolated. Amino acid positions are indicated at the top of the right column. The KEN-box and element 2 are boxed.
The Degradation Signal in Cin8p Is Bipartite and Includes a KEN-Box Plus Downstream Amino Acids
To define amino acids required to specify degradation, we generated random and site-directed mutants in the C terminus of Cin8p. Both types of mutants were subjected to two assays. The first, a colony growth assay, was based on the assumption that a form of Cin8p that is stable because it is missing a cis-acting degradation signal will be toxic when moderately overexpressed (from PGALS), just as moderate overexpression of wild-type Cin8p was toxic to cdh1Δ cells (Figure 2D). The second test was the G1 expression-pulse analysis described above, utilizing mutant forms of 6myc-SV40NLS-Cin8p expressed from PGAL1 (“Degradation assay”). Examples are shown in Figure 5, and all findings are summarized in Figure 6. For the majority of mutants, the two assays resulted in similar conclusions.
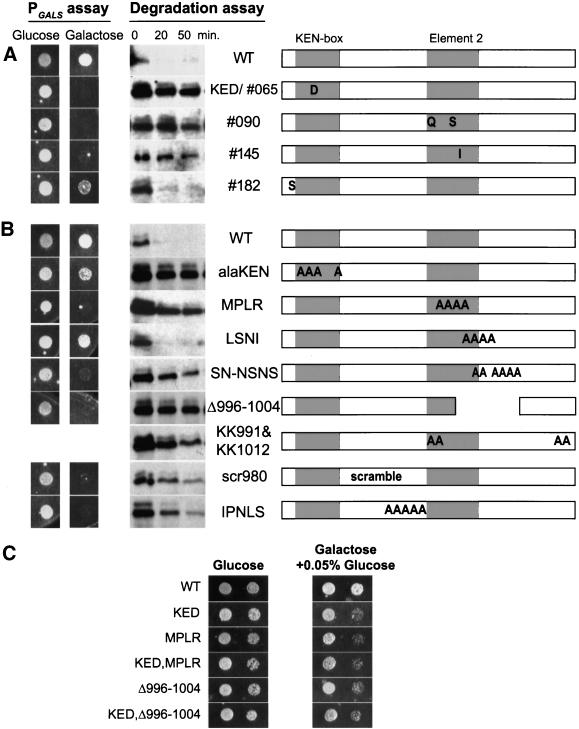
Figure 5.
Moderate overexpression and stability of random and site-directed Cin8p mutants. Select random (A) and site-directed (B) Cin8p mutants were tested for toxicity when moderately overexpressed from PGALS (PGALS assay) and for Cin8p stability in vivo (degradation assay). PGALS assay: Wild-type (MAY2422, WT) cells carrying mutant derivatives of PGALS > CIN8 were spotted onto solid synthetic medium with either glucose or galactose (as in Figure 2D except a single dilution spot is shown). Degradation assay: PGAL1 > 6myc-SV40NLS-CIN8 derivatives of the mutants were tested for stability in strain K1534 by the G1 expression-pulse protocol described in Figure 1C. The figures on the right depict the positions of mutant sites in the amino acid 956-1015 region of Cin8p. The boundaries of element 2 are defined in Figure 8A. Precise sequence changes are described in Figure 6. (C) Wild-type (MAY2422) cells carrying mutant derivatives of PGALS > CIN8 were spotted (as in Figure 2D except two dilution spots are shown) onto solid synthetic medium with either glucose or galactose plus 0.05% glucose as carbon sources.
To generate random mutants we applied mutagenic PCR to the DNA encoding amino acids 956-1015 of Cin8p. A library was created that expressed the Cin8p random mutants from the moderate-level GALS promoter. Expressed from PGALS, wild-type Cin8p does not kill wild-type cells. The library was screened for Cin8p mutants that killed on galactose-containing media. Twenty-eight unique mutants that passed this test were identified. Several mutants were isolated two or more times and most of the mutants contained more than one amino acid change. Nineteen mutants had amino acid changes in the potential KEN-box. The majority of these had changes in the first conserved asparagine (KENxxxN/D) including two mutants where this was the only amino acid changed (043 and KED/065). The KEN-box mutants were very toxic on galactose plates and stable in the degradation assay (e.g., KED/065, Figure 5A).
Interestingly, we recovered nine mutants that had an intact KEN-box (Figure 6B). Seven of these had changes that cluster in the region between amino acids 991 and 998, contained within the region that we had identified earlier as important for the degradation of Cin8p. We are designating this second region “element 2” (borders are defined in Figure 8A). The two mutants with changes in only element 2 (090 and 145, Figure 5A) were very toxic on galactose plates and produced stable proteins in the degradation assay. Three of the non–KEN-box mutants had mutations in the residue just before the KEN-box 084, 122, and 182). Mutant 182, with only one amino acid change, exhibited only weakly stabilized Cin8p. A number of KEN-box and element 2 mutants had other changes located upstream, downstream, or between these elements. It is likely that many of these are silent changes caused by the mutagenesis protocol, but we cannot rule out that some affect Cin8p stability. Taken together, the results from this screen demonstrate two important points: first, the KEN-box is required for Cin8p's degradation, and second, an element downstream from the KEN-box is also required.
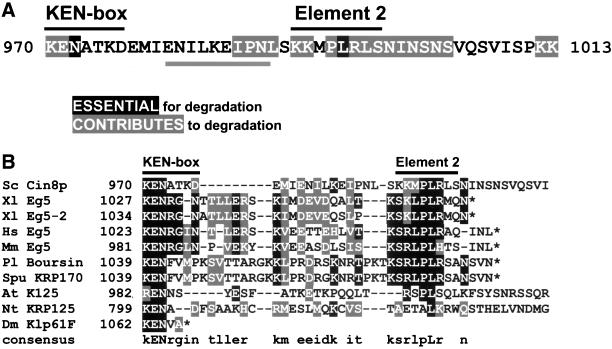
Figure 8.
The destruction sequence in Cin8p and possibly other BimC motors is bipartite. (A) Summary of specific amino acid contributions to Cin8p's degradation signal. The C-terminal side of element 2, amino acid 998, is defined by the shortest truncated form that still supports degradation. The N-terminal side is defined by the positions of the random non–KEN-box mutants (Figure 6B). Note that the borders of element 2 are arbitrary because surrounding amino acids are also important. “Essential” amino acids are those with a single point mutant-stabilized Cin8p. “Contributes” to degradation means the highlighted amino acid had a partial effect when individually mutated or Cin8p was completely stabilized when the residue was mutated in combination with other amino acids. Underlined residues show a region that partially affects Cin8p degradation when deleted or scrambled. (B) Alignment of BimC motors that have a KEN-box–like sequence in the C-terminal domain, after the last coiled-coil. The alignment was performed on sequences starting at the KEN-box (amino acid positions indicated) with the use of the PIMA 1.4 (Smith and Smith, 1992) and BoxShade 3.21 alignment programs and final manual adjustment. Asterisks represent the C terminus of the protein. In the consensus line, capital letters represent identical residues, and lower case letters represent conserved residues. Greater than 40% of sequences must agree for shading. Sc, S. cerevisiae; Xl, Xenopus laevis; Hs, Homo sapiens; Mm, Mus musculus; Pl, Paracentrotus lividus; Spu, Strongylocentrotus purpuratus; At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Dm, Drosophila melanogaster.
To further define the sequence requirements for Cin8p degradation, we generated site-directed mutants in the C terminus of Cin8p and tested their stability (Figures 5B and 6C). Combined mutation of all four critical residues in the KEN-box (KENxxxN/D) to alanine (alaKEN) resulted in a stable Cin8p. However, the extent of toxicity in the PGALS assay was not as strong as other stable mutants, including single KEN-box changes (e.g., KED), perhaps because Cin8p activity was affected. The effects of the random and site-directed KEN-box mutants clearly indicate that it is an essential element targeting Cin8p for degradation.
We made various deletion and alanine mutations downstream of the KEN-box (Figures 5B and 6C). Alanine-scanning mutagenesis identified a patch of residues, MPLR (amino acids 993–996), as essential for degradation. Within this patch, changes to alanine of leucine 995 (L995A) and to a lesser extent proline 994 (P994A) had the greatest stabilizing effect. L995 was also identified in the random mutagenesis screen (145). For both mutants the changes at L995 were conservative, suggesting that very specific recognition of this residue is required for degradation.
One other patch of alanine mutations had a stabilizing effect. Changing a series of six serines and asparagines to alanine between amino acids 998 and1004 (SN-NSNS) partially stabilized Cin8p. A deletion encompassing this region was also stabilized (Δ996-1004). Neither scrambling the amino acids in this region (scr996) nor making changes in one or a few of these residues had much effect (LSNI; S998,1002A; ala996; SNSN). In fact, no individual serine alteration, including a potential CDK1 phosphorylation site (S1010), seemed to affect Cin8p degradation, ruling out the likelihood of a critical phosphorylation event. Therefore, it appears that residues after amino acid 998 contribute to Cin8p's degradation signal, in the context of the full-length protein, even though they are not essential.
We investigated the possibility that the region downstream from the KEN-box contains the ubiquitination target residues. There are two pairs of lysines, which when mutated to alanines in pairs (KK991 and KK1012), had only a mild effect on Cin8p stability. When all four lysines were mutated together, Cin8p was partially stabilized (KK991,KK1012, Figure 5B). Providing four C-terminal lysines alone to an otherwise stable Cin8p truncation (992 plus KK) did not rescue degradation. Therefore, it is possible that one (or more) of these downstream lysines serves as a target for the covalent addition of ubiquitin by the APC, although none appeared to be absolutely required for Cin8p degradation. We could not examine the site of ubiquitin addition, unfortunately, because we were unable to detect ubiquitinated forms of Cin8p with the use of established methods (Willems et al., 1996; Zachariae and Nasmyth, 1996). Because the KEN-box has been demonstrated to be a target signal for ubiquitin addition by APCCdh1, it is likely that a ubiquitinated form of Cin8p is the target for degradation (Pfleger and Kirschner, 2000).
Mutation and deletion of the region between the KEN-box and element 2 also partially stabilized Cin8p (e.g., scr980 and IPNLS, Figure 5B). We have not identified precise residue requirements in this region. It is possible that this region acts as a spacer between the KEN-box and element 2.
Finally, to address the possibility that the KEN-box and element 2 were acting independently and additively, we created double mutants and compared their effects with single mutants. This was difficult, however, because the single mutants exhibited maximum effects in our two assays. Single mutants often appeared fully stable in degradation assays (i.e., compare the −990 single mutant to the −955 double mutant in Figure 4A) and were lethal in the PGALS assay. Therefore, we compared single and double mutants in the PGALS assay in which the level of transcription was reduced by addition of 0.05% glucose to the galactose medium (Figure 5C). Single mutants in the KEN-box (KED) and element 2 (MPLR and Δ996-1004) were slow-growing compared with wild type but not lethal on galactose/0.05% dextrose medium. Double mutants (KED,MPLR and KED,Δ996–1004) were affected to the same extent as the single mutants.
The C Terminus of Cin8p Is Necessary and Sufficient to Target Degradation
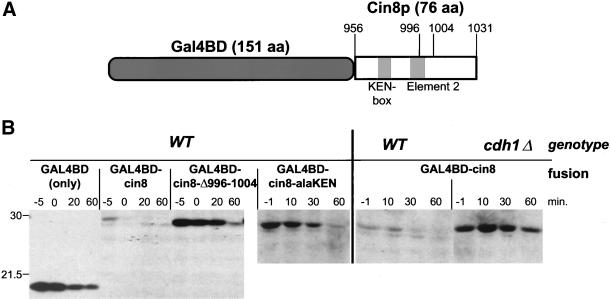
One hallmark of destruction signal sequences is that they can confer instability to a heterologous, normally stable polypeptide. We investigated whether the C-terminal region of Cin8p possessed this property by fusing it to the nuclear-localized DNA-binding domain of Gal4p (Silver et al., 1984). Amino acids 956-1031 of Cin8p, encompassing the region after the fourth coiled-coil domain and up to but not including Cin8p's NLS, were fused to Gal4BD (Figure 7A).
Figure 7.
The C terminus of Cin8p destabilizes a heterologous protein. (A) Schematic of Gal4BD-Cin8p fusion (to scale). (B) The C terminus of Cin8p is necessary and sufficient for degradation. Wild-type (K1534, WT) or cdh1Δ (MAY5353) strains carrying plasmids expressing Gal4BD (PGAL1 > GAL4BD) or Gal4BD fused to a C-terminal portion of wild-type Cin8p (PGAL1 > GAL4BD-cin8), Cin8-Δ996-1004p (PGAL1 > GAL4BD-cin8-Δ996-1004), or Cin8-alaKENp (PGAL1 > GAL4BD-cin8-alaKEN) were subjected to the to the G1 expression-pulse protocol described in Figure 1C. Samples were collected 1–5 min before the end of the galactose pulse (−1 or −5 min) and at times indicated after the pulse. The wild-type fusion was nearly completely degraded by the time the first time sample was processed. The positions of molecular weight standards are shown on the left. The Western blot was probed with anti-Gal4BD antibody.
The Gal4BD 19-kDa polypeptide was stable (t1/2 ∼ 40 min) when transiently expressed from the GAL1 promoter in the expression-pulse assay (Figure 7B). In contrast, the Gal4BD-Cin8p fusion (26.5 kDa) was unstable (t1/2 < 5 min) in wild-type cells but stabilized in a cdh1Δ strain. Therefore, this 76-amino acid region of Cin8p causes the Gal4BD fusion to be degraded in an APCCdh1-dependent manner.
We next deleted nine residues in the Cin8p portion of the fusion protein, within element 2. Pulse-chase analysis revealed this fusion was stable (t1/2 ∼ 30 min, Figure 7B, Gal4BD-cin8-Δ996-1004). Mutation of the KEN-box also stabilized the fusion protein (t1/2 ∼ 20 min, Gal4BD-cin8-alaKEN). These findings demonstrate that amino acids 956-1031 from the C terminus of Cin8p are sufficient to confer APC-dependent degradation of a heterologous protein and that both the KEN-box and element 2 are important for this degradation signal.
Expression of Stable Cin8p Causes Mild Spindle Defects but Degradation Is Not Essential for Normal Cell Cycle Progression
We predicted that degradation of Cin8p, expressed from its endogenous promoter, would not be an essential process because the regulator Cdh1p is not essential. However, moderate overexpression of stable mutants from PGALS caused lethality, in contrast to wild-type Cin8p (Figure 5). We examined the phenotype of these cells (PGALS-cin8-Δ996-1004 or PGALS-cin8-KED expressed for 3–8 h) and observed that ∼50% of cells accumulated at the large-budded stage (compared with 20% for the PGALS control) with mostly unsegregated nuclei. Approximately 60% of budded cells had no spindle (10% for the control). These observations are consistent with an arrest in mitosis with a failure to build or maintain bipolar spindles. Moderate overexpression of wild-type Cin8p caused a similar but less severe phenotype.
We assessed the phenotypic consequence of stable forms of Cin8p expressed from its endogenous promoter. Stable Cin8p mutants (e.g., cin8-alaKEN; cin8-KED; cin8-Δ996-1004; cin8-MPLR; cin8-KED,Δ996-1004) were fully capable of complementing the cin8Δ defect (see MATERIALS AND METHODS). No differences were observed between wild type and stable forms for their ability to support cell growth over a wide range of temperatures. Furthermore, stable Cin8p forms caused no obvious delay to the cell cycle as assessed by flow cytometric analysis of DNA content and cell morphology (Hildebrandt and Hoyt, unpublished results). There was a subtle difference in spindle morphology, however, when we observed the population of mitotic cells in asynchronous cultures. There were 15–21% fewer large-budded cells with obvious bipolar spindle structures in strains expressing stable mutants of Cin8p (cin8-KED and cin8-Δ996-1004) compared with wild-type Cin8p (Table 2). This reduction in bipolar spindles was attributed to a combined reduction in the fraction of cells with preanaphase spindles (short spindles) and anaphase spindles.
Table 2.
Cells expressing stable Cin8p accumulate mitotic cells with no spindle
| Relevant genotypeb | Large-budded (mitotic) cells witha
|
|||
|---|---|---|---|---|
No spindle
|
Short spindle
|
Anaphase spindle
|
||
| (%) | (%) | (%) | (n) | |
| CIN8 | 21 | 47 | 32 | 383 |
| cin8-KED | 42 | 38 | 20 | 309 |
| cin8-Δ996-1004 | 36 | 42 | 22 | 480 |
The indicated strains were grown to midlog phase, fixed, and stained for microtubules and DNA. Large-budded cells were selected microscopically, and their microtubules were observed by indirect immunofluorescence. Short spindles were defined as a bright bar of microtubule staining approximately half the diameter of the mother cell body (approximately 2–3 μm). Anaphase spindles were defined as a line of microtubule staining extending into both mother and bud and from half to two cell body diameters in length. “No spindle” cells contained no clearly observable bipolar structure but possessed clear arrays of microtubules in either one or both cell bodies.
cin8Δ strain (MAY2063) carrying a low-copy CEN plasmid with CIN8, cin8-KED, or cin8-Δ996-1004.
DISCUSSION
Cin8p's Degradation Signal Has Two Essential Parts: a KEN-Box and a Downstream Element
We have found that Cin8p is degraded in an APCCdh1-dependent manner in the period from late mitosis through G1 phase. We could find no evidence for a functioning canonical d-box sequence in Cin8p. Instead, targeting for degradation appeared to be accomplished exclusively by a C-terminal region that includes a KEN-box. The KEN-box was originally defined by in vitro ubiquitination and proteolysis studies of human Cdc20 (Pfleger and Kirschner, 2000). It's role in targeting proteins for destruction in vivo has been demonstrated for human Cdc6, securin, and Drosophila cyclin A, where it appears to share duties with a closely located d-box (Petersen et al., 2000; Jacobs et al., 2001; Zur and Brandeis, 2001). Our finding that KEN-box–mediated destruction occurs in S. cerevisiae demonstrates that it is conserved across distantly related eukaryotic species.
In the in vitro analysis of hCdc20, a polypeptide terminating in the sequence KEN was able to serve as a substrate for APCCdh1-mediated ubiquitination (Pfleger and Kirschner, 2000). The final N/D residue in the KEN-box consensus (KENxxxN/D) was found to contribute to degradation of only certain experimental substrates. In the cases of human Cdc6, securin, and Drosophila cyclin A, the extent of the sequence required for KEN-box function was not determined. With the use of an in vivo approach to dissect Cin8p's degradation signal we also identified the KEN-box as being critical for APCCdh1–mediated degradation. We found that the KEN-box was necessary but not sufficient to target this protein for destruction in vivo. Additional amino acids located up to 22 residues downstream (element 2, Figure 8A) were required. For example, conservative changes affecting leucine 995 resulted in stabilization of Cin8p to the same extent as KEN-box mutations (Figure 5).
We note that element 2 sequences in Cin8p and other related BimC kinesins (see below and Figure 8B) possess a weak match to part of the d-box consensus (RxxL). Although it remains possible that the KEN-box and element 2 could act additively, as observed for the KEN-box and d-box of human Cdc6, securin, and Drosophila cyclin A, our findings suggest that for Cin8p they act as a single unit specifying degradation. Mutations affecting either element caused effects of similar magnitude. In addition, KEN-box/element 2 double mutants were no more severely affected than either single mutant in our assays (Figure 5C; also, compare truncation mutant Cin8-990p, which has the KEN-box but not element 2, to Cin8-955p, which has neither in Figure 4A). This is consistent with both elements contributing to the same degradative mechanism.
Until there is structural data for how the APC recognizes substrates, we cannot distinguish whether element 2 is a novel destruction signal, a cryptic d-box, or an extension of the KEN-box. We can, however, suggest a few molecular explanations for Cin8p's apparent bipartite destruction sequence. First, the APCCdh1 may recognize the entire region, with the identity of the intervening amino acids being less critical than the specific sequences of the KEN-box and element 2. The less stringent requirement for downstream amino acids noted by Phleger and Kirschner (2000), therefore, may be a consequence of different experimental substrates or systems. Second, APC-targeting signals may need to appear within a particular protein structure for recognition. For example, the sequence with a perfect match to the d-box consensus in the N terminus of Cin8p did not specify degradation (this study) and the d-box in Xenopus cyclin A requires the presence of a downstream region for it to act as a degradation signal (Stewart et al., 1994; King et al., 1996b). Thus, element 2 may contribute to the proper structural context for KEN-box recognition by APCCdh1. Third, element 2 could confer a specific Cin8p function or localization causing it to become susceptible to the actions of APCCdh1. This seems unlikely, however, because element 2 was important for degradation of the Gal4BD-Cin8p fusion, which has no motor activity and presumably is bound to DNA rather than microtubules. Finally, it might confer an additional level of regulation to certain APC substrates. For instance, overexpression of CDH1 reduced Clb2p levels in asynchronous cultures (Schwab et al., 1997; Visintin et al., 1997), implying that Cdh1p is limiting for Clb2p degradation. However, we were unable to see a similar effect on Cin8p levels upon CDH1 overexpression (Hildebrandt and Hoyt, unpublished results). Therefore, additional factors may be limiting for Cin8p degradation.
Some, but not all, BimC motors possess a KEN-box–like sequence 24–196 residues beyond the end of the predicted coiled-coil regions in their C termini (a notable exception is Kip1p). All those with a C-terminal KEN-box also possess conserved element 2 sequences downstream (Figure 8B, with the exception of KLP61F, which may not have a complete KEN-box). Therefore, it seems likely that the combined KEN-box/element 2 sequence is utilized by the APCCdh1 for targeting of other BimC motors as well.
APCCdh1- Versus APCCdc20-mediated Destruction of Cin8p
Some APC substrates are recognized by both APCCcd20 and APCCdh1. Hsl1p has a d-box, which is responsible for binding interactions with Cdc20p and Cdh1p (Burton and Solomon, 2000). Additionally, the APC substrate Clb2p is degraded in a biphasic manner, first by APCCdc20 in early anaphase and then by APCCdh1 during telophase and G1 (Baumer et al., 2000a; Yeong et al., 2000). Despite the similar cell cycle profiles that we observed for Cin8p and Clb2p, Cin8p appeared to be degraded exclusively via the APCCdh1. First, cdc20-1 cells were not sensitive to moderate overexpression of Cin8p, but cdh1Δ cells were. Second, Cin8p was stable in late anaphase (Figure 1D), a stage when Cdc20p is presumably active. Third, Cin8p levels were unaffected by Cdc20p overexpression (Hildebrandt and Hoyt, unpublished results). Fourth, Cin8p does not have a functioning canonical d-box, the only known targeting signal for APCCdc20. Finally, because the Cin8p motor makes the greatest contribution to anaphase spindle elongation (Saunders et al., 1995), it seems unlikely that it should be targeted for destruction as cells initiate anaphase. Nevertheless, our studies cannot rule out the possibility that a small fraction of Cin8p degradation is mediated by the APCCdc20 pathway.
Cin8p Must be Nuclear for Degradation
Cin8p's NLS and destruction signal were found in its C-terminal tail domain. Another sequence element conserved in the C-terminal domain of most BimC motors (but not Cin8p and Kip1p) is the “BimC-box.” This region contains a recognition sequence for the cdc2 kinase that has been demonstrated to perform an important role in cell cycle-regulated motor localization (Blangy et al., 1995; Sawin and Mitchison, 1995; Sharp et al., 1999). Thus, a conserved property of this motor family may be that determinants of regulation and localization are found in the C-terminal tail.
APC subunits have been found to localize to nuclei and are often associated with spindles, centrosomes, and kinetochores (Tugendreich et al., 1995; Lim et al., 1998; Huang and Raff, 1999; Kurasawa and Todokoro, 1999). In S. cerevisiae, mutations disrupting nucleocytoplasmic transport also disrupt APC-mediated degradation (Loeb et al., 1995; Baumer et al., 2000b). In Schizosaccharomyces pombe, the failure to localize the proteosome to the nucleus was found to inhibit the destruction of securin and a mitotic cyclin (Tatebe and Yanagida, 2000). Therefore, it is expected that APC substrates may be required to enter the nucleus to become destabilized, especially in fungal cells that do not exhibit mitotic nuclear envelope breakdown. Our findings verify this prediction. C-terminal mutations that reduced the ability of Cin8p to enter the nucleus greatly stabilized the protein. The addition of a heterologous NLS to the N terminus restored its susceptibility to the degradative machinery (Figure 3).
Why Does Cin8p Need to Be Degraded?
Unlike APCCdc20, the APCCdh1 degradation pathway is not essential for S. cerevisiae viability. However, cdh1Δ cells display clear growth defects (Schwab et al., 1997; Visintin et al., 1997). Likewise, S. cerevisiae cells are quite sensitive to the level of Cin8p activity. When Cin8p was overexpressed in cells arrested with preanaphase spindles, the spindles overelongated as if in anaphase (Saunders et al., 1997). Moderate overexpression of a nondegradable form of Cin8p was toxic to cells (Figure 5). Expression of stable Cin8p mutants from the endogenous promoter caused no obvious growth defects, but we did observe an increased number of preanaphase and late anaphase cells with no bipolar spindles (Table 2).
Bipolar spindle formation can fail when the balance of spindle motor forces is perturbed. This has been observed both for loss of BimC activity (Enos and Morris, 1990; Hagan and Yanagida, 1990; Hoyt et al., 1992; Roof et al., 1992; Sawin et al., 1992; Heck et al., 1993; Blangy et al., 1995) and when Cin8p is overexpressed, leading to spindle overelongation and breakage (Saunders et al., 1997; and this study). We speculate that one of the main functions of Cin8p degradation is to reset motor activity to a low, premitotic state important for the early stage of spindle assembly. Because the degradation pathway for Cin8p is not essential and expression of stable Cin8p is only slightly detrimental to cell division, there may exist an additional mechanism for regulating Cin8p motor activity in vivo.
ACKNOWLEDGMENTS
We wish to thank Tami Kingsbury for the initial study of Cin8p levels, John Geiser, Doug Koshland, Doug Kellogg, and Eric Schott for strains and reagents, Laura Totis for technical assistance, and Cindy Dougherty and Srinivas Venkatram for critical reading of the manuscript. These studies were supported by National Institutes of Health grant GM40714 to M.A.H. and National Research Service Award fellowship GM18745 to E.R.H.
Abbreviations used:
- APC
anaphase-promoting complex
- d-box
destruction box
- DMSO
dimethyl sulfoxide
- HA
hemagglutinin
- NLS
nuclear localization signal
- ORF
open reading frame
- PGAL1, GAL1 promoter
PCR, polymerase chain reaction
REFERENCES
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Baumer M, Braus GH, Irniger S. Two different modes of cyclin clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett. 2000a;468:142–148. doi: 10.1016/s0014-5793(00)01208-4. [DOI] [PubMed] [Google Scholar]
- Baumer M, Kunzler M, Steigemann P, Braus GH, Irniger S. Yeast Ran-binding protein Yrb1p is required for efficient proteolysis of cell cycle regulatory proteins Pds1p, and Sic1p. J Biol Chem. 2000b;275:38929–38937. doi: 10.1074/jbc.M007925200. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol Cell Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Mutagenic PCR. In: Dieffenbach CW, Dveksler GS, editors. PCR Primer: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 583–589. [Google Scholar]
- Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Gheber L, Miller D, Hoyt MA. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J Cell Biol. 1999;147:335–349. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically-acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint pathway. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast α-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Giet R, Uzbekov R, Cubizolles F, Le Guellec K, Prigent C. The Xenopus laevis Aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J Biol Chem. 1999;274:15005–15013. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Goh PY, Lim HH, Surana U. Cdc20 protein contains a destruction-box but, unlike Clb2, its proteolysis is not acutely dependent on the activity of anaphase- promoting complex. Eur J Biochem. 2000;267:434–449. doi: 10.1046/j.1432-1327.2000.01014.x. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Roof DM. The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J Biol Chem. 1999;274:28779–28786. doi: 10.1074/jbc.274.40.28779. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Heck MMS, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LSB. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Mitotic motors in S. cerevisiae. Biochim Biophys Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene-products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–2195. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Jacobs HW, Keidel E, Lehner CF. A complex degradation signal in cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J. 2001;20:2376–2386. doi: 10.1093/emboj/20.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang Y-L, Huang J, Peters J-M, McLaughlin ME, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters J-M, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996a;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996b;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Todokoro K. Identification of human APC10/Doc1 as a subunit of anaphase promoting complex. Oncogene. 1999;18:5131–5137. doi: 10.1038/sj.onc.1203133. [DOI] [PubMed] [Google Scholar]
- Lim HH, Goh PY, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Pedroni S, Fournier C, Gautero H, Lecomte MC. Method of site-directed mutagenesis using long primer-unique site elimination and exonuclease III. Biotechniques. 1997;22:430–434. doi: 10.2144/97223bm11. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for the APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA, Keegan LP, Ptashine M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci USA. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat BF, Pringle JR. A mutant of yeast defective in cellular morphogenesis. Science. 1978;200:1171–1173. doi: 10.1126/science.349694. [DOI] [PubMed] [Google Scholar]
- Smith RF, Smith TF. Pattern-induced multi-sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for use in comparative protein modeling. Protein Eng. 1992;5:35–41. doi: 10.1093/protein/5.1.35. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Kobayashi H, Harrison D, Hunt T. Destruction of Xenopus cyclins A and B2, but not B1, requires binding to p34cdc2. EMBO J. 1994;13:584–594. doi: 10.1002/j.1460-2075.1994.tb06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Yanagida M. Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr Biol. 2000;10:1329–1338. doi: 10.1016/s0960-9822(00)00773-9. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Uzbekov R, Prigent C, Arlot-Bonnemains Y. Cell cycle analysis and synchronization of the Xenopus laevis XL2 cell line: study of the kinesin related protein XlEg5. Microsc Res Tech. 1999;45:31–42. doi: 10.1002/(SICI)1097-0029(19990401)45:1<31::AID-JEMT3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1, a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zur A, Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20:792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]