Abstract
Objective
The molecular diagnosis of extreme forms of obesity, in which accurate detection of both copy number variations (CNVs) and point mutations, is crucial for an optimal care of the patients and genetic counseling for their families. Whole-exome sequencing (WES) has benefited considerably this molecular diagnosis, but its poor ability to detect CNVs remains a major limitation. We aimed to develop a method (CoDE-seq) enabling the accurate detection of both CNVs and point mutations in one step.
Methods
CoDE-seq is based on an augmented WES method, using probes distributed uniformly throughout the genome. CoDE-seq was validated in 40 patients for whom chromosomal DNA microarray was available. CNVs and mutations were assessed in 82 children/young adults with suspected Mendelian obesity and/or intellectual disability and in their parents when available (ntotal = 145).
Results
CoDE-seq not only detected all of the 97 CNVs identified by chromosomal DNA microarrays but also found 84 additional CNVs, due to a better resolution. When compared to CoDE-seq and chromosomal DNA microarrays, WES failed to detect 37% and 14% of CNVs, respectively. In the 82 patients, a likely molecular diagnosis was achieved in >30% of the patients. Half of the genetic diagnoses were explained by CNVs while the other half by mutations.
Conclusions
CoDE-seq has proven cost-efficient and highly effective as it avoids the sequential genetic screening approaches currently used in clinical practice for the accurate detection of CNVs and point mutations.
Keywords: Augmented whole-exome sequencing, Copy number variation, Intellectual disability, Molecular diagnosis, Next-generation sequencing, Obesity
Highlights
-
•
Whole-exome sequencing (WES) poorly detects CNVs.
-
•
Whole-genome sequencing remains expensive and hard to handle in clinical practice.
-
•
CoDE-seq (based on an augmented WES protocol) accurately detect CNVs.
-
•
CoDE-seq is highly effective for the diagnosis of obesity & intellectual disability.
1. Introduction
A complex, polygenic disorder determined by genetic, epigenetic, and environmental components, obesity has now reached epidemic proportions [1]. The genetic component is far from insignificant as the heritability of obesity or body mass index (BMI) has been estimated around 70% [1]. Rarer and more severe forms of obesity (i.e. Mendelian obesity) can be due to only one genetic event (e.g. point mutation, copy number variation [CNV]) [1]. These monofactorial forms of obesity can be associated with severe clinical features including intellectual disability, as observed in patients with Bardet-Biedl syndrome or Alström syndrome [2]. Furthermore, we and others previously demonstrated that a CNV leading to a deletion in chromosome 16 on the short arm p, in band 11.2 (chr16p11.2) causes a highly penetrant form of severe obesity, often associated with intellectual disability [3], [4]. A molecular diagnosis of these extreme forms of obesity, which is able to accurately detect both CNVs and point mutations, is crucial for the optimal care of the patients and genetic counseling for their families. Indeed, curing extreme obesity can be reached in LEP deficient patients via recombinant leptin therapy [5] and in POMC deficient patients via the MC4R agonist Setmelanotide [6]. Furthermore, we previously demonstrated that bariatric surgery should be avoided in obese patients carrying a rare deleterious mutation in MC4R as these patients have more complications and reoperations than non-carriers [7].
Next-generation sequencing, in particular whole-exome sequencing (WES), has considerably benefited molecular diagnosis in patients presenting with Mendelian disorders, including monofactorial obesity [8]. Lower cost, reduced labor intensity, shorter delay time, and enhanced effectiveness for the detection of point mutations have been the key WES advantages compared to Sanger sequencing [9], [10]. A major caveat of WES is the poor detection rate of CNVs, partly explained by the weak reproducibility of CNV predicting software programs [11]. In addition, probes used for WES only target coding regions that represent 2% of the whole genome in humans, and the distribution of exons is not uniform throughout the genome, limiting further CNV detection sensitivity. Indeed, the gaps between exome probes can be very large (up to 4 Mb in non-telomeric or non-centromeric regions for the NimbleGen SeqCap EZ MedExome enrichment). As point genetic variants in the exome do, CNVs cause many Mendelian disorders including monofactorial obesity (Table A). Therefore, in clinical practice, patients suspected of having such Mendelian disorders are referred to clinical geneticists who first apply a chromosomal DNA microarray for CNV detection, which, if negative, is potentially followed by WES or targeted gene panel sequencing for point mutation identification [9]. When compared to WES, whole-genome sequencing has proven more accurate and sensitive for CNV detection [12], but it remains costly and very hard to handle in clinical practice (particularly for routine computer analyses).
Here, we aimed to develop a new next-generation sequencing strategy (named ‘CoDE-seq’ for Copy number variation Detection and Exome sequencing) enabling the accurate detection of both CNVs and point mutations in only one step, which would be more affordable and easier to handle than whole-genome sequencing, more sensitive for CNV detection than WES, and at least as accurate as chromosomal DNA microarray for CNV detection. For this purpose, we designed a specific capture based on an augmented WES protocol, which has been first assessed in patients for whom array comparative genomic hybridization (aCGH) was also performed. After the full validation of CoDE-seq, 82 children or young adults with suspected Mendelian obesity and/or intellectual disability (as well as their parents when available) were analyzed, and a molecular diagnosis was achieved in more than 30% of them (half due to CNVs, half due to point mutations).
2. Methods
2.1. Participants
We investigated blood DNA samples from 145 participants including:
-
-
Forty patients (24 males/16 females) for whom aCGH (Agilent 60K v2) was performed in order to identify pathogenic CNVs. These patients were referred to the Centre de Génétique Chromosomique in Saint Vincent de Paul hospital (Lille, France) by pediatric neurologists (38 patients under 16 years of age) or by adult neurologists (two patients). These aCGH-based tests were required as these patients had neurological disorders: 13 participants had intellectual deficiency, seven participants had behavioral disorders, 12 participants had learning disabilities, five participants had both intellectual deficiency and behavioral disorders, and three participants had both behavioral disorders and learning disabilities. We also analyzed 28 parents of these patients in order to investigate de novo genetic events or transmission of genetic events.
-
-
Thirty children (15 males/15 females) from Obesity Saint Vincent (OSV) cohort study (Saint Vincent de Paul hospital, Lille, France). This cohort study included obese children (below 18 years of age) with extended international (IOTF) BMI higher than 30, who were recruited between 2015 and 2016 (inclusion is planned to continue until 2020). Inclusion criteria were syndromic obesity (e.g. associated with intellectual disability, eating disorders, behavior disorders, visual and/or auditory impairments, dysmorphic features, malformations, sleep disorders, hypogonadism), isolated obesity (i.e. both parents were not obese), and/or consanguineous obesity. Exclusion criteria were putative common obesity, inability to take blood sample, inability to get consent from the child and both parents, or refusal of one parent to participate. For each child, clinical data were collected: in utero events, birth parameters, neonatal events, psychomotor development, neurological disorders, obesity onset (adiposity rebound), metabolic parameters, treatment, eating habits, physical activity, education, and family history (BMI of both parents and of siblings, potential overweight/obesity onset, potential history of diabetes, socio-economic classification of both parents). Clinical data of the children are reported in Table B. We also analyzed 33 parents of the children in order to investigate de novo genetic events or transmission of genetic events.
-
-
Twelve children or young adults (7 males/5 females) referred to the CNRS unit #8199 (Lille, France). These individuals presented with severe obesity and/or neurological disorders. Clinical data of these individuals are reported in Table B. We also analyzed two parents of one child to investigate de novo genetic events or transmission of genetic events.
2.2. CoDE-seq target enrichment design
We combined a commercially available capture targeting the human whole exome (NimbleGen SeqCap EZ MedExome Enrichment) with our enhanced capture that we designed (NimbleGen SeqCap EZ Choice XL). This specific capture consisted of a backbone of ∼120 bp probes regularly spaced throughout the entire genome. More specifically, probes were placed every 10 kb apart in the genomic regions known to be affected by CNVs (based on the literature, and in particular the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources [DECIPHER] [13] [Table A]), which encompassed 881 Mb (i.e. about 30% of the human genome), and probes were placed every 25 kb in the rest of the genome. The probes targeting whole exome and backbone covered 65 Mb of the genome. The effective coverage after next-generation sequencing reached 91 Mb.
2.3. CoDE-seq target enrichment preparation and next-generation sequencing
Target enrichment was performed according to the manufacturer's protocol (NimbleGen SeqCap EZ) for Illumina sequencing. Briefly, 1 μg DNA was fragmented through sonication (Covaris E220 Focused-ultrasonicator). The fragmented DNA samples were end-repaired and ligated to the Illumina adapters using the KAPA HTP Library Preparation Kits, on the Hamilton Microlab STARlet automate. These samples were subsequently amplified by polymerase chain reaction using primers complementary to the adapters. After size selection and sample quantification (Perkin Elmer LabChip GX), five samples were combined in a single pool of at least 1 μg and hybridized to the biotin-labeled SeqCap EZ probe pool. After 72 h at 47 °C, the captures were purified using the SeqCap Hybridization and Wash Kit on the Agilent Bravo Automated Liquid Handling Platform. Captures were subsequently amplified using the KAPA HiFi HotStart ReadyMix and quantified by both Caliper LifeScience LabChip GX and Qubit fluorometric quantitation assays (Thermo Fisher Scientific). Then, the samples were sequenced on the Illumina HiSeq 4000 system (with a throughput of one pool including five samples per lane), using a paired-end 2 × 150 bp protocol.
2.4. Bioinformatics analyses
The demultiplexing of sequence data (from BCL files generated by Illumina sequencing systems to standard FASTQ file formats) was performed using bcl2fastq Conversion Software (Illumina; version 2.19.1). Subsequently, sequence reads from FASTQ files were mapped to the human genome (hg38/GRCh38) using Burrows-Wheeler Aligner (version 0.7.15) [14]. On average, 145M reads were generated per sample, and 95.7 ± 0.8% of them were accurately mapped. The variant calling was performed using Genome Analysis ToolKit (GATK; version 3.7) [15]. The annotation of point variants (including missense variants, nonsense variants, small insertions and deletions [indels], splice-site variants … ) was performed using the Ensembl Perl Application Program Interfaces (version 89) and other Perl scripts to include data from both dbSNP (version 135) and dbNSFP (version 3.4) databases [16], [17]. Our analysis of rare point variants was focused on genes involved in Mendelian forms of obesity and/or intellectual disability (Table C). We also analyzed rare point variants in genes involved in Mendelian conditions that can be related to obesity (hypercholesterolemia, diabetes and lipodystrophy; Table C). We used standards and guidelines of the American College of Medical Genetics and Genomics for the interpretation of rare point variants [18]. Of note, for the moderate pathogenic criterion PM2, we used the genome aggregation database (gnomAD) browser spanning 123,136 exome sequences and 15,496 whole-genome sequences as control population [19], and for the supporting pathogenic criterion PP3, we used four software programs: PolyPhen-2 (HumDiv), SIFT, MutationAssessor and PhyloP [20], [21], [22], [23]. The point variants that were tagged “pathogenic”, “likely pathogenic” or “variants of uncertain significance” were covered with more than 20 reads and had a Phred quality score of at least 150. The detection of CNVs was performed using the eXome Hidden Markov Model (XHMM; version 1.0) program [24]. This approach uses principal-component analysis to normalize sequence read depth for each of the target (which was calculated via GATK; version 3.7) [15] and a hidden Markov model (HMM) [24]. Among the 40 patients for whom aCGH raw data were available, we compared the detection of CNVs through aCGH, CoDE-seq and WES. For the specific comparison of CNV detection between CoDE-seq and WES, XHMM program was run in the 40 patients, with respectively the browser extensible data (BED) of CoDE-seq or the BED of NimbleGen SeqCap EZ MedExome only, as input.
3. Results
After next-generation sequencing of the 145 DNA samples (from 82 patients with suspected Mendelian obesity and/or intellectual disability, and their parents when available), the 91.5 Mb CoDE-seq target was covered with a mean depth of 120.3 ± 31.5 reads. On average, 99.1 ± 0.4% of the target was covered with at least eight reads, 97.0 ± 2.7% of the target was covered with at least 20 reads and 83.2 ± 11.2% of the target was covered with at least 50 reads.
In order to assess our new method, we analyzed first the 40 patients with neurological disorders for whom the CNV dataset from the aCGH was available. This specific analysis used all captured regions (coding regions and intergenic regions). Second, in all participants, we investigated the genomic regions specifically known to be involved in Mendelian forms of intellectual disability and obesity.
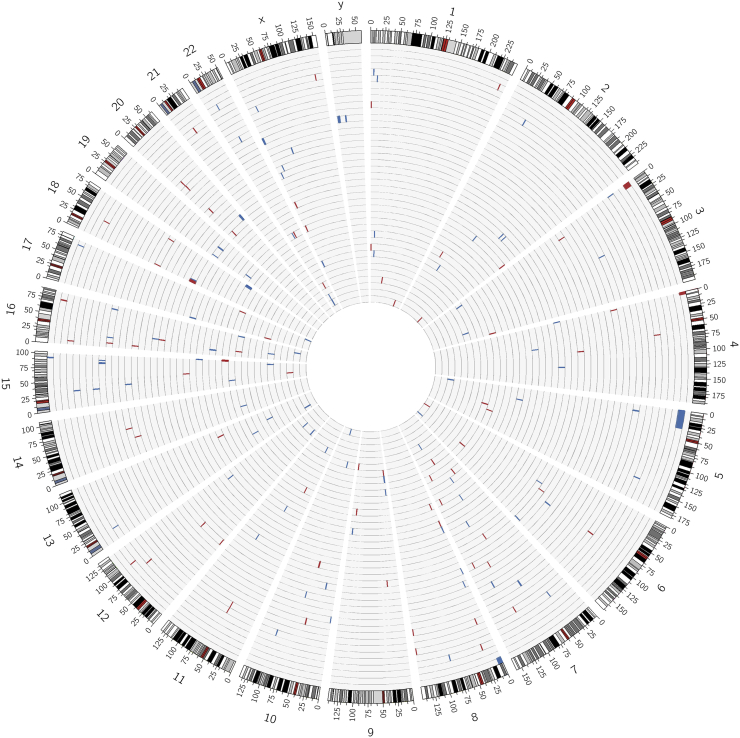
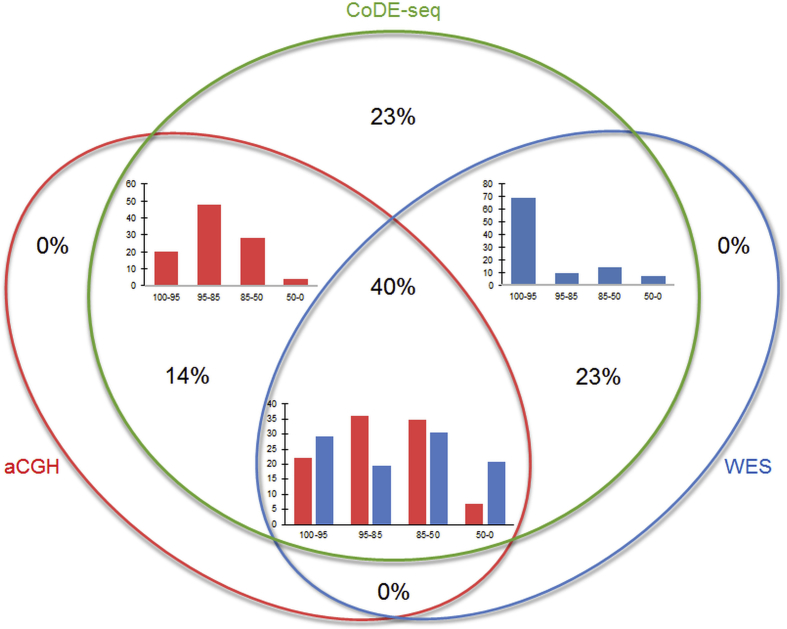
In the 40 patients with neurological disorders for whom aCGH data were available, our CoDE-seq method detected 181 CNVs (102 gains/79 losses) located in all chromosomes (Figure 1), with a mean size of 720 kb (minimum: 1.8 kb; maximum: 31 Mb) (Table D). In contrast, in the same samples, aCGH only detected 97 CNVs (59 gains/38 losses), with a mean size of 1.1 Mb (minimum: 57 kb; maximum: 31 Mb) (Table D). Importantly, all these 97 CNVs were detected by CoDE-seq, but 67% of these shared CNVs looked smaller (±5%) when detected by aCGH compared to the ones detected by CoDE-seq (Figure 2, and Table D), suggesting a better resolution via CoDE-seq. Eighty-four CNVs (46%) detected by CoDE-seq were not identified by aCGH (Figure 2). These newly identified CNVs had a mean size of 162 kb (minimum: 1.8 kb; maximum: 2.5 Mb) (Table D). In the same samples, we subsequently compared the CNV detection through WES versus CoDE-seq or aCGH. Via WES, 114 CNVs (73 gains/41 losses) were detected, with a mean size of 855 kb (minimum: 1.8 kb; maximum: 27 Mb) (Table D). These 114 CNVs were all detected by CoDE-seq, although 55% of these shared CNVs looked smaller (±5%) when detected by WES compared to the ones detected by CoDE-seq (Figure 2, and Table D). Sixty-seven CNVs (37%) detected by CoDE-seq were not identified by WES (Figure 2). These CNVs had a mean size of 197 kb (minimum: 11 kb; maximum: 667 kb) (Table D). Among the 114 CNVs detected by WES, 42 CNVs (37%) were not identified by aCGH, and among the 97 CNVs detected by aCGH, 25 CNVs (26%) were not identified by WES (Figure 2, and Table D). Overall, among the 181 CNVs detected by CoDE-seq, 42 CNVs (23%) were identified neither by aCGH nor by WES (Figure 2), while 24 of these 42 CNVs mapped coding regions (Table D). These 42 CNVs had a mean size of 144 kb (minimum: 11 kb; maximum: 456 kb) (Table D), and their profiles are shown in Figure. A. Therefore, CoDE-seq has proven more sensitive for CNV detection than aCGH and WES.
Figure 1.
Location of the 181 CNVs detected by CoDE-seq throughout the genome (from chromosome 1 to chromosomes X,Y) in the 40 participants for whom aCGH data were available. Each light grey circle depicts a participant's genome (clockwise from chromosome 1 to chromosomes X,Y). CNVs are depicted by a blue bar (copy-gain variant) or a red bar (copy-loss variant). This figure was done using the Circos software package [34].
Figure 2.
CNV detection through CoDE-seq, aCGH or whole-exome sequencing (WES) in the 40 participants for whom aCGH data were available. CoDE-seq detected 181 CNVs; aCGH detected 97 CNVs and whole-exome sequencing detected 114 CNVs. Histograms show the frequency (%) of the CNVs detected by aCGH (red histograms) or by whole-exome sequencing (blue histograms) according to their size (100–95: between 95 and 100% of the size of the CNVs detected by CoDE-seq; 95–85: between 85 and 95% of the size of the CNVs detected by CoDE-seq; 85–50: between 50 and 85% of the size of the CNVs detected by CoDE-seq; 50–0: below 50% of the size of the CNVs detected by CoDE-seq). CNV frequency (%) in each part of the Venn diagram is also shown.
Subsequently, we aimed to analyze pathogenic CNVs and point mutations in the 82 patients with neurological disorders and/or severe obesity. For this purpose, the investigation of point variants and CNVs was focused on a list of genes known to be involved in Mendelian obesity and/or intellectual disability, and in Mendelian conditions associated with obesity (Table C). We also analyzed the CNVs detected in the large genomic regions known to be associated with these conditions (Table A). We found 13 likely pathogenic CNVs (7 gains/6 losses) with a mean size of 3.5 Mb (minimum: 241 kb; maximum: 30.7 Mb) in 13 (15.9%) participants (Table 1). These CNVs were located in chromosomes 5, 7, 15, 16, 20, and X, and they had previously been demonstrated to cause obesity and/or intellectual disability (Table 1) [3], [4], [25], [26], [27], [28], [29], [30], [31]. Notably, six out of the 13 participants carried a CNV chr16p11.2 (Table 1). Among them, three patients had a ∼600 kb proximal chr16p11.2 deletion and presented with both obesity and intellectual disability; one girl had a ∼600 kb proximal chr16p11.2 duplication and presented with intellectual disability, short stature, thinness and dysmorphic feature; and two patients had a ∼200 kb distal chr16p11.2 deletion and presented with intellectual disability for the first child, or seizures, behavioral disorder, hypermetropia, obesity, and dysmorphic features for the second child (Table 1). Of note, we identified 45 CNVs of uncertain significance (27 gains/18 losses) with a mean size of 195 kb (minimum: 9.8 kb; maximum: 718 kb) in 33 participants (Table E). These CNVs mapped genomic regions harboring genes involved in intellectual disability and/or obesity (Table C), or large genomic regions known to be associated with these conditions (Table A) but with a smaller size. In parallel, when investigating point mutations in the 82 patients suffering from neurological disorder and/or severe obesity, the molecular diagnosis involving a Mendelian disease gene related to the clinical phenotype at the time of examination was reported for 14 (17.1%) patients (Table 2). Among the pathogenic or likely pathogenic genetic variants, one non-synonymous mutation (p.Arg18Cys) and one frameshift mutation (p.Ala68Glnfs*2) in the Mendelian obesity MC4R gene were found in two girls with morbid obesity and severe intellectual disability associated with behavioral disorders, dysmorphic features, and/or malformations (Table 2). Moreover, in a morbidly obese boy with large abdominal lipoma, we identified a nonsense mutation (p.Arg130*) in PTEN (Table 2). When investigating the family, we found that the mother who previously developed a thyroid cancer and uterine fibroids, and who presented with morbid obesity, also carried the same nonsense mutation in PTEN (Table 2). In a boy presenting with a very severe and complex clinical presentation of intellectual disability, we reported two molecular diagnoses: one non-synonymous mutation (p.Met74Val) in CASR and two compound heterozygous mutations in MTHFR (p.Val450Leu and p.*657Serext*50; Table 2). A molecular diagnosis was achieved in the following other genes in 10 patients with intellectual disability: ATRX, MECP2, RAI1, EEF1A2, KDM5B, NF1, SLC6A1, TSC1, and NEXMIF/KIAA2022 (Table 2). Of note, in seven patients, we identified eight incidental findings related to monogenic forms of diabetes, hyperinsulinemia, seizure, hypercholesterolemia and intellectual disability (Table F). In nearly all participants, we detected variants of uncertain significance (Table G).
Table 1.
Likely pathogenic CNVs identified in the 82 participants with obesity and/or neurological disorder.
| Patient | Sex* | Chr | Start | End | Size | Probes | Type | Parental origin** | Gene | Phenotype | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 32,996 | F | 16 | 29,568,896 | 30,206,106 | 637.21 | 215 | Gain | M | SPN, QPRT, C16orf54, ZG16, KIF22, MAZ, PRRT2, PAGR1, MVP, CDIPT, SEZ6L2, ASPHD1, KCTD13, TMEM219, TAOK2, HIRIP3, INO80E, DOC2A, C16orf92, FAM57B, ALDOA, PPP4C, TBX6, YPEL3, GDPD3, MAPK3, CORO1A, BOLA2, BOLA2B, SLX1A, SLX1B, SULT1A4, SULT1A3 | Intellectual disability [psychomotor retardation, language delay], short stature, thinness, dysmorphic feature | [25] |
| 36,504 | F | 5 | 41,442 | 31,162,895 | 30,743.13 | 3989 | Gain | Unk | PLEKHG4B, LRRC14B, CCDC127, SDHA, PDCD6, AHRR, EXOC3, SLC9A3, CEP72, TPPP, ZDHHC11B, ZDHHC11, BRD9, TRIP13, NKD2, SLC12A7, SLC6A19, SLC6A18, TERT, CLPTM1L, SLC6A3, LPCAT1, MRPL36, NDUFS6, IRX4, IRX2, C5orf38, IRX1, ADAMTS16, ICE1, MED10, UBE2QL1, NSUN2, SRD5A1, PAPD7, ADCY2, C5orf49, FASTKD3, MTRR, SEMA5A, TAS2R1, FAM173B, CCT5, CMBL, MARCH6, ROPN1L, ANKRD33B, DAP, CTNND2, DNAH5, TRIO, FAM105A, OTULIN, ANKH, FBXL7, MARCH11, ZNF622, RETREG1, MYO10, BASP1, H3.Y, CDH18, CDH10, CDH12, CDH9, PRDM9 | Intellectual disability | [26] |
| 36,981 | F | 15 | 98,507,942 | 101,861,073 | 3353.13 | 513 | Loss | Unk | FAM169B, IGF1R, PGPEP1L, SYNM, TTC23, LRRC28, MEF2A, LYSMD4, ADAMTS17, CERS3, LINS1, ASB7, ALDH1A3, LRRK1, CHSY1, SELENOS, SNRPA1, PCSK6, TM2D3, TARSL2, LOC100128108, OR4F6, OR4F15 | Intellectual disability [psychomotor retardation], failure to thrive, short stature, thinness, microcephaly, dysmorphic feature | [27] |
| 38,213 | F | 20 | 59,032,190 | 64,329,015 | 5296.83 | 1180 | Gain | Unk | ATP5F1E, PRELID3B, ZNF831, EDN3, PHACTR3, SYCP2, FAM217B, PPP1R3D, FAM217B, CDH26, C20orf197, CDH4, TAF4, LSM14B, PSMA7, SS18L1, MTG2, HRH3, OSBPL2, ADRM1, LAMA5, RPS21, CABLES2, RBBP8NL, GATA5, MIR1-1HG, SLCO4A1, NTSR1, MRGBP, OGFR, COL9A3, TCFL5, DPH3P1, DIDO1, GID8, SLC17A9, BHLHE23, YTHDF1, BIRC7, NKAIN4, ARFGAP1, COL20A1, CHRNA4, KCNQ2, EEF1A2, PPDPF, PTK6, SRMS, FNDC11, HELZ2, GMEB2, STMN3, RTEL1, TNFRSF6B, ARFRP1, ZGPAT, LIME1, SLC2A4RG, ZBTB46, ABHD16B, TPD52L2, DNAJC5, UCKL1, ZNF512B, SAMD10, PRPF6, C20orf204, SOX18, TCEA2, RGS19, OPRL1, LKAAEAR1, OPRL1, NPBWR2, MYT1, PCMTD2 | Intellectual disability [psychomotor retardation, spatial disorientation], aggressive behavior, short stature | [31] |
| 38,645 | M | 16 | 28,811,431 | 29,052,826 | 241.4 | 136 | Loss | Unk | ATXN2L, TUFM, SH2B1, ATP2A1, RABEP2, CD19, NFATC2IP, SPNS1, LAT | Intellectual disability [psychomotor retardation, language delay] | [4] |
| 38,839 | M | X | 53,231,708 | 53,878,306 | 646.6 | 185 | Gain | Unk | IQSEC2, SMC1A, RIBC1, HSD17B10, HUWE1 | Intellectual disability [psychomotor retardation], neonatal hypotonia, thinness | [28] |
| 39,409 | F | 7 | 73,316,540 | 74,798,284 | 1481.74 | 339 | Gain | D | TRIM50, FKBP6, FZD9, BAZ1B, BCL7B, TBL2, MLXIPL, VPS37D, DNAJC30, BUD23, STX1A, ABHD11, CLDN3, CLDN4, METTL27, TMEM270, ELN, LIMK1, EIF4H, LAT2, RFC2, CLIP2, GTF2IRD1, GTF2I, NCF1, GTF2IRD2 | Intellectual disability [psychomotor retardation, language delay], behavioral disorder | [29] |
| 39,509 | M | 15 | 31,739,875 | 32,243,158 | 503.28 | 58 | Gain | F | OTUD7A, CHRNA7 | Intellectual disability [language delay], seizures | [30] |
| 39,638 | F | 15 | 31,729,880 | 32,225,059 | 495.18 | 60 | Gain | Unk | OTUD7A, CHRNA7 | Intellectual disability | [30] |
| OSV20 | M | 16 | 29,572,106 | 30,188,122 | 616.02 | 211 | Loss | F | SPN, QPRT, C16orf54, ZG16, KIF22, MAZ, PRRT2, PAGR1, MVP, CDIPT, SEZ6L2, ASPHD1, KCTD13, TMEM219, TAOK2, HIRIP3, INO80E, DOC2A, C16orf92, FAM57B, ALDOA, PPP4C, TBX6, YPEL3, GDPD3, MAPK3, CORO1A | Intellectual disability [language delay], obesity | [3] |
| OSV21 | F | 16 | 29,642,461 | 30,194,811 | 552.35 | 203 | Loss | F | SPN, QPRT, C16orf54, ZG16, KIF22, MAZ, PRRT2, PAGR1, MVP, CDIPT, SEZ6L2, ASPHD1, KCTD13, TMEM219, TAOK2, HIRIP3, INO80E, DOC2A, C16orf92, FAM57B, ALDOA, PPP4C, TBX6, YPEL3, GDPD3, MAPK3, CORO1A, BOLA2, BOLA2B, SLX1A, SLX1B | Intellectual disability [language delay], obesity | [3] |
| OSV46 | F | 16 | 28,811,431 | 29,052,826 | 241.4 | 136 | Loss | Unk | ATXN2L, TUFM, SH2B1, ATP2A1, RABEP2, CD19, NFATC2IP, SPNS1, LAT | Seizures, behavioral disorder, hypermetropia, obesity, dysmorphic feature | [4] |
| U2 | F | 16 | 29,590,876 | 30,188,122 | 597.25 | 209 | Loss | Unk | SPN, QPRT, C16orf54, ZG16, KIF22, MAZ, PRRT2, PAGR1, MVP, CDIPT, SEZ6L2, ASPHD1, KCTD13, TMEM219, TAOK2, HIRIP3, INO80E, DOC2A, C16orf92, FAM57B, ALDOA, PPP4C, TBX6, YPEL3, GDPD3, MAPK3, CORO1A | Intellectual disability [psychomotor retardation], severe obesity | [3] |
All genomic coordinates are based on the hg38 reference sequence.
Chr, Chromosome; D, de novo; F*, female; F**, father; M*, male; M**, mother; Ref, reference; Unk, unknown.
Table 2.
Pathogenic or likely pathogenic point mutations causing obesity and/or neurological disorder in the 82 participants.
| Patient | Sex* | Gene | Inheritance | Mutation | Status | Parental origin** | rs ID | Pathogenicity | ACMG criteria | Phenotype | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 36,711 | M | ATRX | XL | c.6242T > C/p.Ile2081Thr | hem | M | – | Likely pathogenic | PM1, PM2, PP2, PP4 | Intellectual disability [psychomotor retardation], aggressive behavior, attention deficit | – |
| 38,114 | F | MECP2 | XL | c.622C > T/p.Gln208* | het | D | rs61749729 | Pathogenic | PVS1, PS1, PS2, PM2, PP2, PP4 | Intellectual disability [language delay, pervasive developmental disorder], stereotypes | [35] |
| 39,268 | M | RAI1 | AD | c.4685A > T/p.Gln1562Leu | het | Unk | – | Likely pathogenic | PM2, PM5, PP2, PP4 | Intellectual disability [psychomotor retardation], behavioral disorder, heart disorder, seizures | – |
| 39,465 | F | EEF1A2 | AD | c.1375_1383del/p.Gln459_Ala461del | het | M | – | Pathogenic | PS1, PM2, PM4, PP2, PP4 | Intellectual disability [psychomotor retardation, language delay], anxiety, coarse facial features | ClinVar RCV000487209.1 |
| 39,612 | M | KDM5B | AD/AR | c.688C > T/p.Arg230* | het | Unk | rs867831466 | Pathogenic | PVS1, PP2, PP4 | Intellectual disability [psychomotor retardation] | – |
| OSV11 | F | NF1 | AD | c.3989A > C/p.Glu1330Ala | het | Unk | – | Likely pathogenic | PM1, PM2, PP2, PP4 | Intellectual disability [language delay], too friendly, morbid obesity | – |
| OSV37 | F | MC4R | AD | c.52C > T/p.Arg18Cys | het | F | rs749768113 | Pathogenic | PS1, PS3, PM1, PP1, PP2, PP4 | Intellectual disability [language delay], morbid obesity, emotional instability, aggressive behavior, sleep disorder, synophria, hypertrichosis, cross-eye, hypermetropia | [36] |
| OSV52 | M | SLC6A1 | AD | c.715-1G > A/- | het | Unk | – | Pathogenic | PVS1, PM2, PM6, PP2, PP4 | Intellectual disability [language delay], aggressive behavior, grimace, anxiety, eye twitch, obesity | – |
| OSV53 | M | TSC1 | AD | c.2665_2666delinsAT/p.Glu889Ile | hom | Unk | rs587778724 | Likely pathogenic | PM1, PM2, PP2, PP3, PP4 | Intellectual disability [learning difficulties], morbid obesity, hypermetropia | – |
| OSV65 | F | NEXMIF | XL | c.2692del/p.Gln898Argfs*11 | het | D | – | Pathogenic | PVS1, PS2, PM2, PP2, PP4 | Intellectual disability [psychomotor retardation, language delay], obesity, too friendly, impulsivity, hypermetropia | – |
| OSV77 | M | TSC1 | AD | c.827C > A/p.Ser276Tyr | het | F | – | Likely pathogenic | PM1, PM2, PP2, PP3, PP4 | Behavioral disorder, isolated morbid obesity, dysmorphic feature | – |
| OSV9 | F | MC4R | AD | c.202del/p.Ala68Glnfs*2 | het | M | – | Pathogenic | PVS1, PM1, PM2, PP1, PP2, PP4 | Intellectual disability [language delay, dysphasia], morbid obesity, malformation | – |
| U3 | M | PTEN | AD | c.388C > T/p.Arg130* | het | M | rs121909224 | Pathogenic | PVS1, PS1, PS3, PM1, PP1, PP2, PP4 | Morbid obesity, macrocephaly, abdominal lipoma | [37] |
| U5 | M | CASR | AD/AR | c.220A > G/p.Met74Val | het | F | – | Likely pathogenic | PS1, PM2, PP2, PP3, PP4 | Intellectual disability [severe psychomotor retardation, no language], behavioral disorder, seizures, neonatal hypotonia, kyphosis, cross-eye, ataxic gait, sleep disorder | [38] |
| U5 | M | MTHFR | AR | c.1348G > T/p.Val450Leu | comp het | M | – | Pathogenic | PVS1, PM2, PP2, PP4 | Intellectual disability [severe psychomotor retardation, no language], behavioral disorder, seizures, neonatal hypotonia, kyphosis, cross-eye, ataxic gait, sleep disorder | – |
| U5 | M | MTHFR | AR | c.1970G > C/p.*657Serext*50 | comp het | F | rs749490263 | Pathogenic | PVS1, PS1, PS3, PM1, PP2, PP4 | Intellectual disability [severe psychomotor retardation, no language], behavioral disorder, seizures, neonatal hypotonia, kyphosis, cross-eye, ataxic gait, sleep disorder | [39] |
ACMG, American College of Medical Genetics and Genomics; AD, autosomal dominant; AR, autosomal recessive; Chr, Chromosome; comp het, compound heterozygous; D, de novo; F*, female; F**, father; hem, hemizygous; het, heterozygous; hom, homozygous; M*, male; M**, mother; PM, moderate pathogenic criterion; PP, supporting pathogenic criterion; PS, strong pathogenic criterion; PVS, very strong pathogenic criterion; Ref, reference; Unk, unknown; XL, X-linked.
4. Discussion
In the present study, we have firmly demonstrated that CoDE-seq based on an augmented WES protocol enables the accurate and cost-effective detection of both CNVs and point mutations in a single step. In our hands, CoDE-seq not only detected all CNVs identified by aCGH but also found additional CNVs, due to the much higher number of probes used for CoDE-seq, compared to the number of probes used for aCGH (Table D). Moreover, we confirmed that WES leads to poor CNV detection (even in coding regions), which may explain the current clinical use of chromosomal DNA microarray (for CNV detection only) and WES or targeted gene panel sequencing (for point mutation identification). CoDE-seq not only dramatically saves labor time but also money as its cost is only 25% higher than the cost of standard WES (around $600 per sample), which is equivalent to the invoicing of aCGH only. Furthermore, CoDE-seq remains far less expensive than whole-genome sequencing as it still targets less than 3% of the whole genome, and it is significantly easier to analyze.
When we assessed both CNVs and point mutations through CoDE-seq in 82 patients with suspected Mendelian obesity and/or intellectual disability, a likely molecular diagnosis was achieved in more than 30% of the patients. Half of the genetic diagnoses were explained by pathogenic CNVs and the other half were caused by pathogenic point mutations, a finding that has not been made so far, and should encourage to systematically look for both genetic events. Most pathogenic CNVs were located on chr16p11.2. The ∼600 kb proximal chr16p11.2 deletion and the ∼200 kb distal chr16p11.2 deletion cause a highly penetrant form of obesity, often associated with intellectual disability [3], [4], while the ∼600 kb proximal chr16p11.2 duplication causes underweight, microcephaly and intellectual disability [25]. Further efforts are needed to clarify the extend by which CNVs explain these phenotypes; we found many CNVs of potential interest (Table E), but we were unable to address their pathogenicity due to the lack of consensus guidelines and databases with lists and frequencies of CNVs in the general population (like the gnomAD browser for point variants [19]). Regarding point mutations, we surprisingly identified two pathogenic mutations in MC4R (encoding melanocortin 4 receptor) in two girls with morbid syndromic obesity, while it is believed that MC4R mutations represent the most frequent genetic cause of Mendelian non-syndromic obesity [1]. Even if we cannot preclude that in these two girls other mutations/CNVs in novel regions (not specifically analyzed in this study focused on candidate regions only) might cause their syndromic features, this result highlights that genes involved in Mendelian non-syndromic obesity should also be carefully screened in patients presenting with obesity associated with syndromic clinical features. After the identification of the pathogenic mutation in PTEN (encoding phosphatase and tensin homolog) in a morbidly obese boy with large abdominal lipoma and his mother, we re-examined the boy and his mother. Although not presenting with intellectual disability, we suspected that the boy had the Bannayan-Riley-Ruvalcaba syndrome while the mother had Cowden syndrome. In both cases, lifetime risks for a variety of cancers is very high, in particular for breast cancer (85%), thyroid cancer (35%), endometrium cancer (28%), colorectal cancer (9%), kidney cancer (33%), and melanoma (6%) [32]. Therefore, the boy and the mother will be under a life-long comprehensive surveillance by clinicians. The mother has elected to undergo a preventive endometrectomy and mastectomy. In another boy presenting with very severe intellectual disability (no language) associated with behavioral disorder, seizures, neonatal hypotonia, kyphosis, cross-eyedness, incontinence, ataxic gait and sleep disorder, we identified a pathogenic mutation in CASR (encoding calcium sensing receptor), which probably caused his epileptic seizures, and two compound heterozygous mutations in MTHFR (encoding methylene tetrahydrofolate reductase), which probably caused most of the severe clinical phenotypes of the boy. MTHFR deficiency may be treated with high-dose betaine, methionine, pyridoxine, vitamin B12, and folic acid supplements [33].
5. Conclusion
The CoDE-seq approach has proven cost-efficient for the accurate detection of both CNVs and coding point mutations and seems highly effective for the molecular diagnosis of intellectual disability and obesity, reaching more than 30% of success. We believe that in 2018, this method remains far easier to handle and less expensive than whole-genome sequencing, and therefore should be useful in clinical practice.
Acknowledgments
The authors would like to thank the children, young adults, and their families who participated in this study. We thank Frédéric Allegaert and Nicolas Larcher for their contribution to DNA extraction. We are also grateful to Françoise Boidein, Marie-Bertille Dehouck and Christine Vassel for collecting clinical data. This work was supported by grants from the French National Research Agency (ANR-10-LABX-46 [European Genomics Institute for Diabetes] and ANR-10-EQPX-07-01 [LIGAN-PM], to PF), from the European Research Council (ERC GEPIDIAB – 294785, to PF; ERC Reg-Seq – 715575, to AB) and from FEDER (to PF). AB was supported by Inserm.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.05.005.
Contributor Information
Louise Montagne, Email: montagne.louise@ghicl.net.
Philippe Froguel, Email: p.froguel@imperial.ac.uk.
Amélie Bonnefond, Email: amelie.bonnefond@inserm.fr.
Conflict of interest
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Moustafa J.S.El-Sayed, Froguel P. From obesity genetics to the future of personalized obesity therapy. Nature Reviews Endocrinology. 2013;9(7):402–413. doi: 10.1038/nrendo.2013.57. [DOI] [PubMed] [Google Scholar]
- 2.Ramachandrappa S., Farooqi I.S. Genetic approaches to understanding human obesity. The Journal of Clinical Investigation. 2011;121(6):2080–2086. doi: 10.1172/JCI46044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463(7281):671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463(7281):666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farooqi I.S., Jebb S.A., Langmack G., Lawrence E., Cheetham C.H., Prentice A.M. Effects of recombinant leptin therapy in a child with congenital leptin deficiency. New England Journal of Medicine. 1999;341(12):879–884. doi: 10.1056/NEJM199909163411204. [DOI] [PubMed] [Google Scholar]
- 6.Kühnen P., Clément K., Wiegand S., Blankenstein O., Gottesdiener K., Martini L.L. Proopiomelanocortin deficiency treated with a Melanocortin-4 receptor agonist. New England Journal of Medicine. 2016;375(3):240–246. doi: 10.1056/NEJMoa1512693. [DOI] [PubMed] [Google Scholar]
- 7.Bonnefond A., Keller R., Meyre D., Stutzmann F., Thuillier D., Stefanov D.G. Eating behavior, low-frequency functional mutations in the Melanocortin-4 receptor (MC4R) gene, and outcomes of bariatric operations: a 6-year prospective study. Diabetes Care. 2016;39(8):1384–1392. doi: 10.2337/dc16-0115. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England Journal of Medicine. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan T.Y., Dillon O.J., Stark Z., Schofield D., Alam K., Shrestha R. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatrics. 2017;171(9):855–862. doi: 10.1001/jamapediatrics.2017.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnefond A., Durand E., Sand O., De Graeve F., Gallina S., Busiah K. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5(10):e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong C.S., Singh L.N., Mullikin J.C., Biesecker L.G. Assessing the reproducibility of exome copy number variations predictions. Genome Medicine. 2016;8(1):82. doi: 10.1186/s13073-016-0336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. American Journal of Human Genetics. 2016;98(1):58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl Resources. The American Journal of Human Genetics. 2009;84(4):524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Human Mutation. 2016;37(3):235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american College of medical genetics and genomics and the association for molecular pathology. Genetics in Medicine: Official Journal of the American College of Medical Genetics. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nature Protocols. 2016;11(1):1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 22.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Research. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper G.M., Stone E.A., Asimenos G., NISC Comparative Sequencing Program, Green E.D., Batzoglou S. Distribution and intensity of constraint in mammalian genomic sequence. Genome Research. 2005;15(7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromer M., Moran J.L., Chambert K., Banks E., Bergen S.E., Ruderfer D.M. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. American Journal of Human Genetics. 2012;91(4):597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velagaleti G.V., Morgan D.L., Tonk V.S. Trisomy 5p. A case report and review. Annales De Genetique. 2000;43(3–4):143–145. doi: 10.1016/s0003-3995(00)01030-3. [DOI] [PubMed] [Google Scholar]
- 27.van Duyvenvoorde H.A., Lui J.C., Kant S.G., Oostdijk W., Gijsbers A.C.J., Hoffer M.J.V. Copy number variants in patients with short stature. European Journal of Human Genetics: European Journal of Human Genetics. 2014;22(5):602–609. doi: 10.1038/ejhg.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Froyen G., Belet S., Martinez F., Santos-Rebouças C.B., Declercq M., Verbeeck J. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. American Journal of Human Genetics. 2012;91(2):252–264. doi: 10.1016/j.ajhg.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castiglia L., Husain R.A., Marquardt I., Fink C., Liehr T., Serino D. 7q11.23 microduplication syndrome: neurophysiological and neuroradiological insights into a rare chromosomal disorder. Journal of Intellectual Disability Research: Journal of Intellectual Disability Research. 2017 doi: 10.1111/jir.12457. [DOI] [PubMed] [Google Scholar]
- 30.Gillentine M.A., Berry L.N., Goin-Kochel R.P., Ali M.A., Ge J., Guffey D. The cognitive and behavioral phenotypes of individuals with CHRNA7 duplications. Journal of Autism and Developmental Disorders. 2017;47(3):549–562. doi: 10.1007/s10803-016-2961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in Medicine: Official Journal of the American College of Medical Genetics. 2011;13(9):777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan M.-H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2012;18(2):400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levin B.L., Varga E. MTHFR: addressing genetic counseling dilemmas using evidence-based literature. Journal of Genetic Counseling. 2016;25(5):901–911. doi: 10.1007/s10897-016-9956-7. [DOI] [PubMed] [Google Scholar]
- 34.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vacca M., Filippini F., Budillon A., Rossi V., Mercadante G., Manzati E. Mutation analysis of the MECP2 gene in British and Italian Rett syndrome females. Journal of Molecular Medicine (Berlin, Germany) 2001;78(11):648–655. doi: 10.1007/s001090000155. [DOI] [PubMed] [Google Scholar]
- 36.Vaisse C., Clement K., Durand E., Hercberg S., Guy-Grand B., Froguel P. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. The Journal of Clinical Investigation. 2000;106(2):253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelen M.R., van Staveren W.C., Peeters E.A., Hassel M.B., Gorlin R.J., Hamm H. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Human Molecular Genetics. 1997;6(8):1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 38.Vargas-Poussou R., Mansour-Hendili L., Baron S., Bertocchio J.-P., Travers C., Simian C. Familial hypocalciuric hypercalcemia types 1 and 3 and primary hyperparathyroidism: similarities and differences. Journal of Clinical Endocrinology & Metabolism. 2016;101(5):2185–2195. doi: 10.1210/jc.2015-3442. [DOI] [PubMed] [Google Scholar]
- 39.Tonetti C., Saudubray J.-M., Echenne B., Landrieu P., Giraudier S., Zittoun J. Relations between molecular and biological abnormalities in 11 families from siblings affected with methylenetetrahydrofolate reductase deficiency. European Journal of Pediatrics. 2003;162(7–8):466–475. doi: 10.1007/s00431-003-1196-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.