Abstract
Lag1p and Lac1p are two homologous transmembrane proteins of the endoplasmic reticulum in Saccharomyces cerevisiae. Homologous genes have been found in a wide variety of eukaryotes. In yeast, both genes, LAC1 and LAG1, are required for efficient endoplasmic reticulum-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. In this study, we show that lag1Δlac1Δ cells have reduced sphingolipid levels due to a block of the fumonisin B1-sensitive and acyl-CoA–dependent ceramide synthase reaction. The sphingolipid synthesis defect in lag1Δlac1Δ cells can be partially corrected by overexpression of YPC1 or YDC1, encoding ceramidases that have been reported to have acyl-CoA–independent ceramide synthesis activity. Quadruple mutant cells (lag1Δlac1Δypc1Δydc1Δ) do not make any sphingolipids, but are still viable probably because they produce novel lipids. Moreover, lag1Δlac1Δ cells are resistant to aureobasidin A, an inhibitor of the inositolphosphorylceramide synthase, suggesting that aureobasidin A may be toxic because it leads to increased ceramide levels. Based on these data, LAG1 and LAC1 are the first genes to be identified that are required for the fumonisin B1-sensitive and acyl-CoA–dependent ceramide synthase reaction.
INTRODUCTION
The fate of eukaryotic cells is supervised by a tightly networked signaling system, which comprises a variety of molecular sensors. Different stimuli are exactly perceived and subsequently initiate defined signal transduction cascades, which modulate the molecular and general status of the cell. By such mechanisms, the cell responds to stress or changes in its environment and adapts to the new situation.
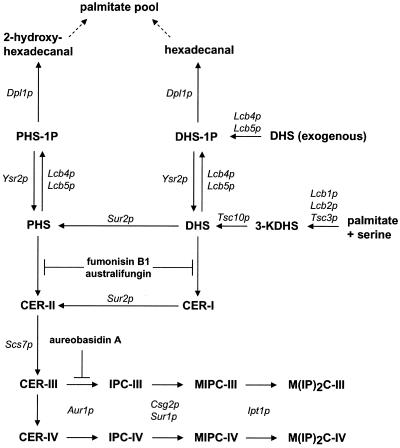
Recently, it has been shown that sphingolipid metabolism plays a role in signal transduction in mammalian cells (Mathias et al., 1998) as well as in the baker's yeast S. cerevisiae (Dickson, 1998). These compounds are involved in the response to a variety of extracellular signals and physiological situations. Our knowledge of sphingolipid metabolism in yeast has greatly increased over the past years (Dickson and Lester, 1999; see Figure 1). In this pathway, ceramide is a central molecule. Structurally essential for cell growth, ceramide also mediates different cellular events, such as apoptosis, growth arrest, and stress response (Mathias et al., 1998; Perry and Hannun, 1998). As a signaling molecule, its turnover has to be tightly regulated. Nevertheless, the regulation of the level of ceramide in cells is still poorly understood. Ceramide is synthesized mainly from fatty acyl-CoA and PHS or DHS by a CoA-dependent ceramide synthase. Recently, two genes, YPC1 and YDC1, have been shown to have a minor ceramide synthase activity due to their reverse ceramidase action (Mao et al., 2000a, 2000b). This ceramide synthase activity is independent of CoA and is not inhibited by fumonisin B1 (FB1). However, to date, no genes involved in the CoA-dependent ceramide synthase reaction have been identified.
Figure 1.
Sphingolipid metabolism in yeast. This scheme presents a summary of the general sphingolipid metabolism in yeast as described in the literature. The important steps are mentioned in the text. The Roman numerals I–IV indicate the ascending order of hydroxylation of the ceramide backbone (i.e., ceramide containing (I) DHS and C26:0, (II) PHS and C26:0, (III) PHS and C26:0-OH, and (IV) PHS and C26:0–2OH. For detailed information see Dickson (1998). DHS, dihydrosphingosine; DHS-1P, DHS-1-phosphate; IPC, inositolphosphorylceramide; KDHS, keto-DHS; MIPC, mannosylinositolphosphorylceramide; M(IP)2C, mannosyldiinositolphosphorylceramide; PHS, phytosphingosine; PHS-1P, PHS-1-phosphate. The site of action of the inhibitors FB1, australifungin, and aureobasidin A are shown.
Barz and Walter (1999) recently described two highly homologous ER membrane proteins, Lag1p and Lac1p, of S. cerevisiae. Sequence homologues of LAG1 and LAC1 were found in numerous eukaryotes including humans (Jiang et al., 1998; Brandwagt et al., 2000), but nothing is known about the molecular function of the encoded proteins. The single deletion of either gene in yeast had no obvious phenotype, whereas the double deletion revealed very interesting features. The absence of both LAG1 and LAC1 resulted in a serious defect in the cell growth, a thickened cell wall, and a strongly reduced transformation ability. Interestingly, a lag1Δlac1Δ strain showed an ∼50% reduction in the rate of transport of glycosylphosphatidylinositol (GPI)-anchored proteins to the Golgi, whereas the maturation of non–GPI-anchored proteins was unaltered. Because transport of GPI-anchored proteins requires ongoing sphingoid base synthesis (Horvath et al., 1994; Muniz et al., 2001), we decided to analyze sphingolipid biosynthesis in the lag1Δlac1Δ deletion strain. By combination of in vivo and in vitro approaches, we demonstrate that Lag1p and Lac1p are essential for the acyl-CoA–dependent and FB1-sensitive ceramide synthase reaction.
MATERIALS AND METHODS
Miscellaneous
The strains of S. cerevisiae used in this study and their genotypes are given in Table 1. Except for the experiments in Figures 5 and 6 and growth in presence of aureobasidin A, all experiments were performed in the W303 genetic background. RH4838, RH2881, RH5308, and RH 5515 were used for the former experiments. Cultivation of yeast as well as molecular and biochemical standard techniques were performed as described previously (Barz and Walter, 1999). PCR was performed in a GeneAmp PCR System 9700 (PE Applied Biosystems, Norwalk, CT) with the use of TaKaRa LA Taq DNA polymerase (TaKaRa Biomedicals, Tokyo, Japan) according to the manufacturer's directions. PCR products were subsequently cloned into the pCR2.1 vector with the use of the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA) and sequenced. For the determination of protein concentration the BCA protein assay (Pierce, Rockford, IL) was used. Authentic yeast sphingolipid standards were a kind gift of Robert L. Lester (University of Kentucky, Lexington, KY). Radioisotopes, ortho-[32P]phosphate (carrier free, ∼8000 Ci/mmol), myo-[2-3H]inositol (10–20 Ci/mmol), [9,10(n)-3H]palmitate (PA; 40–60 Ci/mmol), l-[U-14C]serine (150 mCi/mmol) were obtained from Amersham (Uppsala, Sweden), except for d-erythro-[4,5-3H]dihydrosphingosine (30–60 Ci/mmol; [3H]DHS), which was obtained from BioTrend Chemikalien GmbH (Cologne, Germany). Unless otherwise indicated, chemicals and enzymes were obtained from Roche (Mannheim, Germany), ICN (Indianapolis, IN) or Sigma Chemical Co. (St. Louis, MO). Aureobasidin A and australifungin were obtained from Takara Shozu Co. (Otsu, Japan), and Merck (Rahway, NJ), respectively.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference/Source |
|---|---|---|
| W303-1A | MATa ade2-1 his3-11 leu2-3,-112 trp1-1 ura3-1 can1-100 | Ref. 38 |
| W303-1B | MATα ade2-1 his3-11 leu2-3,-112 trp1-1 ura3-1 can1-100 | Ref. 38 |
| WBY286 | W303-1A lac1Δ::ADE2 | Ref. 7 |
| WBY616 | W303-1B lac1Δ::ADE2 lag1Δ::HIS3 | Ref. 7 |
| WBY743 | W303-1A lac1Δ::ADE2 lag1Δ::HIS3 [pWB96] | Ref. 7 |
| WBY777 | W303-1B lac1Δ::ADE2 lag1Δ::HIS3 [pWB98] | Ref. 7 |
| WBY783 | W303-1B lac1Δ::ADE2 lag1Δ::HIS3 [pWB783] | This study |
| WBY824 | W303-1A lac1Δ::ADE2 lag1Δ::HIS3 [pWB824] | This study |
| WBY822 | W303-1A lac1Δ::ADE2 lag1Δ::HIS3 ypc1Δ::LEU2 ydc1Δ::TRP1 | This study |
| STY18 | W303-1A lac1Δ::ADE2 lag1Δ::HIS3 [pWB96, pWB98] | This study |
| RH5308 | MATa lac1::LEU2 lag1::TRP1 his3 trp1 ura3 leu2 bar1 AUR1::3HA::HIS3 | This study |
| RH5309 | MATa lac1::LEU2 lag1::URA3 his3 trp1 ura3 leu2 bar1 | This study |
| RH5515 | plag1-1TS transformed in RH 5309 | This study |
| RH2881 | MATa his3 trp1 ura3 leu2 bar1 | This study |
| RH4838 | MATa his3 leu2 ura3 trp1 bar1 AUR1::3HA::HIS3 | K. Funato |
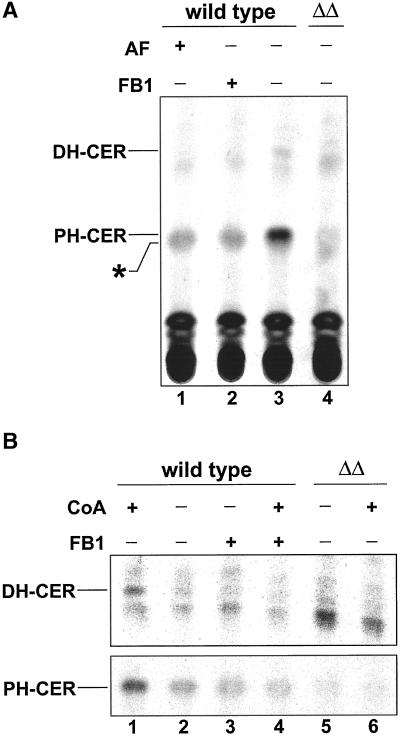
Figure 5.
In vitro ceramide synthase assay. (A) Enriched microsomal membranes derived from wild-type or lag1Δlac1Δ (ΔΔ) cells were preincubated with wild-type cytosol, a mixture of cold and labeled dihydrosphingosine (0.5 μCi), an ATP-regenerating system, and in the presence of australifungin (lane 1) or fumonisin B1 (lane 2) for 15 min at 10°C. The ceramide synthase reaction was started by adding CoA and a liposomal mixture of hexacosanoic acid and phosphatidylinositol. After a further 120 min of incubation at 24°C, the reaction was stopped with chloroform/methanol 1:1 [vol/vol], and the lipids were hydrolyzed with mild base and desalted. Equal amounts were spotted onto HPTLC plates and resolved in solvent C. (B) CoA dependence. Experiments are performed at the same conditions as in A, except for omitting CoA (lanes 2, 3, and 5). Lipids were visualized and quantified with the use of tritium-sensitive screens and a Cyclone phosphorimager. As dihydroceramide and phytoceramide spots had very different signal intensities, a shorter exposure time for phytoceramide is shown. Abbreviations: AF, australifungin; DH-CER, dihydroceramide; FB1, fumonisin B1; PH-CER, phytoceramide. An unidentified lipid is denoted with an asterisk.
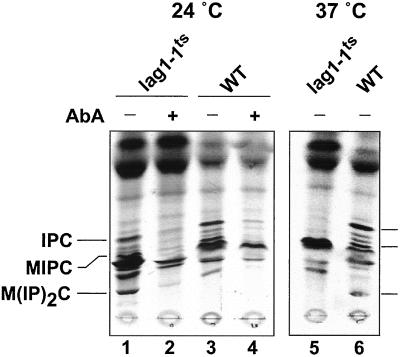
Figure 6.
A temperature-sensitive allele of LAG1. The lag1Δlac1Δ strain was transformed with a plasmid carrying a temperature-sensitive allele of LAG1 (lag1–1TS). Cells were grown overnight at 24°C in SDYE medium till early logarithmic phase, resuspended in SD medium to an absorbance of 10 OD/ml. Then, 0.5 ml of cells were labeled with [3H]DHS (4 μCi, final concentration 0.15 μM) at the indicated temperature for 2 h after prior incubation at 24°C or 37°C for 1 min in the presence of aureobasidin A (AbA; lanes 2 and 4). Further energy-dependent metabolism was stopped by adding NaN3/NaF, and the labeled lipids were extracted, hydrolyzed with mild base, and analyzed on TLC plates with the use of solvent B. Lipids were visualized and quantified with the use of tritium-sensitive screens and a Cyclone phosphorimager.
High-copy Suppressor Screen
The genomic LEU2-library (Nasmyth and Reed, 1980) was shuffled into WBY777 (grown in YPD) with the use of a standard protocol. Fifteen agar plates each having a lawn of colonies were replica plated onto plates containing fluoroorotate as counterselective agent and incubated at 30°C for 3–4 d. One hundred thirty-five differently sized colonies were streaked to single colonies on SC-LEU plates followed by colony PCR to identify plasmid-encoded LAG1 or LAC1, respectively. Negative clones were taken to recover the containing plasmids, which were then shuffled into WBY777. After counterselection against pWB98 on fluoroorotate agar plates, the resulting clones were tested for their growth ability. YBR183w with its flanking sequences was subcloned into pRS425 (Christianson et al., 1992) with the use of the native EagI and BglII restriction sites to yield pWB783. YPL087w was amplified with its flanking sequences with the use of genomic PCR, and the resulting 1.9-kb fragment cloned into pRS425 with the use of the EagI restriction sites to yield pWB824.
Disruption of YBR183w and YPL087w
The chromosomal deletion alleles ybr183w1Δ::LEU2 and ypl087wΔ::TRP1 were created by replacing the entire coding sequences of YBR183w and YPL087w with yeast-integrative plasmids. To disrupt YBR183w, the noncoding flanking regions of this gene (400–600 bp) were amplified by genomic PCR with the use of oligonucleotides that incorporated XhoI and XbaI restriction sites at the ends of the fragments proximal to YBR183w and HindIII sites at the outside ends of the fragments distant from YBR183w. These fragments were then digested with the relevant restriction enzymes and cloned in a three-way ligation into XbaI- and XhoI-cut pRS405 (Sikorski and Hieter, 1989). The resulting plasmid, pWB754, was linearized with HindIII and used to transform WBY286 (Barz and Walter, 1999) cells to Leu+ prototrophy. One of the Leu+ transformants was purified and designated WBY759 (lac1Δ::ADE2 ybr183wΔ::LEU2). Correct chromosomal deletion of YBR183w was confirmed by extensive genomic PCR. A similar strategy was used to replace the entire coding region of YPL087w with a yeast-integrative vector pRS404 (Sikorski and Hieter, 1989). The 5′- and 3′-flanking sequences of YPL087w (400–600 bp) were amplified by genomic PCR and cloned in a three-piece ligation into BamHI- and XhoI-digested pRS404. The resulting plasmid, pWB755, was linearized with EcoRI and used to transform WBY283 (Barz and Walter, 1999) cells to Trp+ prototrophy. Correct replacement of YPL087w was confirmed in one Trp+ transformant (termed WBY758; lag1Δ::HIS3 ypl087wΔ::TRP1) by genomic PCR analysis. WBY758 and WBY759 were mated to construct a diploid that is heterozygous for LAG1, LAC1, YBR183w, and YPL087w. The resulting His+Ade+Leu+Trp+ diploid (designated WBY796) was sporulated in liquid sporulation medium and asci were dissected into tetrads. One haploid His+Ade+Leu+Trp+ (lag1Δ::HIS3 lac1Δ::ADE2 ybr183wΔ::LEU2 ypl087wΔ::TRP1) clone was designated WBY822.
Lipid Analysis
Pulse Labeling.
Pulse labeling of lipids with [3H]inositol and lipid extraction was performed as previously described (Puoti et al., 1991), except that the yeast cells were grown in YPD or selective SC media. Pulse labeling with [3H]DHS or [3H]PA was performed with cells of early logarithmic phase that were resuspended to an absorbance of 1 OD600 in 1 ml of fresh SC media. Before labeling, the cells were preincubated with 100 μM FB1 (Calbiochem, Bad Soden, Germany) for 2 h at 30°C as indicated. Subsequently, 2 μCi of [3H]DHS or 5 μCi of [3H]PA were added, and the cells were further incubated for the indicated time span.
Long-term Labeling.
For long-term labeling of lipids, cells of early logarithmic phase were diluted in 5 ml media containing the relevant labeled precursor to an absorbance of 0.1 OD600 and grown at 30°C overnight. The labeling was performed as follows: 50 μCi [3H]inositol in SC media lacking inositol (Culbertson and Henry, 1975), 5 μCi [14C]serine in SC media lacking serine, 10 μCi [3H]DHS in SC media, 500 μCi [32P]phosphate in YPD. Before harvesting the cell density was determined by absorbance of a 1:20 dilution at 600 nm.
Extraction, Hydrolysis, Desalting, and TLC.
Generally, preparation of whole cell lipid extracts was performed according to the procedure IIIB of Hanson and Lester (1980). If necessary, the extracted lipids were subjected to mild alkaline methanolysis (Becker and Lester, 1980) and partitioned between butanol and water as described (Krakow et al., 1986). The pooled organic phases were dried in a speedvac and resuspended in equivalent volumes corresponding to absorbance at 600 nm, that is, the cell number, of CMW (chloroform/methanol/water, 10:10:3 [vol/vol]). Lipids were subjected to high-performance TLC plates (Kieselgel 60; Merck, Darmstadt, Germany) and resolved in either solvent A (chloroform/methanol/0.25% KCl, 55:45:10 [vol/vol]), solvent B (chloroform/methanol/4.2 N NH4OH, 9:7:2 [vol/vol]), or solvent C (chloroform/acetic acid, 9:1 [vol/vol]) as indicated. For visualization of the 3H- or 14C-labeled lipids, the plates were treated with EN3HANCE (NEN Life Science Products, Boston, MA) before exposition to XOMAT films (Eastman Kodak, Rochester, NY). Quantification of the [32P]phosphate-labeled lipids was performed with the use of a Fujifilm FLA2000 phosphorimager (Fuji Photo Film, Tokyo, Japan). All lipids were identified with the use of authentic standards except for phytosphingosine-1-phosphate (PHS-1P), which was assigned by its relative mobility on TLC compared with dihydrosphingosine-1-phosphate (DHS-1P) and mannosyldiinositolphosphorylceramide (M(IP)2C; Mao et al., 2000a; Nagiec et al., 1998).
In vitro Ceramide Synthase Assay
For microsomal membrane preparation, a 2-liter culture grown at 24°C to a density of ∼5 × 107 cells/ml was harvested, washed with 100 mM Tris/HCl, pH 9.4, and incubated 20 min at a density of 20 OD600/ml in 20 mM DTT, 100 mM Tris-HCl, pH 9.4, at room temperature. The cells were resuspended (2 ml/g cells) in 0.7 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1.5% peptone, 0.75% yeast extract, 0.5% glucose containing zymolyase 20T (2.5 mg/g of cells; Seigagaku Corp., Tokyo, Japan) and incubated for 45 min at 24°C. The resulting spheroplasts were overlaid onto 15 ml of 20 mM HEPES, pH 7.4, 0.8 M sucrose, 1.5% ficoll 400 and spun, and the pellet was washed with 0.7 M sorbitol, 20 mM HEPES, pH 7.4. The spheroplasts were disrupted by resuspension in 0.1 M sorbitol, 20 mM HEPES, pH 7.4, 150 mM potassium acetate, 2 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 μg/ml protein inhibitors and were homogenized with a Potter-Elvehjem homogenizer on ice. Subsequently, cell debris was removed by centrifugation at 4°C at 1000 × g, and microsomal membranes were collected by centrifugation at 4°C at 100,000 × g. The pellet was washed and resuspended in B88 (20 mM HEPES-KOH, pH 6.8, 150 mM potassium acetate, 5 mM magnesium acetate, 250 mM sorbitol) and stored at −80°C. The preparation of wild-type cytosol was performed essentially as described (Salama et al., 1993).
The in vitro assay was performed as follows. In a total volume of 50 μl of B88, microsomal membranes (200 μg), cytosol (100 μg), ATP-regenerating system (1 mM ATP, 40 mM phosphocreatine, 0.2 mg/ml creatine phosphokinase), GDP-mannose (50 μM), and a mix of unlabeled and labeled DHS (10 pmol of [3H]DHS and 40 pmol of DHS, 0.5 μCi) were firstly incubated for 15 min at 10°C. Then, 50 μM CoA and a mix of liposomes containing hexacosanoic acid (C26) and PI (50 μM/250 μM) were added. The samples were incubated for 2 h at 24°C. The reaction was stopped by adding 333 μl of chloroform/methanol (1:1 [vol/vol]). The lipids were submitted to a mild-alkaline treatment, extracted by butanol as described above, and analyzed by TLC with the use of solvent C to identify ceramide (Morell and Radin, 1970), which was visualized and quantified with the use of tritium-sensitive screens and a Cyclone phosphorimager (Packard, Meriden, CT).
RESULTS
The transport of GPI-anchored proteins from the ER to the Golgi requires ongoing sphingoid base and/or ceramide synthesis (Horvath et al., 1994; Skrzypek et al., 1997; Sutterlin et al., 1997). Barz and Walter (1999) raised the possibility that the reduction in kinetics of GPI-anchored protein transport to the Golgi compartment in the lag1Δlac1Δ deletion strain could be due to an alteration in sphingolipid biosynthesis. Thus, we decided to analyze the sphingolipid composition of lag1Δlac1Δ cells in comparison to wild-type cells.
The lag1Δlac1Δ Strain Has a Strongly Reduced Level of Sphingolipids
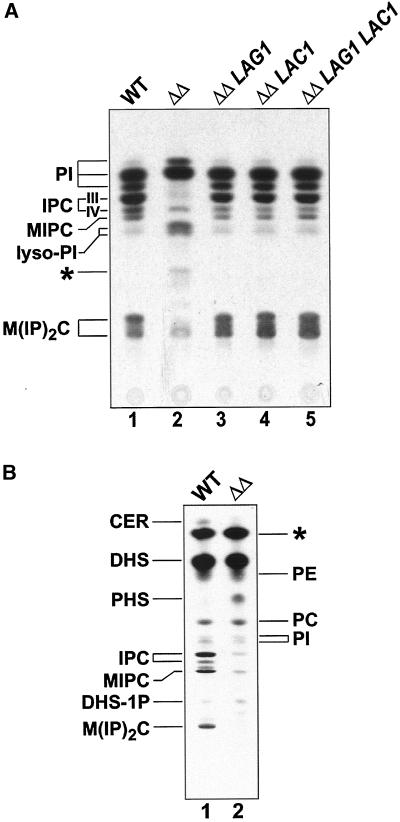
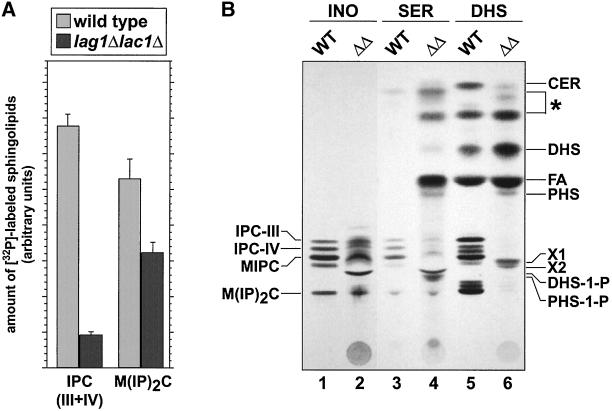
In yeast, the incorporation of inositol into sphingolipids occurs by the addition of inositolphosphate, derived from phosphatidylinositol (PI), to the ceramide moiety (Becker and Lester, 1980). The resulting inositolphosphorylceramide (IPC) is then mannosylated to yield mannosylinositolphosphorylceramide (MIPC), which serves as substrate for the synthesis of the mature sphingolipid M(IP)2C (see also Figure 1). The biosynthetic rates of these complex sphingolipids were investigated using the technique of in vivo pulse-chase labeling with the use of [3H]inositol as the radioactive marker. Compared with wild-type cells, lag1Δlac1Δ cells exhibited a completely altered synthesis pattern of inositol-containing lipids (Figure 2A, lanes 1 and 2). The bands corresponding to IPC-IV (containing phytoceramide with C26:0–2OH) and M(IP)2C were much less intense, whereas IPC-III (containing phytoceramide with C26:0-OH) and MIPC were not detectable. The synthesis rate of PI remained high but appeared to be qualitatively altered in favor of species with elongated fatty acids (faster migrating PI species). The amount of lyso-PI varied but was always higher than in the wild-type cells. Interestingly, lag1Δlac1Δ cells showed a band that was absent in the wild-type cells (Figure 2A, denoted with an asterisk; see below).
Figure 2.
Synthesis rate of inositol-containing sphingolipids is altered in lag1Δlac1Δ cells. (A) [3H]Inositol labeling. Cells of wild-type (lane 1), lag1Δlac1Δ (lane 2), lag1Δlac1Δ expressing LAG1 (lane 3), lag1Δlac1Δ expressing LAC1 (lane 4), and lag1Δlac1Δ expressing LAG1 and LAC1 (lane 5) were pulse labeled with [3H]inositol for 20 min followed by a chase of 60 min. Further energy-dependent metabolism was stopped by adding NaN3/NaF, and the labeled lipids were extracted and analyzed on HPTLC plates, resolved in solvent A, and subjected to fluorography. (B) [3H]DHS labeling. Cells of wild-type (lane 1) and lag1Δlac1Δ (lane 2) were pulse labeled with [3H]DHS for 2 h, and the labeled lipids were extracted and analyzed on HPTLC plates, resolved in solvent B, and subjected to fluorography. CER, ceramide; DHS, dihydrosphingosine; DHS-1P, DHS-1-phosphate; IPC, inositolphosphorylceramide; MIPC, mannosylinositolphosphorylceramide; M(IP)2C, mannosyldiinositolphosphorylceramide; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PHS, phytosphingosine; PI, phosphatidylinositol; unidentified lipids are marked with an asterisk.
The deficiency of lag1Δlac1Δ cells with respect to inositol incorporation into sphingolipids could be completely corrected when the double deletion strain was complemented with either LAG1 or LAC1 on a centromeric plasmid (Figure 2, lanes 3 and 4). The complementation with both LAG1 and LAC1 (lane 5) did not increase the synthesis of sphingolipids compared with the single complementation experiments. A similar effect was observed for the ability of Lag1p or Lac1p to revert the growth defect of lag1Δlac1Δ (Barz and Walter, 1999) and underlines the redundant functions of Lag1p and Lac1p.
To further analyze sphingolipid synthesis in the lag1Δlac1Δ strain, we repeated the in vivo labeling with [3H]DHS. A similar drastically altered pattern of complex sphingolipids was observed (Figure 2B). Almost no IPC, MIPC, and M(IP)2C were detectable. Moreover, an accumulation of PHS and DHS-1P was observed. It has been shown that exogenous DHS is first phosphorylated upon uptake by the long-chain base kinases Lcb4p and Lcb5p (Nagiec et al., 1998) and then dephosphorylated by Ysr2p or Ysr3p before it can be efficiently converted to ceramide or PHS (Mao et al., 1997; Mandala et al., 1998; see also Figure 1). From our [3H]DHS labeling, the phosphorylation of sphingoid bases and the subsequent dephosphorylation appears to be unaltered because lag1Δlac1Δ cells exhibited wild-type amounts of DHS-1P and accumulated PHS. These labelings suggest that the lag1Δlac1Δ strain is deficient at a step after synthesis or incorporation of sphingoid bases.
Pulse-chase labeling reflects de novo synthesis of sphingolipids. To have an overview of the relative pool of sphingolipids in the lag1Δlac1Δ strain, we decided to study their levels by long-term labeling in order to reach an equilibrium of the label in the cell. The cells were grown in the presence of radiolabeled inositol, serine, DHS, or phosphate for at least six generations. Figure 3 shows a comparison between mild base-treated lipid extracts that were typical for wild-type and lag1Δlac1Δ cells. Consistent with the reduced synthesis rate, the amount of sphingolipids was generally reduced in the deletion strain, although not as dramatically as observed in the pulse-chase experiments. These data suggest that the lag1Δlac1Δ strain can adapt to the reduced rate of sphingolipid synthesis to accumulate a low level of sphingolipids, perhaps by an alternative mechanism. Quantification of phosphate-labeled lipids showed that IPC is reduced to ∼13% and M(IP)2C to ∼75% of the wild-type amount (Figure 3A). MIPC was not quantified, because two extra bands, X1 and X2, which accumulated in the mutant, migrated slightly below MIPC (Figure 3B, lane 2, 4, 6) and were labeled by [32P]phosphate, [3H]inositol, [14C]serine, and [3H]DHS, but not by [3H]mannose (unpublished results). The origin of these lipids as well as of the ones that accumulated in the mutant and migrated between ceramide and DHS (Figure 3B, lane 4, 6; denoted with an asterisk) will be discussed. Deacylated lipid extracts of mutant cells long-term labeled with either [3H]inositol or [14C]serine always showed a reduced labeling in the TLC in the region where IPC/MIPC would be expected, reflecting a block in the sphingolipid biosynthesis pathway. In addition, PHS and DHS seemed to accumulate as well as their phosphorylated counterparts (Figure 3B, lane 4). The presence of free fatty acid in lipid extracts from serine-labeled mutant cells (Figure 3B, lane 4) indicates an accumulation of phosphorylated sphingoid bases, which can be broken down to hexadecanals and phosphoethanolamine by the lyase Dpl1p (Saba et al., 1997; see also Figure 1).
Figure 3.
Comparison of lipid extracts long-term labeled with different precursors of sphingolipids. Wild type (WT) and lag1Δlac1Δ (ΔΔ) were labeled overnight with either (A) [32P]phosphate, or (B) [3H]inositol (lanes 1 and 2), [14C]serine (lanes 3 and 4), [3H]dihydrosphingosine (lanes 5 and 6). Whole cell lipid extracts were prepared, deacylated, and analyzed on HPTLC plates resolved in solvent B. Error bars in B indicate two independent experiments with three determinations each. Abbreviations: see also legend to Figure 2; FA, fatty acid; unidentified lipids are marked with an asterisk. X1 and X2 are previously undescribed alkali-stable lipids, which were present in all labeling experiments. Phosphate-containing sphingolipids in A were quantified with the use of a phosphoimager.
Localization of the Sphingolipid Synthesis Defect in Double Mutant Cells to Acyl-CoA–dependent Ceramide Synthesis
The pulse-chase and long-term labelings suggest that the lag1Δlac1Δ strain is deficient at a step after synthesis of sphingoid bases namely ceramide synthase, transport of ceramide from ER to Golgi, or IPC synthase. The activity of the IPC synthase, encoded by AUR1, was tested in vivo by [3H]inositol labeling in presence of exogeneous C6-ceramide. In these conditions, the lag1Δlac1Δ strain is able to make C6-IPC (unpublished results). This result suggests that the deficient step of the lag1Δlac1Δ strain may be the synthesis of ceramide.
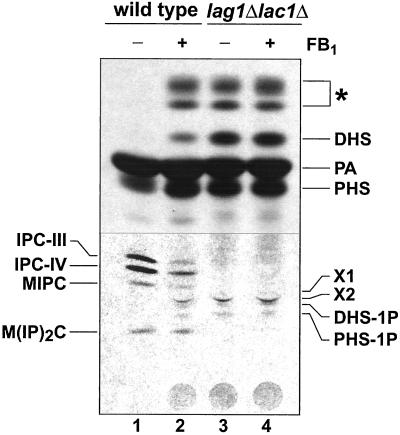
The lag1Δlac1Δ strain is resistant to Fumonisin B1, an inhibitor of the acyl-CoA–dependent ceramide synthesis (unpublished results). This result and our data prompted us to compare the pattern of de novo synthesized sphingolipids of lag1Δlac1Δ and wild-type cells treated with this inhibitor. When cells were pulse-labeled for 60 min with [3H]palmitate, labeled sphingolipids could be recovered from wild-type cells, but not from lag1Δlac1Δ cells (Figure 4). Beside the different sphingoid base species and X1/X2, the mutant cells accumulated some unusual lipids that were not normally found in the wild-type cells (Figure 4; denoted with an asterisk; see DISCUSSION). After a 2-h preincubation with FB1, the same unusual lipids were labeled in wild-type cells. The lipid profile from the mutant cells, in contrast, remained unchanged when the cells were treated with the ceramide synthase inhibitor. These observations strongly suggest that the altered lipid profile present in the lag1Δlac1Δ deletion strain results from a block in the FB1-sensitive ceramide synthase reaction.
Figure 4.
Lipid extracts of lag1Δlac1Δ cells resemble those from FB1 treated wild-type cells. Overnight cultures of wild-type (lanes 1 and 2) and lag1Δlac1Δ (lanes 3 and 4) were resuspended in 1 ml of SC-media to a density of ∼107 cells, preincubated for 120 min in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 100 μM fumonisin B1 (FB1) and subsequently labeled with 5 μCi [3H]palmitate for 60 min. Whole cell lipid extracts were analyzed on HPTLC plates resolved in solvent B. A short exposure of the top of the TLC and a long exposure of the bottom of the TLC plate are shown. Abbreviations: see legend to Figure 3. PA, palmitate. Note that the lipids, which accumulate in the mutant, are also present in FB1 treated wild-type lipid extracts (marked with an asterisk).
The above data suggest that Lag1p and Lac1p are required for ceramide synthesis in vivo via the FB1-sensitive ceramide synthase. Therefore, we decided to measure ceramide synthase more directly with the use of an in vitro assay. In vitro ceramide synthase activity with [3H]DHS as substrate requires a microsomal fraction, ATP (Funato and Riezman, unpublished results), CoA, and cytosol providing a factor different from the sphingoid base kinases (Funato and Riezman, unpublished results). By this approach, wild-type membranes typically synthesized both dihydroceramide and phytoceramide (Figure 5A, lane 3). These two forms of ceramide were quite well separated, with two weak unidentified bands, running slightly below phytoceramide and below dihydroceramide, respectively, when an acidic solvent system was used. The relative contribution of the two forms of ceramide to the overall amount was variable, depending on the batch of membranes. The level of complex sphingolipids made in this assay was very low (unpublished results), allowing us to assume that the amount of ceramide we detect reflects the amount made during the assay, although we cannot rule out ceramide hydrolysis by ceramidases. When the assay was performed in the presence of the potent ceramide synthase inhibitors australifungin (Mandala et al., 1995; lane 1) or FB1 (Wang et al., 1991; lane 2), no bands corresponding to ceramide were detectable, but an unidentified lipid denoted with an asterisk remained unaltered or even accumulated. Therefore, this band is not a product of the FB1-sensitive ceramide synthase. The pattern of the lag1Δlac1Δ strain resembled that of wild-type cells treated with FB1: almost no dihydro- and phytoceramide were detectable. This demonstrates that Lag1p and Lac1p are essential for the FB1-sensitive ceramide synthase reaction.
Recently, Mao and coworkers (Mao et al., 2000a, 2000b) have identified two genes, YPC1 and YDC1, involved in the FB1-resistant and acyl-CoA–independent ceramide synthase. These data prompted us to test the CoA dependence of the LAG1/LAC1-dependent ceramide synthase activity. With the use of the in vitro assay, ceramide synthase activity was tested in the presence or absence of CoA. When CoA was omitted from the assay, wild-type membranes showed a strongly reduced level of ceramide, to levels that were similar to amounts found in the presence of FB1 (Figure 5B, lanes 1–4). On the other hand, the synthesis pattern in the lag1Δlac1Δ double mutant strain was unaffected by CoA (Figure 5B, lanes 5 and 6). These results demonstrate that CoA-dependent ceramide synthesis requires Lag1p and Lac1p and that the low level of ceramide synthesis present in the double mutant cells is CoA-independent.
To further characterize the ceramide synthase activity of Lag1p and Lac1p, we decided to overexpress either one or both proteins in wild-type cells and to measure the level of ceramide in these cells by the in vitro assay. These experiments did not give definite results in the sense that overexpression of these proteins resulted in only a slight increase of the total ceramide level (unpublished results). However, these results are not so surprising. Ceramide is a signaling molecule, so its level has to be tightly regulated. Slight changes of its concentration have been shown to induce cell responses (Perry and Hannun, 1998). For example, when yeast cells are submitted to heat shock, the level of ceramide only increases threefold, whereas other molecules become much more abundant (C20-PHS and C20-DHS increase by 100-fold; Dickson et al., 1997). Alternatively, it may be that LAG1 and LAC1 are not the only genes that need to be overexpressed to increase ceramide synthesis (see DISCUSSION).
It is possible that the defect in ceramide synthesis in the double deletion strain is an indirect consequence of another defect present that is directly due to the deletions. If this is the case, then one would expect to encounter a lag in the appearance of the ceramide synthesis defect in a strain carrying a conditional allele of one of the genes. To begin to address this issue, the lag1Δlac1Δ strain was transformed with a plasmid carrying a temperature-sensitive allele of LAG1 (lag1–1TS). At the permissive temperature (24°C), this strain grows at a wild-type rate, whereas at the nonpermissive temperature (37°C) the growth rate is strongly reduced to a level of the same range as the one of the lag1Δlac1Δ strain (unpublished results). At the permissive temperature the sphingolipid patterns of the lag1Δlac1Δ-plag1–1TS and wild-type strains were similar as judged by in vivo [3H]DHS labeling (Figure 6, lanes 1–4). In contrast, the incorporation of [3H]DHS into IPC was immediately blocked when the cells were shifted at the nonpermissive temperature. Even after a 1-min incubation at 37°C, no IPC synthesis was detectable in our labelings (Figure 6, lanes 5 and 6). These results suggest that LAG1, and most likely LAC1, may be directly involved in ceramide synthesis. However, because it takes a considerable amount of labeling time to obtain enough incorporation into IPC to obtain quantifiable results, we cannot rule out that, in fact, the loss of ceramide synthase activity required a longer time. Interestingly, when the temperature-sensitive cells were pregrown and labeled at the nonpermissive temperature, sphingolipids were slightly detectable as in the double deletion mutant (unpublished results), showing that the strain can adapt to a loss of acyl-CoA–dependent ceramide synthesis.
Overproduction of the Ceramidases Ypc1p and Ydc1p in the lag1Δlac1Δ Strain Partially Restore Growth and Sphingolipid Synthesis
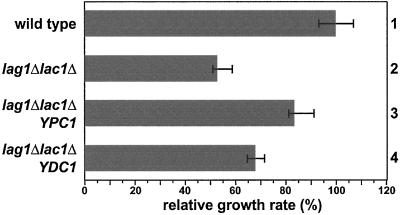
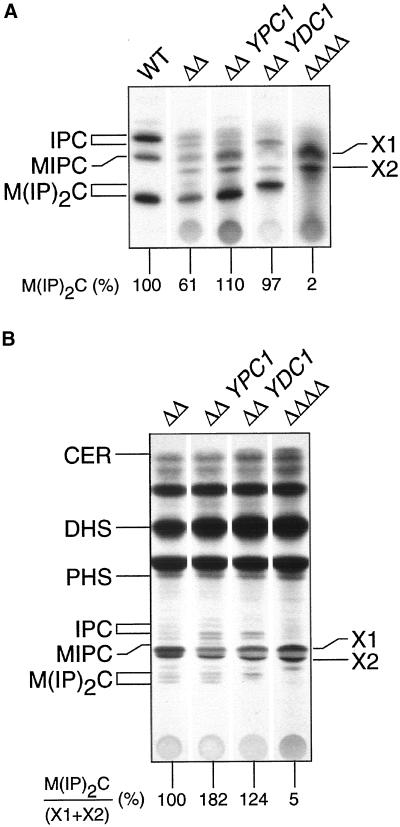
Before starting the investigations on sphingolipid biosynthesis we intended to characterize the function of Lag1p and Lac1p by finding suppressors of the lag1Δlac1Δ deletion phenotype. To identify proteins that function together with Lag1p and Lac1p, a high-copy suppressor screen was performed in the deletion background. With the use of a high-copy genomic library, 135 colonies were selected that carried plasmids suppressing the growth defect of the double deletion strain. One hundred fifteen of these suppressor strains grew at rates identical to wild type and were found by colony PCR to contain plasmids carrying LAG1 or LAC1. The remaining 20 suppressors restored growth to ∼80% of the wild-type rate. The plasmids of 8 of these mutants carried overlapping inserts with 2 previously undescribed genes YBR183w and YBR184w. Subcloning showed that the overexpression of YBR183w alone was sufficient for partial reversion of the growth defect (Figure 7, row 3). PCR with the remaining 12 plasmids revealed that all plasmids had inserts containing YBR183w. The overall amino acid sequence showed no significant homology to Lag1p or Lac1p. However, it was 71% identical to another putative membrane protein that is encoded by the gene YPL087w. Even though YPL087w was not identified in the high-copy suppressor screen, the cloning and overexpression of this gene with the use of the plasmid pWB824 in the lag1Δlac1Δ deletion strain restored growth to ∼65% of the wild-type rate (Figure 7, row 4).YBR183w has been found to encode an alkaline phytoceramidase with reverse action and was named YPC1 (Mao et al., 2000a). YPL087w was reported to encode a dihydroceramidase named Ydc1p (Mao et al., 2000b). Like Ypc1p, Ydc1p exhibits a reverse ceramidase action but to a lower extent than Ypc1p, which explains why we did not detect it in our screening. Consistent with our findings, the authors originally identified YPC1 in a high-copy suppressor screen for plasmids that endowed resistance to FB1. This property was due to the FB1-resistant ceramide synthetic activity of the ceramidases and showed that YPC1 and YDC1 encode CoA-independent and FB1-insensitive ceramide synthase activity. To clarify whether the rescue of the growth defect is a consequence of restoration of sphingolipid biosynthesis, we analyzed the levels of sphingolipids by long-term phosphate labeling in the suppressor strains. Overexpression of YPC1 or YDC1 in lag1Δlac1Δ cells increased the amount of M(IP)2C to at least wild-type levels (Figure 8A), and different forms of this sphingolipid appeared depending on the gene overexpressed. This is consistent with differences in the specificity of the two ceramidases for phytoceramide and dihydroceramide (Mao et al., 1997, 2000b) with the upper band being M(IP)2C with dihydroceramide and the lower band with phytoceramide. Similar observations came from [3H]DHS long-term labeling (Figure 8B). Overexpression of YPC1 or YDC1 in lag1Δlac1Δ cells increased the level of M(IP)2C but to a lesser extent when compared with phosphate labeling. In fact, in the double knock-out background, most of the exogenous DHS was incorporated into the two extra lipids, X1 and X2, perhaps an explanation for this difference. However, the rise in the ratio of M(IP)2C/(X1+X2) highlights the action of YDC1 and YPC1. Therefore, the suppression of the mutant cell growth phenotype most likely results from the introduction of an alternative pathway to make ceramides.
Figure 7.
Overexpression of YPC1 and YDC1 partially rescues the growth defect of lag1Δlac1Δ cells. A high copy suppressor screen was performed to identify suppressors of the lag1Δlac1Δ growth defect as described in MATERIALS AND METHODS. High-copy plasmids encoding YPC1 (row 3) and YDC1 (row 4), respectively, were retransformed into the lag1Δlac1Δ background and the relative growth rate determined in comparison to the wild-type (row 1, set to 100%) and lag1Δlac1Δ (row 2).
Figure 8.
YPC1 and YDC1 control sphingolipid levels in lag1Δlac1Δ mutant cells. Cells of wild-type (WT), lag1Δlac1Δ cells (ΔΔ), lag1Δlac1Δ cells overexpressing YPC1 (ΔΔ YPC1), lag1Δlac1Δ cells overexpressing YDC1 (ΔΔ YDC1), or the quadruple deletion strain lag1Δlac1Δypc1Δydc1Δ (ΔΔΔΔ) were grown overnight in the presence of either (A) [32P]phosphate or (B) [3H]dihydrosphingosine. Whole cell lipid extracts were hydrolyzed with mild base, desalted, and analyzed on HPTLC plates resolved in solvent B. Labeled lipids were quantified with the use of a phosphorimager. Abbreviations: see legend to Figure 3. Note that in lag1Δlac1Δypc1Δydc1Δ strain the sphingolipids IPC and M(IP)2C are completely absent.
No Sphingolipids Are Detectable in the Quadruple lag1Δlac1Δypc1Δydc1Δ Mutant
Because Lag1p and Lac1p appear to be required for the normal production of ceramide via CoA-dependent and FB1-sensitive ceramide synthase, we wondered whether the observed low amounts of sphingolipid-bound ceramide in lag1Δlac1Δ cells were synthesized by the ceramidases, via their reverse activity. We constructed deletion strains of YPC1 and YDC1, respectively, and thus obtained different combinations of all four deletions by mating and sporulation. The double deletion of YPC1 and YDC1 had no obvious defect and a lipid pattern similar to wild type (unpublished results). The quadruple deletion strain was not lethal, but had some additional defects in comparison to the lag1Δlac1Δ double deletion. Although it grew in liquid media with a rate comparable to lag1Δlac1Δ, the formation of single colonies on agar plates was strongly reduced (unpublished results). Surprisingly, investigation of the occurrence of sphingolipids in the lag1Δlac1Δypc1Δydc1Δ strain by in vivo labeling with phosphate and quantification with the use of a phosphorimager revealed a complete absence of M(IP)2C (Figure 8A). Instead, X1 and X2, which migrated exactly like the lipids found in lag1Δlac1Δ cells, were the most prominent bands after mild alkaline hydrolysis. Figure 8B shows base-treated whole cell lipid extracts that were steady state labeled with [3H]DHS. A comparison with the phosphate-labeled extracts (Figure 8A) of the same strains under similar conditions clearly demonstrates the defective incorporation of exogenous DHS into ceramide in the absence of LAG1 and LAC1 and demonstrates the increase in severity of this phenotype when the ceramidase genes are deleted.
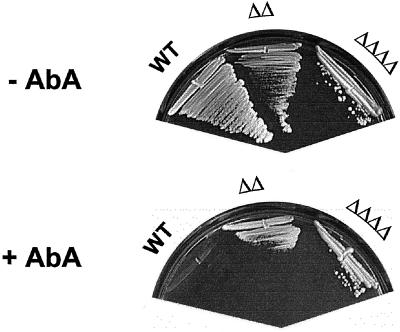
Sphingolipid synthesis has been suggested to be essential because aureobasidin A, an inhibitor of inositolphosphorylceramide synthase, is lethal and because the gene (AUR1) encoding this enzyme is essential (Nagiec et al., 1997). At first, this is difficult to reconcile with our results, especially with the total absence of sphingolipids in the lag1Δlac1Δypc1Δydc1Δ strain. However, an alternative explanation for the toxicity of this compound and the lethality of the aur1Δ mutation could be that the lethality is caused by an accumulation of ceramide rather than by the absence of sphingolipids. To test this, we examined the growth of our mutant strains on YPUAD plates containing ∼5 μg/ml aureobasidin A (Figure 9). Indeed, the double and the quadruple mutant grew at this concentration, but isogenic wild-type cells did not. These data imply that ceramide production derives mainly from the CoA-dependent ceramide synthase activity of Lac1p and Lag1p, whereas the CoA-independent ceramide production of Ypc1p and Ydc1p is only minor and not sufficient to reach a level of ceramide required to inhibit colony formation.
Figure 9.
Aureobasidin-independent growth of lag1Δlac1Δ and lag1Δlac1Δypc1Δydc1Δ cells. Aureobasidin A was spread onto YPUAD plates to a concentration of approximately 5 μg/ml. The designated strains were streaked onto the plates and incubated at 24°C for 2 d. Abbreviations: WT, wild-type; ΔΔ, lag1Δlac1Δ; ΔΔΔΔ, lag1Δlac1Δypc1Δydc1Δ.
These results suggest that the lethality caused by aureobasidin A is due to accumulation of ceramide rather than to the absence of sphingolipids.
DISCUSSION
The major finding of this study is that the homologous ER membrane proteins, Lag1p and Lac1p, are required for the CoA-dependent and FB1-sensitive ceramide synthase reaction. This conclusion is based on several findings. First, the double deletion strain lag1Δlac1Δ has strongly reduced levels of inositol-containing sphingolipids. This biosynthetic defect was shown to occur at the stage of ceramide synthase reaction as judged by in vivo and in vitro labeling with several precursors to sphingolipids. Second, the lipid composition of the lag1Δlac1Δ strain was not influenced by the ceramide synthase inhibitor FB1 and wild-type lipid extracts after treatment with FB1 resemble those of the mutant. Third, the overexpression of genes encoding the ceramidases, Ypc1p or Ydc1p, partially restored growth and sphingolipid synthesis in lag1Δlac1Δ cells. This finding is similar to the observation that overexpression of YPC1 confers FB1 resistance to wild-type cells (Mao et al., 2000a). By their reverse activity, the ceramidases are thus able to suppress both defects in ceramide synthase, one caused by mutation of LAG1 and LAC1 and the other by FB1 treatment. Fourth, the remaining amount of normal sphingolipids in lag1Δlac1Δ cells was completely absent in cells that carry the additional deletions of YPC1 and YDC1. Therefore, the residual ceramide synthesizing activity in the absence of LAG1 and LAC1 is accomplished by the reversal of the action of ceramidases, demonstrating a lack of acyl-CoA–dependent ceramide synthase activity in lag1Δlac1Δ cells. This was also shown more directly by an in vitro assay for CoA dependence of ceramide synthesis with the use of wild-type and mutant membranes.
The analysis of the quadruple deletion strain lag1Δlac1Δypc1Δydc1Δ provides another very interesting finding as this strain apparently can live without making detectable ceramides or normal sphingolipids. This may seem surprising at first because sphingolipids have been reported to be essential for the viability of yeast (Dickson, 1998). However, mutants with a deletion in the LCB1 gene can survive, but not at high temperature, if they carry another suppressor mutation in SLC1, which leads to accumulation of a novel lipid with a long-chain fatty acid, mannose, and inositol (Patton et al., 1992; Nagiec et al., 1993). Similar to the findings with the suppressed lcb1 strain, lag1Δlac1Δ cells can adapt and also synthesized novel lipids, which may be able to substitute for the essential functions of ceramide and normal sphingolipids. There are different possibilities concerning the nature of these lipids. In various experiments, two lipids became apparent in the mutant or FB1-treated wild-type extracts that migrate on TLC slightly below ceramide (Figure 3B, denoted with asterisks). Because of its hydrophobic nature, the upper of these bands could be a fatty acid derivative like methyl-palmitate, from methylation of the palmitate that results from DHS-1P hydrolysis (Zatz et al., 1981; see Figure 1), or like hexadecanal, as a direct product of the sphingoid base phosphate lyase (Saba et al., 1997). As in mammalian cells, the corresponding lipid could also be a derivative of DHS, e.g., N,N-dimethyl-DHS (Igarashi and Hakomori, 1989) or N-acetyl-DHS (Lee et al., 1996). Consequently, the lower of the two bands could result from breakdown or derivation of PHS because it is more polar. Especially dimethyl- or acetyl-DHS (-PHS) appear to be reasonable candidates, as they could serve as the lipid moiety of X1 and X2, which were base-stable, labeled by inositol, phosphate, PA, DHS, and serine, but not mannose (unpublished results), and therefore different from the lipids previously detected in the suppressed LCB1-deleted strain. Alternatively, the labeled lipids could be lyso-PI species with a long-chain fatty acid in an ether or alkyl linkage to the glycerol backbone.
In any event, it should be noted that even although the double, lag1Δlac1Δ, and quadruple, lag1Δlac1Δypc1Δydc1Δ, deletion strains survive with strongly reduced or undetectable synthesis of normal ceramide or sphingolipids, these strains are not healthy. They multiply much slower than wild-type cells and have difficulties in forming colonies from single cells (unpublished results). This suggests that ceramide and sphingolipids are very important, albeit nonessential for viability. Moreover, mutant lag1Δlac1Δ cells also have problems surviving at 37°C or after heat shock (unpublished results). Until now, it was claimed that only the sphingoid bases and their phosphorylated counterparts are required for the full heat shock response in yeast (Mandala et al., 1998; Skrzypek et al., 1999). From our results it is conceivable that ceramide is also necessary for survival under these stress conditions.
Finally, our inability to overproduce ceramide by overexpression of LAG1 and LAC1 could be explained in three ways. First, ceramide production may be tightly regulated and the rate of ceramide accumulation may be controlled independently of ceramide synthase protein levels. This would be consistent with the hypothesis that the lethality due to the deletion of AUR1 or application of aureobasidin A is due to the overproduction of ceramide. Cells that overproduced ceramide because of overproduction of LAG1 and LAC1 would not have grown and would not been recovered. Second, ceramide synthesis requires numerous factors, such as sphingoid bases, C26-fatty acids, CoA, Acyl-CoA binding protein, and these compounds/factors or the enzymes synthesizing them may be rate limiting for ceramide synthesis. In this case, overexpression of ceramide synthase activity would not necessarily increase the amount of ceramide production detectably. Third, LAG1 and LAC1 could encode one functional subunit of ceramide synthase that is required for, but not sufficient for ceramide synthesis. In this case overexpression of the missing subunits would be necessary to increase activity of the enzyme.
The simplest explanation is that these proteins are subunits of the CoA-dependent ceramide synthase. With the use of tagged versions of these proteins, we should be able to purify and characterize the ceramide synthase in order to characterize this important enzyme in greater detail. Finally, during the processing of this article another study has appeared implicating LAG1 and LAC1 in ceramide synthesis (Guillas et al., 2001). This study also provides some further characterization of the novel lipids that accumulate in the lag1Δlac1Δ mutant cells.
ACKNOWLEDGMENTS
We express our esteem and gratitude to R. Wiemeyer for excellent technical assistance, A. Milbradt for help on the suppressor analysis, R.L. Lester for yeast sphingolipid standards, K. Funato for the in vitro ceramide synthesis protocol, and E. Hartmann for the lag1–1TS allele. This work was supported by research fellowships from the Fonds Der Chemischen Industrie (to S.S.), FEBS (to B.V.), and a grant from the Bundesamt für Bildung und Wissenschaft (EC network grant HPRN-CT-2000–00077 on Sphingolipids; to H.R.).
Abbreviations used:
- DHS
dihydrosphingosine
- DHS-1P
DHS-1-phosphate
- ER
endoplasmic reticulum
- FB1
fumonisin B1
- GPI
glycosylphosphatidylinositol
- IPC
inositolphosphorylceramide
- KDHS
keto-DHS
- MIPC
mannosylinositolphosphorylceramide
- M(IP)2C
mannosyldiinositolphosphorylceramide
- PA
palmitic acid
- PHS
phytosphingosine
- PHS-1P
PHS-1-phosphate
- PI
phosphatidylinositol
- SPT
serine:palmitoyl transferase
REFERENCES
- Barz WP, Walter P. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol Biol Cell. 1999;10:1043–59. doi: 10.1091/mbc.10.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker GW, Lester RL. Biosynthesis of phosphoinositol-containing sphingolipids from phosphatidylinositol by a membrane preparation from Saccharomyces cerevisiae. J Bacteriol. 1980;142:747–754. doi: 10.1128/jb.142.3.747-754.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandwagt BF, Mesbah LA, Takken FL, Laurent PL, Kneppers TJ, Hille J, Nijkamp HJ. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc Natl Acad Sci USA. 2000;97:4961–4966. doi: 10.1073/pnas.97.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Culbertson MR, Henry SA. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975;80:23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1438:305–321. doi: 10.1016/s1388-1981(99)00068-2. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids are potential heat stress signals in Saccharomyces. J Biol Chem. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BA, Lester RL. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J Lipid Res. 1980;21:309–315. [PubMed] [Google Scholar]
- Horvath A, Sutterlin C, Manning-Krieg U, Movva NR, Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Hakomori S. Enzymatic synthesis of N,N-dimethyl-sphingosine: demonstration of the sphingosine: N-methyltransferase in mouse brain. Biochem Biophys Res Commun. 1989;164:1411–1416. doi: 10.1016/0006-291x(89)91827-5. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- Krakow JL, Hereld D, Bangs JD, Hart GW, Englund PT. Identification of a glycolipid precursor of the Trypanosoma brucei variant surface glycoprotein. J Biol Chem. 1986;261:12147–12153. [PubMed] [Google Scholar]
- Lee TC, Ou MC, Shinozaki K, Malone B, Snyder F. Biosynthesis of N-acetylsphingosine by platelet-activating factor: sphingosine CoA-independent transacetylase in HL-60 cells. J Biol Chem. 1996;271:209–217. doi: 10.1074/jbc.271.1.209. [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci USA. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala SM, Thornton RA, Frommer BR, Curotto JE, Rozdilsky W, Kurtz MB, Giacobbe RA, Bills GF, Cabello MA, Martin I, Pelaez F, Harris GH. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot (Tokyo) 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- Mao C, Wadleigh M, Jenkins GM, Hannun YA, Obeid LM. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000a;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem. 2000b;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- Mathias S, Pena LA, Kolesnick RN. Signal transduction of stress via ceramide. Biochem J. 1998;335:465–480. doi: 10.1042/bj3350465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell P, Radin NS. Specificity in ceramide biosynthesis from long chain bases and various fatty acyl coenzyme A's by brain microsomes. J Biol Chem. 1970;245:342–350. [PubMed] [Google Scholar]
- Muniz M, Morsomme P, Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Skrzypek M, Nagiec EE, Lester RL, Dickson RC. The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J Biol Chem. 1998;273:19437–19442. doi: 10.1074/jbc.273.31.19437. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- Nasmyth KA, Reed SI. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JL, Srinivasan B, Dickson RC, Lester RL. Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J Bacteriol. 1992;174:7180–7184. doi: 10.1128/jb.174.22.7180-7184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry DK, Hannun YA. The role of ceramide in cell signaling. Biochim Biophys Acta. 1998;1436:233–243. doi: 10.1016/s0005-2760(98)00145-3. [DOI] [PubMed] [Google Scholar]
- Puoti A, Desponds C, Conzelmann A. Biosynthesis of mannosylinositolphosphoceramide in Saccharomyces cerevisiae is dependent on genes controlling the flow of secretory vesicles from the endoplasmic reticulum to the Golgi. J Cell Biol. 1991;113:515–525. doi: 10.1083/jcb.113.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- Salama NR, Yeung T, Schekman RW. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek M, Lester RL, Dickson RC. Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae. J Bacteriol. 1997;179:1513–1520. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Doering TL, Schimmoller F, Schroder S, Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J Biol Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- Zatz M, Dudley PA, Kloog Y, Markey SP. Nonpolar lipid methylation. Biosynthesis of fatty acid methyl esters by rat lung membranes using S-adenosylmethionine. J Biol Chem. 1981;256:10028–10032. [PubMed] [Google Scholar]