Abstract
Cell-substrate interactions play a crucial role in the design of better biomaterials and integration of implants with the tissues. Adhesion is the binding process of the cells to the substrate through interactions between the surface molecules of the cell membrane and the substrate. There are several factors that affect cell adhesion including substrate surface chemistry, topography, and stiffness. These factors physically and chemically guide and influence the adhesion strength, spreading, shape and fate of the cell. Recently, technological advances enabled us to precisely engineer the geometry and chemistry of substrate surfaces enabling the control of the interaction cells with the substrate. Some of the most commonly used surface engineering methods for eliciting the desired cellular responses on biomaterials are photolithography, electron beam lithography, microcontact printing, and microfluidics. These methods allow production of nano- and micron level substrate features that can control cell adhesion, migration, differentiation, shape of the cells and the nuclei as well as measurement of the forces involved in such activities. This review aims to summarize the current techniques and associate these techniques with cellular responses in order to emphasize the effect of chemistry, dimensions, density and design of surface patterns on cell-substrate interactions. We conclude with future projections in the field of cell-substrate interactions in the hope of providing an outlook for the future studies.
Keywords: Microfabrication, Micropattern, Cell-material interaction, Differentiation, Cell adhesion
Graphical abstract
Highlights
-
•
This article aims to provide insight into cell-material interactions and importance of topography on cellular processes.
-
•
Reviewed current progress of nano and micropatterning techniques.
-
•
Review mainly focused on fabrication of nano-micropatterns and interaction with cells.
-
•
Discussed the relation between surface topography and cell adhesion, morphology, proliferation and fate.
-
•
Reviewed the literature on biomaterial substrate modification.
1. Introduction
In the history of the biomaterials field since the ancient Egyptians, the achievement of biocompatibility was the main concern because it is the most critical property for a typical biomaterial. As the scientific and technological tools evolved it became evident that the architecture and topography of an implant is just as central to the implant design as the chemistry. The ultimate goal of research on cell-substrate interactions is to study the relationship between the substrate surface and the cell response it evokes. There are numerous studies involving substrates with different topography and chemistry, targeting different tissues, cells and cell responses. However, there is still no universal rule of thumb applicable to all situations. The outstanding question in implant surface design is to identify the best design for a target tissue and how it can be achieved with the current knowledge base. The proper approach would be the determination of ideal surface properties for each specific application while taking into consideration the contribution of surface topography to the performance of the implant. Today, with the vast variety of the scientific tools available, response of a cell to any stimulus can be controlled and studied in detail.
Cell shape, adhesion, migration, and fate are controlled by the properties of the substrate. These are topography (the surface architecture), stiffness, and bioactive cell adhesive cues such as peptides and proteins. In 1912, Harrison was first to show the effect of solid support materials on cell movement and morphogenesis by using spider web as the substrate [1]. He demonstrated that the cell shape and migration were correlated with the substrate organization and topography. Later, Paul Weiss (1947) showed that cells move and migrate by contact guidance [2]. Curtis and Varde were the first researchers to take advantage of these findings and employed topographical cues for the control of cell behavior [3]. The microfabrication techniques, initially developed for the electronic industry, came into use for the study of the behavior of cells on micro- and nanopatterned surfaces nearly three decades ago [4]. Since then, many studies were conducted to elucidate cell-substrate interactions on engineered surfaces prepared using many different materials, architectures and cells [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]]. All of these studies showed that cellular functions are affected and in some cases improved by the substrates mimicking the extracellular matrix (ECM) topography. The substrate is not just a cell support but also a guide for adhesion, proliferation, morphology, and spreading by providing physical and chemical signals [11,22]. Substrate topography can affect the cellular functions differently depending on the cell type, pattern size and geometry, stiffness and chemical properties of the substrate material [20]. Sub-micron- to -nanoscale topographies affect cells directly since they have the similar size with ECM proteins such as fibronectin, collagen and laminin. These tissue subunits contain a large number of cells so, sub-millimeter topography can also affect cell-cell interactions, cell-cell signaling, and other cellular activities [23]. In order to understand the influence of surface topography on shape and other properties of a cell, the cell adhesion and mechanotransduction should be studied thoroughly.
Although the relationship between substrate topography and cell behavior has been studied by many authors, the main difficulty in this area is the abundance and complexity of the substrates which have different mechanical properties, sizes, shapes, distribution and chemistry of topographical cues in addition to the cell and tissue types used. There are several reviews in the literature which focus on topography especially concentrating on feature size and tissue type [24,25]. There are others on production methods of substrates [[26], [27], [28]], on the biological aspects of the interaction [29,30] and on the comparison of physical and chemical cues [31,32]. The current review aims to provide an integrative perspective on the biology, production methodology and the cell-topography relationships presented according to the cellular responses evoked with special emphasis on feature size.
This review summarizes the cell adhesion process in order to provide a framework to understand the mechanisms of cell-substrate interaction in the context of tissue engineering and the methods for fabricating such substrates. It provides a comprehensive survey of the current literature to reveal the role of surface pattern chemistry, dimensions, density and design on cell adhesion, alignment, migration, differentiation. It also evaluates nano- and microfabricated substrates as tools for controlling cell and nuclear shape, as well as measurement of the cellular forces.
2. Cell adhesion
Adhesion is a fundamental cellular process in tissue formation. It is about the binding of a cell to the extracellular matrix, surface or another cell through use of certain surface proteins [33]. This binding achieved through cell adhesion proteins, results in two particular mechanisms for the intracellular signal generation: originating a force on cytoskeletal elements which is transmitted throughout the cytosol to the nuclear lamina, and activation of signaling pathways and messengers [34]. These two processes are not mutually exclusive. Binding to a surface via adhesion molecules and transmission of a signal to the cytoskeleton involves a specific process called “focal adhesion”. Focal adhesion points are intersection nodes where the environmental mechanical signals received are transduced to intracellular forces and chemical signals through cytoskeletal connections and signaling proteins [35]. After the generation of a traction force on the cytoskeleton, this force is transmitted by the cytoskeletal elements to various structures in the cell in which nucleus holds distinct importance [36]. Following cell adhesion, many cellular events, including differentiation, apoptosis, and changes in the gene expression profiles, are indirectly affected from the forces generated on nuclear lamina [36,37].
Cells adhere to each other, to ECM or any substrate with the structures called cell junctions which are tight junctions (zonula occludens), intermediate junction (zonula adherens), and desmosomes (macula adherens). Zonula occludens (tight junction) is formed by the fusion of adjacent cell membranes, zonula adherens (intermediate junction) is a ∼200 Å intercellular space occupied by homogeneous amorphous material, and macula adherens (desmosome) is a ∼240 Å intercellular space with a central dense disc [38]. Adhesion of cells to substrates was first studied using the interference reflection microscopy, and showed that cell-substrate interactions took place at the adhesions (100 Å) while rest of the cell surface was further away [39]. It was shown that a surface treatment that generates hydroxyl groups on polystyrene resulted in enhanced cell attachment [40]. Therefore it was proven that for the cells to adhere to a surface, there must be specific chemical groups available on the substrate [40]. This was later explained by the presence of specific cell surface proteins called Cell Adhesion Molecules (CAMs) [41]. These molecules are classified under integrin (receptor) family, immunoglobulin superfamily, selectins and cadherins. Integrins are made up of α and β subunits and are responsible for cell-cell and cell-matrix interactions. Immunoglobulin superfamily consists of CD2, CD58, intercellular adhesion molecules (ICAMs), vascular cell adhesion molecule-1 (VCAM-1), platelet-endothelial cell adhesion molecule-1 (PE-CAM-1), and MAdCAM-1. Selectins are a group of cell adhesion molecules that are expressed on the surface of endothelial cells, leucocytes and platelets and have three subfamilies: E-selectin, P-selectin and L-selectin. Cadherins are a superfamily of Ca++ dependent cell adhesion molecules which are important in cell-cell interactions [42].
Early studies revealed two distinct structures in cell adhesions: close contacts and focal contacts which are separated by 30 nm and 10–15 nm from the substrate, respectively, in fibroblasts [43]. In recent studies, these contacts are classified as focal complexes, focal adhesions and fibrillary adhesions [44]. Focal complexes are located at the edge of a lamellipodium and are constituted of paxillin, vinculin, and tyrosine-phosphorylated proteins. Focal adhesions are located at the cell periphery and constitute of α5 integrin, paxillin, vinculin, α actinin, talin, focal adhesion kinase (FAK), and tyrosine-phosphorylated proteins. Fibrillary adhesions are located in the central regions of cells and are made of α5 integrin and tensin [44].
3. Adhesion of cells to ECM and mechanotransduction
Interactions of cells with the ECM and the neighboring cells elicit responses that have an essential role in the regulation of the behavior and fate of the cell. ECM constitutes a physical and chemical microenvironment, a site for anchorage of cells, and guides cell migration during embryonic development and wound repair. Therefore, it plays a key role in tissue morphogenesis. The ECM also acts as a carrier for the transmission of environmental signals to cells influencing proliferation, differentiation, and apoptosis [45]. Cells must sense, respond, and adapt to their physical environments at every level (molecular, cellular, tissue, organ and organism). Cells respond to mechanical cues by initiating signals that result in adaptations in cytoskeletal architecture and gene expression [46]. Adhesion to the ECM is achieved by all types of adherent cells. The adhesion of the cells is achieved by specific cell surface proteins called integrins, and these proteins affect the organization of the cytoskeleton [34,47,48]. The cell adhesion molecules have affinity for a variety of extracellular ligands including but not limited to fibronectin, vitronectin and various types of collagens. The best characterized adhesions are the focal adhesions [45]. Integrins are responsible for mediating cell-matrix adhesions. 24 known integrin αβ pairs are constituted of 18α and 8β subunits. These α and β subunits are type I transmembrane glycoproteins with large extracellular domains, single spanning transmembrane domains and, with the exception of β4, short cytoplasmic domains [49]. Extracellular domains of integrins bind to ECM ligands and divalent cations. After binding of the extracellular domain, a cascade of intracellular events succeeds. This is called the ‘outside-in’ signaling. However, integrin function is not unidirectional. There also exists an ‘inside-out’ signaling which uses integrins as bi-directional passageways of mechanochemical information. Besides working as substrate anchors, they are also linked to the actin cytoskeleton (except α6β4, which makes connections to intermediate filaments) [50].
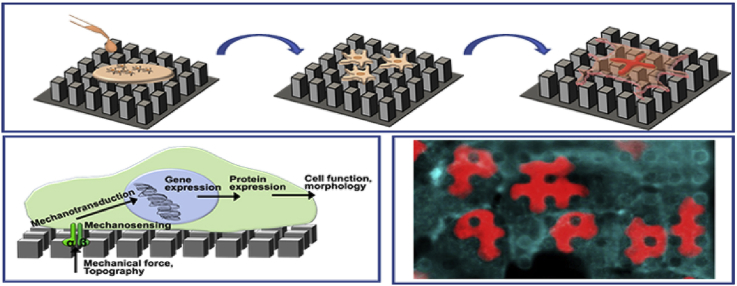
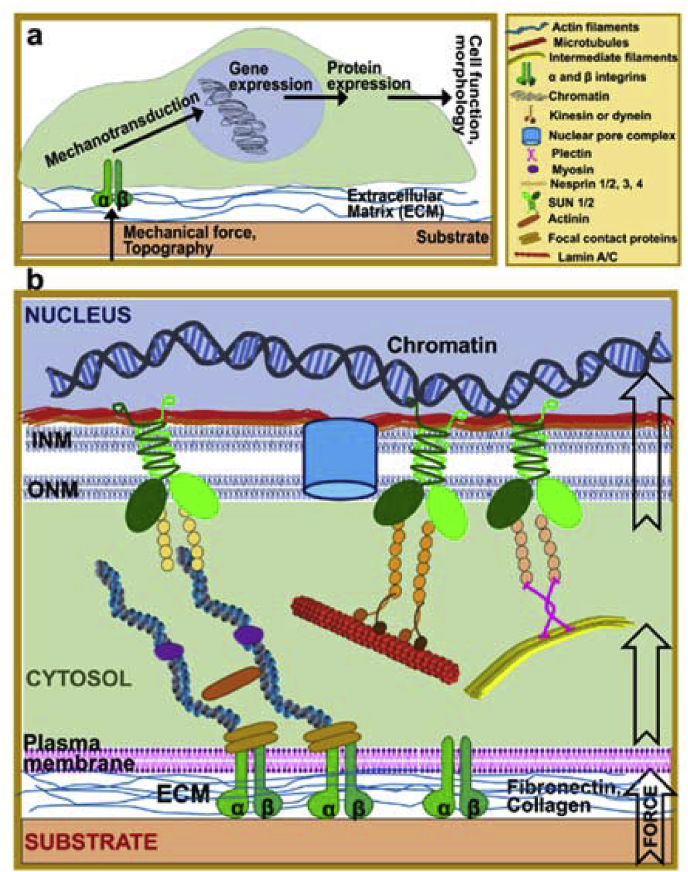
Cell exert traction forces to the substrate during adhesion. These forces are generated by cytoskeletal proteins. To explain the mechanics of the forces generated through cellular cytoskeletal elements cellular tensegrity theory was proposed [51]. This theory constructs cells from interconnected continuous series of tension elements and discontinuous series of compression resistant elements. The cell exerts a centripetal tension force on adhesion points, while the underlying ECM resists this continuous cytoskeletal tension and act as a discontinuous compression resistant element [51]. This theory was later grounded the mechanotransduction concept. Mechanotransduction is the process by which cells convert mechanical stimuli into biochemical signals (Fig. 1a) [35]. It enables cells to sense their physical environment and to respond by adjusting their structure and function. Mechanotransduction has roles in the regulation of blood pressure, remodeling of bone, maintenance of muscle and perception of touch and sound. Cell growth, migration and gene expression are influenced by mechanotransduction in most cell types [10]. Cells utilize a variety of mechanosensitive elements to sense applied forces and substrate stiffness including conformational changes in proteins at focal adhesions and inside the cytoskeleton [52].
Fig. 1.
Mechanotransduction pathways. a- Signaling in mechanotransduction. b- Detail of mechanotransduction from substrate to nucleus.
Mechanotransduction process is not just limited to outside to cytosol, but it proceeds all the way to the nucleus (Fig. 1b). The nuclear envelope separates the nuclear and cytoplasmic compartments and serves as a mechanosensory element regulating both biochemical and physical interaction of the nucleus with cell cytoskeleton, cell membrane and ECM [46]. A central role in this mechanosensory process has been attributed to lamins. It is proposed that tensile properties of the lamina would radiate through the cytoskeleton to the plasma membrane, generating a mechanotransduction signaling capability in the cell that links the extracellular matrix to the inside of the nucleus, considering the connectedness of nuclear lamina proteins [36,37]. Lamins are nuclear intermediate filaments, and they are the major components of the nuclear lamina which is a dense protein network underlying the inner nuclear membrane. They are extended parts of the LINC (linker of nucleoskeleton and cytoskeleton) complex which enables force transmission across the nuclear envelope [47]. The LINC complex itself is composed of two protein families: SUN domain proteins at the inner nuclear membrane and KASH domain proteins at the outer nuclear membrane. Many lamin binding proteins also interact with chromatin, particularly with silenced heterochromatic form, and the lamins have been shown to bind to DNA directly. These interactions complete a continuous physical linkage through which traction forces can be transmitted from the cell exterior to chromatin.
4. Fabrication of micro-nanostructures and substrates for 2D structures
Developments in surface patterning technologies at micro and nano-scale paved the way to novel cell-surface interaction studies. The production of surface topographies with geometrical micro and nanopatterns, like channels, pillars, and pits with controlled dimensions was made possible through the use of various methods [14]. In the last few decades engineering topographies for generating better cell-substrate attachments, cell differentiation and proliferation for tissue engineering has been studied in detail in the recent years [6,15,19,[53], [54], [55], [56], [57]].
4.1. Photolithography
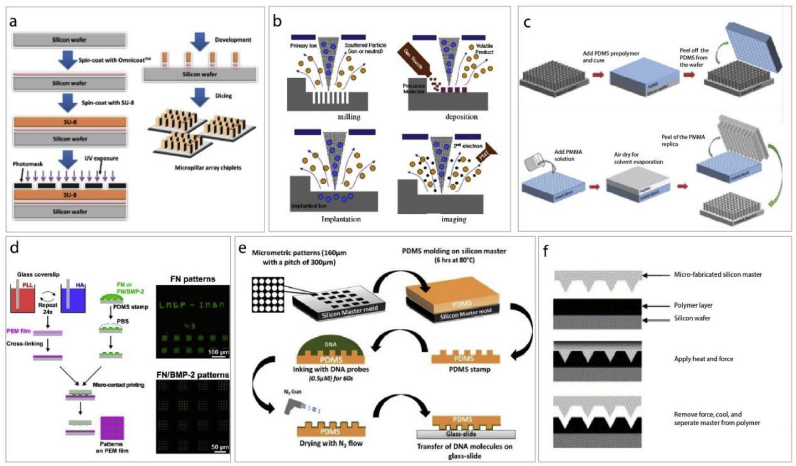
Photolithography is formation or removal of a pattern on a substrate by UV exposure of a photo mask coated with a resin (Fig. 2a). The substrate used is generally a silicon wafer, and the photo mask is a film that allows transmission of the UV light through the unmasked regions [63]. High intensity UV is used to transfer the pattern onto the template or image of the mask to the surface of the substrate [63,64]. Photomasks are transparent glass plates and patterns (or their inverse) are mostly metals that block UV transmission [65].
Fig. 2.
Micro- and nano fabrication methods. a- Production scheme of micropatterned substrates using photolithography [58]. b- Electron beam lithography consists of milling, deposition, implantation and imaging steps [59]. c- Solvent casting is a soft lithography method used for the production of micro and nano patterned substrates [60] (modified with permission). d- Layer-by-layer coating combined with microcontact printing [61]. e− Direct microcontact printing methods scheme [62]. f- Hot embossing process [9].
In the photolithography process, there are 4 fundamental steps: positioning of the mask on the substrate, exposure with UV, and development of the resin, and finally, the etching of the silicon layer. The positive resists used in coating the substrate become soluble in the developer solution as a result of exposure to UV while the negative resists become stabilized or crosslinked and become insoluble in the developer after exposure to UV. In the development step, the soluble parts of the resist are washed away [65]. When this method is applied to thick resists, the resultant resist can also be used in the fabrication of thick tiny reactors for making micro channels [65].
4.2. Electron beam lithography
Electron beam lithography (EBL) can produce very tiny patterns with dimensions of up to 3–5 nm. It needs an electron source and a scanning electron microscope to perform transfer design (Fig. 2b). Electrons from the source are accelerated in an electron field, where the electron beam is focused on a narrow spot (2–5 nm) by passing through the lenses. EBL is a process similar to photolithography; photolithography can expose a whole wafer at once but EBL makes it in a series of exposures, so it takes a long time. Also, the stage movement, calibration and settle times are slow and long. Fabrication procedures of EBL and photolithography are also similar in that both use a resist to coated on a substrate. However, EBL needs an electron sensitive coat instead of a light sensitive one. This polymer coat is either degraded or crosslinked upon exposure. Then in the development step, exposed patterns are revealed. In the EBL process, conducting substrates or metallic films coated non-conducting substrates are used [66].
4.3. Soft lithography and microcontact printing
Micro and nanostructures can be constructed using soft lithography and employ polymers such as polydimethylsiloxane, PDMS as templates (soft lithography) or stamps (microcontact printing) (Fig. 2c–e). Soft lithography has the advantages of low cost and ease and is frequently used in the production of micropatterned substrates from various materials [16,58,60,67,68]. Curved surfaces can also be used successfully due to the flexibility of PDMS while photolithography is not suitable for such surfaces [69]. Microcontact printing is a method similar to image transfer like a regular stamp and ink. It is used to pattern molecules on a surface for cell behavior and protein adsorption applications [61,62]. In microcontact printing, PDMS is used to produce a stamp by replica molding. Then the stamp is inked to adsorb cells, proteins and other molecules to transfer them onto the substrate. After the stamp is pressed, the solvent is evaporated, and molecules are transferred, the stamp is removed revealing the patterns. Microcontact printing can be used for large areas, and multiple copies of the patterns can be obtained by a single stamp by applying it repeatedly. High aspect ratio patterns is not possible with this technique.
4.4. Hot embossing
Hot embossing is another technique widely used for micropatterning [9]. Thermoplastic polymers are used in this method and heat is applied to polymer to raise it above its transition temperature and the polymer softens (Fig. 2f) [9,16]. Then the master is pressed onto the warm polymer. When cold, the master is remove from the polymer replica. It is a straightforward and short procedure [66].
4.5. Microfluidics
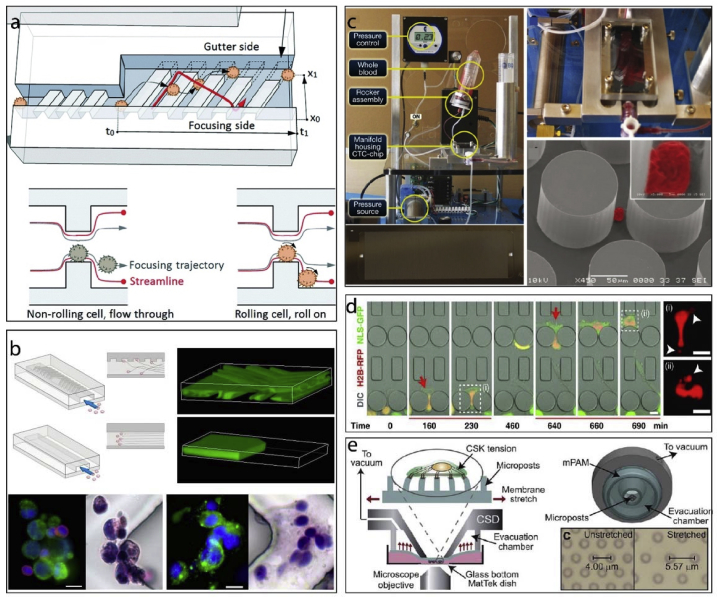
Microfluidics use small amounts of fluids with the help of channels with dimensions of 10–100 μm. They use small quantities of samples and carry out separations and detections with high resolution and sensitivity, low cost, and short analysis times [70]. Microfluidic chips are often used to create gradients of proteins, growth factors, drugs or their combination [71]. By manipulating the flow rates of the input channels, gradients (diffusion profile) with variable intensities can be formed. Aside from rapidly changing the fluidic environment around a cell, microfluidics can also be used to study patterned substrates (Fig. 3) [[72], [73], [74], [75], [76]]. In a study, cells were forced to roll over an obstacle course combining both effects of the fluid shear and physical obstruction of micropatterned substrates (Fig. 3a) [72]. Microfabricated culture systems are advantageous as they offer control of the culture environment with high reproducibility at the level of single cells. For example, such a device can be used to capture circulating tumor cells (CTCs) in blood samples (Fig. 3b–c) [73,74]. Thus, a high control of the cell culture environment can be obtained by tightly regulating cell shape, dimensionality, adhesive surfaces/ligands, amount of cell-cell contacts, and the level and nature of provided soluble factors. Denais et al. studied the nuclear rupture mechanics in confined environments using a micropatterned microfluidics device (Fig. 3d) [75]. Similarly, another microfluidics device was used for pulling the micropatterned substrates to study cell stretching (Fig. 3e) [76]. In general, these systems enable the researchers to study the behavior of cells in cancer and also have great potential in personalized treatments as diagnostic tools [77].
Fig. 3.
Microfluidics applications of micro and nano patterning technologies. a- Cell rolling cytometer [72] (modified with permission). b- A microfluidics device with herringbone micropatterns for capturing CTCs [73] (Copyright (2010) National Academy of Sciences. Modified with permission). c- CTCs were isolated from whole blood samples by using a microfluidics device [74]. d- Cell confinement microfluidics device to study DNA rupture [75] (Modified with permission). e− A cell stretching device to study cell contractile response [76] (Modified with permission).
5. Applications for tissue engineering and Cell material interactions
There are several applications for tissue engineering and cell-material interactions of the substrates produced using the methods mentioned in the previous sections. These applications include adhesion, alignment, migration and differentiation of the cells, as well as tools for studying the nuclear elasticity, deformation, control of cell shape and measuring cellular forces. Substrates produced using these methods have grooves and channels, pillars, wells and pits, and proteins immobilized on the surfaces creating controlled patterns. The examples of the produced surfaces, surface pattern dimensions, cells and tissues tested with them and the cell responses are summarized in Table 1.
Table 1.
Summary of the substrates properties and the cellular responses they evoked.
| Grooves/Channels | |||||
| Material | Dimension | Cell/tissue | Remarks | Ref. | |
| Width/Diameter | Depth/Height | ||||
| PMMA | 10, 25, 100 μm | 330 nm | HOBs | Decreased nanogroove widths led to increased contact guidance, decreased adhesion and increased angiogenic gene expressions | 78,79 |
| 2, 3, 6, 12 μm | 0.2, 0.5, 1.1, 1.9 μm | Baby hamster kidney cells, | Increased alignment with increased depth and decreased with increased width. | 21,80 | |
| PDMS | 5, 10, 20, 60 μm | 25 μm | Human neural stem cell | Increased alignment and induced neurite growth with decreased micropattern dimensions. Increased neuron density but altered neurite alignment with increased micropattern dimesions. | 81 |
| 30 μm | 10 μm | VEC | Orientation along grooves; changes in gene expression | 82,83 | |
| 20–60 μm | 11 μm | VSMC | Enhanced alignment of cell/nuclei on narrow grooves | 83,84 | |
| 20, 50, 80 μm | 5, 12 μm | VSMC | Enhanced cell/nuclei aspect ratio and cell alignment | 83,85 | |
| 3.5 μm | 0.2–5 μm | VEC | Orientation along grooves; no change in proliferation | 83,86 | |
| 2–10 μm | 50–200 nm | VSMC or VEC | Cell orientation and migration along grooves; enhanced cell elongation | 19,83 | |
| 1200 nm | 600 nm | VEC | Increased cell elongation, alignment and migration along grooves and reduced cell proliferation | 83,87 | |
| 600 nm | 600 nm | Human embryonic stem cells | Reduced cell proliferation | 21,87 | |
| PS | 10 μm | 3 μm | Rat astrocytes | Low adhesion, strong alignment | 21,88 |
| 1–10 μm | 0.5–1.5 μm | Rat bone marrow cells | On large grooves, focal adhesions cover the surface, On narrow grooves, focal adhesions are only on edges |
21,89 | |
| 1, 2, 5, 10 μm | 0.5, 1, 1.5 μm | Rat bone marrow cells | Better mineralization with 1 μm depth and 1–2 μm width | 89 | |
| 20–1000 nm | 5–530 nm | Fibroblasts | No alignment for depths ˂ 35 nm or widths ˂ 100 nm | 21,90 | |
| Polyimide | 4 μm | 5 μm | Osteoblasts | Strong alignment, no change in adhesion | 9,21 |
| PDLA | 10 μm | 3 μm | Schwann cells (nerve cells) |
Strong alignment | 21,91 |
| PHBV | 1–10 μm | 5–30 μm | Rat mesenchymal stem cell-derived osteoblasts |
Increased osteoblast adhesion and alignment | 79,92 |
| PLGA | 350, 700, 1050 nm | 500 nm | VEC | Enhanced adhesion strength, Increased cell alignment along grooves |
83,93 |
| Ti | 750 nm- 100 μm | 200 nm | VEC | Increased cell alignment along grooves and higher cell density on grooves with width < 10 μm | 83,94 |
| Quartz | 0.5,5,10, 25 μm | 0.5, 5 μm | Murine macrophage | Increased orientation | 21,95 |
| 12.5 μm | 5 μm | Fibroblasts | Change in gene expression profile | 21,96 | |
| 1, 4 μm | 1.1 μm | MSC | Alignment better in the wider grooves | 21,97 | |
| 2–10 μm | 30–280 nm | Murine macrophage | Higher phagocytotic activity when topography size is equal to collagen fiber size | 21,98 | |
| TCPS | 5, 45 μm | 5 μm | Primary (glioma) and metastatic (lung and colon) tumors | Induced migration of primary and metastatic brain, lung and colon cancer cells | 99 |
| HA on PET | 5, 25 μm | – | Articular knee chondrocytes | Induced adhesion, migration, alignment, and differentiation of chondrocytes | 79,100 |
| Collagen | 27 μm | 12 μm | Mesenchymal osteoprogenitor cells | Cell alignment and enhanced bone formation | 53 |
| 10 μm | 30 μm | Human corneal keratocytes and D407 | Higher mechanical properties on patterned collagen films | 101 | |
| 650, 500, 332.5 nm | 300, 250, 200 nm | HMEC | No change in proliferation and has a minimal effect on cell alignment, enhanced cell retention under flow-shear conditions | 67 | |
| Collagen coated with fibrinogen | 27 μm | 12 μm | Rat bone marrow osteoblast cells | Enhanced cell orientation and bone formation | 102 |
| Ti— coated Si |
15 μm |
200 nm |
T24 |
Less round and smaller cell shape |
21,103 |
|
Pillars | |||||
|
Material |
Dimension |
Cell/tissue |
Remarks |
Ref. |
|
| Width/Diameter |
Depth/Height |
||||
| PMMA | 4, 8, 16 μm | 8 μm | DPSC | Control of fate of the stem cells | 60 |
| 100 nm | 160 nm | Fibroblasts | Smaller, less organized actin cytoskeleton | 21,104 | |
| 100 nm | 160 nm | Fibroblasts | Less spreading | 21,105 | |
| PLGA | 4, 8, 16 μm | 8 μm | Saos-2, L929, SH-SY5Y, MCF7, hOB | Nuclear deformation in cancer cells (Saos-2, MCF-7, SH-SY5Y) not in noncancerous cells (hOB, L929) | 58 |
| 3 μm | 7 μm | MSC | Geometry of cell nuclei responds to the micropillar array | 106 | |
| 3 μm | 5 μm | BMSC | Severe nucleus deformation, no change in proliferation and differentiation | 107 | |
| PLGA and PDMS | 30 μm | 4, 9 μm | NIH 3T3 fibroblasts | On PDMS, 3T3 cells on stiffer (longer) pillar area. No such effect on PLGA. | 108 |
| PDMS | 1–5.6 μm | 1–8 μm | VEC | Enhanced cell alignment and elongation on PDMS pillars | 109 |
| PDMS coated with fibronectin | 10 μm | 10 μm | MCF-10 A, MDA-MB-231 | Epithelial to mesenchymal transition of the breast cancer cells within enclosed micropillar arrays | 110 |
| PLLA | 2–20 μm with interpillar spaces 2–20 μm |
5–6 μm | Saos-2, MG-63, OHS4 | Nuclear deformation in cancer cells (Saos-2) higher than in healthy counterparts (OHS-4 and MG-63). Saos-2 cells deformed severely on pillars with 5–10 μm spacing. | 111 |
| PLLA, PLLA: PLGA blend | 200 nm | 900 nm | Saos-2, BMSC | Saos-2 cells populated fields with pillars 1 μm apart but not on pillar-free surfaces. BMSCs avoided fields with interpillar distances <2 μm. | 112 |
| Collagen and PLGA | 8, 16 μm | 8 μm | Saos-2 | Increased proliferation and ALP production on collagen micropillars. Increased nuclear deformation on PLGA micropillars | 113 |
| Ti | 21 nm | 15 nm | BMSC and hBMHCs | Improved bone deposition on nanopillars | 114 |
| Alumina |
110 nm |
– |
Mouse bone marrow stem cells |
Increased proliferation and differentiation |
21,115 |
|
Wells/Pits | |||||
|
Material |
Dimension |
Cell/tissue |
Remarks |
Ref. |
|
| Width/Diameter |
Depth/Height |
||||
| PMMA | 120 nm | 100 nm | MSC | Stimulated differentiation and production of bone mineral in vitro | 21,116 |
| 35, 75, 120 nm | – | Fibroblasts | Reduced adhesion, orientation and distinction of symmetries | 21,[117], [118], [119] | |
| PDMS | 2, 5, 10 μm | – | Human fibroblasts | 2 and 5 μm showed better proliferation 10 μm showed no effect |
21,120 |
| PC | 7, 25, 50 μm | 0.5, 1.5, 2.5 μm | Fibroblasts | No orientation | 21,121 |
| Titanium | 100, 30, 10 μm | – | MG63 | Cell attachment, growth, aggregation and morphology depends on the presence and dimension of the micropatterns | 79,122 |
| PCL | 30 μm | 80, 220, 333 nm | BMSC | Optimal adhesion on 80 nm deep pits, inductive capability on 220 nm deep pits | 123 |
| 150 nm | 80 nm | Fibroblasts | Less focal contacts and vinculin F-actin cytoskeleton less developed |
21,124 | |
*Cells (hOB: primary human osteoblasts, MSC: mesenchymal stem cells, VEC: vascular endothelial cells, VSMC: vascular smooth muscle cells, DPSC: human dental pulp mesenchymal stem cells, HMEC: human microvascular endothelial cells, BMSC: bone marrow stem cells, Saos-2: osteosarcoma cells, MCF-10 A: mammary epithelial cells, MDA-MB-231: breast adenocarcinoma cells, MG63: osteoblast like cells, OHS4: human osteosarcoma cells, L929: mouse fibroblast cell, SH-SY5Y: human neuroblastoma cells, T24: human bladder carcinoma, D407: retinal pigment epithelial cells, hBMHCs: human bone marrow hematopoietic cells).
**Polymers (HA: hyaluronic acid, PET: polyethylene terephthalate, PLLA: poly(l-lactic acid), PMMA: poly(methylmethacrylate), PDMS: polydimethyl siloxane, PLGA: polylactic acid-co-glycolic acid, PCL: polycaprolactone, PC: polycarbonate, PS: polystyrene, PDLA: poly(D,l-lactic acid), PHBV: poly(hydroxybutyrate-co-hydroxyvalerate), TCPS: tissue culture polystyrene).
5.1. Cell adhesion and alignment
Cell adhesion depends on substrate properties, and it is a multistep process. First, proteins from the culture medium are adsorbed onto the surface, and followed by cell adhesion. Then the cells release compounds involved in signaling, ECM deposition, cell proliferation and differentiation. These all depend on the chemical interaction of the cell and the substrate, and therefore, the chemical composition of the surface is among the most important properties affecting the cell-substrate interaction. Chemical features of a substrate include surface charges, wettability, and protein adsorption ability [125]. Studies show that moderately hydrophilic surfaces lead to more cell adhesion and good spreading, proliferation, and differentiation on a surface [126,127]. Cell adhesion was found to be decreased with increasing contact angle (from 0° to 106°) in studies with osteoblasts, and the highest adhesion was observed between 60° and 80° in studies with fibroblasts [18,128]. Protein adsorption is another event occurring just before cell-substrate contact. Many proteins such as immunoglobulins, vitronectin, fibronectin, and fibrinogen adsorb on substrate surfaces [127]. Studies show that ions on surfaces can improve their biocompatibility and the cell affinity to surface and differentiation [129]. As an example, attachment and spreading of osteoblast and fibroblast were enhanced when the surface was positively charged compared to negative and neutral [130].
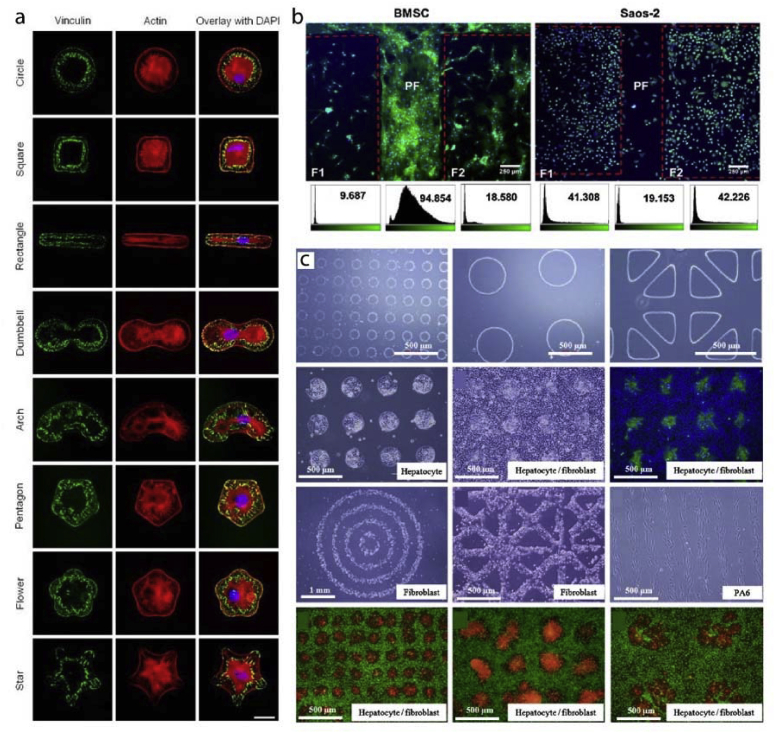
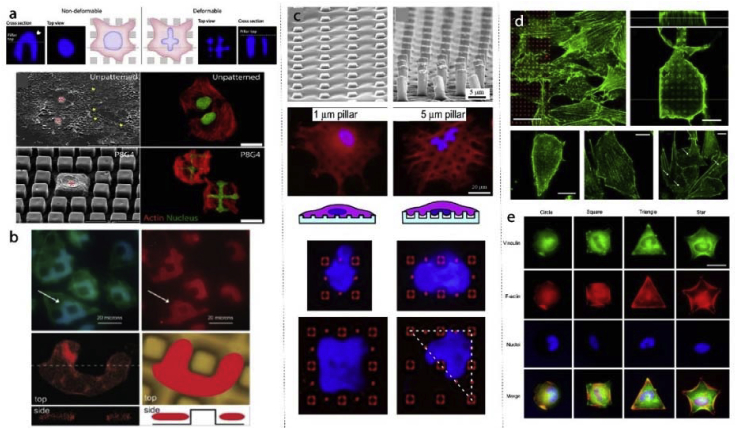
The idea of adsorbing proteins to the surface can be further advanced with protein micropatterns. For example, in a study fibronectin patterns spanning the area of a cell were produced [131]. Cells conformed to the fibronectin patterns in the form of circles, squares, rectangles, arcs, pentagons, flowers, and stars (Fig. 4a). A similar cell patterning approach was achieved by using micro- and nanopillars. Ozcelik et al. showed different cells prefer different surface roughness and topography (Fig. 4b) [112]. In this study, stem cells crowded smooth surfaces while osteosarcoma cells preferred surfaces decorated with nanotopography. In another study, micropatterns were created using several ECM proteins: hyaluronic acid, fibronectin and collagen in a layer-by-layer fashion [132]. These multi-material patterns allowed coculturing hepatocytes, fibroblasts and embryonic stem cells in a patterned manner when different cells adhered to different surfaces (Fig. 4c). These examples demonstrate the versatility of chemical and physical micropatterns for studying cell adhesion.
Fig. 4.
Cell adhesion to micro and nano patterned substrates. a- Fibronectin surface micropatterns influence cell shape and actin cytoskeleton organization of the cells [131] (Modified with permission). b- Nanopatterned regions and unpatterned aisles were used to differentially attach stem cells and osteosarcoma cells to the surfaces [112]. c- Hyaluronic acid, fibronectin and collagen micropatterns were used to study cocultures of hepatocytes and fibroblasts [132].
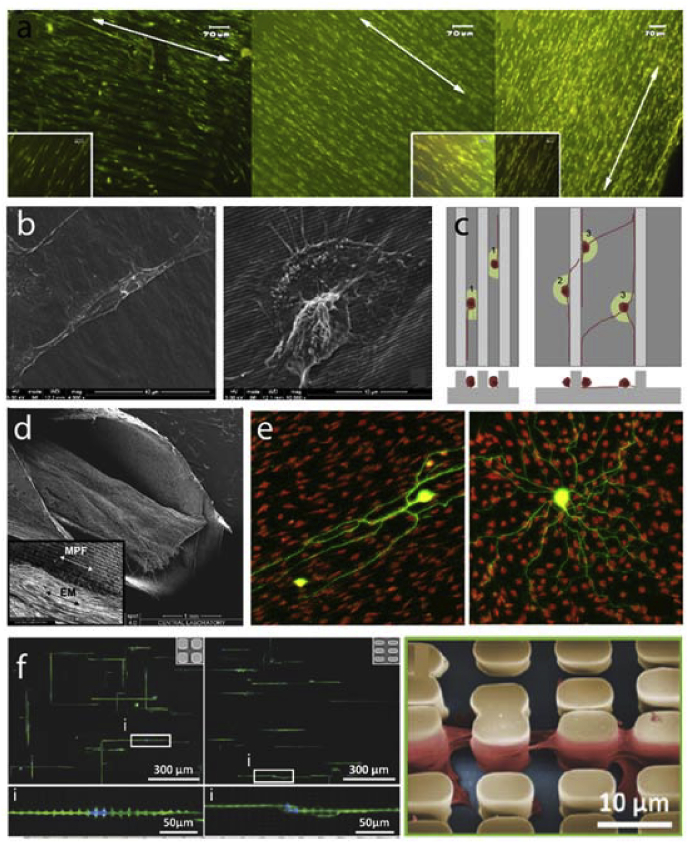
Tissues have predetermined architectures. In some cases, these architectures require alignment of cells. To mimic the cell alignment and tissue architecture, micro and nanopatterns and grooves were employed for tissue engineering applications [11,13,60,67,68,81,101]. Vrana et al. achieved aligned corneal keratinocytes and retinal pigmented epithelial cells mimicking the corneal architecture (Fig. 5a) [101]. Peterbauer et al. showed that two different mammalian cells (CHO and rat myoblasts) were aligned on nano-grooved substrates (154 nm) [133]. Lu et al. fabricated nano and micro patterned titanium surfaces (random and 750 nm, 2 μm, 5 μm, 25 μm, 75 μm, 100 μm grooves) and showed increased endothelial cell density as the pattern dimension decreased to nano leading to enhanced spreading and alignment of cells on nanogrooves [94]. Endothelial cells grown on nanopatterned collagen films also showed alignment (Fig. 5b) [67]. In other studies, stems cells were differentiated on micro-grooved substrates to align nerve cells (Fig. 5c–d) [68,81]. A similar alignment effect was also achieved within a coculture system where Schwann cells were aligned on laminin micropatterns leading to aligned growth of neurites (Fig. 5e) [13].
Fig. 5.
Cell alignment on micro and nanopatterned substrates. a- Alignment of human corneal keratocytes on patterned collagen films [101]. b- Endothelial cells were aligned on top of nanopatterned grooves [67] (Modified with permission). c- Schematic illustration of elongation and alignment of stems cells differentiated into nerve cells [81]. d- Nerve guidance conduit design with micro-grooved inner surface architecture [68] (Modified with permission). e− Schwann cells were aligned to laminin patterns in the absence of other guidance cues which resulted in aligned neurite growth on top [13] (Modified with permission).
5.2. Cell migration
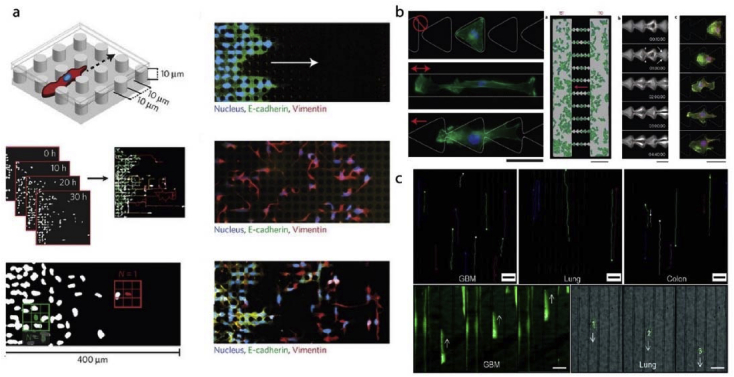
Cell migration is a complex, multistep process and integral for embryonic development, wound healing, ossification, and in immunity [134]. It is also an essential contributor to disease processes including cancer [135]. Many studies seek the answer to the problem how cells migrate in an obstacle course. Since micro-nanopatterned substrates are promising candidates to mimic tissue architecture, there are several studies on the subject [17,99,110,[136], [137], [138], [139], [140]]. In an example, migration of breast cells was studied within an enclosed micropillar array (Fig. 6a) [110]. Migration rate and length of non-malignant and malignant breast cells were different from one another. In the study of Mahmud et al. migration of breast cancer, melanoma and embryonic stem cells were studied on disconnected triangles, straight lines, and triangular ratchets (Fig. 6b) [17]. They showed that cells prefer certain topographies for migration; ratchets had more cells to migrate than straight lines. In another study, primary and metastatic cancer cells exhibited directional migration on microgrooves (Fig. 6c) [99]. Using microgrooves, cell migration patterns were classified as aligned, confined and the combination of the two. All these studies demonstrated the usefulness of engineered microtopographies.
Fig. 6.
Cell migration on micropatterned substrates. a- Migration and epithelial to mesenchymal transition of the breast cancer cells were investigated within an enclosed micropillar array [110] (Modified with permission). b- Disconnected triangles, straight line and triangular ratchet micropatterns were employed to study cancer and embryonic cell migration [17]. c- Migration of primary and metastatic brain, lung and colon cancer cells were studied on microgrooved substrates [99] (Modified with permission).
5.3. Differentiation
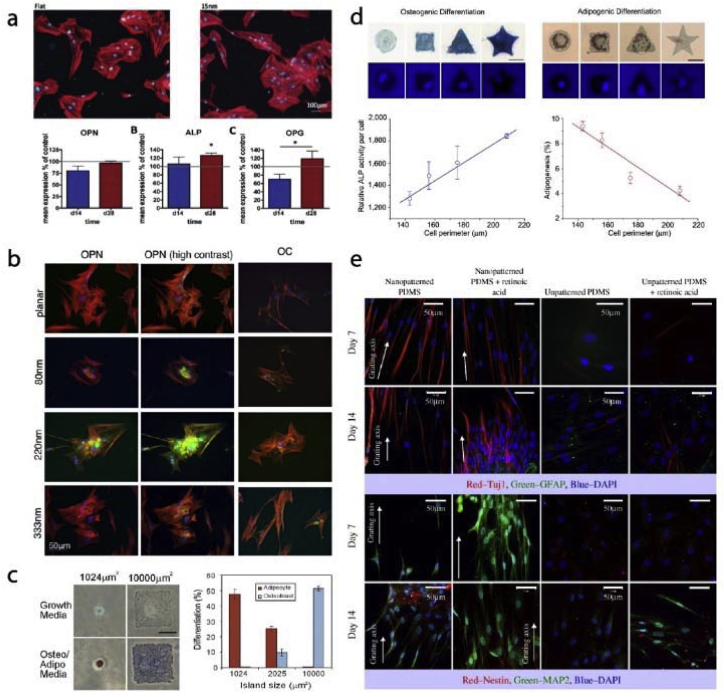
Tissue engineering and regenerative medicine disciplines integrate chemical, architectural and physical cues of the scaffolds to control the fate of cells by inducing differentiation for targeted tissue regeneration. Substrate topography at nano or micro level is one of the determinants of cell fate. In a study, it was shown that ALP activity, proliferation, and expression of calvaria, number of osteoblastic cells were higher on rough surfaces with average roughness of up to 0.8 μm than on smooth ones [141]. Human osteoblastic cells were also shown to have increased spreading and proliferation on rough surfaces [142]. Surface roughness can be macro- (100 μm – mm), micro- (100 nm–100 μm) or nano-level (less than 100 nm). Cells respond differently to different level of roughness [143]. Similarly, the engineered topography is just as influential on cell fate as random roughness (Fig. 7). In one study, 15 nm pillars were shown to upregulate osteogenic markers of the stem cells and induce differentiation towards both osteoclasts and osteoblasts (Fig. 7a) [114]. Similarly, Davison et al. demonstrated higher osteogenic marker expression on nanopits compared to planar substrates (Fig. 7b) [123]. Chaubey et al. produced micropatterned (grooves with 7.3 μm separation) films to study the effects of 2D modified substrates on adipogenic differentiation of stem cells [144]. Micropatterned surfaces increased adipogenic phenotype for up to 10 days. Cell confinement and cell shape control were achieved by using fibronectin micropatterns. For example, McBeath et al. showed the size of the cell confinement area is the critical factor for cell fate determination (Fig. 7c) [145]. In a set up where cells were supplemented with a combination of osteo- and adipo-differentiation medium and were either confined to a very small area or a 10x larger area determined the cell fate. Cells restricted to small patterns differentiated towards adipocytes while cells on larger patterns showed osteogenic tendencies. Peng et al. made a similar observation (Fig. 7d) [146]. Cells were constrained into specific shapes with different perimeters. As the perimeter of the cell increased, osteogenic marker ALP production increased while decreased perimeter led to adipogenic cell increase. These studies are not limited to bone tissue engineering. Yim et al. studied neurogenic differentiation of stem cells on nanopatterns and showed increased expression of early neurogenic markers on nanopatterned substrates (Fig. 7e) [147].
Fig. 7.
Cell differentiation on micro and nanopatterned substrates. a- Titanium nanopatterns induce ostegenic marker expression in stem cells [114] (Modified with permission). b- Stem cells cultured on 220 nm deep pits showed higher osteogenic marker intensity compared to planar substrate [123]. c- Stem cells confined to 1024 μm and 10000 μm fibronectin islands. Adipogenic differentiation is inversely related to island size while osteogenic differentiation is directly proportional to it [145] (Modified with permission). d- Differentiation of stem cells on different sized and shaped fibronectin islands [146]. e− Neurogenic differentiation and expression of nerve markers of stem cells on nanopatterned substrates [147].
5.4. Nuclear elasticity and deformation and control of cell shape
Forces applied on the cell surfaces resulted in cell responses including the reorganization of cytoskeletal elements, actin microfilaments, intermediate filaments, microtubules and nuclear structures [148]. Rigidity, or deformability, of a cell nucleus is determined primarily by: (1) chromatin as well as the nucleoskeletal organization, and (2) expression and assembly of lamins as part of the nuclear lamina [149]. Micro and nanopatterns induce mechanotransduction and cause distortions in the cell shape. The topographical cues not only distort and deform cells but also affect the nuclei within. Recent studies demonstrated that particular cell types are more prone the nuclear deformations (Fig. 8a) [58]. In the study of Davidson et al. it was shown that cell nuclei deform under stress generated by patterned substrates (Fig. 8b) [16]. It was shown that nuclei of cancer cells extensively deform on patterned substrates unlike the healthy cells [150]. This suggests the presence of a difference in both the mechanotransduction and signaling between cancer and healthy cells and opens the door to the possibility of discrimination of cancer cells through the substrate. A recent study showed nuclear shape deformations with bone marrow stem cells (BMSCs) (Fig. 8c) [107]. In the study of Badique et al. invasion capacity of a cancer cell was correlated with the deformation capacity and this could be measured by designing a patterned surface which can deform cell nucleus [111].
Fig. 8.
Deformation of cells and cell nuclei on micropatterned substrates. a- Nuclear deformability, which reflects nuclear elasticity, is different in different cell types. Micro and nanopatterned substrates could be used to reveal and visualize the elasticity differences of cells [58] (Modified with permission). b- Osteosarcoma cells show nuclear deformations on micropatterned substrates [16] (Modified with permission). c- Stem cell nuclei also demonstrated deformations on micropatterns and cell nuclei were confined into interpillar spaces [107]. d- Actin reorganization and proliferation was studied on nanopillar arrays [107]. e− Control of cell shape can be achieved with cell adhesive micropatterns produced from RGD peptides [146].
Micro and nanopatterns are also used for the control of cell shape. Buch-Manson et al. used nanopillar arrays with different pillar densities to study cytoskeletal remodeling of fibroblasts (Fig. 8d) [151]. They showed that the interface between the nanopatterned and planar regions had crucial importance to study actin remodeling and proliferation. In another study, arginine–glycine–aspartic acid (RGD) micropatterns were produced using photolithography. Stem cells were successfully confined to these RGD islands and took predetermined shapes (circle, square, triangle or star) (Fig. 8e) [146]. They showed that by controlling cell shape, cell fate could be influenced indirectly.
5.5. Quantification of cellular forces
Cell-material interactions differ in disease and health state of the cells. The interaction between the diseased cells and surfaces with microtopographical features have significant importance in the study of some diseases since traction forces of the cells on these surfaces can be quantified. This would help reveal the intrinsic differences between various conditions.
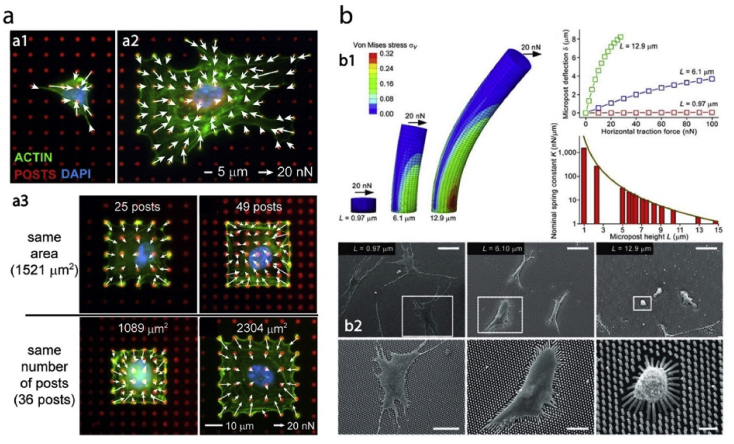
Micropost arrays are being used to measure cellular traction forces for nearly a decade [[152], [153], [154], [155], [156]]. In the study of Han et al. cells were seeded on surfaces with different micropost densities and coated with a hydrophobic polymer and fibronectin (Fig. 9a) [153]. Their results showed that force generation by a cell through focal adhesions can be modulated by substrate stiffness, spread area, and post density. Understanding the effect of stiffness and spreading on traction forces generated by cells would provide insight into the formation of focal adhesions. The response of stem cells to changing micropost rigidity was studied by Fu et al. (Fig. 9b) [155]. They showed that traction forces were strongly correlated with cell spreading regardless of micropost rigidity. They postulated that rigidity sensing occurs at micron scale rather than nanoscale.
Fig. 9.
Quantification of cellular forces using micro and nanopatterned substrates. a- Traction forces of the endothelial cells were measured using a micropost array. Cells were either interacted with the microposts freely, or confined to an area with predetermined number of posts or post density [153] (Modified with permission). b- Cell traction forces of the stem cells were measured using elastomeric micropost with controlled stiffness [155].
6. Conclusion
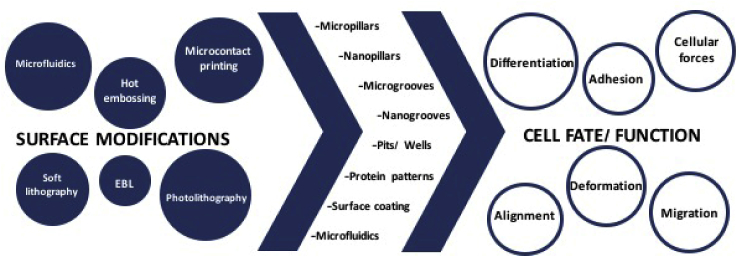
Cell-substrate interaction is an integral part of tissue engineering and implant design. It is also a very well-studied discipline. Nano and microfabrication technologies present a series of novel approaches for the control of substrate topography and chemistry. With the use of these technologies, cell processes like attachment, alignment, migration, and differentiation could be modulated (Fig. 10).
Fig. 10.
Surface modifications and resultant effects on cells.
There are several methods used in the fabrication of surfaces with desired properties including photolithography, EBL, soft lithography, microcontact printing and hot embossing. Photolithography uses a light source to transfer the design onto the resin, while in EBL an electron beam is employed. Soft lithography and microcontact printing methods are used for chemical patterning of surfaces where the desired molecule is stamped onto the surface. Hot embossing, on the other hand, is a convenient method for the fabrication of replicas of micro-nano surfaces. All these methods could be integrated into microfluidics devices in order to control the cell microenvironment further. With the use of these novel technologies, the study of cell-substrate interactions and implant design were improved. Cell adhesion is a very complex process that requires specific chemical, physical and mechanical interactions with the substrate. In tissue engineering and regenerative medicine, control of substrate properties allows control of differentiation, achieving better tissue integration and on the whole, improved biomaterials performance. Devices produced using these methods could also make detection and screening of diseased cells or quantification of the forces generated during cell-material interactions possible. In conclusion, it can be stated that physical and chemical surface modifications at the nano- and micro levels have proven their worth in the biomedical device design.
In this review, we have presented the application of the afore mentioned methods to the creation of micro- and nanostructured surfaces to present a general perspective to the readers on cell-substrate interactions. It was shown that the geometry and the size of topographical features affect substantially the cell adhesion, alignment, migration, differentiation and morphology; however, no universal correlation was found between the size of the topographical features (nano-to-micro) and the induced cell responses. Nevertheless, in a case by case basis, promising results are observed in controlling the responses of the cells through the use of these novel approaches. Tuning of the surface properties of a substrate with patterns created by photolithographic approaches and causing the reprogramming the cells towards a specific fate, instead of conventional chemical induction, is one such example.
On the other hand, all the methods and tools that are mentioned here are valid for cell monolayers studied under in vitro conditions. The adaptation of these 2D methods to 3D microenvironments and assessment of their results are still being debated.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Harrison R.G. The cultivation of tissues in extraneous media as a method of morpho-genetic study. Anat. Rec. 1912;6:181–193. [Google Scholar]
- 2.WEISS P. The problem of specificity in growth and development. Yale J. Biol. Med. 1947;19:235–278. http://www.ncbi.nlm.nih.gov/pubmed/20284280 [PMC free article] [PubMed] [Google Scholar]
- 3.CURTIS A.S., VARDE M. Control of cell behavior: topological factors. J. Natl. Cancer Inst. 1964;33:15–26. http://www.ncbi.nlm.nih.gov/pubmed/14202300 [PubMed] [Google Scholar]
- 4.Voldman J., Gray M.L., Schmidt M.A. Microfabrication in biology and medicine. Annu. Rev. Biomed. Eng. 1999;1:401–425. doi: 10.1146/annurev.bioeng.1.1.401. http://rleweb.mit.edu/biomicro/reprints/jvoldman_annrev.pdf [DOI] [PubMed] [Google Scholar]
- 5.Curtis A., Wilkinson C. Topographical control of cells. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. http://www.ncbi.nlm.nih.gov/pubmed/9613804 [DOI] [PubMed] [Google Scholar]
- 6.Rajnicek A., Britland S., McCaig C. Contact guidance of CNS neurites on grooved quartz: influence of groove dimensions, neuronal age and cell type. J. Cell Sci. 1997;110(2):2905–2913. doi: 10.1242/jcs.110.23.2905. http://www.ncbi.nlm.nih.gov/pubmed/9359873 [DOI] [PubMed] [Google Scholar]
- 7.Hutmacher D.W. Scaffold design and fabrication technologies for engineering tissues–state of the art and future perspectives. J. Biomater. Sci. Polym. Ed. 2001;12:107–124. doi: 10.1163/156856201744489. http://www.ncbi.nlm.nih.gov/pubmed/11334185 [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Pépin A. Nanofabrication: conventional and nonconventional methods. Electrophoresis. 2001;22:187–207. doi: 10.1002/1522-2683(200101)22:2<187::AID-ELPS187>3.0.CO;2-0. doi:10.1002/1522-2683(200101)22:2<187::AID-ELPS187>3.0.CO;2–0. [DOI] [PubMed] [Google Scholar]
- 9.Charest J.L., Bryant L.E., Garcia A.J., King W.P. Hot embossing for micropatterned cell substrates. Biomaterials. 2004;25:4767–4775. doi: 10.1016/j.biomaterials.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Dalby M.J. Topographically induced direct cell mechanotransduction. Med. Eng. Phys. 2005;27:730–742. doi: 10.1016/j.medengphy.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Hasirci V., Kenar H. Novel surface patterning approaches for tissue engineering and their effect on cell behavior. Nanomedicine. 2006;1:73–90. doi: 10.2217/17435889.1.1.73. [DOI] [PubMed] [Google Scholar]
- 12.Curtis A.S., Dalby M., Gadegaard N. Cell signaling arising from nanotopography: implications for nanomedical devices. Nanomedicine. 2006;1:67–72. doi: 10.2217/17435889.1.1.67. [DOI] [PubMed] [Google Scholar]
- 13.Thompson D.M., Buettner H.M. Neurite outgrowth is directed by Schwann cell alignment in the absence of other guidance cues. Ann. Biomed. Eng. 2006;34:161–168. doi: 10.1007/s10439-005-9013-4. [DOI] [PubMed] [Google Scholar]
- 14.Hasirci V., Vrana E., Zorlutuna P., Ndreu A., Yilgor P., Basmanav F.B., Aydin E. Nanobiomaterials: a review of the existing science and technology, and new approaches. J. Biomater. Sci. Polym. Ed. 2006;17:1241–1268. doi: 10.1163/156856206778667442. [DOI] [PubMed] [Google Scholar]
- 15.Zorlutuna P., Hasirci N., Hasirci V. Nanopatterned collagen tubes for vascular tissue engineering. J. Tissue Eng. Regen. Med. 2008;2:373–377. doi: 10.1002/term.99. [DOI] [PubMed] [Google Scholar]
- 16.Davidson P.M., Özçelik H., Hasirci V., Reiter G., Anselme K. Microstructured surfaces cause severe but non-detrimental deformation of the cell nucleus. Adv. Mater. 2009;21:3586–3590. [Google Scholar]
- 17.Mahmud G., Campbell C.J., Bishop K.J.M., Komarova Y.A., Chaga O., Soh S., Huda S., Kandere-Grzybowska K., Grzybowski B.A. Directing cell motions on micropatterned ratchets. Nat. Phys. 2009;5:606–612. [Google Scholar]
- 18.Wei J., Igarashi T., Okumori N., Igarashi T., Maetani T., Liu B., Yoshinari M. Influence of surface wettability on competitive protein adsorption and initial attachment of osteoblasts. Biomed. Mater. 2009;4:45002. doi: 10.1088/1748-6041/4/4/045002. [DOI] [PubMed] [Google Scholar]
- 19.Biela S.A., Su Y., Spatz J.P., Kemkemer R. Different sensitivity of human endothelial cells, smooth muscle cells and fibroblasts to topography in the nano–micro range. Acta Biomater. 2009;5:2460–2466. doi: 10.1016/j.actbio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Bettinger C.J., Langer R., Borenstein J.T. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew. Chem. Int. Ed. 2009;48:5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez E., Engel E., Planell J.A., Samitier J. Effects of artificial micro- and nano-structured surfaces on cell behaviour. Ann. Anat. - Anat. Anzeiger. 2009;191:126–135. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Hasirci V., Pepe-Mooney B.J. Understanding the cell behavior on nano-/micro-patterned surfaces. Nanomedicine. 2012;7:1375–1389. doi: 10.2217/nnm.12.7. [DOI] [PubMed] [Google Scholar]
- 23.Wang L. Biomimetic topography: bioinspired cell culture substrates and scaffolds. Adv. Biomimetics. 2011 InTech. [Google Scholar]
- 24.Martínez E., Engel E., Planell J.A., Samitier J. Effects of artificial micro- and nano-structured surfaces on cell behaviour. Ann. Anat. - Anat. Anzeiger. 2009;191:126–135. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Lim J.Y., Donahue H.J. Cell sensing and response to micro- and nanostructured surfaces produced by chemical and topographic patterning. Tissue Eng. 2007;13:1879–1891. doi: 10.1089/ten.2006.0154. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson C.D., Riehle M., Wood M., Gallagher J., Curtis A.S. The use of materials patterned on a nano- and micro-metric scale in cellular engineering. Mater. Sci. Eng. C. 2002;19:263–269. [Google Scholar]
- 27.Norman J.J., Desai T.A. Methods for fabrication of nanoscale topography for tissue engineering scaffolds. Ann. Biomed. Eng. 2006;34:89–101. doi: 10.1007/s10439-005-9005-4. [DOI] [PubMed] [Google Scholar]
- 28.Engel E., Michiardi A., Navarro M., Lacroix D., Planell J.A. Nanotechnology in regenerative medicine: the materials side. Trends Biotechnol. 2008;26:39–47. doi: 10.1016/j.tibtech.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Tay C.Y., Irvine S.A., Boey F.Y.C., Tan L.P., Venkatraman S. Micro-/Nano-engineered cellular responses for soft tissue engineering and biomedical applications. Small. 2011;7:1361–1378. doi: 10.1002/smll.201100046. [DOI] [PubMed] [Google Scholar]
- 30.Streicher R.M., Schmidt M., Fiorito S. Nanosurfaces and nanostructures for artificial orthopedic implants. Nanomedicine. 2007;2:861–874. doi: 10.2217/17435889.2.6.861. [DOI] [PubMed] [Google Scholar]
- 31.Desai T.A. Micro- and nanoscale structures for tissue engineering constructs. Med. Eng. Phys. 2000;22:595–606. doi: 10.1016/s1350-4533(00)00087-4. [DOI] [PubMed] [Google Scholar]
- 32.Craighead H., James C., Turner A.M. Chemical and topographical patterning for directed cell attachment. Curr. Opin. Solid State Mater. Sci. 2001;5:177–184. [Google Scholar]
- 33.Ruoslahti E., Pierschbacher M.D. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 34.Mitra S.K., Hanson D.A., Schlaepfer D.D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 35.Wang N., Tytell J.D., Ingber D.E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 36.Hutchison C.J. Lamins: building blocks or regulators of gene expression? Nat. Rev. Mol. Cell Biol. 2002;3:848–858. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- 37.Hutchison C.J., Worman H.J. A-type lamins: guardians of the soma? Nat. Cell Biol. 2004;6:1062–1067. doi: 10.1038/ncb1104-1062. [DOI] [PubMed] [Google Scholar]
- 38.Farquhar M., Palade G. Junctional complexes in various epithelia. J. Cell Biol. 1963:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.curtis a.s. The mechanism of adhesion of cells to glass. a study by interference reflection microscopy. J. Cell Biol. 1964;20:199–215. doi: 10.1083/jcb.20.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis A., Forrester J.V., McInnes C., Lawrie F. Adhesion of cells to polystyrene surfaces. J. Cell Biol. 1983;97:1500–1506. doi: 10.1083/jcb.97.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelman G.M. Cell adhesion molecules. Science (Wash. D C) 1983;219 doi: 10.1126/science.6823544. http://science.sciencemag.org/content/219/4584/450.abstract 450 LP-457. [DOI] [PubMed] [Google Scholar]
- 42.Elangbam C.S., Qualls C.W., Dahlgren R.R. Cell adhesion molecules–update. Vet. Pathol. 1997;34:61–73. doi: 10.1177/030098589703400113. [DOI] [PubMed] [Google Scholar]
- 43.Izzard C.S., Lochner L.R. Formation of cell-to-substrate contacts during fibroblast motility: an interference-reflexion study. J. Cell Sci. 1980;42:81–116. doi: 10.1242/jcs.42.1.81. [DOI] [PubMed] [Google Scholar]
- 44.Geiger B., Bershadsky A., Pankov R., Yamada K.M. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 45.Geiger B., Bershadsky A., Pankov R., Yamada K.M. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 46.Uzer G., Rubin C.T., Rubin J. Cell mechanosensitivity is enabled by the LINC nuclear complex. Curr. Mol. Biol. Reports. 2016;2:36–47. doi: 10.1007/s40610-016-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isermann P., Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsimbouri P. Adult stem cell responses to nanostimuli. J. Funct. Biomater. 2015;6:598–622. doi: 10.3390/jfb6030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo B.-H., Carman C.V., Springer T. a. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berrier A.L., Yamada K.M. Cell-matrix adhesion. J. Cell. Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 51.Ingber D.E. Cellular tensegrity: defining new rules of biological design that govern the cytoskeleton. J. Cell Sci. 1993;104(3):613–627. doi: 10.1242/jcs.104.3.613. http://www.ncbi.nlm.nih.gov/pubmed/8314865 [DOI] [PubMed] [Google Scholar]
- 52.McGregor A.L., Hsia C.-R., Lammerding J. Squish and squeeze — the nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 2016;40:32–40. doi: 10.1016/j.ceb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ber S., Torun Köse G., Hasırcı V. Bone tissue engineering on patterned collagen films: an in vitro study. Biomaterials. 2005;26:1977–1986. doi: 10.1016/j.biomaterials.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Charest J.L., García A.J., King W.P. Myoblast alignment and differentiation on cell culture substrates with microscale topography and model chemistries. Biomaterials. 2007;28:2202–2210. doi: 10.1016/j.biomaterials.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 55.Karuri N.W., Nealey P.F., Murphy C.J., Albrecht R.M. Structural organization of the cytoskeleton in SV40 human corneal epithelial cells cultured on nano- and microscale grooves. Scanning. 2008;30:405–413. doi: 10.1002/sca.20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu X., Leng Y. Comparison of the osteoblast and myoblast behavior on hydroxyapatite microgrooves. J. Biomed. Mater. Res. B Appl. Biomater. 2008;90B:438–445. doi: 10.1002/jbm.b.31304. [DOI] [PubMed] [Google Scholar]
- 57.Franco D., Klingauf M., Bednarzik M., Cecchini M., Kurtcuoglu V., Gobrecht J., Poulikakos D., Ferrari A. Control of initial endothelial spreading by topographic activation of focal adhesion kinase. Soft Matter. 2011;7:7313. [Google Scholar]
- 58.Ermis M., Akkaynak D., Chen P., Demirci U., Hasirci V. A high throughput approach for analysis of cell nuclear deformability at single cell level. Sci. Rep. 2016;6:36917. doi: 10.1038/srep36917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim C.-S., Ahn S.-H., Jang D.-Y. Review: developments in micro/nanoscale fabrication by focused ion beams. Vacuum. 2012;86:1014–1035. [Google Scholar]
- 60.Hasturk O., Sivas A., Karasozen B., Demirci U., Hasirci N., Hasirci V. Quantification of type, timing, and extent of cell body and nucleus deformations caused by the dimensions and hydrophilicity of square prism micropillars. Adv. Healthc. Mater. 2016;5:2972–2982. doi: 10.1002/adhm.201600857. [DOI] [PubMed] [Google Scholar]
- 61.Fitzpatrick V., Fourel L., Destaing O., Gilde F., Albigès-Rizo C., Picart C., Boudou T. Signal mingle: micropatterns of BMP-2 and fibronectin on soft biopolymeric films regulate myoblast shape and SMAD signaling. Sci. Rep. 2017;7:41479. doi: 10.1038/srep41479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fredonnet J., Foncy J., Lamarre S., Cau J.-C., Trévisiol E., Peyrade J.-P., François J.M., Séverac C. Dynamic PDMS inking for DNA patterning by soft lithography. Microelectron. Eng. 2013;111:379–383. [Google Scholar]
- 63.Berkowski K.L., Plunkett K.N., Yu Q., Moore J.S. Introduction to photolithography: preparation of microscale polymer silhouettes. J. Chem. Educ. 2005;82:1365. [Google Scholar]
- 64.Subramani K. Emerg. Nanotechnologies Manuf. Elsevier; 2010. Fabrication of hydrogel micropatterns by soft photolithography; pp. 261–276. [Google Scholar]
- 65.Nguyen N.-T., Nguyen N.-T. Micromixers. Elsevier; 2012. Fabrication technologies; pp. 113–161. [Google Scholar]
- 66.McMurray R., J M., Gadegaar N. Nanopatterned surfaces for biomedical applications. Biomed. Eng. Trends Mater. Sci. 2011 InTech. [Google Scholar]
- 67.Zorlutuna P., Rong Z., Vadgama P., Hasirci V. Influence of nanopatterns on endothelial cell adhesion: enhanced cell retention under shear stress. Acta Biomater. 2009;5:2451–2459. doi: 10.1016/j.actbio.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 68.Yucel D., Kose G.T., Hasirci V. Polyester based nerve guidance conduit design. Biomaterials. 2010;31:1596–1603. doi: 10.1016/j.biomaterials.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 69.W.L.I.U W., Chen Z.-L., Jiang X.-Y. Methods for cell micropatterning on two-dimensional surfaces and their applications in biology. Chin. J. Anal. Chem. 2009;37:943–949. [Google Scholar]
- 70.Whitesides G.M. The origins and the future of microfluidics, Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 71.Huda S., Pilans D., Makurath M., Hermans T.M., Kandere-Grzybowska K., Grzybowski B.A. Microfabricated systems and assays for studying the cytoskeletal organization, micromechanics, and motility patterns of cancerous cells. Adv. Mater. Interfaces. 2014;1:1400158. doi: 10.1002/admi.201400158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi S., Levy O., Coelho M.B., Cabral J.M.S., Karp J.M., Karnik R. A cell rolling cytometer reveals the correlation between mesenchymal stem cell dynamic adhesion and differentiation state. Lab a Chip. 2014;14:161–166. doi: 10.1039/c3lc50923k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stott S.L., Hsu C.-H., Tsukrov D.I., Yu M., Miyamoto D.T., Waltman B.A., Rothenberg S.M., Shah A.M., Smas M.E., Korir G.K., Floyd F.P., Gilman A.J., Lord J.B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L.V., Lee R.J., Isselbacher K.J., Maheswaran S., Haber D.A., Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A., Ryan P., Balis U.J., Tompkins R.G., Haber D.A., Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., Te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mann J.M., Lam R.H.W., Weng S., Sun Y., Fu J. A silicone-based stretchable micropost array membrane for monitoring live-cell subcellular cytoskeletal response. Lab a Chip. 2012;12:731–740. doi: 10.1039/c2lc20896b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Håkanson M., Cukierman E., Charnley M. Miniaturized pre-clinical cancer models as research and diagnostic tools. Adv. Drug Deliv. Rev. 2014;69–70:52–66. doi: 10.1016/j.addr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Biggs M.J.P., Richards R.G., McFarlane S., Wilkinson C.D.W., Oreffo R.O.C., Dalby M.J. Adhesion formation of primary human osteoblasts and the functional response of mesenchymal stem cells to 330nm deep microgrooves. J. R. Soc. Interface. 2008;5:1231–1242. doi: 10.1098/rsif.2008.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coutinho D., Costa P., Neves N., Gomes M.E., Reis R.L. Tissue Eng. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. Micro- and nanotechnology in tissue engineering; pp. 3–29. [Google Scholar]
- 80.Clark P., Connolly P., Curtis A.S., Dow J.A., Wilkinson C.D. Topographical control of cell behaviour: II. Multiple grooved substrata. Development. 1990;108:635–644. doi: 10.1242/dev.108.4.635. http://www.ncbi.nlm.nih.gov/pubmed/2387239 [DOI] [PubMed] [Google Scholar]
- 81.Béduer A., Vieu C., Arnauduc F., Sol J.-C., Loubinoux I., Vaysse L. Engineering of adult human neural stem cells differentiation through surface micropatterning. Biomaterials. 2012;33:504–514. doi: 10.1016/j.biomaterials.2011.09.073. [DOI] [PubMed] [Google Scholar]
- 82.Huang N.F., Lai E.S., Ribeiro A.J.S., Pan S., Pruitt B.L., Fuller G.G., Cooke J.P. Spatial patterning of endothelium modulates cell morphology, adhesiveness and transcriptional signature. Biomaterials. 2013;34:2928–2937. doi: 10.1016/j.biomaterials.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greiner A.M., Sales A., Chen H., Biela S.A., Kaufmann D., Kemkemer R. Nano- and microstructured materials for in vitro studies of the physiology of vascular cells. Beilstein J. Nanotechnol. 2016;7:1620–1641. doi: 10.3762/bjnano.7.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glawe J.D., Hill J.B., Mills D.K., McShane M.J. Influence of channel width on alignment of smooth muscle cells by high-aspect-ratio microfabricated elastomeric cell culture scaffolds. J. Biomed. Mater. Res. 2005;75A:106–114. doi: 10.1002/jbm.a.30403. [DOI] [PubMed] [Google Scholar]
- 85.Sarkar S., Dadhania M., Rourke P., Desai T.A., Wong J.Y. Vascular tissue engineering: microtextured scaffold templates to control organization of vascular smooth muscle cells and extracellular matrix. Acta Biomater. 2005;1:93–100. doi: 10.1016/j.actbio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 86.Uttayarat P., Chen M., Li M., Allen F.D., Composto R.J., Lelkes P.I. Microtopography and flow modulate the direction of endothelial cell migration. Am. J. Physiol. Cell Physiol. 2008;294:H1027–H1035. doi: 10.1152/ajpheart.00816.2007. [DOI] [PubMed] [Google Scholar]
- 87.Gerecht S., Bettinger C.J., Zhang Z., Borenstein J.T., Vunjak-Novakovic G., Langer R. The effect of actin disrupting agents on contact guidance of human embryonic stem cells. Biomaterials. 2007;28:4068–4077. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Recknor J.B., Recknor J.C., Sakaguchi D.S., Mallapragada S.K. Oriented astroglial cell growth on micropatterned polystyrene substrates. Biomaterials. 2004;25:2753–2767. doi: 10.1016/j.biomaterials.2003.11.045. [DOI] [PubMed] [Google Scholar]
- 89.Matsuzaka K., Walboomers X.F., de Ruijter J.E., Jansen J.A. The effect of poly-L-lactic acid with parallel surface micro groove on osteoblast-like cells in vitro. Biomaterials. 1999;20:1293–1301. doi: 10.1016/s0142-9612(99)00029-0. http://www.ncbi.nlm.nih.gov/pubmed/10403047 [DOI] [PubMed] [Google Scholar]
- 90.loesberg w., teriet j., vandelft f., schon p., figdor c., speller s., vanloon j., walboomers x., jansen j. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials. 2007;28:3944–3951. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 91.Miller C., Shanks H., Witt A., Rutkowski G., Mallapragada S. Oriented Schwann cell growth on micropatterned biodegradable polymer substrates. Biomaterials. 2001;22:1263–1269. doi: 10.1016/s0142-9612(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 92.Kenar H., Kocabas A., Aydinli A., Hasirci V. Chemical and topographical modification of PHBV surface to promote osteoblast alignment and confinement. J. Biomed. Mater. Res. 2008;85A:1001–1010. doi: 10.1002/jbm.a.31638. [DOI] [PubMed] [Google Scholar]
- 93.Kwon H., Choi D.-H., Bae J.-H., Kim J.-H., Kim Y.-S. mRNA expression pattern of insulin-like growth factor components of granulosa cells and cumulus cells in women with and without polycystic ovary syndrome according to oocyte maturity. Fertil. Steril. 2010;94:2417–2420. doi: 10.1016/j.fertnstert.2010.03.053. [DOI] [PubMed] [Google Scholar]
- 94.Lu J., Rao M.P., MacDonald N.C., Khang D., Webster T.J. Improved endothelial cell adhesion and proliferation on patterned titanium surfaces with rationally designed, micrometer to nanometer features. Acta Biomater. 2008;4:192–201. doi: 10.1016/j.actbio.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 95.Wójciak-Stothard B., Madeja Z., Korohoda W., Curtis A., Wilkinson C. Activation of macrophage-like cells by multiple grooved substrata. Topographical control of cell behaviour. Cell Biol. Int. 1995;19:485–490. doi: 10.1006/cbir.1995.1092. [DOI] [PubMed] [Google Scholar]
- 96.Dalby M.J., Riehle M.O., Yarwood S.J., Wilkinson C.D.W., Curtis A.S.G. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp. Cell Res. 2003;284:274–282. doi: 10.1016/s0014-4827(02)00053-8. http://www.ncbi.nlm.nih.gov/pubmed/12651159 [DOI] [PubMed] [Google Scholar]
- 97.Wood A. Contact guidance on microfabricated substrata: the response of teleost fin mesenchyme cells to repeating topographical patterns. J. Cell Sci. 1988;90(Pt 4):667–681. doi: 10.1242/jcs.90.4.667. http://www.ncbi.nlm.nih.gov/pubmed/3075621 [DOI] [PubMed] [Google Scholar]
- 98.Wójciak-Stothard B., Curtis A., Monaghan W., Macdonald K., Wilkinson C. Guidance and activation of murine macrophages by nanometric scale topography. Exp. Cell Res. 1996;223:426–435. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- 99.Gallego-Perez D., Higuita-Castro N., Denning L., DeJesus J., Dahl K., Sarkar A., Hansford D.J. Microfabricated mimics of in vivo structural cues for the study of guided tumor cell migration. Lab a Chip. 2012;12:4424. doi: 10.1039/c2lc40726d. [DOI] [PubMed] [Google Scholar]
- 100.Barbucci R., Torricelli P., Fini M., Pasqui D., Favia P., Sardella E., d'Agostino R., Giardino R. Proliferative and re-defferentiative effects of photo-immobilized micro-patterned hyaluronan surfaces on chondrocyte cells. Biomaterials. 2005;26:7596–7605. doi: 10.1016/j.biomaterials.2005.05.090. [DOI] [PubMed] [Google Scholar]
- 101.Vrana N.E., Elsheikh A., Builles N., Damour O., Hasirci V. Effect of human corneal keratocytes and retinal pigment epithelial cells on the mechanical properties of micropatterned collagen films. Biomaterials. 2007;28:4303–4310. doi: 10.1016/j.biomaterials.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 102.Ber S., Torun Köse G., Hasırcı V. Bone tissue engineering on patterned collagen films: an in vitro study. Biomaterials. 2005;26:1977–1986. doi: 10.1016/j.biomaterials.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 103.Andersson A.-S., Bäckhed F., von Euler A., Richter-Dahlfors A., Sutherland D., Kasemo B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials. 2003;24:3427–3436. doi: 10.1016/s0142-9612(03)00208-4. http://www.ncbi.nlm.nih.gov/pubmed/12809771 [DOI] [PubMed] [Google Scholar]
- 104.Dalby M.J., Riehle M.O., Sutherland D.S., Agheli H., Curtis A.S.G. Changes in fibroblast morphology in response to nano-columns produced by colloidal lithography. Biomaterials. 2004;25:5415–5422. doi: 10.1016/j.biomaterials.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 105.Dalby M.J., Riehle M.O., Sutherland D.S., Agheli H., Curtis A.S.G. Fibroblast response to a controlled nanoenvironment produced by colloidal lithography. J. Biomed. Mater. Res. 2004;69A:314–322. doi: 10.1002/jbm.a.20138. [DOI] [PubMed] [Google Scholar]
- 106.Liu X., Liu R., Gu Y., Ding J. Nonmonotonic self-deformation of cell nuclei on topological surfaces with micropillar array. ACS Appl. Mater. Interfaces. 2017;9:18521–18530. doi: 10.1021/acsami.7b04027. [DOI] [PubMed] [Google Scholar]
- 107.Pan Z., Yan C., Peng R., Zhao Y., He Y., Ding J. Control of cell nucleus shapes via micropillar patterns. Biomaterials. 2012;33:1730–1735. doi: 10.1016/j.biomaterials.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 108.Wei J., Shi J., Wang B., Tang Y., Tu X., Roy E., Ladoux B., Chen Y. Fabrication of adjacent micropillar arrays with different heights for cell studies. Microelectron. Eng. 2016;158:22–25. [Google Scholar]
- 109.Dickinson L.E., Rand D.R., Tsao J., Eberle W., Gerecht S. Endothelial cell responses to micropillar substrates of varying dimensions and stiffness. J. Biomed. Mater. Res. 2012;100A:1457–1466. doi: 10.1002/jbm.a.34059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wong I.Y., Javaid S., Wong E.A., Perk S., Haber D.A., Toner M., Irimia D. Collective and individual migration following the epithelial–mesenchymal transition. Nat. Mater. 2014;13:1063–1071. doi: 10.1038/nmat4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Badique F., Stamov D.R., Davidson P.M., Veuillet M., Reiter G., Freund J.-N., Franz C.M., Anselme K. Directing nuclear deformation on micropillared surfaces by substrate geometry and cytoskeleton organization. Biomaterials. 2013;34:2991–3001. doi: 10.1016/j.biomaterials.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 112.Özçelik H., Padeste C., Hasirci V. Systematically organized nanopillar arrays reveal differences in adhesion and alignment properties of BMSC and Saos-2 cells. Colloids Surfaces B Biointerfaces. 2014;119:71–81. doi: 10.1016/j.colsurfb.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 113.Antmen E., Ermis M., Demirci U., Hasirci V. Engineered natural and synthetic polymer surfaces induce nuclear deformation in osteosarcoma cells. J. Biomed. Mater. Res. B Appl. Biomater. 2018 doi: 10.1002/jbm.b.34128. [DOI] [PubMed] [Google Scholar]
- 114.Silverwood R.K., Fairhurst P.G., Sjöström T., Welsh F., Sun Y., Li G., Yu B., Young P.S., Su B., Meek R.M.D., Dalby M.J., Tsimbouri P.M. Analysis of Osteoclastogenesis/Osteoblastogenesis on Nanotopographical Titania surfaces. Adv. Healthc. Mater. 2016;5:947–955. doi: 10.1002/adhm.201500664. [DOI] [PubMed] [Google Scholar]
- 115.Popat K.C., Chatvanichkul K.-I., Barnes G.L., Latempa T.J., Grimes C.A., Desai T.A. Osteogenic differentiation of marrow stromal cells cultured on nanoporous alumina surfaces. J. Biomed. Mater. Res. 2007;80A:955–964. doi: 10.1002/jbm.a.31028. [DOI] [PubMed] [Google Scholar]
- 116.Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P., Wilkinson C.D.W., Oreffo R.O.C. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 117.Dalby M.J., Gadegaard N., Riehle M.O., Wilkinson C.D., Curtis A.S. Investigating filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int. J. Biochem. Cell Biol. 2004;36:2005–2015. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 118.Curtis A.S., Casey B., Gallagher J.O., Pasqui D., Wood M.A., Wilkinson C.D. Substratum nanotopography and the adhesion of biological cells. Are symmetry or regularity of nanotopography important? Biophys. Chem. 2001;94:275–283. doi: 10.1016/s0301-4622(01)00247-2. [DOI] [PubMed] [Google Scholar]
- 119.Curtis A.S.G., Gadegaard N., Dalby M.J., Riehle M.O., Wilkinson C.D.W., Aitchison G. Cells React to nanoscale order and symmetry in their Surroundings. IEEE Trans. NanoBioscience. 2004;3:61–65. doi: 10.1109/tnb.2004.824276. [DOI] [PubMed] [Google Scholar]
- 120.Green A.M., Jansen J.A., van der Waerden J.P.C.M., Von Recum A.F. Fibroblast response to microtextured silicone surfaces: Texture orientation into or out of the surface. J. Biomed. Mater. Res. 1994;28:647–653. doi: 10.1002/jbm.820280515. [DOI] [PubMed] [Google Scholar]
- 121.Hunt J.A., Williams R.L., Tavakoli S.M., Riches S.T. Laser surface modification of polymers to improve biocompatibility. J. Mater. Sci. Mater. Med. 1995;6:813–817. [Google Scholar]
- 122.Zinger O., Zhao G., Schwartz Z., Simpson J., Wieland M., Landolt D., Boyan B. Differential regulation of osteoblasts by substrate microstructural features. Biomaterials. 2005;26:1837–1847. doi: 10.1016/j.biomaterials.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 123.Davison M.J., McMurray R.J., Smith C.-A., Dalby M.J., Meek R.D. Nanopit-induced osteoprogenitor cell differentiation: the effect of nanopit depth. J. Tissue Eng. 2016;7 doi: 10.1177/2041731416652778. 2041731416652778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gallagher J.O., McGhee K.F., Wilkinson C.D.W., Riehle M.O. Interaction of animal cells with ordered nanotopography. IEEE Trans. NanoBioscience. 2002;1:24–28. doi: 10.1109/tnb.2002.806918. http://www.ncbi.nlm.nih.gov/pubmed/16689218 [DOI] [PubMed] [Google Scholar]
- 125.Ishikawa J., Tsuji H., Sato H., Gotoh Y. Ion implantation of negative ions for cell growth manipulation and nervous system repair. Surf. Coating. Technol. 2007;201:8083–8090. [Google Scholar]
- 126.Goddard J.M., Hotchkiss J.H. Polymer surface modification for the attachment of bioactive compounds. Prog. Polym. Sci. 2007;32:698–725. [Google Scholar]
- 127.Xu L.-C., Siedlecki C.A. Effects of surface wettability and contact time on protein adhesion to biomaterial surfaces. Biomaterials. 2007;28:3273–3283. doi: 10.1016/j.biomaterials.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tamada Y., Ikada Y. Effect of Preadsorbed proteins on cell adhesion to polymer surfaces. J. Colloid interface Sci. 1993;155:334–339. [Google Scholar]
- 129.Bet M.R., Goissis G., Vargas S., Selistre-de-Araujo H.S. Cell adhesion and cytotoxicity studies over polyanionic collagen surfaces with variable negative charge and wettability. Biomaterials. 2003;24:131–137. doi: 10.1016/s0142-9612(02)00270-3. http://www.ncbi.nlm.nih.gov/pubmed/12417186 [DOI] [PubMed] [Google Scholar]
- 130.Schneider G.B., English A., Abraham M., Zaharias R., Stanford C., Keller J. The effect of hydrogel charge density on cell attachment, Biomaterials. 2004;25:3023–3028. doi: 10.1016/j.biomaterials.2003.09.084. [DOI] [PubMed] [Google Scholar]
- 131.Eisenberg J.L., Beaumont K.G., Takawira D., Hopkinson S.B., Mrksich M., Budinger G.R.S., Jones J.C.R. Plectin-containing, centrally localized focal adhesions exert traction forces in primary lung epithelial cells. J. Cell Sci. 2013;126:3746–3755. doi: 10.1242/jcs.128975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takahashi S., Yamazoe H., Sassa F., Suzuki H., Fukuda J. Preparation of coculture system with three extracellular matrices using capillary force lithography and layer-by-layer deposition. J. Biosci. Bioeng. 2009;108:544–550. doi: 10.1016/j.jbiosc.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 133.Peterbauer T., Yakunin S., Siegel J., Hering S., Fahrner M., Romanin C., Heitz J. Dynamics of spreading and alignment of cells cultured in vitro on a grooved polymer surface. J. Nanomater. 2011;2011:1–10. [Google Scholar]
- 134.Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 135.Franz C.M., Jones G.E., Ridley A.J. Cell migration in development and disease. Dev. Cell. 2002;2:153–158. doi: 10.1016/s1534-5807(02)00120-x. [DOI] [PubMed] [Google Scholar]
- 136.Jiang X., Bruzewicz D.A., Wong A.P., Piel M., Whitesides G.M. Directing cell migration with asymmetric micropatterns. Proc. Natl. Acad. Sci. U. S. A. 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lehnert D., Wehrle-Haller B., David C., Weiland U., Ballestrem C., Imhof B.A., Bastmeyer M. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J. Cell Sci. 2004;117:41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 138.Kumar G., Ho C.-C., Co C.C. Guiding cell migration using one-way micropattern arrays. Adv. Mater. 2007;19:1084–1090. [Google Scholar]
- 139.Kim D.-H., Seo C.-H., Han K., Kwon K.W., Levchenko A., Suh K.-Y. Guided cell migration on microtextured substrates with variable local density and anisotropy. Adv. Funct. Mater. 2009;19:1579–1586. doi: 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raghavan S., Desai R.A., Kwon Y., Mrksich M., Chen C.S. Micropatterned Dynamically adhesive substrates for cell migration. Langmuir. 2010;26:17733–17738. doi: 10.1021/la102955m. [DOI] [PubMed] [Google Scholar]
- 141.Hatano K., Inoue H., Kojo T., Matsunaga T., Tsujisawa T., Uchiyama C., Uchida Y. Effect of surface roughness on proliferation and alkaline phosphatase expression of rat calvarial cells cultured on polystyrene. Bone. 1999;25:439–445. doi: 10.1016/s8756-3282(99)00192-1. [DOI] [PubMed] [Google Scholar]
- 142.Lim J.Y., Hansen J.C., Siedlecki C.A., Runt J., Donahue H.J. Human foetal osteoblastic cell response to polymer-demixed nanotopographic interfaces. J. R. Soc. Interface. 2005;2:97–108. doi: 10.1098/rsif.2004.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vagaská B., Bacáková L., Filová E., Balík K. Osteogenic cells on bio-inspired materials for bone tissue engineering. Physiol. Res. 2010;59:309–322. doi: 10.33549/physiolres.931776. http://www.ncbi.nlm.nih.gov/pubmed/19681662 [DOI] [PubMed] [Google Scholar]
- 144.Chaubey A., Ross K.J., Leadbetter R.M., Burg K.J.L. Surface patterning: Tool to modulate stem cell differentiation in an adipose system. J. Biomed. Mater. Res. B Appl. Biomater. 2008;84B:70–78. doi: 10.1002/jbm.b.30846. [DOI] [PubMed] [Google Scholar]
- 145.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S., Shape Cell, Tension Cytoskeletal, Regulate RhoA. Stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 146.Peng R., Yao X., Ding J. Effect of cell anisotropy on differentiation of stem cells on micropatterned surfaces through the controlled single cell adhesion. Biomaterials. 2011;32:8048–8057. doi: 10.1016/j.biomaterials.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 147.Yim E.K.F., Pang S.W., Leong K.W. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dahl K.N., Ribeiro A.J.S., Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Krause M., Wolf K. Cancer cell migration in 3D tissue: Negotiating space by proteolysis and nuclear deformability. Cell Adhes. Migrat. 2015;9:357–366. doi: 10.1080/19336918.2015.1061173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Davidson P.M., Fromigué O., Marie P.J., Hasirci V., Reiter G., Anselme K. Topographically induced self-deformation of the nuclei of cells: dependence on cell type and proposed mechanisms. J. Mater. Sci. Mater. Med. 2010;21:939–946. doi: 10.1007/s10856-009-3950-7. [DOI] [PubMed] [Google Scholar]
- 151.Buch-Månson N., Spangenberg A., Gomez L.P.C., Malval J.-P., Soppera O., Martinez K.L. Rapid Prototyping of polymeric nanopillars by 3D direct laser Writing for controlling cell behavior. Sci. Rep. 2017;7:9247. doi: 10.1038/s41598-017-09208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li B., Xie L., Starr Z.C., Yang Z., Lin J.-S., Wang J.H.-C. Development of micropost force sensor array with culture experiments for determination of cell traction forces. Cell Motil. Cytoskeleton. 2007;64:509–518. doi: 10.1002/cm.20200. [DOI] [PubMed] [Google Scholar]
- 153.Han S.J., Bielawski K.S., Ting L.H., Rodriguez M.L., Sniadecki N.J. Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys. J. 2012;103:640–648. doi: 10.1016/j.bpj.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sniadecki N.J., Chen C.S. Microfabricated silicone elastomeric post arrays for measuring traction forces of adherent cells. Meth. Cell Biol. 2007;83:313–328. doi: 10.1016/S0091-679X(07)83013-5. [DOI] [PubMed] [Google Scholar]
- 155.Fu J., Wang Y.-K., Yang M.T., Desai R.A., Yu X., Liu Z., Chen C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Br. J. Pharmacol. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ricart B.G., Yang M.T., Hunter C.A., Chen C.S., Hammer D.A. Measuring traction forces of Motile Dendritic cells on micropost arrays. Biophys. J. 2011;101:2620–2628. doi: 10.1016/j.bpj.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]