Abstract
Background
Red ginseng is a popularly used traditional medicine with antiaging effects in Asian countries. The present study aimed to explore the changes in protein expression underlying the mechanisms of life span extension and antiaging caused by red ginseng extract (RGE) in Drosophila melanogaster.
Methods
A proteomic approach of two-dimensional polyacrylamide gel electrophoresis (2-DE) was used to identify the differential abundance of possible target proteins of RGE in D. melanogaster. The reliability of the 2-DE results was confirmed via Western blotting to measure the expression levels of selected proteins. Proteins altered at the expression level after RGE treatment (1 mg/mL) were identified by matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry and by searching against the National Center for Biotechnology nonredundant and Uniprot protein databases. The differentially expressed proteins were analyzed using bioinformatics methods.
Results
The average survival life span of D. melanogaster was significantly extended by 12.60% with RGE treatment (1 mg/mL) compared to untreated flies. This followed increased superoxide dismutase level and decreased methane dicarboxylic aldehyde content. Based on the searching strategy, 23 differentially expressed proteins were identified (16 up-regulated and 7 down-regulated) in the RGE-treated D. melanogaster. Transduction pathways were identified using the Kyoto Encyclopedia of Genes and Genomes database, and included the hippo and oxidative phosphorylation pathways that play important roles in life span extension and antiaging process of D. melanogaster.
Conclusion
Treatment with RGE in D. melanogaster demonstrated that mechanisms of life span extension and antiaging are regulated by multiple factors and complicated signal pathways.
Keywords: antiaging, life span extension, proteome, red ginseng extract
1. Introduction
Aging, a spontaneous and complex process with the passage of time, is one of the most important risk factors for increasing susceptibility to multiple diseases [1]. It involves much morphological and functional deterioration in biological systems, which are associated with various molecular, cellular, and organic changes [2]. Aging features a progressive accumulation of oxidative agents associated with decreased efficiency of antioxidant defense mechanisms. Related to this is a progressive decrease in immune activity or immune senescence. As animals age, their mitochondrial function and thus cellular metabolism systematically decline, leading to a loss of cellular homeostasis and the occurrence of multiple disorders [3], [4]. Thus, a number of efforts have been made to elucidate the mechanisms of the processes of aging and to discover new compounds that retain antiaging activities.
Ginseng (Panax ginseng Meyer, Araliaceous) is an important medicinal herb that has long been used to treat various diseases in Asian countries (i.e., Korea, China, and Japan) [5]. Among several kinds of P. ginseng products, red ginseng, produced by steaming and drying fresh ginseng, is a main active ginseng. During this process, ginsenosides undergo chemical changes that have the potential to create special physiologic activities in vivo [6]. This is known to have various physiological activities, including—but not limited to—antioxidant, anticancer, tonic, and antiaging effects. Red ginseng improved learning and memory of the mice [7], and Korean Red Ginseng tonic extended the life span of Drosophila melanogaster [8]. However, there is very limited information on the proteomic mechanisms underlying the antiaging effect and life span extension of red ginseng on D. melanogaster or other animal models, because existing studies have primarily focused on pharmacology and/or pharmacodynamics association studies [9], [10].
In this study, D. melanogaster, an internationally recognized model for studying life span that is widely used in antiaging medical research, was used to elucidate this issue in vivo. A proteomic approach of two-dimensional polyacrylamide gel electrophoresis (2-DE) was used to investigate changes in age-related specific indicators and relative abundance of proteins in expression caused by feeding with red ginseng extract (RGE). Our findings could be helpful in revealing the mechanisms and addressing biological questions underlying the effects of red ginseng on life span and antiaging.
2. Materials and methods
2.1. Plant materials and reagents
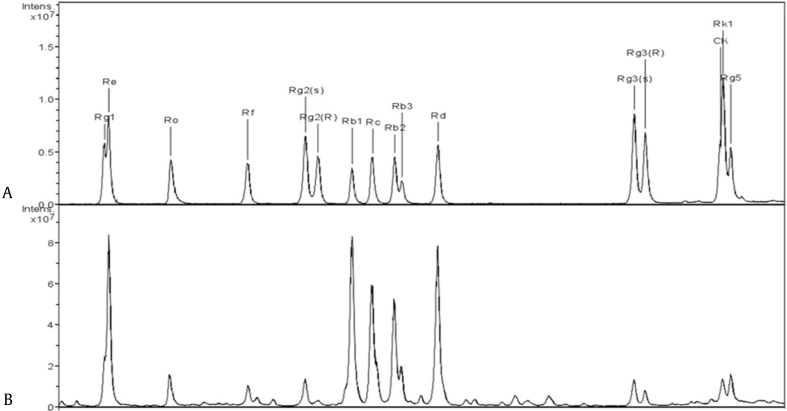
Fresh ginseng (6 yr) was obtained from Jilin City, Jilin Province, China. Red ginseng and its extract (RGE) were processed and provided by the Medicinal Plant Analysis laboratory of the Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences (Jilin, China), according to the national standard of China (Technical specifications for red ginseng processing, NO. NY/T 2784-2015). The panaxoside content (Fig. 1, Table S1) in the RGE was determined as previously described [11], using high-performance liquid chromatography, as follows (all in mg/g): Rb1 30.13, Rb2 14.67, Rb3 8.23, Rc 20.45, Rd 15.57, Re 16.44, Rf 1.83, Rg1 4.47, Rg2(s) 1.96, Rg2(r) 0.42, Rg3(s) 2.06, Rg3(r) 1.18, Rg5 3.31, Rk1 1.07, and Ro 4.09.
Fig. 1.
Panaxoside content of red ginseng extract (RGE) determined by high-performance liquid chromatography (HPLC). (A) HPLC chromatogram of ginseng saponin standard. (B) HPLC chromatogram of RGE.
Urea, 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS), Pharmalyte, dithiothreitol (DTT), and iodoacetamide were obtained from AMRESCO (Solon, OH, USA). ReadyPrep 2-D cleanup kit and reagents used for 2 DE and Western blotting were purchased from Bio-Rad (Richmond, CA, USA). The primary antibodies for Western blotting included rabbit anti-14-3-3 zeta (1433Z) polyclonal antibody, rabbit antivacuolar-type H(+)-ATPase (VATA) polyclonal antibody, rabbit antiheat-shock protein 70 (HSP70) polyclonal antibody, rabbit antiglycerol-3-phosphate dehydrogenase (GPDA) polyclonal antibody, mouse antiubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) monoclonal antibody mouse antifructose-bisphosphate aldolase (ALF) monoclonal antibody, and mouse antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (dilution 1:1000; Proteintech, Chicago, IL, USA). The secondary antibody used in this study was horseradish peroxidase-conjugated goat antirabbit/mouse IgG (dilution 1:2,000; Proteintech).
2.2. Life span analysis of D. melanogaster
Wild type D. melanogaster (Oregon K) was a gift from Jilin Agricultural University and was used throughout the experiment. Single populations (200 flies each) of males were housed in a biochemical incubator with 12-h dark/light cycle at 25°C, 60% humidity, and with free access to basal food (water, 1.2% agar, 2.2% sucrose, 1.8% yeast extra powder, 8% corn extract, and 6.25 μL/L propionic acid). For RGE treatment, basal food was supplemented with RGE at a final w/v concentration of 1 mg/mL optimized by our previous study (unpublished). The food was changed every 2–3 d.
To examine the effects of RGE on the life span of flies, survival was observed and documented at 8:00 p.m. when transferring to fresh basal food or basal food supplemented with RGE every 2–3 d. Survivorships were scored regularly and eliminated the unnatural death flies, and death flies were removed out of the cage when possible. Prism 5 (GraphPad Software Inc., San Diego, CA, USA) was used for data analysis, and statistical significance was tested with log-rank statistical methods. Mean (average value of all values) and maximum (average values of the last 20 dead flies) values were presented to compare results.
2.3. Methane dicarboxylic aldehyde and superoxide dismutase assessment in whole tissue
Flies of wild type and RGE-treated groups were anesthetized and collected at the age of 21 d. Flies (whole tissue) were homogenized and centrifuged at 4,000g for 10 min. Supernatants were collected gently and kept at −80°C until analysis measurements.
The extent of lipid peroxidation in whole tissue was estimated by measurement of methane dicarboxylic aldehyde (MDA) formation [12]. The MDA assay was performed using an MDA kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the manufacturer's instructions. In brief, 0.2 mL thiobarbituric acid and antioxidant reagents were respectively pipetted into a microcentrifuge tube containing 0.1 mL of wild type or RGE-treated homogenate and MDA standard solution. Tubes were vortexed and then incubated at 95°C for 40 min and centrifuged at 4,000g for 10 min. Then, 0.3 mL of the supernatant from each of the tubes was transferred to the quartz sampling cell, and the absorbance was measured at 532 nm using a spectrophotometer (NanoDrop Lite; Thermo-Fisher, Waltham, MA, USA). The content of MDA was expressed as nmol MDA/mg homogenate protein. The total protein concentration was determined by the Bradford method, using bovine serum albumin as the standard.
Whole tissue total superoxide dismutase (SOD) activity was measured using enzyme-linked immunosorbent assay kits (Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer's instructions. Briefly, 0.1 mL of whole tissue homogenate was mixed with 0.9 mL of physiological saline (0.9% NaCl), and then 0.1 mL of 15mM pyrogallic acid was added. The activity was measured at 420 nm for 3 min using a multidetection microplate reader (Infinite 200; Tecan, Männedorf, Switzerland) and expressed as U/mg protein. One unit was determined as the amount of enzyme that inhibited the oxidation of pyrogallic acid by 50%. Experiments for MDA content and SOD activity were performed in triplicate in this study.
2.4. Sample preparation and protein extraction
Both wild type and RGE-treated D. melanogaster (21 d old, about 20 flies each) were resuspended in 600 μL lysis buffer (7M Urea, 2M Thiourea, 4% CHAPS, and 1% phenylmethylsulfonyl fluoride (PMSF)). Bullet Blender (Next Advance, Averill Park, NY, USA) was used to break the tissue as described previously [13]. Supernatants were collected after sharking in ice for 2 h and centrifugation at 12,000g at 4°C for 10 min. Concentrations of proteins were detected using the RC DC Protein Assay (Bio-Rad, Hercules, CA, USA). The protein samples were purified using the 2D Cleanup kit (Bio-Rad, Hercules, CA, USA) following the manufacturer's instructions. Experiments in this study were performed in triplicate gels (n = 3).
2.5. Two-dimensional gel electrophoresis
Proteins (both wild type and RGE-treated) were first separated with isoelectric focusing using linear precast immobilized pH gradient (IPG) strips (17 cm, 3–10 linear pH gradients; Bio-Rad, Hercules, CA, USA). IPG strips with 1.2 mg of proteins were rehydrated for 12 h and focused at 15,000 Vhs (Bio-Rad, Hercules, CA, USA). First-dimension strips were stored at −80°C or equilibrated immediately in first equilibration solution [75nM Tris–HCL, pH 8.8, 6M urea, 2M thiourea, 30% glycerol, 2% sodium dodecyl sulfate (SDS), and 0.002% bromophenol blue] with 100mM DTT for 15 min, followed by the second equilibration with 250mM iodoacetamide for 15 min. 2-DE was performed using 12.5% polyacrylamide gels at 90 V for 30 min and 140 V for 4–5 h at 20°C in Bio-Rad systems (Bio-Rad, Hercules, CA, USA).
2.6. Staining and image analysis
Gels (six in total) were stained using Pierce Silver Stain Kit (Thermo Fisher, MA, USA) as per the manufacturer's instructions. The stained gels were scanned using an image scanner (UMAX, Dallas, TX, USA) at 600 dpi. All detected spots were automatically matched by gel-to-gel comparison using the PDquest software version 8.0 (Bio-Rad, Hercules, CA, USA). Areas of all detected spots were normalized. Those protein spots with reproducible and statistically significant changes in abundance were considered to be differentially expressed. Only those spots with quantitative changes >1.5-fold in abundance, p value < 0.05, were considered for subsequent analyses.
2.7. Mass spectrometry and database search
Protein spots were manually excised from the preparative gel, digested with trypsin, and analyzed using matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry (MALDI-TOF MS/MS) with a 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA). The peptide mass fingerprint was analyzed with GPS (Applied Biosystems) MASCOT (Matrix Science, London, UK). The identified proteins were named according to the corresponding annotations in the National Center for Biotechnology (NCBI, http://www.ncbi.nlm.nih.gov/) and/or Universal Protein Resource (Uniprot, http://www.uniprot.org/). For proteins without functional annotation in the databases, homologs of these proteins were searched against the NCBI nonredundant protein database with BLASTP (http://blast.ncbi.nlm.nih.gov/) to annotate these identities. The experimental molecular mass of each protein spot was estimated by comparison with molecular weight standards, whereas the experimental isoelectric point (pI) was determined by the migration of protein spots on linear IPG strips.
2.8. Bioinformatics analysis
Functional annotation analysis was performed for up-regulated and down-regulated differentially expressed proteins using STRING (version 10.0) (http://string-db.org/). The associated Gene Ontology (GO) terms (i.e., molecular function, biological process, and cell component) were annotated based on the database. Additionally, to further investigate the signaling pathways of the identified proteins, protein–protein interactions analyzed by available experimental evidence, and statistical enrichment tests were carried out for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations.
2.9. Validation by Western blotting
Six differentially expressed proteins (VATA, GPDA, HSP70, 1433Z, ALF, and UCHL1) were randomly selected for further verification using Western blotting. Total protein from both wild type and RGE-treated flies were extracted using lysis buffer and concentrations determined (as described above). Equal amounts of proteins (40–50 μg) were resolved on 12% polyacrylamide–SDS gels and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA) using standard methods blocked with 5% (w/v) nonfat dry milk (NFDM; BD Biosciences, San Diego, CA, USA) freshly made in TBS-T [Tween-Tris buffered saline: 0.1% Tween-20 in 100mM Tris–HCL (pH 7.5), 0.9% NaCl], rock on a rotating shaker for 2 h at room temperature or overnight at 4°C. The membranes were probed with specific primary antibodies (dilution 1:1,000 in TBS-T/1% NFDM) for 2 h at room temperature or overnight at 4°C, washed three times of 15 min each with TBS-T, and then incubated for an additional 1.5 h with horseradish-peroxidase-conjugated secondary antibodies (dilution 1:2000 in TBS-T/1% NFDM) at 37°C. The levels of selected proteins were visualized using an ECL system (GE Healthcare, Waukesha, WI, USA). The Western blots were performed in triplicate, and the target protein bands were quantified by scanning densitometry using Image J (version 1.50) (National Institutes of Health, MD, USA) processing software and normalized to the signal intensity of GAPDH.
2.10. Statistical analysis
Values in the figures are expressed as the mean ± standard deviation. Statistical analysis was carried out with three replicates for proteomic and biochemical analyses. The results of the spot intensities and physiological data were analyzed using Student t test to determine the significant difference between group means. A p value <0.05 was considered statistically significant (SPSS for Windows, version 12.0; SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Effects of life span extension and antioxidant index induced by RGE in D. melanogaster
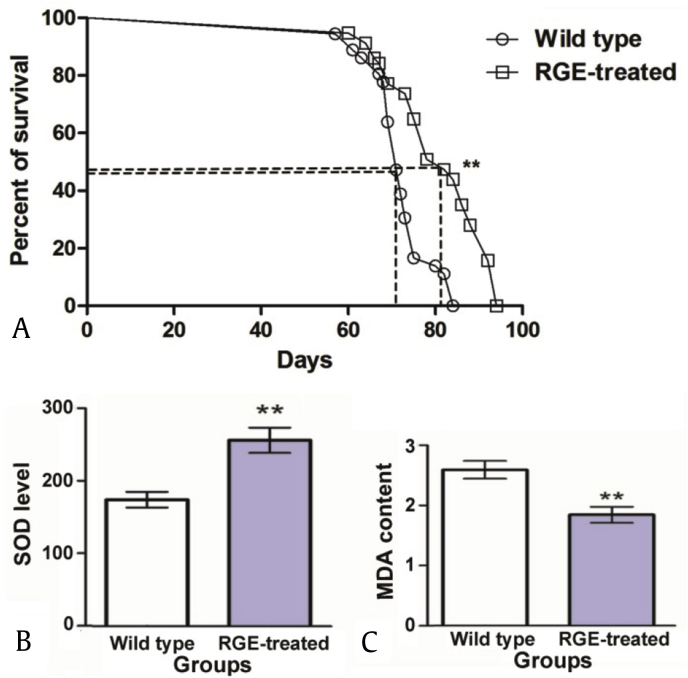
In this study, 200 males (20 flies per cage) of wild type and RGE-treated flies were used for longevity screening. RGE extended D. melanogaster life span and generated significantly lower mortality in older population groups (Fig. 2A). All wild type flies had died by Day 84, whereas the RGE-treated flies survived until Day 94 (Fig. 2A). RGE-treated flies showed significantly higher mean and maximum longevities compared with wild type flies (p < 0.01). In comparison to the wild type, RGE-treated flies had a 12.60% and 11.90% increase in mean and maximum life span, respectively.
Fig. 2.
Effects on life span, superoxide dismutase (SOD) level and methane dicarboxylic aldehyde (MDA) content in Drosophila melanogaster treated with or without red ginseng extract (RGE). (A) Life span curves of male flies feed with RGE. Wild type (circles): mean survival life span, 71.67 d; maximum life span, 79.8 d; RGE-treated (squares): mean survival life span, 80.7 d; maximum life span 93.8 d. In comparison to the wild type, RGE-treated flies had a 12.60% and 11.90% increase in mean and maximum life span, respectively. Life span curve was constructed with Prism 5 software and statistical significance was determined with log-rank test. Bar graphs of average level of SOD and average content of MDA were repeated three times and analyzed by Student t test. (B) Average level of SOD. (C) Average content of MDA. Data in this figure are shown as mean ± standard deviation. Statistical significance: **p < 0.01 (RGE-treated vs. wild type).
In addition, the SOD level and the MDA content were highly significantly changed by RGE treatment. Total SOD enzyme activity (U/mg protein) was significantly increased in RGE-treated flies (p < 0.01) compared to the wild type (Fig. 2B), whereas the MDA content (nmol/mg protein) was significantly lower in RGE-treated flies (p < 0.01) compared to wild type flies (Fig. 2C).
3.2. Proteomic analysis of wild type and RGE-treated D. melanogaster
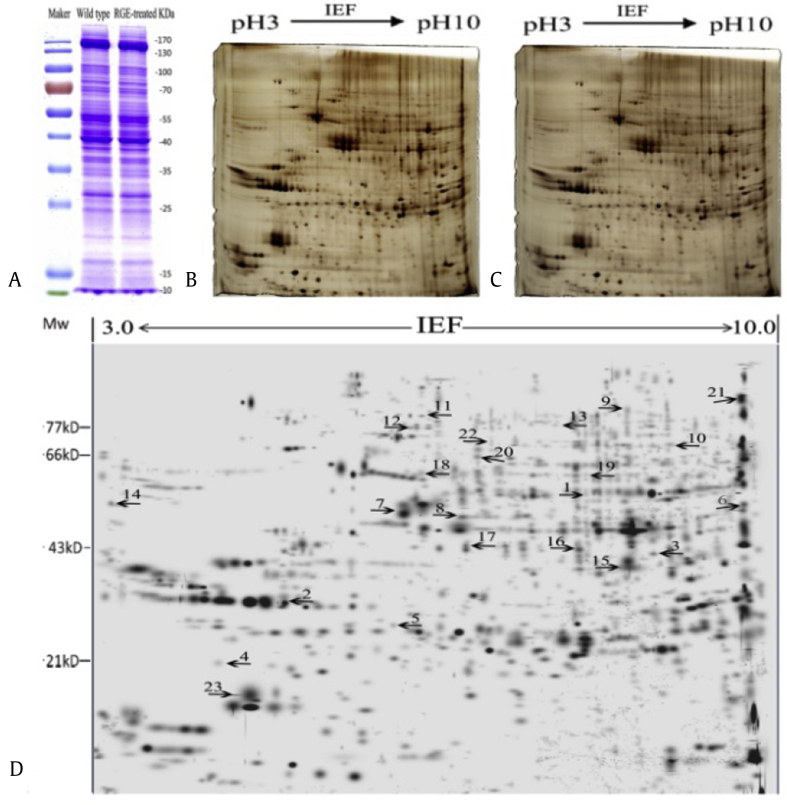
A 2-DE-based comparative proteomics analysis was performed to investigate the possible target-related proteins of RGE in D. melanogaster. Extracted proteins were separated by using SDS polyacrylamide gel electrophoresis (SDS–PAGE) to detect the quality of protein extraction (Fig. 3A). The proteins were further separated by using 2-DE and silver stained to evaluate their relative abundance levels by analyzing all protein spots from three independent replicates using an imaging software (Fig. 3B and C). Among the 65 differentially abundant protein spots on the 2-DE gels, 38 spots were successfully identified by using MALDI-TOF MS/MS (Table S2). Based on the information found in the NCBI, Uniprot, and GO protein databases, 23 (16 up-regulated and 7 down-regulated) differentially accumulated protein spots were identified as related to RGE treatment (Fig. 3D; Table 1).
Fig. 3.
Proteome maps (two-dimensional polyacrylamide gel electrophoresis images; representative of six gels) of wild type and red ginseng extract (RGE)-treated Drosophila melanogaster. (A) Coomassie Brilliant Blue-stained gels of protein extracts from wild type (left) and RGE-treated (right) flies. Proteins (40 μg in total) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) to detect the quality of extracted protein. Silver stained gels (n = 3) of extracted protein: (B) Wild type flies. (C) RGE-treated flies. Proteins (1.2 mg) were resolved in 17 cm linear, immobilized pH (3–10) gradient gels and then separated by 12.5% SDS–PAGE. All spots were matched and normalized by gel-to-gel comparison using the PDquest software version 8.0 (Bio-Rad, Hercules, CA, USA), and only those spots with reproducible changes (quantitative changes > 1.5-fold in abundance, p < 0.05) were considered for subsequent analyses. (D) Spot numbers indicated on the master gel are differentially expressed proteins related to RGE treatment that were subjected to matrix-assisted laser desorption/ionization-time of flight/time of flight mass spectrometry. IEF, isoelectrofocusing.
Table 1.
Protein identifications of differentially expressed proteins using MALDI-TOF MS/MS
| Entry | Accession no. | Protein name | Score | Mass ku/pI | Molecular function | Change |
|---|---|---|---|---|---|---|
| 1 | Q9VN95 | Enolase-phosphatase E1 (ENOPH) | 159 | 54.6/8.68 | Acireductone synthase activity | Increase |
| 2 | P29310 | 14-3-3 protein zeta (1433Z) | 100 | 28.3/4.77 | Protein heterodimerization activity | Increase |
| 3 | P07764 | Fructose-bisphosphate aldolase (ALF) | 245 | 45.8/7.08 | Fructose-bisphosphate aldolase activity | Increase |
| 4 | P35122 | Ubiquitin carboxyl-terminal hydrolase (Uch; also named UCHL1) | 80 | 100.0/9.37 | Thiol-dependent ubiquitin-specific protease activity | Increase |
| 5 | P13395 | Spectrin alpha chain (SPTCA) | 85 | 481.2/6.02 | Actin binding | Increase |
| 6 | P84029 | Cytochrome c-2 (CYC2) | 715 | 45.6/9.47 | Electron carrier activity | Increase |
| 7 | P83967 | Actin, indirect flight muscle (ACT6) | 628 | 42.1/5.29 | Structural constituent of cytoskeleton | Increase |
| 8 | P02574 | Actin, larval muscle (ACT4) | 580 | 42.2/5.3 | Structural constituent of cytoskeleton | Increase |
| 9 | P48610 | Arginine kinase (KARG) | 122 | 40.1/6.04 | Arginine kinase activity | Increase |
| 10 | P48611 | 6-Pyruvoyl tetrahydrobiopterin synthase (PTPS) | 81 | 20.1/6.07 | 6-Pyruvoyltetrahydropterin synthase activity | Increase |
| 11 | P29844 | Heat shock 70 kDa protein (HSP70) | 586 | 72.3/5.22 | ATP binding | Increase |
| 12 | Q27331 | V-type proton ATPase catalytic subunit A (VATA) | 464 | 68.5/5.23 | Proton-transporting ATPase activity | Increase |
| 13 | Q94511 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial (NDUS1) | 135 | 79.5/6.43 | Metal ion binding | Increase |
| 14 | P53501 | Actin-57B (ACT3) | 128 | 42.2/5.23 | Structural constituent of cytoskeleton | Increase |
| 15 | P13706 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic (GPDA) | 396 | 39.2/6.17 | NAD binding | Decrease |
| 16 | Q9VWH4 | Probable isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial (IDH3A) | 202 | 38.9/6.96 | Isocitrate dehydrogenase (NAD+) activity | Decrease |
| 17 | Q9W0T1 | Nucleosome-remodeling factor subunit NURF301 (NU301) gene E(bx) | 291 | 32.7/5.58 | Ligand-dependent nuclear receptor binding | Decrease |
| 18 | P10981 | Actin-87E (ACT5) | 163 | 38.2/5.36 | ATP binding | Decrease |
| 19 | P00334 | Alcohol dehydrogenase (ADH) | 187 | 57.3/6.37 | Protein homodimerization activity | Decrease |
| 20 | Q9VPH2 | DNA primase large subunit (PRI2) | 78 | 65.5/6.38 | DNA primase activity | Decrease |
| 21 | Q9V3R1 | Accessory gland protein Acp36DE (A36DE) | 676 | 84.5/9.18 | Hormone activity | Decrease |
| 22 | P08171 | Esterase-6 (EST6) | 88 | 61.9/5.78 | Carboxylic ester hydrolase activity | Decrease |
| 23 | Q9VA73 | Calcium binding protein 1 (CABP1) | 217 | 21.5/4.71 | Calcium ion binding | Decrease |
Mass ku (kDa)/pI, molecular mass and isoelectric point; score corresponding with two-dimensional gel electrophoresis gel as shown in Fig. 2 or calculated by mass spectrum identification or PDquest software. In the table, increase means up-regulation in RGE-treated, compared to the wild type; decrease means down-regulation in RGE-treated, compared to the wild type
RGE, red ginseng extract; MALDI-TOF MS/MS, matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry
3.3. Functional classification of the differentially expressed proteins
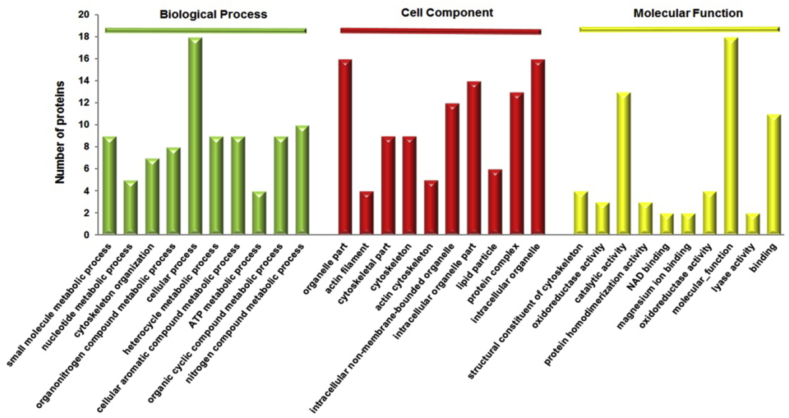
To investigate the function roles of RGE in D. melanogaster, the altered proteins were subjected to GO-based enrichment analysis using STRING version 10.0 (Table S3). The most significantly enriched terms in each GO category were included for “biological process,” “cell component,” and “molecular function” (Fig. 4). The biological process category showed that most proteins were associated with small molecule metabolic process (p = 1.56 × 10−7), nucleotide metabolic process (p = 1.69 × 10−5), and cytoskeleton organization (p = 1.98 × 10−5). In addition, most of the differentially expressed metabolic related-proteins (such as CYC2, 1433Z, GPDH, VATA, and ND75) were found to be up-regulated in RGE-treated D. melanogaster. The “cell component” annotation revealed a major spectrum of organelle part (p = 3.53 × 10−9), actin filament (5.14 × 10−9), and cytoskeletal part (2.21 × 10−8). Proteins that participated in the “molecular function” were related to the structural constituent of cytoskeleton (p = 2.72 × 10−7), oxidoreductase activity (p = 1.51 × 10−4), and catalytic activity (p = 2.25 × 10−4). Most proteins enriched within these GO terms were up-regulated in the RGE-treated D. melanogaster compared to the wild type.
Fig. 4.
Gene Ontology (GO) term annotation analysis of the proteins altered by red ginseng extract. The differentially expressed proteins are categorized into molecular function, biological process, and cell component by using GO annotation software (STRING version 10.0). ATP, adenosine triphosphate; NAD, nicotinamide adenine dinucleotide.
3.4. Enrichment of pathways in which the differentially expressed proteins participated
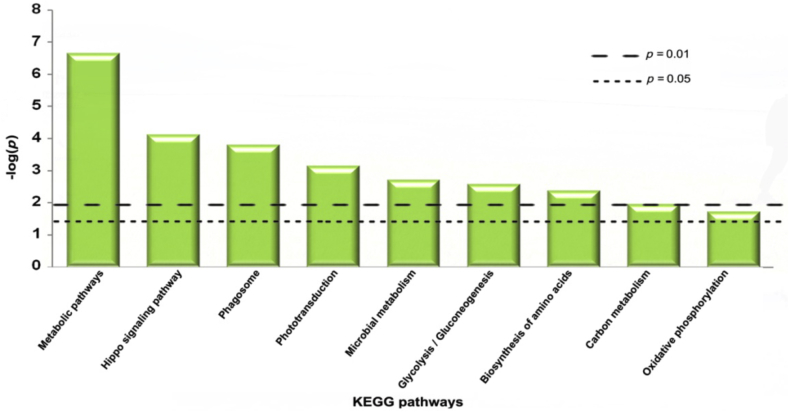
Nine KEGG pathways (p < 0.05) were enriched from the dataset using STRING software (version 10.0) (Fig. 5). The most significant pathways for the differentially expressed proteins were metabolic pathways (10 proteins, p = 2.11 × 10−7) and hippo signaling pathway (3 proteins, p = 7.36 × 10−5), which are involved in potentiation of the antiaging process in flies. As to the “metabolic” pathways, six of 10 enriched proteins (ALF, CYC2, PTPS, VATA, ND75, and ENPD) were found to be up-regulated in RGE-treated D. melanogaster, whereas EST6, IDH3A, PRI2, and ADH were down-regulated. Regarding the “hippo” signaling pathway, most of the enriched proteins (1433Z and ACT6) were up-regulated in RGE-treated flies.
Fig. 5.
Distribution of KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways in which the differentially expressed proteins participate. KEGG pathways are arranged in ascending order according to the p values. Dash line represents p = 0.01 or p = 0.05.
3.5. Validation of the selected differentially expressed proteins
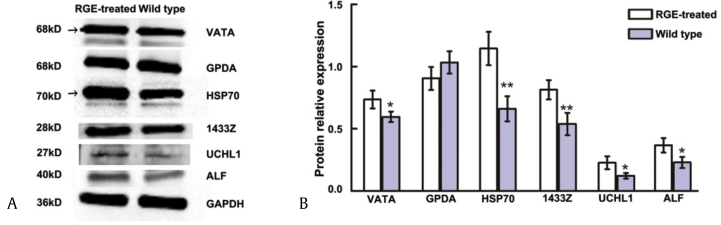
Expression levels of VATA, GPDA, HSP70, 1433Z, UCHL1, and ALF were confirmed using Western blotting (Fig. 6). Expression levels of VATA, HSP70, 1433Z, UCHL1, and ALF were significantly up-regulated in RGE-treated D. melanogaster versus wild type. Although GPDA expression appeared to be lower in RGE-treated flies, this was not significant. These results of Western blotting are consistent with those of the 2-DE MALDI-TOF MS/MS analysis.
Fig. 6.
Relative expression levels of VATA, GPDA, HSP70, 1433Z, UCHL1, and ALF by Western blot analysis. (A) Western blotting in this study was performed in triplicate and the target protein bands were quantified by scanning densitometry using Image J processing software and normalized to the signal intensity of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (B) The data are expressed as mean ± standard deviation and analyzed using Student t test. Statistical significance: *p < 0.05; **p < 0.01 (RGE-treated vs. wild type). 1433Z, 14-3-3 zeta; ALF, fructose-bisphosphate aldolase; GPDA, glycerol-3-phosphate dehydrogenase; HSP70, heat shock 70 kDa protein; UCHL1, ubiquitin carboxyl-terminal hydrolase; VATA, V-type proton ATPase catalytic subunit A.
4. Discussion
Proteomic technology has become an indispensable and efficient tool in modern bioscience research, and this technology has been successfully used to study the mechanisms of traditional medicines by comparing the protein expression profiles. To our knowledge, this study is the first proteomic analysis for revealing the mechanisms of life span extension and antiaging on D. melanogaster treated with RGE.
In this study, D. melanogaster treated with RGE (1 mg/mL) extended the life span significantly compared to the wild type (p < 0.01). Further results demonstrated that the effects of life span extension of RGE on D. melanogaster might be attributed to elevation of SOD levels, reduction in lipid peroxidation of MDA content, and a range of differently expressed proteins. Proteomic analysis detected 23 differentially expressed proteins in the RGE-treated D. melanogaster versus wild type: 16 up-regulated and 7 down-regulated. These proteins were found to involved in multiple biological processes and signaling pathways, such as hippo and oxidative phosphorylation signaling. Taken together, the results provided a scientific basis for the further exploitation of mechanisms of longer life span and/or antiaging altered by RGE in D. melanogaster.
Age-associated neurodegenerative disorders including Alzheimer's disease (AD) and Parkinson's disease (PD) are associated with multifactorial etiology [14]. In this study, UCHL1, a ubiquitin ligase exclusively expressed in neurons, is up-regulated in RGE-treated D. melanogaster versus wild type. UCHL1 was shown to be involved in the pathophysiology of PD and AD [15] and is elevated in AD patients [16], AD mice [17], and senescent C57BL/6J mouse pituitaries [18] compared to controls. However, UCHL1 overexpression has been shown to delay AD progression in vivo [19], and low levels have also been reported in postmortem brain tissue from AD patients [20]. The increased expression level of UCHL1 protein by RGE treatment suggested that it may play important roles in antiaging and life span extension, and potentially neurodegenerative disorders. HSP70, which also showed increased expression in RGE-treated D. melanogaster, plays important protective roles in various neurodegenerative disorders [21]. HSP70 is up-regulated in AD model mice compared to normal mice [17], [22]. Further studies are required to explore these mechanisms during the process of neurodegeneration, and RGE may offer new avenues for novel therapeutic approaches for the treatment of chronic neurodegenerative disorders that currently have no effective medication.
Oxidation impairs protein function as the proteins are unfolded leading to an increase in protein hydrophobicity. This often leads to the formation of toxic aggregates that interfere with normal cellular functions, which is a sign of the aging process and many age-related diseases. In this study, changes in the expression levels of HSP70 and UCHL1 (both up-regulated in RGE-treated D. melanogaster) could be a response to general age-derived cellular alterations, such as increasing oxidative burden, accumulation of damaged proteins, and elevated protein synthesis. At the molecular level, to ensure survival, cells have developed antioxidants to counteract this oxidative damage accumulation [23]. It is wildly believed that HSP70 and UCHL1, which are responsible for the removal of damaged proteins, counteract the oxidative stress damage of proteins [24]. HSP70 has also been shown to confer protection against the harmful effects of oxidative stress, as well as modulate the inflammatory status [23]. Furthermore, the overexpression of HSP70 in mice muscle eliminated the age-related increase in posttranslational modifications of proteins [25].
Ginseng can increase long-term resistance to stress and disease and therefore affects life span [26]. Our study demonstrated an elevation of SOD and a reduction in lipid peroxidation of MDA content in D. melanogaster treated with RGE. In addition, three antioxidant-related proteins—HSP70, UCHL1, and ALF [27], [28], [29]—in 2-DE results were identified by Western blotting and elevated in D. melanogaster induced by RGE. Previous studies reported that Korean Red Ginseng ameliorated oxidative tissue injury, provided an antiaging effect, and suppressed age-related renal injury [9]. In addition, ginsenoside Rg1 improved cognitive ability and promoted neurogenesis by enhancing the antioxidant and anti-inflammatory capacity in the hippocampus [30]. Therefore, the higher expression of the antioxidants HSP70 and UCHL1 in RGE-treated flies may facilitate slower aging progression and prolong life span.
HSP70 is an important molecular chaperone, which plays a key role in cell protection by assisting with protein homeostasis or protein quality control [31], [32]. In addition, aging has been related to impairment in activity and availability of HSP70 induction. Previous reports revealed that exogenous HSP70 protein can significantly enhance mouse life span [33] and stimulate the growth of aged mesenchymal stem cells in vitro [34]. Ginseng treatment increased expression of HSP70 in rats, which showed protected functions of neurological responses and memory ability [35]. In the present study, we found an increased level of HSP70 in RGE-treated D. melanogaster, suggesting a protective role in aging and life span extension. Therefore, RGE might be considered a potential source to identify compounds that delay aging.
The hippo pathway is a signaling cascade that plays an evolutionarily conserved role in organ size control from Drosophila to humans by regulating cell proliferation, apoptosis, stem cell/progenitor cell fate determination, self-renewal control, and tissue architecture and renewal [36], [37], [38]. Mutations of these genes lead to a common phenotype of tissue overgrowth and enlarged organ size in Drosophila eyes and wings [39]. Hippo pathway components also mediated crosstalk with other pathways, for example, Wnt, TGF, and BMP pathways [40]. In the present study, the differentially expressed proteins, 1433Z and ACT6, of the hippo pathway were up-regulated in D. melanogaster treated by RGE versus wild type. This may indicate a role in maintaining cellular health and tissue turnover, induced by RGE treatment.
V-type ATPase (VATA), an up-regulated protein in RGE-treated D. melanogaster, is categorized as a rotary ATP synthase/ATPase complex [41]. ATP synthase is a key enzyme of mitochondrial energy conversion, and inhibition of ATP synthase may cause energy deprivation and increased reactive oxygen species production. A high reactive oxygen species content induces cellular necrosis and/or apoptosis [42], [43]. Because the oxidative phosphorylation pathway is induced (via both VATA and ND75 up-regulation) by RGE treatment, this may provide the energy for the antiaging process and life span extension.
5. Conclusions
This is the first comprehensive study of D. melanogaster treated with RGE using a 2-DE-based quantitative proteomic approach. In this study, we found that RGE had longevity-enhancing effects on D. melanogaster by altering the expression of antioxidants and regulating by multiple pathways. Further studies are warranted to explore the novel molecular mechanisms underlying the antiaging effects and life span extension by treatment with red ginseng.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by the Project of the National Science Foundation for Youths of China (No. 31401606), Jilin Science & Technology Development Plan (20140101125JC and 201627), the special fund for agro-scientific research in the public interest (201303111), and the International Science & Technology Cooperation Program of China (2015DFA31290). The authors also thank Dr. Eric Arthur Lord (University of Otago) and Dr. Yali Li for kindly reading and revising through the manuscript.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2017.04.006.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Ghimire S., Kim M.S. Jujube (Ziziphus Jujuba Mill.) fruit feeding extends lifespan and increases tolerance to environmental stresses by regulating aging-associated gene expression in Drosophila. Biogerontology. 2017;18:263–273. doi: 10.1007/s10522-017-9686-8. [DOI] [PubMed] [Google Scholar]
- 2.Ahangarpour A., Lamoochi Z., Fathi Moghaddam H., Mansouri S.M. Effects of Portulaca oleracea ethanolic extract on reproductive system of aging female mice. Int J Reprod Biomed (Yazd) 2016;14:205–212. [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg S., Feller C., Forne I., Schiller E., Sévin D.C., Schauer T., Regnard C., Straub T., Prestel M., Klima C. Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. EMBO Rep. 2016;17:455–469. doi: 10.15252/embr.201541132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghimire S., Kim M.S. Defensive behavior against noxious heat stimuli is declined with aging due to decreased pain-associated gene expression in Drosophila. Biomol Ther (Seoul) 2015;23:290–295. doi: 10.4062/biomolther.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong W.Y., Farooqui T., Koh H.L., Farooqui A.A., Ling E.A. Protective effects of ginseng on neurological disorders. Front Aging Neurosci. 2015;7:129. doi: 10.3389/fnagi.2015.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim G.N., Lee J.S., Song J.H., Oh C.H., Kwon Y.I., Jang H.D. Heat processing decreases Amadori products and increases total phenolic content and antioxidant activity of Korean Red Ginseng. J Med Food. 2010;13:1478–1484. doi: 10.1089/jmf.2010.1076. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y., Oh S. Administration of red ginseng ameliorates memory decline in aged mice. J Ginseng Res. 2015;39:250–256. doi: 10.1016/j.jgr.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M.S. Korean Red Ginseng tonic extends lifespan in D. melanogaster. Biomol Ther (Seoul) 2013;21:241–245. doi: 10.4062/biomolther.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S., Kim C.S., Min J., Lee S.H., Jung Y.S. A high-fat diet increases oxidative renal injury and protein glycation in d-galactose-induced aging rats and its prevention by Korea red ginseng. J Nutr Sci Vitaminol (Tokyo) 2014;60:159–166. doi: 10.3177/jnsv.60.159. [DOI] [PubMed] [Google Scholar]
- 10.Kang T.H., Park H.M., Kim Y.B., Kim H., Kim N., Do J.H., Kang C., Cho Y., Kim S.Y. Effects of red ginseng extract on UVB irradiation-induced skin aging in hairless mice. J Ethnopharmacol. 2009;123:446–451. doi: 10.1016/j.jep.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Han J., Li P., Cai W., Shao X. Fast determination of ginsenosides in ginseng by high-performance liquid chromatography with chemometric resolution. J Sep Sci. 2014;37:2126–2130. doi: 10.1002/jssc.201400403. [DOI] [PubMed] [Google Scholar]
- 12.Huang X., Wang X., Lv Y., Xu L., Lin J., Diao Y. Protection effect of kallistatin on carbon tetrachloride-induced liver fibrosis in rats via antioxidative stress. PLoS One. 2014;9:e88498. doi: 10.1371/journal.pone.0088498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Z., Ba H., Zhang W., Coates D., Li C. iTRAQ-based quantitative proteomic analysis of the potentiated and dormant antler stem cells. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17111778. pii: E1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erdő F., Denes L., de Lange E. Age-associated physiological and pathological changes at the blood–brain barrier: a review. J Cereb Blood Flow Metab. 2016 doi: 10.1177/0271678X16679420. pii: 0271678X16679420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeffer M., Plenzig S., Gispert S., Wada K., Korf H.W., Von Gall C. Disturbed sleep/wake rhythms and neuronal cell loss in lateral hypothalamus and retina of mice with a spontaneous deletion in the ubiquitin carboxyl-terminal hydrolase L1 gene. Neurobiol Aging. 2012;33:393–403. doi: 10.1016/j.neurobiolaging.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Öhrfelt A., Johansson P., Wallin A., Andreasson U., Zetterberg H., Blennow K., Svensson J. Increased cerebrospinal fluid levels of ubiquitin carboxyl-terminal hydrolase L1 in patients with Alzheimer's disease. Dement Geriatr Cogn Dis Extra. 2016;6:283–294. doi: 10.1159/000447239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan A.T., Dobson R.J., Sattlecker M., Kiddle S.J. Alzheimer's disease: are blood and brain markers related? A systematic review. Ann Clin Transl Neurol. 2016;3:455–462. doi: 10.1002/acn3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzban G., Grillari J., Reisinger E., Hemetsberger T., Grabherr R., Katinger H. Age-related alterations in the protein expression profile of C57BL/6J mouse pituitaries. Exp Gerontol. 2002;37:1451–1460. doi: 10.1016/s0531-5565(02)00117-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M., Cai F., Zhang S., Zhang S., Song W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer's progression in vivo. Sci Rep. 2014;4:7298. doi: 10.1038/srep07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J., Levey A.I., Weintraub S.T., Rees H.D., Gearing M., Chin L.S., Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 21.Tsai Y.C., Lee Y.M., Lam K.K., Lin J.F., Wang J.J., Yen M.H., Cheng P.Y. The role of heat shock protein 70 in the protective effect of YC-1 on β-amyloid-induced toxicity in differentiated PC12 cells. PLoS One. 2013;8:e69320. doi: 10.1371/journal.pone.0069320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y., Zhao D., Pan B., Wang J., Cui Y., Shi F., Wang C., Yin X., Zhou X., Yang L. Proteomic analysis of protein expression throughout disease progression in a mouse model of Alzheimer's disease. J Alzheimers Dis. 2015;47:915–926. doi: 10.3233/JAD-150312. [DOI] [PubMed] [Google Scholar]
- 23.Martínez de Toda I., De la Fuente M. The role of Hsp70 in oxi-inflamm-aging and its use as a potential biomarker of lifespan. Biogerontology. 2015;16:709–721. doi: 10.1007/s10522-015-9607-7. [DOI] [PubMed] [Google Scholar]
- 24.Di Domenico F., Coccia R., Cocciolo A., Murphy M.P., Cenini G., Head E., Butterfield D.A., Giorgi A., Schinina M.E., Mancuso C. Impairment of proteostasis network in Down syndrome prior to the development of Alzheimer's disease neuropathology: redox proteomics analysis of human brain. Biochim Biophys Acta. 2013;1832:1249–1259. doi: 10.1016/j.bbadis.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broome C.S., Kayani A.C., Palomero J., Dillmann W.H., Mestril R., Jackson M.J., Mc Ardle A. Effect of lifelong overexpression of HSP70 in skeletal muscle on age-related oxidative stress and adaptation after nondamaging contractile activity. FASEB J. 2006;20:1549–1551. doi: 10.1096/fj.05-4935fje. [DOI] [PubMed] [Google Scholar]
- 26.Bittles A.H., Fulder S.J., Grant E.C., Nicholls M.R. The effect of ginseng on lifespan and stress responses in mice. Gerontology. 1979;25:125–131. doi: 10.1159/000212330. [DOI] [PubMed] [Google Scholar]
- 27.Goren P., Reznick A.Z., Reiss U., Gershon D. Isoelectric properties of nematode aldolase and rat liver superoxide dismutase from young and old animals. FEBS Lett. 1977;84:83–86. doi: 10.1016/0014-5793(77)81062-4. [DOI] [PubMed] [Google Scholar]
- 28.Choi S., Park K.A., Lee H.J., Park M.S., Lee J.H., Park K.C., Kim M., Lee S.H., Seo J.S., Yoon B.W. Expression of Cu/Zn SOD protein is suppressed in hsp 70.1 knockout mice. J Biochem Mol Biol. 2005;38:111–114. doi: 10.5483/bmbrep.2005.38.1.111. [DOI] [PubMed] [Google Scholar]
- 29.Coulombe J., Gamage P., Gray M.T., Zhang M., Tang M.Y., Woulfe J., Saffrey M.J., Gray D.A. Loss of UCHL1 promotes age-related degenerative changes in the enteric nervous system. Front Aging Neurosci. 2014;6:129. doi: 10.3389/fnagi.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J., Mu X., Zeng J., Xu C., Liu J., Zhang M., Li C., Chen J., Li T., Wang Y. Ginsenoside Rg1 prevents cognitive impairment and hippocampus senescence in a rat model of d-galactose-induced aging. PLoS One. 2014;9:e101291. doi: 10.1371/journal.pone.0101291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen C.J., Choi Y.H., Ing N.H., Kraemer D.C., Vogelsang M.M., Hinrichs K. Heat shock protein 70 gene expression in equine blastocysts after exposure of oocytes to high temperatures in vitro or in vivo after exercise of donor mares. Theriogenology. 2010;74:374–383. doi: 10.1016/j.theriogenology.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 32.Reeg S., Jung T., Castro J.P., Davies K.J., Henze A., Grune T. The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic Biol Med. 2016;99:153–166. doi: 10.1016/j.freeradbiomed.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobkova N.V., Evgen'ev M., Garbuz D.G., Kulikov A.M., Morozov A., Samokhin A., Velmeshev D., Medvinskaya N., Nesterova I., Pollock A. Exogenous Hsp70 delays senescence and improves cognitive function in aging mice. Proc Natl Acad Sci USA. 2015;112:16006–16011. doi: 10.1073/pnas.1516131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreeva N.V., Zatsepina O.G., Garbuz D.G., Evgen'ev M.B., Belyavsky A.V. Recombinant HSP70 and mild heat shock stimulate growth of aged mesenchymal stem cells. Cell Stress Chaperones. 2016;21:727–733. doi: 10.1007/s12192-016-0691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang G., Liu A., Zhou Y., San X., Jin T., Jin Y. Panax ginseng ginsenoside-Rg2 protects memory impairment via anti-apoptosis in a rat model with vascular dementia. J Ethnopharmacol. 2008;115:441–448. doi: 10.1016/j.jep.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Pesce M., Messina E., Chimenti I., Beltrami A.P. Cardiac mechano-perception: a life-long story from early beats to aging and failure. Stem Cells Dev. 2017;26:77–90. doi: 10.1089/scd.2016.0206. [DOI] [PubMed] [Google Scholar]
- 37.Yu F.-X., Zhao B., Guan K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder M.C., Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol. 2012;23:803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Q., Li L., Zhao B., Guan K.L. The Hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116:1431–1447. doi: 10.1161/CIRCRESAHA.116.303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D., Lian T., Tu J., Gaur U., Mao X., Fan X., Li D., Li Y., Yang M. Lnc RNA mediated regulation of aging pathways in Drosophila melanogaster during dietary restriction. Aging (Albany NY) 2016;8:2182–2203. doi: 10.18632/aging.101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forgac M. Vacuolar ATPases: rotary protonpumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2006;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 42.Numoto N., Hasegawa Y., Takeda K., Miki K. Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 2009;10:1228–1234. doi: 10.1038/embor.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skorokhod O.A., Davalos-Schafler D., Gallo V., Valente E., Ulliers D., Notarpietro A., Mandili G., Novelli F., Persico M., Taglialatela-Scafati O. Oxidative stress-mediated antimalarial activity of plakortin, a natural endoperoxide from the tropical sponge Plakortis simplex. Free Radic Biol Med. 2015;89:624–637. doi: 10.1016/j.freeradbiomed.2015.10.399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.