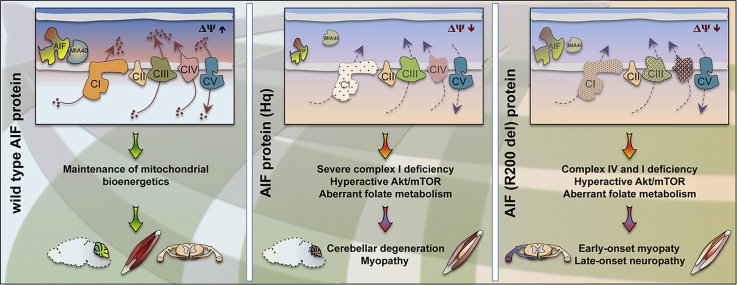
Abstract
Objective
Mutations in the AIFM1 gene have been identified in recessive X-linked mitochondrial diseases. Functional and molecular consequences of these pathogenic AIFM1 mutations have been poorly studied in vivo.
Methods/results
Here we provide evidence that the disease-associated apoptosis-inducing factor (AIF) deletion arginine 201 (R200 in rodents) causes pathology in knockin mice. Within a few months, posttranslational loss of the mutant AIF protein induces severe myopathy associated with a lower number of cytochrome c oxidase-positive muscle fibers. At a later stage, Aifm1 (R200 del) knockin mice manifest peripheral neuropathy, but they do not show neurodegenerative processes in the cerebellum, as observed in age-matched hypomorphic Harlequin (Hq) mutant mice. Quantitative proteomic and biochemical data highlight common molecular signatures of mitochondrial diseases, including aberrant folate-driven one-carbon metabolism and sustained Akt/mTOR signaling.
Conclusion
Our findings indicate metabolic defects and distinct tissue-specific vulnerability due to a disease-causing AIFM1 mutation, with many pathological hallmarks that resemble those seen in patients.
Keywords: Akt/mTOR signaling, Apoptosis-inducing factor (AIF), 1C metabolism, Mitochondria, Mitochondrial diseases, Oxidative phosphorylation
Graphical abstract
Highlights
-
•
Aifm1 (R200 del) mutation causes early-onset myopathy in knockin mice.
-
•
Aifm1 (R200 del) knockin animals do not develop cerebellar degeneration.
-
•
Expression of mutant AIF alters mitochondrial function and Akt/mTOR activity.
-
•
AIF deficiency induces aberrant folate-driven one-carbon metabolism.
-
•
Rapamycin treatment improves metabolic dysfunction in AIF deficient mice.
1. Introduction
Apoptosis-inducing factor (AIF) was originally described as a pro-death molecule that is released from mitochondria during caspase-dependent and independent cell death [1], [2], [3]. Upon proteolysis of the membrane-tethered precursor, AIF translocates into the nucleus, where it participates in chromatin condensation and DNA degradation (reviewed in [4], [5], [6]). Apart from its contribution to various cell death pathways, AIF has a fundamental housekeeping role in mitochondrial bioenergetics (reviewed in [5], [6]). In an evolutionarily conserved manner, lack of AIF alters the expression of several respiratory complex subunits at the posttranscriptional level, resulting in aberrant oxidative phosphorylation (OXPHOS) [7]. Since AIF is a FAD- and NADH-binding low-turnover oxidoreductase [4], [8], it was initially proposed that AIF could act as a broad range antioxidant enzyme in the mitochondrial intermembrane space [9]. However, recent insights provide a clearer molecular mechanism underlying AIF regulation of the OXPHOS system. In patient-derived fibroblasts as well as in various cells from transgenic mice, AIF deficiency causes decreased expression of the coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHD4) (orthologue of the yeast Mia40). As a result of its binding to CHCHD4, AIF contributes indirectly to mitochondrial import and oxidative folding pathways, thereby regulating the assembly of the respiratory complexes [10], [11], [12]. Although these coherent findings seem to provide conclusive molecular mechanisms, the complex pleiotropic effects of AIF deficiency may alter mitochondrial bioenergetics through additional pathways. For instance, among the newly identified interacting partners, AIF and CHCHD4 are both targets of thioredoxin-like proteins, which regulate the redox status of the cell [12]. Moreover, AIF directly binds the tumor suppressor PTEN (phosphatase and tensin homog on chromosome ten) and prevents its oxidative inactivation [13], thereby altering Akt activity and, consequently, overall metabolism. Therefore, we believe that a substantial number of AIF-binding partners may participate in the stepwise-regulated structural assembly and correct maintenance of the OXPHOS system.
Over the past few years, several pathogenic mutations in the AIFM1 locus have been causally implicated in a set of X-linked recessive human disorders (reviewed in [6]). Although clinical manifestations are extremely variable, in most of the reported cases patients exhibited metabolic changes and altered mitochondrial bioenergetics, with many associated features commonly observed in multisystemic diseases generally known as human mitochondrial disorders [14], [15], [16], [17]. The spectrum of AIFM1-related disorders includes many syndromes with various clinical symptoms and clear signs of degenerative processes. The first documented cases of AIFM1 mutations were two male infants with severe encephalomyopathy [18]. In the AIFM1 locus, an ablation of an entire nucleotide triplet coding for the arginine residue 201 (R201 del) results in a mutant AIF protein. Biochemical and structural studies of recombinant wild type and mutant AIF proteins demonstrate aberrant folding and FAD incorporation [18], [19], while patient-derived fibroblasts show altered expression of complex III (CIII) and complex IV (CIV) subunits [18]. Since the first reported case, additional pathogenic mutations in the AIFM1 gene have been described in individuals with complex multisystem disorders featuring a wide range of clinical manifestations, including prominent neurological deficits, progressive muscular wasting and ataxia, hearing loss, optic atrophy, retinopathy, neuropathy, hypomyelination, and spondylometaphyseal dysplasia [18], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]. Notably, additional variants in the AIFM1 gene seem to co-segregate with certain forms of childhood-onset hearing loss, further expanding the spectrum of AIFM1-linked diseases [23]. Most of the annotated pathogenic mutations have structural and functional consequences on AIF properties [18], [19]. However, these in vitro findings correlate only partially with the clinical profiles of patients, making the interpretation of their physiological relevance extremely difficult, especially in the context of drug development.
AIF deficiency has been widely studied in a few animal models, such as the hypomorphic Harlequin (Hq) mutant mice [9]. Contrary to the observed embryonic lethality of Aifm1 knockouts [30], Hq mutant mice are viable and show significant temporal variability in terms of phenotypic abnormalities and degenerative lesions [9], [31]. At the biochemical level, Hq mutant mice show an 80% reduction of AIF expression that leads to a compromised expression of respiratory complex subunits in the optic nerve, retina, brain and skeletal muscle [7], [31]. In an apparent discrepancy, the OXPHOS system is basically unaffected in the heart and liver of Hq mutant mice, whereas it is considerably impaired in organ-specific Aifm1 knockout mice [7], [31], [32], [33]. Throughout different tissues, a certain degree of correlation exists between the residual complex I (CI) activity, pathology onset and progression. However, body weight, growth retardation, fur anomalies and neurological symptoms vary considerably among individual mice. In terms of pathology, Hq mutant animals show extensive neurodegeneration in the cerebellum and retina and, consequently, develop progressive ataxia and become blind between 4 and 7 months of age [9], [31], [34]. Despite these caveats, hypomorphic Hq mutant mice, along with tissue-specific Aifm1 knockout animals, have been widely used to study AIFM1-mediated mitochondrial dysfunction. Nevertheless, since these transgenic mice are models of AIFM1 gene disruption rather than AIF dysfunction, they may not recapitulate the exact molecular pathogenesis of AIFM1 mutations, resulting in animals with pathophysiological profiles and phenotypic traits different from those observed in human patients. Consistent with this view, Hq mutant mice are generally accepted as valuable in vivo models of mitochondrial disorders associated with CI deficiency [32], [35], [36].
Given the clinical heterogeneity of AIFM1-linked disorders and the unclear relationship between the genetic etiology and clinical outcomes, the study of AIFM1-related diseases is extremely challenging, with impaired mitochondrial bioenergetics as the only common denominator. As a consequence, there is a lack of reliable tractable models that recapitulate most of the relevant features observed in patients, undermining preclinical drug testing and the development of future therapies. Here, we provide the first unequivocal evidence that a disease-associated mutation in the Aifm1 gene causes pathology in vivo in mice. In a newly developed Aifm1 (R200 del) knockin mouse model, we show that Aifm1 mutant allele has an mRNA expression pattern similar to the wild type one. Across different tissues, the mutant AIF polypeptide is consistently downregulated in a posttranslational manner. Importantly, the residual amount of the AIF (R200 del) variant is comparable to the wild type AIF protein in Hq mice. As in patients, Aifm1 (R200 del) mice display OXPHOS deficiency in the skeletal muscle, including accumulation of nemaline rod-like structures, reduced COX activity and consequent muscular weakness. At 3 and 6 months of age, Aifm1 (R200 del) animals do not exhibit cerebellar degeneration, but instead show consistent and evident signs of peripheral neuropathy (i.e., swelling and demyelination of axons in the sciatic nerve) between 6 and 12 months of age. In line with previously published findings [7], [9], [10], [11], [31], [32], we show that AIF deficiency causes mitochondrial dysfunction in a tissue-specific fashion. As in other mouse models of mitochondrial diseases, we provide evidence that the AIF (R200 del) variant leads to hyperactive Akt/mTOR signaling, aberrant folate-driven one-carbon (1C) metabolism [37], [38], [39] and potential cataplerotic adaptations linked to enhanced glycolysis. Together, our findings describe molecular and metabolic signatures associated with a distinct disease-causing mutation in the Aifm1 gene, highlighting further the complex etiology of AIFM1-linked mitochondrial diseases.
2. Materials and methods
2.1. Animals
Hq mutant (JAX stock number: 000501) breeding pairs were obtained from The Jackson Laboratory (Bar Harbor, Maine, USA). All mice were housed in groups of two to four under a 12/12 h light/dark cycle (lights on at 6:00 am) with free access to food (ssniff® V1534-300) and tap water. All experiments were approved and performed in conformity to the guidelines of the State Agency for Nature, Environment and Consumer Protection in North Rhine Westphalia. Taconic Biosciences GmbH (Cologne, Germany) generated the targeting vector and performed transfection of ES cells, homologous recombination, in vitro removal of neomycin and puromycin cassettes, ES cells injection in the blastocysts, selection of chimeric mice, and heterozygous transgenic female founders.
2.2. Antibodies
The following antibodies were used: mouse anti-actin (Abcam ab14128), rabbit anti-AIF (Cell Signaling 5318), rabbit anti-Akt (Cell Signaling 4685), rabbit anti-phospho-Akt (Cell Signaling 4058), rabbit anti-βIII tubulin (Cell Signaling 5666), rabbit anti-calbindin (Swant CD 68), rabbit anti-Diablo (Abcam ab8115), rabbit anti-GAPDH (Cell Signalling 2118), mouse anti-GFAP (Cell Signaling 3670), rabbit anti-Iba1 (Wako 019–19741), rabbit anti-GSR (Abcam, ab16801), rabbit anti-MAP2 (Abcam ab 40390), rabbit anti-MBP (Abcam ab40390), rabbit anti-MIA40 (CHCHD4, Protein Tech 21090-1 AP), rabbit anti-MTHFD2 (Abcam ab151447), mouse anti-NDUFA9 (Abcam, ab14713), rabbit anti-NDUFB7 (Protein Tech 14912-1 AP), mouse anti-NDUFS3 (Abcam ab14711), mouse anti-Neurofilament (SMI 312, BioLegend 837904) mouse anti-OXPHOS rodent cocktail (Mitosciences MS604), rabbit anti-p70-S6K (Cell Signaling 9202), rabbit anti-phospho-p70-S6K (Cell Signaling 9205), rabbit anti-PCK1 (Cell Signaling 12940), rabbit anti-PCK2 (Cell Signaling 6924), rabbit anti-PKM1 (Cell Signaling 7067), rabbit anti-PKM2 (Cell Signaling 4053), rabbit anti-PRAS40 (Cell Signaling 2691), rabbit anti-phospho-PRAS40 (Cell Signaling 13175), rabbit anti-RPS6 (Cell Signaling 2217), rabbit anti-phospho-RPS6 (Cell Signaling 4858), rabbit anti-TOM20 (SantaCruz sc-11415), goat anti-rabbit and anti-mouse HRP-conjugated secondary antibodies (Promega), goat anti-mouse and anti-rabbit Alexa Fluor 488 and 568-conjugated secondary antibodies (Life Technologies).
2.3. Cell culture
Primary cortical neurons were isolated from E16.5/17.5 embryos and maintained in Neurobasal medium (Invitrogen) supplemented with 2% glutamax and 2% B27 supplement (Invitrogen) as previously described [40], [41]. Cells were kept in culture for 5 days and then either collected for biochemical analyses or fixed with 4% PFA for immunocytochemistry. Mouse embryonic fibroblasts (MEFs) were isolated from E14.5 embryos and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). Cells were collected for biochemistry 24 h after plating.
2.4. COX and SDH analysis
COX/SDH staining was performed on 10 μm cross-sections of fresh frozen quadriceps previously cut on a cryostat and collected on microscope slides. Briefly, sections were incubated for 60 min at 37 °C with COX (4 mM 3,3-diaminobenzidine tetrahydrochloride, 100 μM cytochrome c and 20 μg/ml catalase in 0.05 M sodium phosphate buffer, pH 7,4) and SDH (1,875 mM nitroblue tetrazolium, 1 mM sodium azide, 200 μM phenazine methosulphate, 130 mM sodium succinate, in 0.1 M phosphate buffer) reaction solution, dehydrated in ascending ethanol series, cleared with Xylene, and coverslipped with DePeX® (VWR).
2.5. Imaging
Brightfield images were obtained using an EPI-SCOPE1 (Carl Zeiss) equipped with a 10x objective. Image processing, such as stitching of low magnification photographs, was done with the ZEN 2012 Blue edition software (Carl Zeiss). Fluorescent images were taken using a LSM700 Zeiss confocal microscope with a 20x Plan-Apochromat objective. Stack images were collected by sequential scanning and maximum intensity projections of acquired z-stacks was done in ImageJ. Image analyses were performed in a blinded manner using ImageJ.
2.6. Immunohistochemistry
Tissue sections were blocked for 1 h at room temperature in blocking buffer containing 10% normal goat serum and 0.1% Triton X-100. Following incubation with primary antibodies overnight at 4 °C, sections were washed and incubated with appropriate Alexa Fluor-conjugated secondary antibodies for 2 h at room temperature. After counterstaining with Hoechst-33342, sections were mounted and coverslipped with fluorescence mounting medium (DAKO).
2.7. Modified Gomori trichrome staining
Fresh-frozen quadriceps were cross-sectioned at 10 μm on a cryostat, stained with hematoxylin and eosin (Sigma–Aldrich) and ultimately incubated for 20 min in Gomori staining solution (0.6% chromotrop 2R, 0.3% Fast Green FCF, 0.6% phosphotungstic acid, 1% acetic acid in distilled water). Following dehydration in ascending ethanol series, sections were cleared with Xylene and coverslipped with DePeX® (VWR). Three non-adjacent sections and at least 60 muscle fibers per animal were analyzed, and the number of fibers showing myopathic features was counted manually.
2.8. Oxygen consumption rate
Oxygen consumption rate (OCR) of cells was measured with an XF24 Extracellular Flux Analyzer via the XF Cell Mito Stress Kit (Seahorse Bioscience). Cells were seeded at 4 × 104 cells/well 24 h before the assay. One hour before the measurement, cells were incubated at 37 °C in a CO2-free incubator. Baseline OCR was measured for 15–20 min, while changes in OCR were assessed following addition of oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) and rotenone/anitmycin A (final concentrations were 1, 0.5 and 0.5 μM respectively). After recording, cells were detached with trypsin and harvested immediately. The number of cells per well was counted and used to normalize the corresponding OCR.
2.9. Proteomic analysis (TMT/iTRAQ scan)
Snap-frozen quadriceps muscle samples from three 6 month-old Aifm1KI/y mice and three control littermates were used for proteomic analysis. DC Biosciences Ltd (Scotland, UK) performed lysis of the samples, protein precipitation, digestion and labeling with a TMT™ Mass Tag Labeling kit. Peptide samples were reconstituted, fractionated, and subjected to LC-MS/MS/MS analysis. Raw MS data were analyzed in MaxQuant. Data were then further reanalyzed, renormalized, and requantified in R.
2.10. Rapamycin treatment and grip strength
Rapamycin (LC Laboratories #R-5000) was dissolved in physiological saline solution supplemented with 5% PEG-400 and 5% Tween-80. Mice were weighed daily and injected intraperitoneally (i.p.) with a dose of 8 mg/kg per day. Control animals received daily vehicle injections at a volume of 66 μl/10 g body weight. Treatment started when the animals were between 12 and 14 weeks of age and lasted for a total of 14 days. Twenty-four hours after the last injection, vehicle and rapamycin-treated mice were tested for grip strength to assess muscle force. Here, animals were held at the tail, allowed to grab onto the bar of a grip strength meter (TSE) with their forepaws, and then gently pulled backwards until they released their grip. Each mouse was tested three times and the average of these measurements was used.
2.11. Real-time PCR
Total RNA extraction was performed on snap frozen tissue samples using QIAzol, QIAshredder, and RNeasy mini kit (Qiagen). DNase I digestion was carried out with the RNase-free DNase kit (Qiagen) and RNA concentrations were determined by spectrometry (Nano Drop, Thermo Fisher). RNA extracts (100 ng) were retrotranscribed using the qScript cDNA SuperMix (Quanta Bioscience). Gene expression analysis was conducted with FastSYBR Green Master Mix (Applied Biosystems) on a Step One Plus Real Time PCR System (Applied Biosystems). Primers used for RT-PCR were as follows: Aifm1 (exon 1–2) F 5′-gacggtgtacgtccagagg-3′ R 5′-gaggaacacgccattgct-3’; Aifm1 (exon 6–7) F 5′-tcagacagtggaatggaaaaga-3′ R 5′-taggcaggtcctgagcaga; Aifm1 (exon 12–13) F 5′-tccaagcacgttctaacatctg-3′ R 5′-gccttcgacccaactttatatc-3’; Asns F 5′-gagaaactcttcccaggcttg-3′ R 5′-caagcgtttcttgatagcgttgt-3’; β-actin F 5′-ctaaggccaaccgtgaaaag-3′ R 5′-accagaggcatacagggaca-3’; Fgf21 F 5′-ctggggtctaccaagcata-3′ R 5′-cacccaggatttgaatgacc-3; Gdf15 F 5′-caaccagagccgagaggac-3′ R 5′-tgcacgcggtaggcttc-3’; Mthfd2 F 5′-catggggcgtgtgggagataat-3′ R 5′-ccgggccgttcgtgagc-3’; Phgdh F 5′-atggccttcgcaaatctgc-3′ R 5′-agttcagctatcagctcctcc-3’; Psat1 F 5′-cagtggagcgccagaatagaa-3′ R 5′-cctgtgccccttcaaggag-3’; Psph F 5′-gtcctcctgctgatggctttc-3′ R 5′-gtaccacttggcgttgtcct-3’; Shmt2 F 5′- tggcaagagatactacggagg-3′ R 5′- cgaggtccaaccccatgat-3’; ΔΔCt values were normalized to β-actin and represented as fold change compared to control mice.
2.12. SDS gel electrophoresis and western blot analysis
Cell pellets and snap frozen tissues were lysed and sonicated in ice-cold RIPA buffer (Sigma Aldrich) supplemented with protease and phosphatase inhibitors (Roche). Proteins were resolved on a 10–12% acrylamide gel and transferred onto nitrocellulose membranes (Bio-Rad). Following 1 h of blocking, membranes were incubated with primary antibodies overnight at 4 °C. On the next day, membranes were washed and incubated with appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. Immunoblots were developed using the chemiluminescent analyzer Chemidoc imaging system (Bio-Rad) and quantified by densitometry (ImageLab software, Bio-Rad).
2.13. Statistics
Data are expressed as mean ± S.E.M. and statistical analyses were performed with Graph Pad Prism software. Unpaired t-tests were used to compare differences between genotype groups (e.g. Aifm1wt/Y vs. Aifm1KI/Y; wt vs. Hq). Data from rapamycin and vehicle treated mice were analyzed using a two-way analysis of variance (ANOVA) with genotype as the within-group and treatment as the between-groups factor. In the case of significant main effects, Bonferroni post-hoc pairwise comparisons were performed. The statistical significance was defined as p < 0.05.
2.14. Tissue preparation
Animals were sacrificed via cervical dislocation. Tissue samples were rapidly removed, snap frozen in liquid nitrogen and stored at −80 °C until further processing. Alternatively, mice were anaesthetized with an overdose of Ketamine:Xylazine and transcardially perfused with phosphate buffered saline (PBS) followed by 4% paraformaldehyde (PFA)-containing PBS. Brains were removed from the scull, post-fixed for 24 h in 4% PFA, and stored in 30% sucrose solution until further processing. A series of four to six 40 μm sagittal brain sections were cut on a cryostat (CryoStar NX700) and collected in PBS. One series was mounted immediately and Nissl-stained with Cresyl Violet (ScyTek); the other series were kept in a cryoprotective solution and stored at −20 °C until further processing. Sciatic nerves were fixed in 4% PFA, paraffin-embedded and cut at a thickness of 5 μm thickness.
3. Results
3.1. Aifm1 (R200 del) knockin mice develop early-onset myopathy
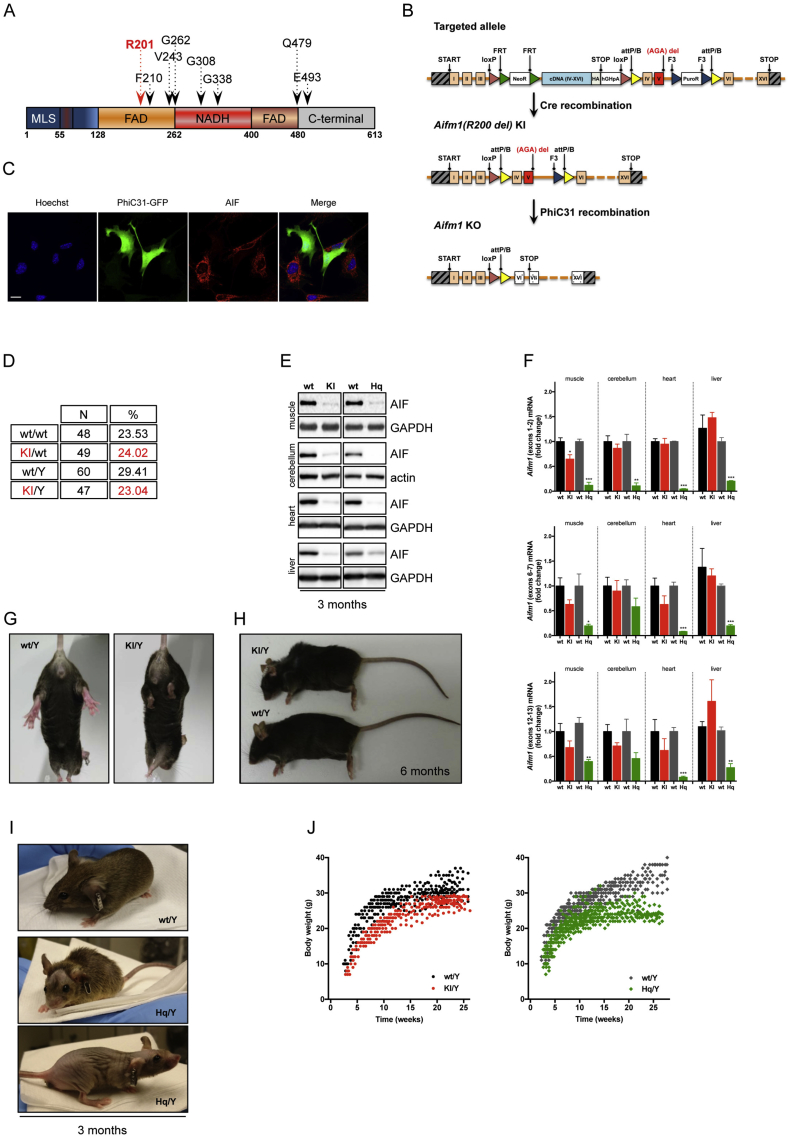
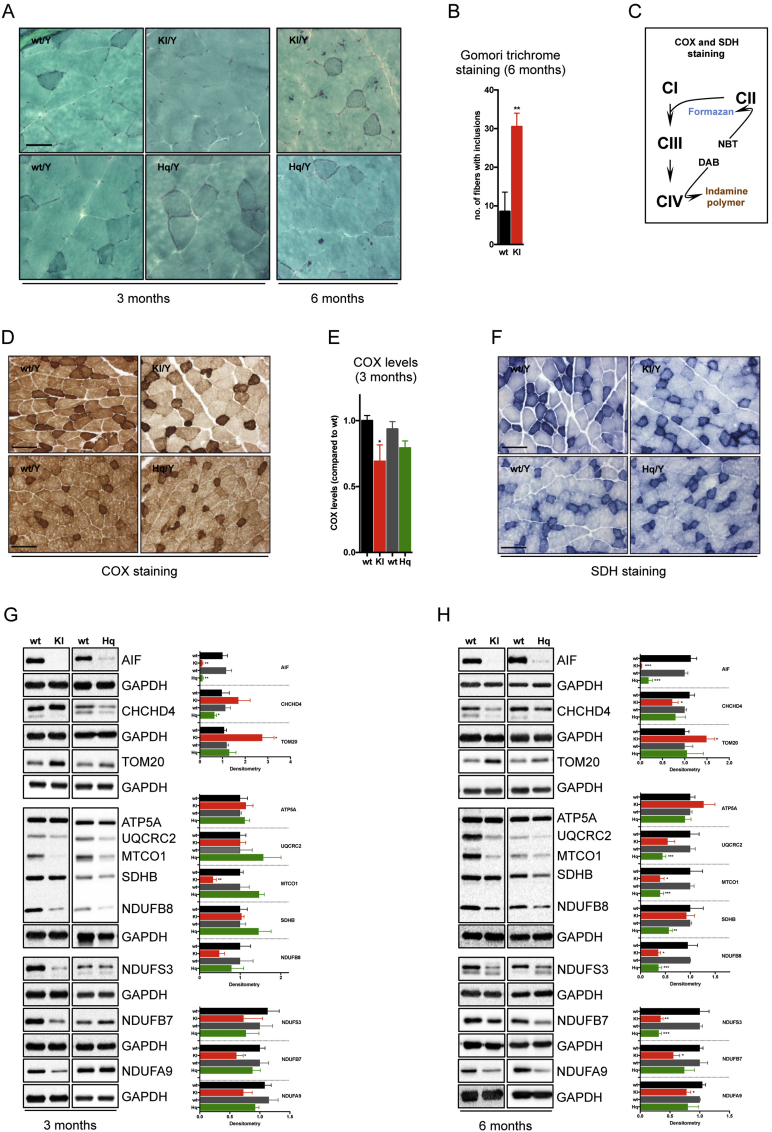
Several disease-causing mutations in the AIFM1 gene have been identified in patients (Figure 1A). Based on prior biochemical analyses, recombinant human AIF protein lacking R201 shows conformational instability and altered redox properties [18]. To study the pathological implications of this mutation in vivo in rodents, we designed a targeting strategy that allowed both the generation of conditional Aifm1 (R200 del) knockin as well as Aifm1 knockout mice. The targeting vector included a loxP-flanked sequence encoding cDNA for exon 4 to 16, the sequence of 3 HA-tags and an additional polyadenylation tail (Figure 1B). The construct was inserted into intron 3 of the native Aifm1 locus, theoretically allowing the expression of a HA-tagged AIF protein. Exon 5 of the native Aifm1 gene carried a deletion for the amino acid R200 (R200 del). Both exon 4 and the mutated exon 5 were flanked with attP/attB sites, which allow the knockout of the Aifm1 gene upon PhiC31-driven recombination (Figure 1C). After transfection in male C57BL/6NTac ES cells, homologous recombinant clones were isolated using double positive Neomycin and Puromycin selection markers flanked by FRT and F3 sites, respectively. The correct genetic recombination was assessed by Southern blot analysis (data not shown). We tested the expression of HA-tagged AIF protein in the resulting positive ES cell clones. Surprisingly, we could not detect AIF expression at the protein and mRNA levels, suggesting that the mRNA splicing and/or maturation were somehow compromised (data not shown). Nevertheless, targeted ES cells were injected after recombinase-mediated in vitro deletion of the selection markers, and several chimeras were obtained. Ultimately, two females with a germline transmission for the transgene were identified as colony founders. Upon breeding with wild type mice, we obtained a very limited number of heterozygous transgenic females, indicating an unexpected mortality of the mutation carriers despite the remaining wild type Aifm1 allele. We hypothesized that the inserted sequence affected mouse survival, perhaps interfering in trans with the maturation or translation of wild type Aifm1 mRNA. Thus, few original female founders were crossed with a male ubiquitously expressing the bacterial Cre-recombinase. Germline expression of the Cre recombinase resulted in genetic recombination of the floxed Aifm1 locus and expression of mutated Aifm1 (R200 del) (Figure 1B). DNA sequencing of Aifm1 exon 5 confirmed the correct recombination (data not shown). From further breeding, we recovered a certain number of viable heterozygous Aifm1 (R200 del) females and hemizygous Aifm1 (R200 del) males in a ratio slightly lower than the expected Mendelian inheritance pattern (Figure 1D). We tested AIF protein expression levels in muscle, cerebellum, heart, and liver tissues from 3 month-old wild type, Hq mutant and Aifm1 (R200 del) males. Likewise, we measured Aifm1 mRNA expression across different organs using a set of validated primers for quantitative RT-PCR. Since Hq mutant animals and Aifm1 (R200 del) mice have different background strains, the relative wild type littermates were used as control animals in all experimental set-ups. Compared to wild type mice, both Hq mutant and Aifm1 (R200 del) males exhibited a similar reduction of the mature ∼62 kDa AIF protein across different tissues (Figure 1E). However, Aifm1 mRNA expression did not differ between Aifm1 (R200 del) and wild type males, contrary to Hq mutant animals, where a significant reduction of Aifm1 transcripts occurred in all tested tissues (Figure 1F). In line with prior in vitro evidence [18], these findings further support that the loss of mutant AIF protein occurs posttranslationally due to its high instability in mammalian cells. As part of our in vivo characterization, we went on with phenotypical and biochemical assessments of Aifm1 (R200 del) mice, using wild type and Hq mutant animals as comparison. While Hq mutant males were highly variable in terms of body weight and baldness, Aifm1 (R200 del) males had normal fur, weighed less than wild type animals but with only slight differences across individuals of the same cohort (Figure 1G–J). Importantly, around 4–5 months of age, Aifm1 (R200 del) homozygous females and hemizygous males displayed hind limb clasping and developed kyphosis (Figure 1G–H), indicating potential muscle atrophy and innervation defects. To provide additional evidence of abnormalities in the skeletal muscle, we performed a modified Gomori trichrome staining on quadriceps muscle sections. Consistent with the early development of myopathy-like features, Aifm1 (R200 del) males exhibited an increased number of nemaline rod-like structures in skeletal muscle fibers between 3 and 6 months of age (Figure 2A–B). Given these observations, we sought to define the biochemical and physiological effects of the mutant AIF protein. Cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) staining (Figure 2C) is a widely used method to assess defects of mitochondrial respiratory complexes in model organisms as well as for diagnostic purposes in patients [42]. Compared to wild type animals, Aifm1 (R200 del) as well as Hq mutant mice exhibited a significantly decreased formation of the brown indamine polymer product in skeletal muscle sections. Notably, Aifm1 (R200 del) mice showed a more pronounced loss of COX-positive muscle fibers compared to Hq mutant animals at 3 months of age (Figure 2D–E), indicating reduced CIV activity. SDH-mediated reduction of nitroblue tetrazolium to blue formazan was relatively similar throughout the different genotypes (Figure 2F). Since AIF deficiency induces tissue-dependent OXPHOS impairment according to the genetic manipulation [7], [31], [32], [33], we performed comprehensive immunoblot analyses using a wide range of validated antibodies. Over time, we found that AIF loss altered CHCHD4 expression in skeletal muscle (Figure 2G–H). At 3 months of age, Aifm1 (R200 del) animals exhibited a clear trend towards a decreased expression of CI and CIV subunits, which became even more significant at 6 months of age (Figure 2G–H). Of note, Hq mutant mice displayed a milder reduction of CI and CIV subunits at 3 months of age, whereas many other respiratory complex subunits were altered significantly at a later stage (Figure 2G–H). Together, our in vivo data indicate that mutant AIF protein causes mitochondrial dysfunction and early-onset myopathy.
Figure 1.
Aifm1 (R200 del) mice have reduced AIF protein levels across organs and display homogenous phenotypic traits. (A) Schematic representation of the human AIF protein. MLS indicates the mitochondrial localization sequence; FAD and NADH are the FAD-binding and NADH-binding motifs, respectively. Numbers define distinct domains within the AIF protein. Arrows indicate mutated residues identified in patients (in red, R201 deletion). (B) Schematic representation of the genetic targeting strategy, enabling the generation of conditional Aifm1 (R200 del) knockin (KI) as well as Aifm1 knockout (KO) mice. (C) Overexpression of PhiC31-GFP in Aifm1 (R200 del) MEFs resulted in AIF knockout. Green-positive cells displayed loss of AIF staining (red). Hoechst-33342 (blue) was used to visualize nuclei. Scale bar = 20 μm. (D) Aifm1 (R200 del) knockin (KI) mice were born at almost the expected Mendelian ratio. (E) Immunoblot and (F) RT-PCR analyses for Aifm1 expression levels in wt, Aifm1 (R200 del) KI and Hq mutant mice across different organs (for RT-PCR: mean ± SEM, Student's t-test, n = 3–5 per genotype, ***p < 0.001, **p < 0.01, *p < 0.05 compared to littermates). (G–H) Aifm1 (R200 del) KI mice developed (G) hind limb clasping and (H) kyphosis around 6 months of age. (I–J) Hq mutant mice showed high variability in phenotypic traits, such as (I) fur loss and (J) body weight changes (n = 15–20 per genotype); (J) compared to littermate controls, Aifm1 (R200 del) KI mice had a consistent decrease in body weight (n = 18–20 per genotype).
Figure 2.
Aifm1 (R200 del) mice show pathological features associated with OXPHOS deficiency in skeletal muscle. (A) Modified Gomori trichrome staining on transverse quadriceps muscle sections in Aifm1 (R200 del) KI, Hq mutant, and respective control animals. Scale bar = 50 μm. (B) At 6 months of age, Aifm1 (R200 del) KI mice showed a significantly higher number of muscle fibers with cytosolic inclusions compared to age-matched wt littermates (mean ± SEM, Student's t-test, n = 4–5 per genotype, **p < 0.01). (C) Schematic representation of the COX and SDH staining. Visualization of COX activity is based on the use of 3,3′-diaminobenzidine (DAB) as electron donor. The reaction product on oxidation of DAB occurs as brown pigmentation corresponding to the distribution of mitochondria. Visualization of SDH activity is based on the use of nitro blue tetrazolium (NBT) as an electron acceptor. (D) Histochemical analysis and (E) relative quantification showed a reduction of COX-positive fibers in quadriceps muscle sections of 3 month-old Aifm1 (R200 del) KI mice compared to controls. Scale bar = 50 μm. (F) SDH staining showed similar CII activity in skeletal muscle of wt and AIF deficient mice. Fibers relying on glycolytic metabolism stain light blue while highly oxidative fibers, that contain more mitochondria, show a dark blue staining. Scale bar = 50 μm. (G–H) Immunoblot analyses were performed using quadriceps muscles from wt, Aifm1 (R200 del) KI and Hq mutant mice at (G) 3 and (H) 6 months of age. Densitometry is relative to wt littermates and reported as mean ± SEM, Student's t-test, number of animals = 4–9 per genotype, ***p < 0.001, **p < 0.01, *p < 0.05. Color code is: black = wt (for KI); red = KI; dark grey = wt (for Hq); green = Hq.
3.2. Aifm1 (R200 del) knockin mice do not exhibit cerebellar neurodegeneration
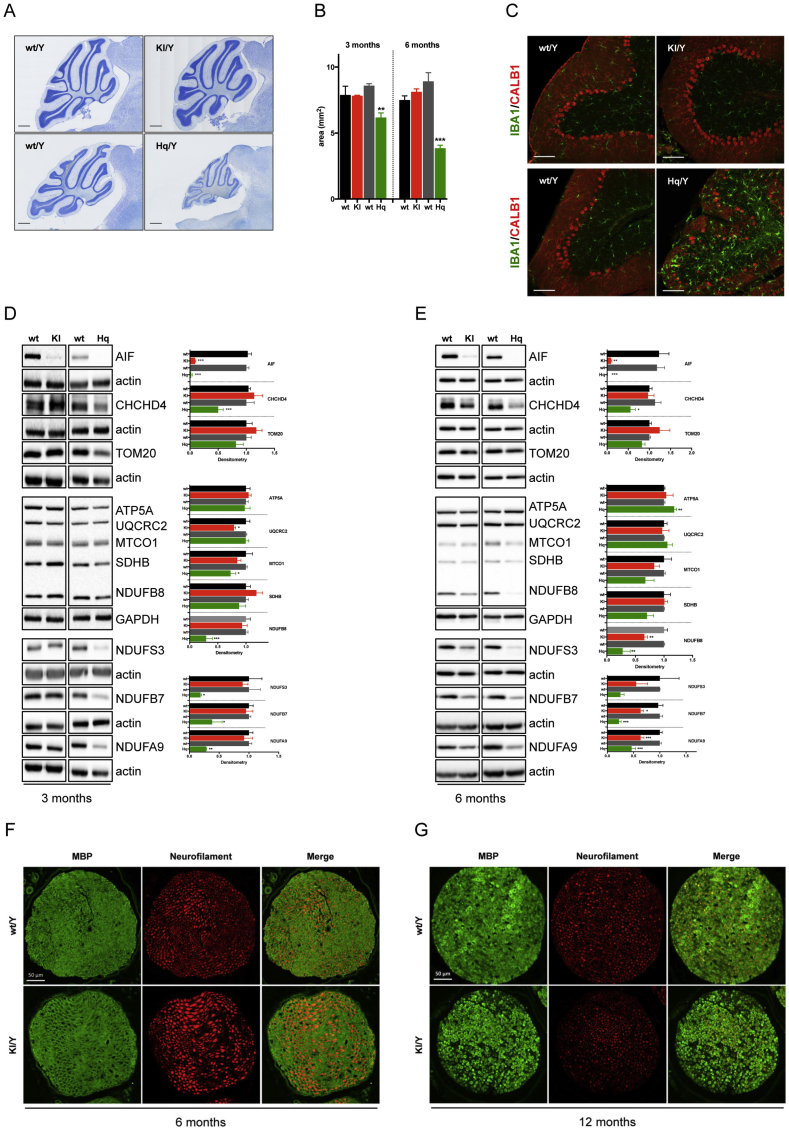
Hq mutant animals develop ataxia between 3 and 7 months of age [9]. Although tissue vulnerability in Hq mutant mice remains a matter of debate, the general consensus is that decreased Aifm1 expression and the consequent loss of AIF protein causes neurodegeneration within the cerebellum in an OXPHOS-dependent manner [9]. Since Aifm1 (R200 del) animals showed reduced cerebellar AIF expression levels akin to Hq mutant mice, we reasoned that related neurodegenerative processes might occur in the brains of our knockin animals. Thus, we performed immunohistochemical analysis and measured the cerebellar size in a group of 3 and 6 month-old wild type, Hq mutant and Aifm1 (R200 del) mice. Surprisingly, and contrary to our original hypothesis, Aifm1 (R200 del) animals did not show loss of Purkinje cells, nor did they exhibit any obvious signs of neurodegeneration within the cerebellum (Figure 3A–C). At 6 months of age, Hq mutant brains displayed a clear increase in immunoreactivity for the microglial marker IBA1, indicating a potential age-dependent microgliosis probably linked to inflammatory processes (Figure 3C). Conversely, the Aifm1 (R200 del) cerebella showed a limited number of IBA1-positive cells (Figure 3C). Despite the significant loss of the full length AIF protein in various brain areas (Figure 3D–E and Supplemental Fig. S1A), Aifm1 (R200 del) cerebella did not exhibit a significant decrease of CHCHD4 protein (Figure 3D–E), which instead was evident in Hq mutant brains as previously reported [10]. While Aifm1 (R200 del) mice showed a trend toward a reduction of mitochondrial CI subunits at 6 months of age, Hq mutant animals presented a marked CI and CIV deficiency at a much younger age, with a clear correlation between CHCHD4 loss and mitochondrial defects in the Hq cerebella (Figure 3D–E).
Figure 3.
Mutant AIF protein does not cause cerebellar degeneration but induces peripheral neuropathy. (A) Representative images of Nissl-stained sagittal brain sections and (B) corresponding quantification of the cerebellar size at 3 and 6 months of age (scale bar = 500 μm, mean ± SEM, Student's t-test, n = 3–4 per genotype, ***p < 0.001, **p < 0.01). (C) Representative images of cerebellar brain sections double-immunostained with calbindin (CALB1; red) and IBA1 (green). At 6 months of age, Hq mutant mice showed Purkinje cell degeneration and greatly enhanced IBA1 immunoreactivity. Scale bar = 100 μm. (D–E) Immunoblot analyses were performed using cerebella from wt, Aifm1 (R200 del) KI and Hq mutant mice at (D) 3 and (E) 6 months of age. Densitometry is relative to wt littermates and reported as mean ± SEM, Student's t-test, n = 4–9 per genotype, ***p < 0.001, **p < 0.01, *p < 0.05). Color code is: black = wt (for KI); red = KI; dark grey = wt (for Hq); green = Hq. (F–G) Cross-sections of the sciatic nerve from (F) 6 and (G) 12 month-old animals were stained with antibodies against Myelin Basic Protein (MBP, green) and Neurofilament (pan axonal, red). Scale bar = 50 μm.
Next, we sought to recapitulate some of our molecular findings in cultured cells. In primary cortical neurons (CNs), decreased AIF expression resulted in diminished CHCHD4 expression and partial OXPHOS impairment, with a significant downregulation of the CIII subunit UQCRC2 (Supplemental Figs. S1B–S1C). Morphologically, Aifm1 (R200 del) knockin CNs displayed aberrant dendritic length; however, they did not show any difference in the number of dendrites compared to wild type cells (Supplemental Fig. S1D). It is worth noting that we also tested the effect of mutant AIF protein in mouse embryonic fibroblast (MEFs). Unlike patient-derived fibroblasts [10], knockin MEFs did not show any difference in terms of OXPHOS subunits nor oxygen consumption rate (Supplemental Figs. S1E–S1F), although they exhibited enhanced glycolysis as revealed by an increased extracellular acidification rate (Supplemental Fig. S1G). It is not surprising that, under standard experimental conditions, cultured primary cells adopt a different metabolism, heavily relying on glycolysis that sufficiently compensates for subtle mitochondrial lesions.
Since Aifm1 (R200 del) knockin mice displayed hind limb clasping starting from around 5 months of age (Figure 1G), we thought that this might be partially due to innervation defects. Therefore, we performed immunohistochemical analyses of myelin basic protein (MBP) and neurofilament in sciatic nerves from 6 to 12 month-old wild type and knockin mice. We observed axonal swelling around 6 months of age (Figure 3F), followed by an obvious loss of neurofilament and MBP staining in 12 month-old Aifm1 (R200 del) knockin mice (Figure 3G). These data suggest possible axonal neuropathy in adulthood as a secondary effect of muscle wasting. Together, our data indicate that mutant AIF protein induces OXPHOS impairment in a tissue-specific and age-dependent manner. Moreover, the onset and progression of the pathogenic processes are clearly diverse in Hq mutant and Aifm1 (R200 del) mice.
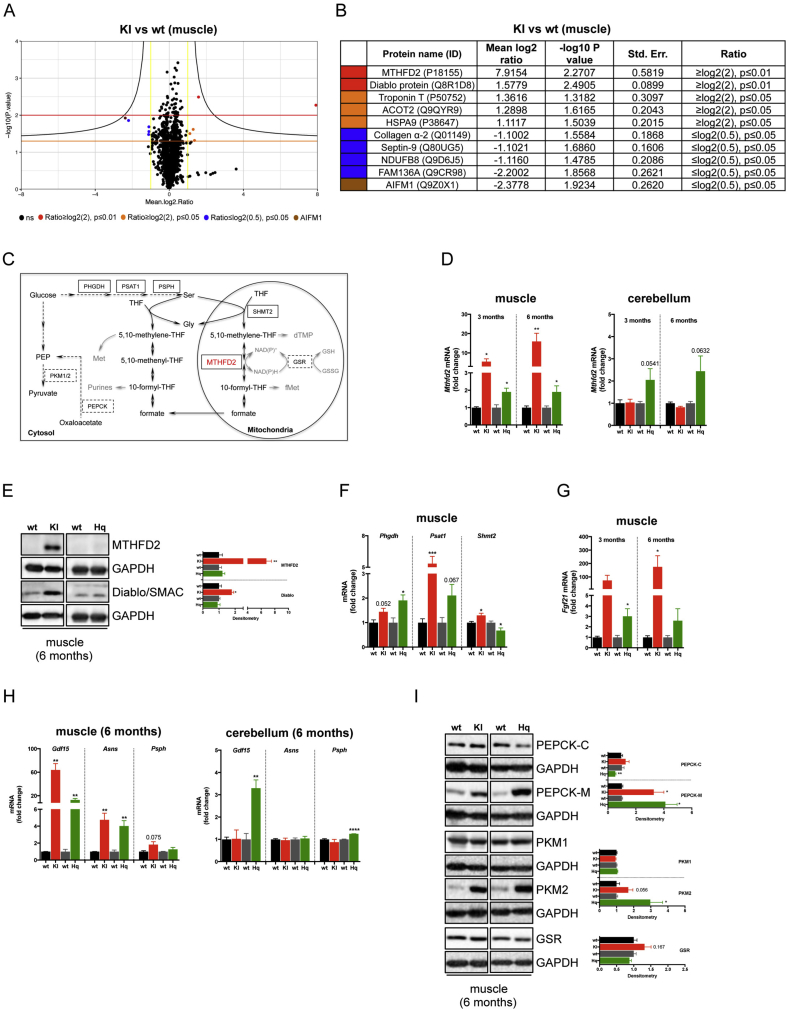
3.3. AIF deficiency induces MTHFD2 upregulation and aberrant glucose metabolism
To gain insights into the molecular processes associated with the expression of mutant AIF (R200 del) protein, we performed proteomic profiling using isobaric tags (TMT/iTRAQ). Compared to wild type, Aifm1 (R200 del) muscles exhibited a significant dysregulation of ten proteins, with the expected diminished levels of AIF and NDUFB8 supporting the quality of our analysis (Figure 4A–B). We found that Collagen α-2, cytoskeletal-associated GTPase Septin-9 and mitochondrial FAM136A were downregulated, whereas mitochondrial stress-70 protein (HSPA9), Acyl-CoA thioesterase 2 (ACOT2), mitochondrial Diablo/Smac and mitochondrial bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase (MTHFD2) were upregulated in Aifm1 (R200 del) mice. MTHFD2 is a mitochondrial protein regulating folate-mediated 1C metabolism (Figure 4C) [43], which has recently attracted much attention in the field of molecular medicine since it is highly upregulated in various models of mitochondrial diseases [38], [39], [44]. We set out to confirm this intriguing result using qRT-PCR and immunoblot analyses. In line with our proteomics, we found that MTHFD2 was significantly upregulated at both the transcriptional and protein levels in muscles from Aifm1 (R200 del) knockin and, to a much lower extent, in muscles and cerebella of Hq mice (Figure 4D–E). In the muscle of 6 month-old animals, the enhanced MTHFD2 expression was accompanied by an upregulation of phosphoglycerate dehydrogenase (Phgdh), phosphoserine aminotransferase (Psat1), and serine hydroxymethyltransferase (Shmt2), three enzymes involved in serine biosynthesis and, as a consequence, in folate-driven 1C metabolism (Figure 4C,F). Next, we tested the expression of fibroblast growth factor 21 (Fgf21), a hormone generally secreted in response to fasting, which is markedly upregulated in disorders resulting from mitochondrial lesions [45], [46]. Consistent with other mouse models of mitochondrial diseases [39], we found Fgf21 overexpression in AIF deficient muscles starting from a young age (Figure 4G). Along with Mthfd2, Phgdh, Psat1, Shmt2, and Fgf21 [46], [47], [48], [49], [50], [51], [52], additional ATF4 target genes were differentially regulated in AIF deficient mice. Specifically, we found that growth differentiation factor 15 (Gdf15), asparagine synthetase (Asns) and phosphoserine phosphatase (Psph) mirrored Mthfd2 upregulation in affected organs of Aifm1 (R200 del) knockin and Hq mutant mice (Figure 4H), further supporting a tissue-specific regulation of ATF4 activity. Since mitochondrial lesions alter serine and folate-driven 1C metabolism, we assessed the expression levels of enzymes that contribute to compensatory cataplerotic reactions [53], [54]. In the muscle of AIF deficient animals, we observed a significant upregulation of the mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) and the cytosolic pyruvate kinase (PKM2) (Figure 4I). These data suggest an increased conversion of the tricarboxylic acid (TCA) cycle intermediate oxaloacetate to phosphoenolpyruvate to pyruvate (Figure 4C), probably as a result of an enhanced glycolytic rate, ATP production and NADH oxidation through aerobic glycolysis. Finally, we tested the expression of glutathione reductase (GSR), an enzyme that converts oxidized glutathione (GSSH) to its reduced form (GSH) (Figure 4C). Our preliminary evidence indicates a tendency toward an enhanced glutathione synthesis (Figure 4I), which may contribute to the oxidative stress tolerance of mitochondrial deficient muscle fibers. Overall, our findings reveal that AIF deficiency alters folate-driven 1C metabolism, induces cataplerotic reactions and stimulates gene expression patterns associated with the transcription factor ATF4.
Figure 4.
AIF deficient mice exhibit aberrant 1-C metabolism. (A) Proteomic profiling using isobaric tags (TMT/iTRAQ) was performed in quadriceps muscle tissues from 6 month-old Aifm1 (R200del) KI mice and littermates (n = 3 per genotype). Volcano plot shows the median log2 ratio against –log10 (p-value). Dots represent individual proteins. Black dots are not significant; colored dots indicate differently expressed proteins. (B) Table of proteins differently expressed in KI mice compared to littermates. (C) Schematic representation of folate-driven 1C metabolism in mammalian cells. In the mitochondria, the reversible conversion of 5,10-methylene-THF to 10-formyl-THF is mediated by MTHFD2 (red) in a NAD(P)+-dependent manner. Enzymes involved in serine metabolism are in black solid box, whereas other enzymes analyzed in this study are in black dashed box. (D) RT-PCR analyses showing Mthfd2 expression levels in muscle and cerebellar tissues from 3 to 6 month-old mice (mean ± SEM, Student's t-test, n = 3–5 per genotype, **p < 0.01, *p < 0.05 compared to littermates). (E) Immunoblot analysis for MTHFD2 and Diablo/SMAC protein expression levels in muscles from 6 month-old animals. (F) RT-PCR analyses for Phgdh, Psat1, and Shmt2 expression in muscle samples from 6 month-old Aifm1 (R200 del) KI and Hq mice (mean ± SEM, Student's t-test, n = 4–6 per genotype, ***p < 0.001, *p < 0.05 compared to littermates). (G) RT-PCR showing expression levels of Fgf21 in Aifm1 (R200 del) KI and Hq mice compared to littermate controls. Quadriceps muscle tissues from 3 to 6 month-old animals were used (mean ± SEM, Student's t-test, n > 3 per genotype, *p < 0.05). (H) RT-PCR analyses for the ATF4 target genes Gdf15, Asns, and Psph in quadriceps muscle and cerebellar tissues from 6 month-old Aifm1 (R200 del) KI and Hq mice compared to littermate controls (mean ± SEM, Student's t-test, n = 4–6 per genotype, ***p < 0.001, **p < 0.01). (I) Immunoblot analysis of muscles from 6 month-old Aifm1 (R200 del) KI and Hq mice showing expression levels of the following enzymes: PEPCK-C, PEPCK-M, PKM1, PKM2, and GSR (densitometry is mean ± SEM, Student's t-test, n = 4–7 per genotype, *p < 0.05 compared to littermate controls). GAPDH was used as internal loading control. Color code is: black = wt (for KI); red = KI; dark grey = wt (for Hq); green = Hq.
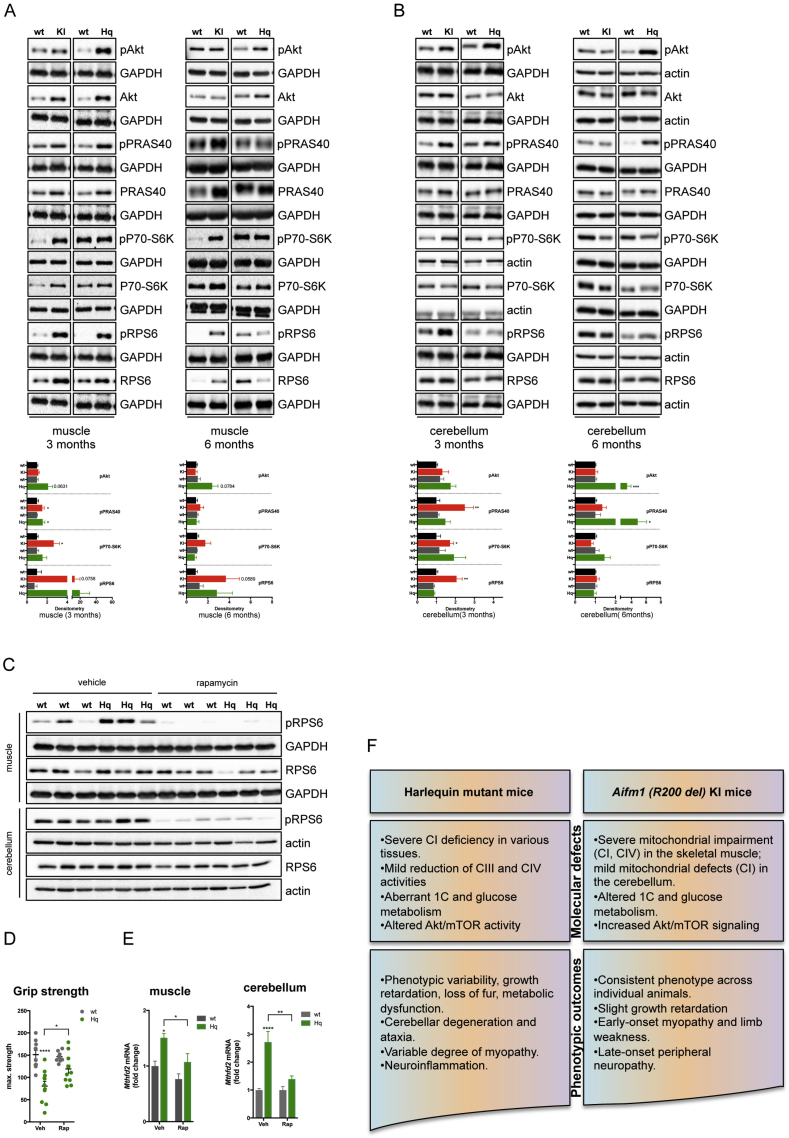
3.4. Loss of AIF leads to sustained Akt/mTOR activity
Since serine metabolism and Mthfd2 expression is linked to mTOR activity [39] and since impaired mitochondrial function stimulates insulin/IGF-1 signaling [37], [55], we tested the phosphorylation status of Akt and mTOR targets in AIF deficient mice and control littermates. We performed immunoblot analyses for the Akt target 40-kDa proline-rich Akt substrate (PRAS40) as well as mTOR substrates p70 S6 kinase (P70-S6K) and ribosomal protein S6 (RPS6). In the muscle and cerebellum, decreased AIF expression correlated with increased Akt and mTOR activity (Figure 5A–B). To further support our findings and address a current knowledge gap in the field, we sought to correlate mTOR activity with Mthfd2 expression in one of our mouse models of AIF deficiency. Similarly to other mitochondrial mutant mice [37], [55], [56], rapamycin treatment attenuated mTOR activity and improved muscular strength in Hq mice (Figure 5C–D). Moreover, rapamycin-mediated inhibition of mTOR signaling led to a significant downregulation of Mthfd2 expression in the muscle and cerebellum of Hq mutant mice compared to vehicle-treated control littermates (Figure 5E), further supporting the link between Akt/mTOR hyperactivity and aberrant 1C metabolism [38], [39], [47]. Taken together, AIF-dependent mitochondrial impairment stimulates Akt/mTOR signaling in a tissue-dependent manner, ultimately leading to a range of molecular defects and disease manifestations in mice (Figure 5F).
Figure 5.
AIF deficiency stimulates Akt and mTOR activity. (A–B) Immunoblot analyses were performed on (A) muscle and (B) cerebellar tissues from 3 to 6 month-old animals. Densitometry is relative to wt littermates and reported as mean ± SEM. Antibodies against total and phosphorylated forms of Akt, 40-kDa proline-rich Akt substrate (PRAS40), p70 S6 kinase (P70-S6K), ribosomal Protein S6 (RPS6) were used (n = 4–9 per genotype). GAPDH and actin were used as loading control (***p < 0.001, **p < 0.01, *p < 0.05). Color code is: black = wt (for KI); red = KI; dark grey = wt (for Hq); green = Hq. (C) Immunoblot analyses on muscle and cerebellar tissue from Hq mice and littermate controls following 2 weeks of rapamycin or vehicle treatment. Compared to vehicle-treated mice, animals of the rapamycin-group showed reduced mTOR activity, as revealed by decreased pRPS6. (D) Grip strength test was performed in wt and Hq mice injected with vehicle or rapamycin. Compared to mock treatment, rapamycin improved muscular performance in Hq animals (n = 9–10 per group). (E) RT-PCR analysis of Mthfd2 expression in wt and Hq mice treated with rapamycin. Compared to vehicle-treated animals, rapamycin-injected mice displayed a significant decrease in Mthfd2 mRNA expression within the quadriceps muscle and cerebellum (n = 5–6 per group). (F) Schematic comparison of molecular and phenotypic defects in AIF deficient animals. Compared to Harlequin mutant mice, Aifm1 (R200 del) knockin animals have a more homogeneous phenotype, with common pathological traits with AIFM1-linked human patients, such as COX defects associated with early-onset myopathy. In Aifm1 (R200 del) knockin animals, molecular defects include CI and CIV impairment, altered carbon metabolism and aberrant Akt/mTOR activity.
4. Discussion
We describe here the tissue-dependent impairment of mitochondrial function in the first knockin mouse model of AIFM1-related mitochondrial disease. Mice carrying the disease-segregating deletion R200 display consistent phenotypes across individual animals. Pathologically, Aifm1 (R200 del) mice progressively accumulate nemaline rod-like structures in muscle fibers, show evident signs of muscle weakness, and develop severe myopathy within a few months. While at 6 months of age Hq mutant mice show extensive cerebellar neurodegeneration, Aifm1 (R200 del) knockin animals do not exhibit obvious brain pathology, loss of Purkinje cells or evidence of neuroinflammation. Only at an advanced age, we observed defects of peripheral motor neurons (i.e., reduced size and myelination of the sciatic nerve). As in Hq mutant mice, Aifm1 (R200 del) knockin animals develop histological signs of progressive retinal degeneration (data not shown). Biochemically, Aifm1 (R200 del) muscle and cerebellar tissues show decreased expression of mutant AIF protein. In a tissue-, cell-, and time-dependent fashion, CHCHD4 deficiency seems to correlate with the status of the OXPHOS system. Our mouse data indicate that the expression of a mutant AIF protein leads to significant loss of CI and CIV subunits as the most pronounced defects. We are rather surprised that Hq mutant and knockin animals do not show similar pathological profiles, given the comparable levels of residual full-length AIF protein. One explanation is that AIF expression varies considerably in Hq mutant cells during development until adulthood, leading to an irreversible compromised status that causes cerebellar degeneration. Alternatively, it is possible that other molecular mechanisms are sufficient to support mitochondrial bioenergetics in the brain, despite the expression of a mutant AIF (R200 del) protein. Finally, it may be that compensatory factors, which are lacking in Hq mutant mice due to their mixed background, contribute to the maintenance of the OXPHOS system in Aifm1 (R200 del) mice. This would also explain the high variability between individual Hq mutant animals, as we observed in our animal cohort and in line with previous reports [31]. We believe that further work will be necessary to define an eventual degree of synthetic lethality between AIF deficiency and other deleterious alleles in model organisms, which may also be relevant for understanding the wide spectrum of clinical manifestations linked to AIFM1 mutations [18], [20], [21], [24], [25], [26], [27], [28], [29], [57].
In an effort to gain insights into the mechanisms underlying AIFM1-linked pathologies, we found that Aifm1 (R200 del) knockin mice exhibit typical molecular hallmarks as observed in other forms of mitochondrial diseases. In this regard, an unbiased proteomic analysis supports that AIF deficiency induces an aberrant 1C metabolism, as revealed by a significantly increased expression of MTHFD2 in affected tissues. Folate-driven 1C metabolism is a series of evolutionarily conserved enzymatic reactions that support a large range of biosynthetic processes, including purine and thymidine synthesis, amino acid (e.g., glycine, serine and methionine) metabolism [43] and mitochondrial OXPHOS through the translation of mitochondrial DNA-encoded proteins [58]. Folate metabolism comprises distinct reactions that occur in the cytosol and in the mitochondria, with only a few intermediates that bridge the two compartments. Apart from glycine and serine, oxidized formate diffuses to the cytosol, whereas reduced tetrahydrofolate (THF) is shuttled into the mitochondria [43]. While the relationship between MTHFD2, ATF4, and mTOR is well established in physiological conditions [47], [49], [50], it remains unclear whether folate-dependent 1C metabolism is a driver of mitochondrial diseases or simply a consequence of aberrant signaling cascades [38], [39]. In cultured cells [49] and in transgenic mice [38], isotope tracing indicates that OXPHOS defects lead to an increased abundance of methylene-THF and de-novo serine biosynthesis, parallel to a decreased production of formate. Based on this line of evidence, it may be that MTHFD2 upregulation stimulates the flux of 1C units toward reduced intermediates (i.e., THF) and serine production. In cells lacking a functional OXPHOS system, disturbed 1C metabolism alters serine-dependent formate synthesis and the usage of essential metabolites through cataplerotic reactions (e.g., conversion of oxaloacetate to phosphoenolpyruvate to pyruvate), which ultimately stimulate alternative routes for purine, methionine biosynthesis, and, eventually, ATP production. As part of an adaptive metabolic stress response, MTHFD2 upregulation depends on the mitochondrial deficiency-induced hyperactive Akt/mTOR signaling, which stimulates the transcriptional activity of ATF4, a stress response that seems to ameliorate proteostasis in cells carrying mitochondrial lesions [50]. Based on our in vivo data using rapamycin, we confirm that mTOR participates in the transcriptional regulation of Mthfd2 in our models. In the future, it will be important to define which of these aforementioned processes are epigenetic adaptations that counteract mitochondrial dysfunction. This may lead to the identification of molecular targets relevant for the development of therapeutic options.
Impaired mitochondrial bioenergetics is a common feature of inherited and sporadic forms of mitochondrial diseases. Over the past years, remarkable achievements in molecular medicine have helped shed light on mitochondrial disease pathophysiology. Yet, patient management and treatment options remain extremely challenging due to the poor correlation between genetic etiology and the wide spectrum of clinical manifestations, organ involvement, disease onset, and progression [16], [17], [59]. Given the heterogeneous clinical patterns of these disorders, it is not surprising that the development of effective therapies lags behind. In this regard, the use of mice is a critical aspect for proof-of-principle assessments of novel treatments and preclinical approaches, with some caveats that are worth mentioning. Among them, there is a growing awareness that findings from these model organisms need to be carefully evaluated due to the different pathophysiology of rodents compared to humans. Also, an increasing body of evidence has challenged the initial assumption that mice carrying OXPHOS lesions are sufficient to mimic a wide spectrum of mitochondrial diseases. While traditional constitutive knockout mouse models have been undoubtedly instrumental for establishing the biological function of a specific targeted gene, their further characterization demonstrated that some of them do not fully recapitulate the biochemical traits and organ defects observed in human patients (reviewed in [60], [61]). As in the case of the Hq mutant mice, striking phenotypic differences often exist across individual animals of the same model, perhaps due to their mixed genetic background or the incomplete penetrance of the hypomorphic mutation. In terms of drug development, such variability may undermine the feasibility even of hypothesis-driven screens of chemical compounds, since they would be extremely laborious and difficult to interpret in terms of their biological effects and meaning. Ultimately, it would be more helpful to compare drug candidates across different mouse models with an established disease progression, rather than a single one with unpredictable pathogenesis. The same argument is also valid in the case of treatments that should be broadly protective from various forms of mitochondria-associated diseases. In our opinion, pathogenic processes underlying distinct human syndromes may be better deciphered through the use of innovative tractable mouse models carrying specific disease-associated mutations. Thus, we believe that our new knockin Aifm1 (R200 del) mouse model is an additional, valuable tool that will positively influence research in molecular medicine and drug development.
5. Conclusion
Our study emphasizes the tissue-specific vulnerability due to a mutant AIF protein and delineates a pathological profile essentially different from the one linked to hypomorphic AIF expression. Mechanistically, AIF deficiency alters respiratory complexes, induces hyperactive Akt/mTOR signaling, and affects anabolic and catabolic processes, including folate-driven 1C-metabolism. Finally, our new knockin Aifm1 (R200 del) mouse model shows many pathological hallmarks that resemble those observed in patients.
Author contribution
Conceptualization, LW and DB; Validation, LW, AG, AP; Formal Analysis, LW, AG, AP; Investigation, LW, AG, DSB, AP, MS; Resources, LW, DSB, MS; Writing, LW and DB; Visualization, LW and DB; Supervision, LW and DB; Project Administration, DB; Funding Acquisition, PN and DB.
Conflict of interests
The authors declare no competing interests.
Acknowledgements
We wish to thank Dr. Adriano Flora (Taconic Biosciences GmbH, Cologne, Germany) for the generation of the Aifm1 (R200 del) mouse model; Dr. Walker Jackson (DZNE, Bonn) for his constructive input; Ms Christiane Bartling-Kirsch (DZNE, Bonn) for her technical assistance; Dr. Katharina Meyer (DZNE, Bonn; Harvard Medical School, Boston, USA) for her preliminary experiments. This research was supported by the DZNE institutional budget and the Helmholtz cross-program topic “Metabolic Dysfunction” and “Aging and Metabolic Programming (AMPro)”. This project was also supported in Germany through the “Bundesministerium für Bildung und Forschung” (BMBF) under the aegis of the EU Joint Programme-Neurodegenerative Disease Research (JPND-www.jpnd.eu; Cellular Bioenergetics of Neurodegenerative Diseases, CeBioND). LW, AG, AP, PN and DB are members of the DFG Cluster of Excellence ImmunoSensation and of the innovation programme under the Marie Skłodowska-Curie grant agreement No 676144 (Synaptic Dysfunction in Alzheimer Disease, SyDAD).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.05.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 2.Yu S.W., Wang H., Poitras M.F., Coombs C., Bowers W.J., Federoff H.J. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 3.Daugas E., Susin S.A., Zamzami N., Ferri K.F., Irinopoulou T., Larochette N. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. The FASEB Journal. 2000;14(5):729–739. [PubMed] [Google Scholar]
- 4.Sevrioukova I.F. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxidants and Redox Signaling. 2011;14(12):2545–2579. doi: 10.1089/ars.2010.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hangen E., Blomgren K., Benit P., Kroemer G., Modjtahedi N. Life with or without AIF. Trends in Biochemical Sciences. 2010;35(5):278–287. doi: 10.1016/j.tibs.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Bano D., Prehn J.H.M. Apoptosis-inducing factor (AIF) in physiology and disease: the tale of a repented natural born killer. EBioMedicine. 2018;30:29–37. doi: 10.1016/j.ebiom.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vahsen N., Cande C., Briere J.J., Benit P., Joza N., Larochette N. AIF deficiency compromises oxidative phosphorylation. The EMBO Journal. 2004;23(23):4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sevrioukova I.F. Redox-linked conformational dynamics in apoptosis-inducing factor. Journal of Molecular Biology. 2009;390(5):924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein J.A., Longo-Guess C.M., Rossmann M.P., Seburn K.L., Hurd R.E., Frankel W.N. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419(6905):367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 10.Meyer K., Buettner S., Ghezzi D., Zeviani M., Bano D., Nicotera P. Loss of apoptosis-inducing factor critically affects MIA40 function. Cell Death & Disease. 2015;6:e1814. doi: 10.1038/cddis.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hangen E., Feraud O., Lachkar S., Mou H., Doti N., Fimia G.M. Interaction between AIF and CHCHD4 regulates respiratory chain biogenesis. Molecular Cell. 2015;58(6):1001–1014. doi: 10.1016/j.molcel.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Nakao L.S., Everley R.A., Marino S.M., Lo S.M., de Souza L.E., Gygi S.P. Mechanism-based proteomic screening identifies targets of thioredoxin-like proteins. Journal of Biological Chemistry. 2015;290(9):5685–5695. doi: 10.1074/jbc.M114.597245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen S.M., Guo M., Xiong Z., Yu Y., Zhao X.Y., Zhang F.F. AIF inhibits tumor metastasis by protecting PTEN from oxidation. EMBO Reports. 2015;16(11):1563–1580. doi: 10.15252/embr.201540536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMauro S., Schon E.A., Carelli V., Hirano M. The clinical maze of mitochondrial neurology. Nature Reviews Neurology. 2013;9(8):429–444. doi: 10.1038/nrneurol.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koopman W.J., Distelmaier F., Smeitink J.A., Willems P.H. OXPHOS mutations and neurodegeneration. The EMBO Journal. 2012 doi: 10.1038/emboj.2012.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koopman W.J., Beyrath J., Fung C.W., Koene S., Rodenburg R.J., Willems P.H. Mitochondrial disorders in children: toward development of small-molecule treatment strategies. EMBO Molecular Medicine. 2016;8(4):311–327. doi: 10.15252/emmm.201506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorman G.S., Chinnery P.F., DiMauro S., Hirano M., Koga Y., McFarland R. Mitochondrial diseases. Nature Reviews Disease Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 18.Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D'Adamo P. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. The American Journal of Human Genetics. 2010;86(4):639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevrioukova I.F. Structure/function relations in AIFM1 variants associated with neurodegenerative disorders. Journal of Molecular Biology. 2016 doi: 10.1016/j.jmb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Rinaldi C., Grunseich C., Sevrioukova I.F., Schindler A., Horkayne-Szakaly I., Lamperti C. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. The American Journal of Human Genetics. 2012;91(6):1095–1102. doi: 10.1016/j.ajhg.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardissone A., Piscosquito G., Legati A., Langella T., Lamantea E., Garavaglia B. A slowly progressive mitochondrial encephalomyopathy widens the spectrum of AIFM1 disorders. Neurology. 2015;84(21):2193–2195. doi: 10.1212/WNL.0000000000001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger I., Ben-Neriah Z., Dor-Wolman T., Shaag A., Saada A., Zenvirt S. Early prenatal ventriculomegaly due to an AIFM1 mutation identified by linkage analysis and whole exome sequencing. Molecular Genetics and Metabolism. 2011;104(4):517–520. doi: 10.1016/j.ymgme.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 23.Zong L., Guan J., Ealy M., Zhang Q., Wang D., Wang H. Mutations in apoptosis-inducing factor cause X-linked recessive auditory neuropathy spectrum disorder. Journal of Medical Genetics. 2015;52(8):523–531. doi: 10.1136/jmedgenet-2014-102961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettwig M., Schubach M., Zimmermann F.A., Klinge L., Mayr J.A., Biskup S. From ventriculomegaly to severe muscular atrophy: expansion of the clinical spectrum related to mutations in AIFM1. Mitochondrion. 2015;21C:12–18. doi: 10.1016/j.mito.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Diodato D., Tasca G., Verrigni D., D'Amico A., Rizza T., Tozzi G. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. European Journal of Human Genetics. 2016;24(3):463–466. doi: 10.1038/ejhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mierzewska H., Rydzanicz M., Bieganski T., Kosinska J., Mierzewska-Schmidt M., Lugowska A. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation - a novel phenotype of the mitochondrial disease. Clinical Genetics. 2016 doi: 10.1111/cge.12792. [DOI] [PubMed] [Google Scholar]
- 27.Morton S.U., Prabhu S.P., Lidov H.G., Shi J., Anselm I., Brownstein C.A. AIFM1 mutation presenting with fatal encephalomyopathy and mitochondrial disease in an infant. Cold Spring Harbor Molecular Case Studies. 2017 Mar;3(2) doi: 10.1101/mcs.a001560. a001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyake N., Wolf N.I., Cayami F.K., Crawford J., Bley A., Bulas D. X-linked hypomyelination with spondylometaphyseal dysplasia (H-SMD) associated with mutations in AIFM1. Neurogenetics. 2017 doi: 10.1007/s10048-017-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu B., Wang M., Castoro R., Simmons M., Dortch R., Yawn R. A novel missense mutation in AIFM1 results in axonal polyneuropathy and misassembly of OXPHOS complexes. European Journal of Neurology - The Official Journal of the European Federation of Neurological Societies. 2017 doi: 10.1111/ene.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joza N., Susin S.A., Daugas E., Stanford W.L., Cho S.K., Li C.Y. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410(6828):549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 31.Benit P., Goncalves S., Dassa E.P., Briere J.J., Rustin P. The variability of the harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PLoS One. 2008;3(9):e3208. doi: 10.1371/journal.pone.0003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pospisilik J.A., Knauf C., Joza N., Benit P., Orthofer M., Cani P.D. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131(3):476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 33.Joza N., Oudit G.Y., Brown D., Benit P., Kassiri Z., Vahsen N. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Molecular and Cellular Biology. 2005;25(23):10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Ghouzzi V., Csaba Z., Olivier P., Lelouvier B., Schwendimann L., Dournaud P. Apoptosis-inducing factor deficiency induces early mitochondrial degeneration in brain followed by progressive multifocal neuropathology. Journal of Neuropathology & Experimental Neurology. 2007;66(9):838–847. doi: 10.1097/NEN.0b013e318148b822. [DOI] [PubMed] [Google Scholar]
- 35.Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. Understanding mitochondrial complex I assembly in health and disease. Biochimica et Biophysica Acta. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Polster B.M. AIF, reactive oxygen species, and neurodegeneration: a “complex” problem. Neurochemistry International. 2013;62(5):695–702. doi: 10.1016/j.neuint.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson S.C., Yanos M.E., Kayser E.B., Quintana A., Sangesland M., Castanza A. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342(6165):1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikkanen J., Forsstrom S., Euro L., Paetau I., Kohnz R.A., Wang L. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metabolism. 2016;23(4):635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Khan N.A., Nikkanen J., Yatsuga S., Jackson C., Wang L., Pradhan S. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metabolism. 2017;26(2) doi: 10.1016/j.cmet.2017.07.007. 419–428 e5. [DOI] [PubMed] [Google Scholar]
- 40.Bano D., Dinsdale D., Cabrera-Socorro A., Maida S., Lambacher N., McColl B. Alteration of the nuclear pore complex in Ca(2+)-mediated cell death. Cell Death and Differentiation. 2010;17(1):119–133. doi: 10.1038/cdd.2009.112. [DOI] [PubMed] [Google Scholar]
- 41.Ziviani E., Lippi G., Bano D., Munarriz E., Guiducci S., Zoli M. Ryanodine receptor-2 upregulation and nicotine-mediated plasticity. The EMBO Journal. 2011;30(1):194–204. doi: 10.1038/emboj.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sciacco M., Bonilla E. Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods in Enzymology. 1996;264:509–521. doi: 10.1016/s0076-6879(96)64045-2. [DOI] [PubMed] [Google Scholar]
- 43.Ducker G.S., Rabinowitz J.D. One-carbon metabolism in health and disease. Cell Metabolism. 2017;25(1):27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tufi R., Gandhi S., de Castro I.P., Lehmann S., Angelova P.R., Dinsdale D. Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson's disease. Nature Cell Biology. 2014;16(2):157–166. doi: 10.1038/ncb2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suomalainen A., Elo J.M., Pietilainen K.H., Hakonen A.H., Sevastianova K., Korpela M. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. The Lancet Neurology. 2011;10(9):806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyynismaa H., Carroll C.J., Raimundo N., Ahola-Erkkila S., Wenz T., Ruhanen H. Mitochondrial myopathy induces a starvation-like response. Human Molecular Genetics. 2010;19(20):3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Sahra I., Hoxhaj G., Ricoult S.J.H., Asara J.M., Manning B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351(6274):728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Sousa-Coelho A.L., Marrero P.F., Haro D. Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. Biochemical Journal. 2012;443(1):165–171. doi: 10.1042/BJ20111748. [DOI] [PubMed] [Google Scholar]
- 49.Bao X.R., Ong S.E., Goldberger O., Peng J., Sharma R., Thompson D.A. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016;5 doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quiros P.M., Prado M.A., Zamboni N., D'Amico D., Williams R.W., Finley D. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of Cell Biology. 2017;216(7):2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han J., Back S.H., Hur J., Lin Y.H., Gildersleeve R., Shan J. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biology. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeNicola G.M., Chen P.H., Mullarky E., Sudderth J.A., Hu Z., Wu D. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nature Genetics. 2015;47(12):1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. Journal of Biological Chemistry. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 54.Ost M., Keipert S., van Schothorst E.M., Donner V., van der Stelt I., Kipp A.P. Muscle mitohormesis promotes cellular survival via serine/glycine pathway flux. The FASEB Journal. 2015;29(4):1314–1328. doi: 10.1096/fj.14-261503. [DOI] [PubMed] [Google Scholar]
- 55.Ising C., Koehler S., Brahler S., Merkwirth C., Hohne M., Baris O.R. Inhibition of insulin/IGF-1 receptor signaling protects from mitochondria-mediated kidney failure. EMBO Molecular Medicine. 2015;7(3):275–287. doi: 10.15252/emmm.201404916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegmund S., Yang H., Sharma R., Javors M., Skinner O., Mootha V. Low-dose rapamycin extends lifespan in a mouse model of mtDNA depletion syndrome. Human Molecular Genetics. 2017 doi: 10.1093/hmg/ddx341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sancho P., Sanchez-Monteagudo A., Collado A., Marco-Marin C., Dominguez-Gonzalez C., Camacho A. A newly distal hereditary motor neuropathy caused by a rare AIFM1 mutation. Neurogenetics. 2017 doi: 10.1007/s10048-017-0524-6. [DOI] [PubMed] [Google Scholar]
- 58.Morscher R.J., Ducker G.S., Li S.H., Mayer J.A., Gitai Z., Sperl W. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018 doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chinnery P.F. Mitochondrial disease in adults: what's old and what's new? EMBO Molecular Medicine. 2015;7(12):1503–1512. doi: 10.15252/emmm.201505079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torraco A., Peralta S., Iommarini L., Diaz F. Mitochondrial Diseases Part I: mouse models of OXPHOS deficiencies caused by defects in respiratory complex subunits or assembly factors. Mitochondrion. 2015;21:76–91. doi: 10.1016/j.mito.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iommarini L., Peralta S., Torraco A., Diaz F. Mitochondrial diseases part II: mouse models of OXPHOS deficiencies caused by defects in regulatory factors and other components required for mitochondrial function. Mitochondrion. 2015;22:96–118. doi: 10.1016/j.mito.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.