Abstract
Measles viruses derived from the live-attenuated Edmonton-B vaccine lineage are currently investigated as novel anti-cancer therapeutics. In this context, tumor specificity and oncolytic potency are key determinants of the therapeutic index. Here, we describe a systematic and comprehensive analysis of a recently developed post-entry targeting strategy based on the incorporation of microRNA target sites (miRTS) into the measles virus genome. We have established viruses with target sites for different microRNA species in the 3′ untranslated regions of either the N, F, H, or L genes and generated viruses harboring microRNA target sites in multiple genes. We report critical importance of target-site positioning with proximal genomic positions effecting maximum vector control. No relevant additional effect of six versus three miRTS copies for the same microRNA species in terms of regulatory efficiency was observed. Moreover, we demonstrate that, depending on the microRNA species, viral mRNAs containing microRNA target sites are directly cleaved and/or translationally repressed in presence of cognate microRNAs. In conclusion, we report highly efficient control of measles virus replication with various miRTS positions for development of safe and efficient cancer virotherapy and provide insights into the mechanisms underlying microRNA-mediated vector control.
Keywords: measles virus, oncolytic viruses, vector engineering, vector safety, microRNAs, microRNA target sites, post-entry targeting
Introduction
Many different classes of viruses with natural and/or engineered specificity for cancer cells have been investigated as novel cancer therapeutics during the last decades.1 In numerous clinical phase I–III trials, virotherapy has proven to be well tolerated, with first signs of clinically relevant anti-tumor efficacy.2, 3, 4 In October 2015, a first-in-class oncolytic herpes simplex virus (talimogene laherparepvec) was approved by the FDA and, shortly after, by the European Medicines Agency (EMA) for treatment of malignant melanoma.5
Due to their unique mechanism of tumor cell killing, oncolytic viruses (OVs) have the potential to expand current multi-modal treatment regimens and may be efficacious in the treatment of chemo- or radiotherapy-resistant cancers.6 Besides the direct lysis of tumor cells, induction of robust and durable anti-tumor immune responses seems to be of major importance for the therapeutic efficacy of OVs.7, 8
Recently, we and others have demonstrated the feasibility of controlling OV tropism by insertion of synthetic target sequences for differentially expressed microRNAs into the viral genome.9, 10, 11, 12 By selecting microRNA species, which are robustly expressed in healthy tissues but whose expression is downregulated in tumor cells, viral replication, and toxicity can be minimized specifically in off-target tissues. In principle, this post-entry (de-)targeting strategy can be adapted to any given virus irrespective of the nature of its nucleic acid or virus structure. This compares favorably to most other strategies (such as transcriptional targeting or employing antibodies as targeting moieties), which are often restricted to certain virus families. Moreover, the small size of microRNA target sites allows for unimpaired viral replication kinetics (in the absence of the respective microRNA) and application in viruses with tightly restricted genome sizes.13 However, direct comparisons of microRNA-controlled versus, e.g., entry-targeted measles viruses (MVs) have not been reported, yet. We have recently shown that a combination of target sites for different microRNAs inserted into a single viral genome allows for protection of multiple sensitive tissues, which is especially important in systemic virotherapy approaches.14
MV is a single-stranded RNA virus with negative polarity and belongs to the family of Paramyxoviridae. It has been successfully engineered for increased tumor cell specificity and enhanced oncolytic potency and is currently under investigation in several phase I clinical trials.15 Its genome is encapsidated by the nucleocapsid (N) protein, giving rise to a helical nucleocapsid which is protected from cleavage by cellular RNases and not accessible to the RNAi machinery. In line with this, we have previously shown that regulation of MV replication by microRNAs is only effective on the mRNA level but not on the genome level.9 Since the transcription rate of MV genes decreases with distance from the genomic leader sequence (“transcriptional gradient”),16, 17 we hypothesized that insertion of miRTS into different genes will result in different levels of microRNA-based vector control. So far, a systematic analysis of possible miRTS insertion positions within the MV genome and the effect of multiple copies of a defined miRTS box is lacking. However, this knowledge is essential to maximize regulation efficiency and to guide the design of microRNA-controlled MVs for clinical translation. Moreover, our knowledge of the mechanisms of microRNA-based MV control is fragmentary thus far. It remains unclear whether mRNAs containing miRTS are predominantly cleaved by RISC (RNA-induced silencing complex) in presence of cognate microRNA or whether translation of these mRNAs is blocked by the RNAi machinery.
In this study, we established and compared different insertion positions for miRTS within the MV genome, more precisely in the 3′ untranslated regions (3′ UTR) of the nucleocapsid (N), fusion (F), hemagglutinin (H), and large (L) genes. Moreover, we investigated strategies to enhance microRNA-mediated attenuation of MV in a given off-target tissue. To this end, miRTS for several different microRNAs enriched in normal brain tissue (miR-124-3p, miR-125b-5p, and miR-7-5p) and MV carrying miRTS boxes for the same microRNA in several positions were analyzed. Finally, we shed light on the different mechanisms of microRNA-mediated attenuation of oncolytic MV.
The development and gradual optimization of genetic modifications enhancing the safety of OVs paves the way for translation of next-generation virotherapeutics with increasing potency. The microRNA-based vector control system presented here adds another layer of safety to OVs while leaving their tumor-cell-killing properties untouched.
Results
Genetic Design of microRNA-Regulated MVs
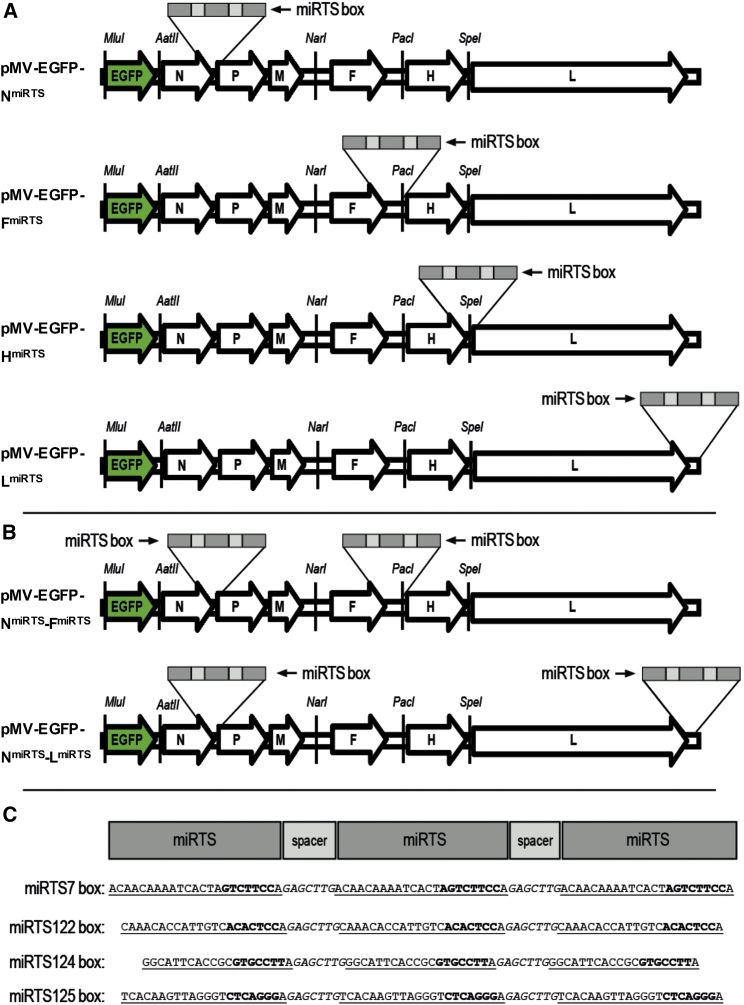
We designed miRTS boxes consisting of three miRTS copies with complete sequence complementarity to the respective cognate mature microRNA (miR-7-5p, miR-122-5p, miR-124-3p, miR-125b-5p; Figure 1C). Identical heptanucleotide spacers were inserted between individual miRTS copies, and additional spacer nucleotides were inserted downstream of the last miRTS copy to ensure an equal length (90 bp) of all miRTS boxes despite different lengths of the mature microRNAs (and thus the respective complementary miRTS). The respective miRTS boxes were inserted into unique restriction sites in the 3′ UTR of the MV N, F, H, or L gene (Figure 1A; see Materials and Methods for details), thereby creating two insertion positions (N and L gene) not reported previously. Additionally, we constructed genomes with miRTS boxes in the 3′ UTRs of two different genes (Figure 1B, N + F or N + L) for analysis of possible additive or synergistic effects on regulation efficiency. Initial experiments were performed with viruses harboring miRTS boxes for miR-7-5p (MV-EGFP-NmiRTS7, -FmiRTS7, -HmiRTS7, -LmiRTS7, -NmiRTS7-FmiRTS7, and -NmiRTS7-LmiRTS7) and verified by using analogous viruses with miRTS boxes for miR-122-5p, miR-124-3p, and miR125b-5p. As described previously,9 we also generated and analyzed respective control viruses with reverse complementary (rc) miRTS boxes, which were invariably unresponsive to the presence of cognate microRNAs (data not shown). All viruses were successfully cloned, rescued, and propagated to high titer stock solutions.
Figure 1.
Genome Structure of Recombinant Measles Viruses
(A) Genome structure of engineered viruses encoding EGFP (enhanced GFP) in an additional transcription unit (ATU) upstream of the N gene as well as a synthetic microRNA target site (miRTS) box in the 3′ UTR of the N, F, H, or L gene. (B) Genome structure of engineered viruses encoding EGFP (enhanced GFP) in an additional transcription unit (ATU) upstream of the N gene as well as synthetic miRTS boxes in the 3′ UTRs of the N and F or N and L genes. (C) Sequences of four different miRTS boxes complementary to miR-7-5p, miR-122-5p, miR-124-3p, and miR-125b-5p, respectively. miRTS are underlined and separated by nonunderlined spacer nucleotides. The miRTS portions that are complementary to the microRNA seed sequences are indicated in bold print.
miRTS Box Insertion Does Not Affect Viral Replication Kinetics
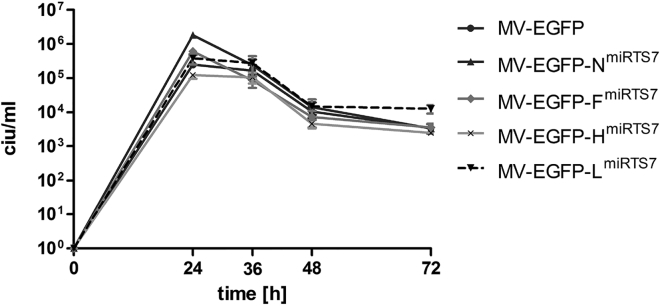
In order to determine possible effects of miRTS box insertion on viral replication kinetics, we performed one-step growth curves on Vero producer cells. As can be seen in Figure 2, all microRNA-controlled viruses (MV-EGFP-NmiRTS7/-FmiRTS7/-HmiRTS7/-LmiRTS7) showed replication kinetics similar to the parental laboratory strain (MV-EGFP). During propagation, titers exceeding 107 cell infectious units per mL (ciu/mL) were achieved for all viruses. Thus, miRTS box insertion into the MV N, F, H, or L gene does not interfere significantly with replication kinetics of MV in Vero cells.
Figure 2.
Replication Kinetics of MV-EGFP-NmiRTS7/-FmiRTS7/-HmiRTS7/-LmiRTS7 in Vero Cells
One-step growth curves of MV-EGFP-NmiRTS7/-FmiRTS7/-HmiRTS7/-LmiRTS7 and MV-EGFP control virus in the producer cell line Vero. Cells were infected at an MOI of 3 and scraped into their medium at 24, 36, 48, 72, and 96 hr post-infection to determine virus progeny titers in cell infectious units (ciu). Error bars represent standard deviation of three technical replicates per sample.
Functional Analysis and Comparison of Different miRTS Box Positions
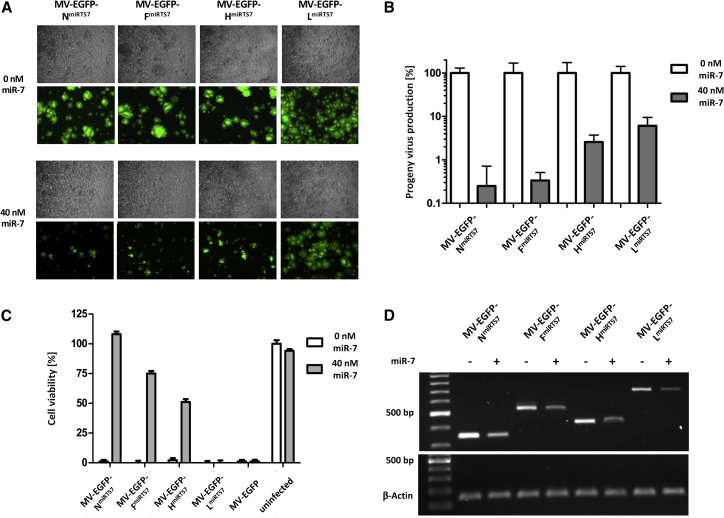
To determine positional effects of miRTS box insertion, we analyzed the responsiveness of recombinant MVs harboring miRTS for miR-7-5p in the 3′ UTR of the N, F, H, or L gene (Figure 1A) toward the presence of cognate miR-7-5p. As shown in Figure 3A, spread of all viruses is attenuated in presence of miR-7-5p, albeit to different degrees. On these fluorescence microscopy images taken at an early time point after infection (34 hr) attenuation of MV-EGFP-NmiRTS7, MV-EGFP-FmiRTS7, and MV-EGFP-HmiRTS7 is pronounced, with a tendency toward the strongest effect for MV-EGFP-NmiRTS7 and MV-EGFP-FmiRTS7. In contrast, MV-EGFP-LmiRTS7 shows only a minor level of attenuation in presence of miR-7-5p.
Figure 3.
Functional Analysis and Comparison of Different miRTS Box Positions
Vero cells were transfected with 0 or 40 nM miR-7-5p mimics and subsequently infected with MV-EGFP-NmiRTS7/-FmiRTS7/-HmiRTS7/-LmiRTS7 or MV-EGFP at an MOI of 0.03. (A) Fluorescence and phase contrast microscopy images 34 hr post-infection, ×50 magnification. (B) Progeny virus determination. Thirty-six hours post-infection, cells were scraped into their medium. Virus progeny titers were determined and are shown with normalization to progeny virus titers in absence of transfected microRNA. Error bars represent SD of three technical replicates per sample. (C) Cell viability of all samples was determined at 96 hr post-infection using a colorimetric (XTT) assay. Error bars represent SD of three technical replicates per sample. (D) Total RNA was isolated 32 hr post infection, subjected to RT-PCR, and cDNA was used for PCR. Gene-specific primer pairs corresponding to regions up- and downstream of the miRTS box insertion sites within the N, F, H, and L ORF, respectively, were used (upper gel), while β-actin-specific primers were used as an input control (lower gel). RT-PCR products were subjected to agarose gel electrophoresis.
In order to quantify these results, progeny production of all viruses was determined in presence or absence of miR-7-5p (Figure 3B). In line with the fluorescence microscopy data, attenuation is most pronounced for MV-EGFP-NmiRTS7 (∼400-fold) and MV-EGFP-FmiRTS7 (∼300-fold), whereas it is weakest for MV-EGFP-LmiRTS7 (∼16-fold). MV-EGFP-HmiRTS7 (∼40-fold) shows an intermediate phenotype.
Next, cell viability of Vero cells infected with the different viruses in presence or absence of the corresponding microRNA was determined. Cell viability assays (XTT) were performed at 70 hr (data not shown) and 98 hr (Figure 3C) post-infection (p.i.). At the earlier time point, attenuation of MV-EGFP-NmiRTS7, MV-EGFP-FmiRTS7, and MV-EGFP-HmiRTS7 was uniformly strong in presence of miR-7-5p, with remaining cell viability of ∼90% compared to uninfected cells. In contrast, viability of Vero cells infected with MV-EGFP-LmiRTS7 was already reduced to ∼35% despite the presence of miR-7-5p. When assessing cell viability 90 hr p.i. (Figure 3C), more subtle differences in the regulation efficiency of the different miRTS box positions became evident. Viability of cells infected with the respective viruses decreased with increasing distance of the miRTS box to the genomic leader sequence.
Gene-specific RT-PCR of viral mRNAs harboring miRTS boxes revealed reduced N, F, H, and L mRNA levels in cells transfected with miR-7-5p mimics for MV-EGFP-NmiRTS7, MV-EGFP-FmiRTS7, MV-EGFP-HmiRTS7, and MV-EGFP-LmiRTS7, respectively (Figure 3D). This indicates that degradation of miRTS box-modified viral mRNAs takes place and contributes to reduced viral titers and cytotoxicity in presence of cognate microRNA.
Regulation Efficiency of Single versus Double miRTS Boxes
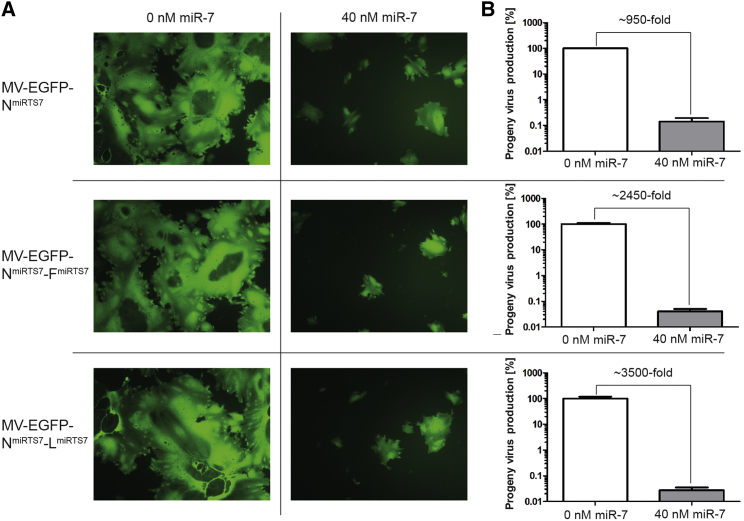
Apart from optimizing the position for miRTS box insertion, increasing the number of miRTS and targeting of multiple viral genes are conceivable ways to enhance the efficiency of microRNA-based virus control. We therefore doubled the amount of functional miRTS in MV-EGFP-NmiRTS7 from three to six (one miRTS box contains three microRNA target sites; Figures 1B and 1C) by insertion of an additional miRTS box within the F- or L-3′ UTR, thereby generating MV-EGFP-NmiRTS7-FmiRTS7 and MV-EGFP-NmiRTS7-LmiRTS7. Moreover, this strategy simultaneously targets two instead of one of the six MV genes. Figure 4 shows viral spread and progeny titers of MV-EGFP-NmiRTS7, MV-EGFP-NmiRTS7-FmiRTS7, and MV-EGFP-NmiRTS7-LmiRTS7 in presence or absence of miR-7-5p. Reduction of viral spread in presence of miR-7-5p was comparable for all viruses (Figure 4A), and progeny virus titers were reduced ∼950-, ∼2,450-, and ∼3,500-fold for MV-EGFP-NmiRTS7, MV-EGFP-NmiRTS7-FmiRTS7, and MV-EGFP-NmiRTS7-LmiRTS7, respectively (Figure 4B).
Figure 4.
Regulation Efficiency of Single versus Double miRTS Box Viruses
Vero cells were transfected with 0 or 40 nM microRNA mimics and subsequently infected with MV-EGFP-NmiRTS7, MV-EGFP-NmiRTS7-FmiRTS7, or MV-EGFP-NmiRTS7-LmiRTS7 at an MOI of 0.03. (A) Forty-one hours post-infection, fluorescence microscopy images were acquired (×100 magnification). (B) Forty-two hours post-infection, cells were scraped into their medium. Virus progeny titers were determined and are shown with normalization to progeny virus titers in absence of transfected microRNA. Error bars represent SD of three technical replicates per sample.
Generation and Characterization of Different Brain-Detargeted Viruses
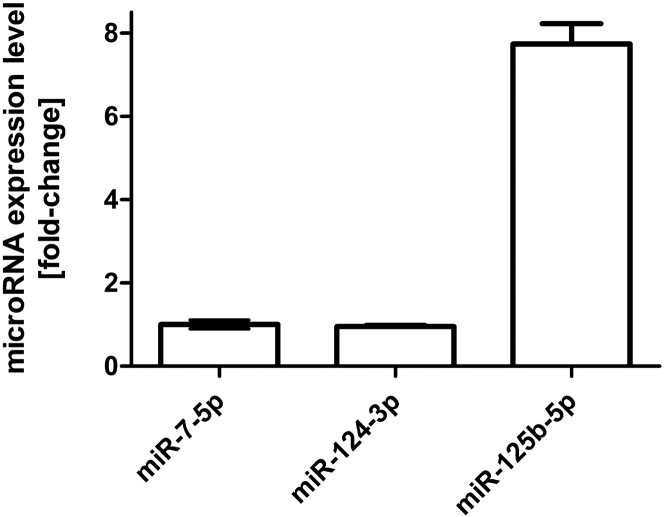
In order to explore microRNA candidates alternative to miR-7-5p that can be exploited for virus attenuation in healthy brain tissue, we analyzed the expression levels of three microRNAs known to be present in human brain tissue.18 The results of qPCR-based relative quantification of microRNA levels in total RNA isolated from human brain tissue are depicted in Figure 5. MiR-7-5p and miR-124-3p are present in similar amounts. In contrast, we determined an ∼8-fold higher expression level of miR-125b-5p compared to miR-7-5p or miR-124-3p.
Figure 5.
MicroRNA Expression Levels in Human Brain
Expression levels of miR-124-3p and miR-125b-5p were determined by qRT-PCR in commercially available human brain total RNA relative to the expression level of miR-7-5p. MicroRNA levels were quantified using a serial dilution standard curve, and fold difference was calculated relative to miR-7-5p. Error bars represent SD of three technical replicates per sample.
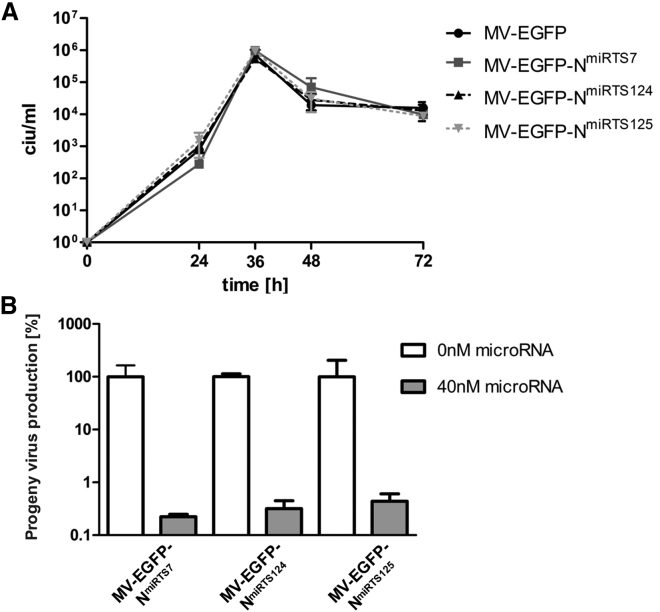
Having verified the expression of miR-124-3p and miR-125b-5p in healthy human brain tissue, we cloned and propagated MV-EGFP-NmiRTS124 and -NmiRTS125 harboring a miRTS box for miR-124-3p or miR-125b-5p, respectively, in the 3′ UTR of the N gene (analogous to MV-EGFP-NmiRTS7; see Figures 1A and 1C). During propagation, titers exceeding 107 ciu/mL were achieved for all viruses. Figure 6A shows the replication kinetics of these viruses compared to MV-EGFP and MV-EGFP-NmiRTS7. All viruses showed similar replication kinetics and replicated to maximum titers of ∼106 ciu/mL.
Figure 6.
Characterization of MV-EGFP-NmiRTS124 and -NmiRTS125
(A) One-step growth curves of MV-EGFP-NmiRTS7/-NmiRTS124/-NmiRTS125 and MV-EGFP control virus in Vero cells. Infection was performed at an MOI of 3. Cells were scraped into their medium at 24, 36, 48, and 72 hr post-infection, and virus progeny titers were determined. Error bars represent SD of three technical replicates per sample. (B) Progeny virus production in the absence or presence of cognate microRNAs. Thirty-eight hours post-infection, cells were scraped in their medium. Virus progeny titers were determined and are shown with normalization to progeny virus titers in absence of transfected microRNA. Error bars represent SD of three technical replicates per sample.
Next, we assessed the level of virus attenuation in presence of 40 nM of the cognate microRNA for all three brain-detargeted viruses. As depicted in Figure 6B, miR-7-5p, miR-124-3p, or miR-125b-5p strongly attenuated MV carrying the respective miRTS box in the N gene 3′ UTR (448-, 315-, and 229-fold reduction in virus progeny production, respectively).
Mechanism of microRNA-Based Vector Control
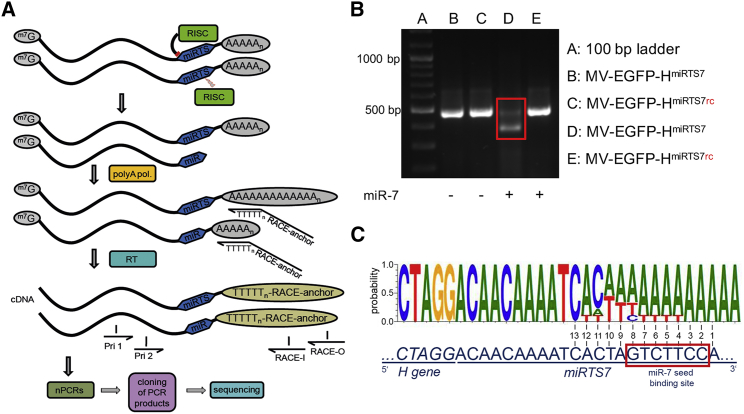
To investigate the mechanistic basis of microRNA-mediated vector control, we amplified and sequenced the 3′ ends of viral mRNAs carrying miRTS boxes using a 3′ RACE (rapid amplification of cDNA ends) approach, which is illustrated in Figure 7A. Vero cells were transfected with 0 or 40 nM miR-7-5p and subsequently infected with miR-7-5p-sensitive MV-EGFP-HmiRTS7 or miR-7-5p-insensitive control virus (MV-EGFP-HmiRTS7rc) harboring a reverse-complementary target site (rc). After isolation of total RNA, a 3′ RACE was performed. Figure 7B shows the gel analysis of the second reaction of a nested PCR (compare Figure 7A). The expected amplicon of ∼480 bp corresponding to the 3′ region of the MV H mRNA was amplified in all samples. However, the band intensity of the ∼480 bp band is reduced specifically in case of MV-EGFP-HmiRTS7 in presence of miR-7-5p. Moreover, under these conditions, a second and more intensive band of ∼380 bp becomes visible. This corresponds well to the estimated size of an amplicon resulting from HmiRTS7 mRNA cleaved within the first miRTS7.
Figure 7.
Mechanism of microRNA-Based Vector Control
To elucidate the mechanism of microRNA-based vector control, a 3′ RACE (rapid amplification of cDNA ends) followed by sequencing approach was applied. Vero cells were transfected with miR-7-5p mimics and subsequently infected with MV-EGFP-HmiRTS7 or MV-EGFP-HmiRTS7rc at an MOI of 0.03. Thirty-five hours p.i., total RNA was isolated. (A) Schematic depiction of the RACE procedure. Total RNA was poly(A) tailed, and cDNA synthesis was performed using a RACE anchor primer. The RACE anchor primer contains a poly-T sequence complementary to the poly(A) tail of poly(A)-tailed RNAs, which is preceded by a nucleotide other than T (V) in order to position the primer at the beginning of the poly(A) sequence of the template. Two reactions of a nested PCR with primers complementary to regions upstream of the miRTS box (Pri 1, Pri 2) and regions within the RACE anchor sequence (RACE-I, RACE-O) were performed. Products of the second reaction of the nested PCR were then subjected to gel electrophoresis and bands containing PCR products of interest were cloned for subsequent Sanger sequencing. (B) Gel electrophoresis image showing products of the second reaction of the nested PCR. PCR products from samples transfected with miR-7-5p and infected with MV-EGFP-HmiRTS7 (red box) were gel purified, cloned, and subjected to sequencing. (C) Sequencing result of cloned RACE-PCR fragments from H mRNA containing a miRTS7 box in presence of miR-7-5p. The upper sequence is the consensus cleavage sequence of the nine RACE-PCR fragments that showed cleavage within the first miRTS. The lower sequence shows the uncleaved sequence of the miRTS for miR-7-5p for comparison. Nucleotides are numbered from 3′ to 5′ starting at the 3′ end of the first microRNA target sequence with the miR-7-5p seed sequence indicated by a red box.
PCR products indicated in Figure 7B (red box; resulting from 3′ RACE of MV-EGFP-HmiRTS7 in presence of miR-7-5p) were cloned and sequenced. Of 24 clones, 11 (46%) were uncleaved, full-length clones and 13 (54%) were of shorter sequence length. Of the 13 shorter clones, 10 (77%) were cleaved within miRTS (nine within the first and one within the second miRTS). Three (23%) clones were cleaved upstream of the first miRTS, probably due to degradation by exonucleases after initial RISC cleavage. Figure S1 shows the distribution and frequency of individual cleavage sites within the cloned and sequenced MV-HmiRTS7 mRNAs (harboring the miRTS box for miR-7-5p) in presence of miR-7-5p. Cleavage within the first miRTS (Figure 7C) preferentially occurred between nucleotides 8 and 11 upstream of the 3′ end of the miRTS, which corresponds to the microRNA region immediately 3′ of the seed sequence. This is evidenced in Figure 7C by the increasing probability of divergent adenines at the 3′ end of the PCR product consensus sequence beginning from position 11 representing the addition of the poly(A) tail during the RACE procedure. To verify these results, we analyzed two additional viruses, MV-EGFP-HmiRTS122 and MV-EGFP-HmiRTS124. Both viruses were attenuated comparably to MV-EGFP-HmiRTS7 in presence of cognate microRNA (data not shown). For MV-EGFP-HmiRTS122, we found that cleavage occurred at the same position within the miRTS box, albeit at a lower frequency when compared to MV-EGFP-HmiRTS7. For MV-EGFP-HmiRTS124, we detected a higher percentage of cleaved mRNA molecules in presence of cognate miR-124-3p. Here, 6 of 26 sequenced clones were full-length (23%), and 20 of 26 (77%) clones were shorter in length, indicating cleavage and degradation. Of those, eight (40%) were cleaved within the first miRTS, one (5%) in the spacer nucleotides between the first and the second miRTS, and 11 (55%) within the H open reading frame (ORF), indicating cleavage and subsequent degradation by exonucleases. The preferred cleavage site within the miRTS was between nucleotides 10 and 11 upstream of the 3′ end of the miRTS (Figure 7C).
Discussion
In this work, we have systematically analyzed different insertion positions for miRTS boxes within the MV genome. miRTS boxes were successfully inserted into the 3′ UTRs of the MV N, F, H, and L genes. The resulting viruses could be propagated to high titers and showed similar replication kinetics. We demonstrated that proximal genomic positions of miRTS boxes are generally more efficient in attenuating virus replication in presence of cognate microRNAs. Interestingly, the decrease in regulation efficiency toward distal genomic positions is paralleled by the decreasing mRNA expression levels toward more distal genomic positions, which is a common theme in the order of Mononegavirales.19 Very high levels of the MV N protein (most proximal MV gene with respect to the viral leader promoter) are needed to encapsidate nascent viral RNA genomes (2,649 copies of the N protein per unmodified genome copy). Accumulation of a sufficient number of N proteins in the infected cell has therefore been proposed to be required for the viral polymerase complex to switch from gene transcription to genome replication.20 This may explain the profound effect on virus replication associated with miRTS box-mediated silencing of the N gene. In contrast, miRTS box-mediated reduction of the viral L protein levels seems to be less detrimental for MV replication. The L gene (most distal MV gene) encodes the catalytic component of the viral polymerase complex and is a priori transcribed at much lower levels than the N gene.16, 17 Our data allow the interpretation that the MV polymerase is present in excess upon infection, and the L protein can therefore be reduced without major slowing of viral replication kinetics, even under experimental conditions allowing for multiple rounds of replication. In contrast, the N, F, and H proteins are structural components of the viral envelope or nucleocapsid, respectively, and thus need to be present at much higher levels.
In an attempt to further increase the attenuation of microRNA-sensitive MV in healthy tissues, we inserted one miRTS box each into the 3′ UTR of the N and F or N and L genes, thereby doubling the number of miRTS (three versus six copies). We hypothesized that the combination of the two most efficient positions (N and F) may result in superior attenuation by the cognate microRNA. With the combination of the N and the L position, we aimed at further reduction of the N protein via microRNA-based reduction of the L protein, which in turn reduces production of all other viral proteins due to its catalytic polymerase function. However, we only obtained 2- to 3-fold differences in regulation efficiency in presence of cognate microRNAs when comparing single- and double-miRTS box viruses, which is within the methodologically inherent variability of the progeny virus quantification assay. In the scenario of efficient suppression of the N gene, all other MV genes should be expressed in excess. Our data suggests that additional suppression of the F or L gene does not reduce expression of these genes to a level that further diminishes MV replication. Another possible explanation could be that the number of available miRTS saturates the RNAi machinery. Even though microRNA-based virus control was not relevantly increased in double-miRTS box viruses, applying this strategy further decreases the likelihood of escape mutants and thereby strengthens the safety profile of MV for clinical application. Additionally, miRTS boxes for several different microRNAs can easily be combined, as we have demonstrated previously.14
Besides the importance of miRTS box positioning, the microRNA species exploited for detargeting is of major significance. Even in equal concentrations, some microRNAs seem to be more potent in silencing viral replication than others.14 Additionally, the “ideal” microRNA for OV control would have a low-to-absent expression in tumor cells and an invariably high expression in healthy tissues. Here, we generated brain-detargeted viruses harboring miR-7-5p, miR-124-3p, or miR-125b-5p target sites and demonstrated unaltered replication kinetics in virus producer cells. All viruses were similarly and strongly attenuated in presence of the respective cognate microRNA. We verified expression of each microRNA in total RNA from nontransformed human brain tissue and found the highest expression levels for miR-125b-5p. As we have previously observed that higher levels of a given microRNA are generally more efficient in attenuating replication of viruses with cognate miRTS (data not shown), we hypothesize that miR-125b-5p may be a highly effective candidate. However, it should be noted that expression levels may vary between individuals and anatomical sites of the brain.21 MiR-125b-5p could be exploited in direct or systemic treatment of various tumor entities, including chronic lymphocytic leukemia, breast cancer, and melanoma, where it is downregulated.22, 23 MiR-7-5p and miR-124-3p are also highly expressed in healthy brain tissue and—in contrast to miR-125b-5p—have been reported to be downregulated in glioblastoma.24, 25, 26, 27 Therefore, miR-7-5p and miR-124-3p expression profiles render them excellent candidates in the context of glioblastoma virotherapy. However, despite similar expression levels in the analyzed total brain RNA, the distribution of candidate microRNAs within distinct anatomical sites of the brain may differ and should be taken into consideration for microRNA selection.
The mechanism of microRNA-based vector control appears to be complex and dependent on more than complementarity between microRNA and miRTS. MicroRNA-targeted mRNAs are either degraded after direct cleavage by Ago proteins of the RISC or their translation into proteins is suppressed.28, 29 In the RACE followed by sequencing analysis using a virus harboring a miRTS box for miR-7-5p, we found cleavage to be the preferential mechanism of silencing. However, it should be noted that direct cleavage and translational repression may not be mutually exclusive but rather act collaboratively to suppress targeted mRNA expression. When we analyzed two additional viruses harboring miRTS boxes for miR-122-5p or miR-124-3p, the rate of cleaved sequences detected differed from the miR-7-5p experiments. As the number of clones assessable by our strategy is very limited, sampling error may be relevant; future studies should therefore include next-generation sequencing to further address this issue. The fact that the viruses were equally attenuated in presence of equimolar concentrations of their cognate microRNAs (data not shown) argues for a varying contribution of direct cleavage and translational repression. Therefore, attempting to predict regulation capability of a certain microRNA based on its ability to mediate cleavage of its complementary miRTS likely falls short. Differences in polyadenylation efficiency of different mRNAs during the RACE procedure might impact these results. However, it has been described that the E. coli poly(A) polymerase used in the presented RACE experiments does not exhibit substantial specificity with regard to the sequence of its RNA substrate.30 Furthermore, slight differences in experimental conditions, the nucleotide sequence of the microRNA itself, or the local sequence context of the miRTS box may influence the mechanism of target mRNA silencing. In our RACE experiments, which represent a snapshot in time, we detected a certain amount of cleaved but nondegraded viral mRNAs by sequencing. This might be surprising since it is generally believed that mRNAs, once cleaved, are rapidly degraded by cytosolic nucleases. Possibly, the fact that we did not find a higher percentage of cleaved viral RNAs despite the substantial downregulation of virus progeny production is indeed due to rapid degradation of cleaved viral RNAs.
In the thriving field of oncolytic virotherapy, the focus is currently shifting from pre-clinical vector engineering to the clinical application of next-generation OVs with increased potency. Increasingly higher doses and combination with established therapy regimens such as chemo- or immunotherapy will increase the likelihood of off-target toxicity. The enhanced microRNA (de-)targeting system presented here may add to the therapeutic index of MV in clinical translation by reducing the probability of off-target replication without sacrificing oncolytic potency. We established two novel insertion positions for miRTS boxes in the MV genome (N and L 3′ UTR) and demonstrated greatest efficiency of microRNA-mediated vector control through miRTS box insertion in leader-proximal genes. The mechanism of microRNA-mediated virus attenuation appears to be multifaceted with varying contributions of mRNA cleavage and other suppressive effects depending on the respective microRNA species exploited for detargeting and warrants further investigation.
Materials and Methods
Cell Culture
Vero (African green monkey kidney) cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Vero cells were cultured in DMEM (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal calf serum (DMEM+). Cells were routinely cultured at 37°C in a humidified atmosphere of 5% CO2. Cell lines were tested negative for mycoplasma contamination using VenorGeM Mycoplasma Detection Kit (Minerva Biolabs, Berlin, Germany).
Recombinant MVs
Custom-designed, complementary DNA oligonucleotides encoding the respective miRTS-boxes (reference microRNA sequences of hsa-miR-7-5p, hsa-miR-122-5p, hsa-miR-124-3p, hsa-miR-125b-5p according to http://www.mirbase.org) or nonresponsive, rc control boxes (rc-boxes) flanked by overhanging ends compatible for insertion into the 3′ UTR of the N, F, H, or L genes were obtained from Eurofins Genomics, Ebersberg, Germany. The sequences of the functional miRTS boxes are shown in Figure 1C. In brief, a cDNA expression plasmid encoding the MV genome (pMV)-EGFP-NmiRTS7 was generated by subcloning the MluI-SbfI fragment from the parental pMV-EGFP31 into pUC29. Via site-directed mutagenesis (QuikChange Lightning kit, Agilent, Santa Clara, USA), a unique AvrII restriction site was inserted into the 3′ UTR of the N gene (forward mutagenesis primer, 5′-AGGTGCGAGAGGCCtAGGACCAGAACAAC-3′; reverse mutagenesis primer, 5′-GTTGTTCTGGTCCTaGGCCTCTCGCACCT-3′). Hybridized complementary DNA oligonucleotides (encoding the miRTS box for miR-7-5p with compatible ends) were cloned into the AvrII restriction site (5′-CTAGGACAACAAAATCACTAGTCTTCCAGAGCTTGACAACAAAATCACTAGTCTTCCAGAGCTTGACAACAAAATCACTAGTCTTCCAGA-3′; 5′- CTAGTCTGGAAGACTAGTGATTTTGTTGTCAAGCTCTGGAAGACTAGTGATTTTGTTGTCAAGCTCTGGAAGACTAGTGATTTTGTTGTC-3′). After insertion of the miRTS7 box, the introduced AvrII site was reverted to its original sequence by a second round of site-directed mutagenesis (forward mutagenesis primer, 5′-TGATTTTGTTGTCCTcGGCCTCTCGCACCTA-3′; reverse mutagenesis primer, 5′- TAGGTGCGAGAGGCCgAGGACAACAAAATCA-3′). Finally, the modified N gene was excised with MluI and SbfI and inserted into the pMV-EGFP vector digested with MluI and SbfI to generate pMV-EGFP-NmiRTS7. pMV-EGFP-FmiRTS7 and -HmiRTS7 were generated by insertion of custom-designed, complementary DNA oligonucleotides encoding the miRTS box for miR-7-5p (with compatible ends) into the 3′ UTR of F via PacI or of H via SpeI as described before.9, 14 To generate pMV-EGFP-LmiRTS7, the 3′ portion of the L gene, including its 3′ UTR, was amplified by high-fidelity PCR (Phusion polymerase, New England Biolabs, Frankfurt am Main, Germany) using primers that introduce artificial BamHI and SalI restriction sites for subcloning into the pUC19 vector (New England Biolabs) (forward primer, 5′-TTTTGGATCCGGCCTTGTCGAACACAGAATGG-3′; reverse primer, 5′-TTTTGTCGACTATGACCATGATTACGCCAAGC-3′). After subcloning, a unique SpeI restriction site was introduced by changing the L stop codon (underlined) from ACTAAT into ACTAGT (forward mutagenesis primer, 5′- CCCTGATTAAGGACTAgTTGGTTGAACTCCGGA-3′; reverse mutagenesis primer, 5′- TCCGGAGTTCAACCAAcTAGTCCTTAATCAGGG-3′). After insertion of the miRTS box for miR-7-5p into the SpeI site, the introduced mutation was reverted to its original sequence via site-directed mutagenesis (forward mutagenesis primer, 5′-CTAGTCTTCCAGACTAaTTGGTTGAACTCCGGA-3′; reverse mutagenesis primer, 5′-TCCGGAGTTCAACCAAtTAGTCTGGAAGACTAG-3′). Finally, the modified portion of the L gene was excised with PmlI and EagI and inserted into the pMV-EGFP vector digested with PmlI and EagI to generate pMV-EGFP-LmiRTS7. Recombinant MV genomic plasmids encoding miRTS boxes for miR-122-5p, miR-124-3p, or miR-125b-5p were generated analogously.
For construction of genomic plasmids containing two miRTS boxes, the modified N gene from pMV-EGFP-NmiRTS7 was excised with MluI and SbfI and exchanged in the recipient plasmids pMV-EGFP-FmiRTS7 and pMV-EGFP-LmiRTS7 digested with MluI and SbfI to generate pMV-EGFP-NmiRTS7-FmiRTS7 and pMV-EGFP-NmiRTS7-LmiRTS7, respectively.
MV genomic plasmids containing miRTS boxes as well as the parental pMV-EGFP were used to generate (rescue) the viruses MV-EGFP, MV-EGFP-NmiRTS7/-FmiRTS7/-HmiRTS7/-LmiRTS7, MV-EGFP-NmiRTS122/-FmiRTS122/-HmiRTS122/-LmiRTS122, MV-EGFP-NmiRTS7-FmiRTS7, MV-EGFP-NmiRTS7-LmiRTS7, MV-EGFP-NmiRTS124, MV-EGFP-HmiRTS124, and MV-EGFP-NmiRTS125 following the procedure described by Radecke et al.32 and Martin et al.33 with slight modifications. All viruses were propagated on Vero cells, and virus stocks from the second or third passage were used for all experiments.
Virus Titration
Virus titrations were carried out in octuplicates in 96-well plates. Virus suspensions were serially diluted 1:10 in 100 μL DMEM+ per well. Vero cells (1.5 × 104 cells/well) were added to each well. Forty-eight hours p.i., syncytia were counted at an appropriate dilution step, and titers were calculated as cell infectious units/mL (ciu/mL). To determine the titers of viral stocks, aliquots were thawed, vortexed briefly, and directly used for titration. To determine virus progeny titers, infected cells were scraped in their medium, frozen in liquid nitrogen, and stored at −80°C. After thawing, samples were vortexed vigorously and centrifuged at 6,000 × g for 5 min (4°C), and the supernatants were used for titration.
Transfection of microRNA Mimics
Vero cells were seeded in 6-well plates at a density of 3 × 105 cells/well and incubated overnight for attachment. MicroRNA mimics (mirVana miRNA Mimics, Life Technologies) were pre-diluted in Opti-MEM (Life Technologies) to achieve a final concentration of 40 nM and were transfected using 8 μL of Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer’s protocol.
Infection Experiments
Assessment of viral spread and replication in presence or absence of exogenous microRNAs was performed as described previously.14 In brief, Vero cells were transfected with microRNA mimics (miR-7-5p/-122-5p/-124-3p/-125b-5p) as described above or mock transfected (Opti-MEM + Lipofectamine 2000 only) in triplicates. Six hours post-transfection, cells were infected at an MOI of 0.03 by aspirating the culture medium and adding the respective viruses diluted in Opti-MEM or adding Opti-MEM only (mock). Viruses were allowed to adsorb to the cells in the incubator for 2 hr before inocula were removed, cells were washed, and standard culture medium was added.
To determine virus progeny production, infected cells were scraped in their culture medium at the designated time points, and samples were titrated as described above.
One-Step Growth Curves
Vero cells were seeded in 12-well plates at a density of 1.5 × 105 cells/well. After incubation overnight, cells were infected in triplicates at an MOI of 3 in Opti-MEM. Viruses were allowed to adsorb for 6 hr before the inoculum was removed, cells were washed, and culture medium was added (600 μL DMEM+). At the designated time points (24, 36, 48, 72, and 96 hr), cells were scraped in their medium, and viral progeny titers were determined as described above.
Cell Viability Assays
Vero cells were seeded in 12-well plates at a density of 2.5 × 105 cells/well and transfected 6 hr later with microRNA mimics as described above. Sixteen hours post-transfection, cells were infected at an MOI of 0.03 or mock infected. Seven hours p.i., medium was exchanged to DMEM+, and the plates were further incubated. At the designated time points, cell viability was determined using the Colorimetric Cell Viability Kit III (XTT) (PromoCell, Heidelberg, Germany) according to the manufacturer’s protocol.
RT-PCR
Vero cells were seeded in 6-well plates at a density of 3.5 × 105 cells/well. After 18 hr, cells were transfected with microRNA mimics as described above. Twenty hours post-transfection, cells were washed with 1 mL PBS (Life Technologies) and infected with MV-EGFP-NmiRTS7, MV-EGFP-FmiRTS7, MV-EGFP-HmiRTS7, or MV-EGFP-LmiRTS7 at an MOI of 0.03. Thirty-two hours p.i., medium was aspirated, cells were washed with 1 mL PBS, and total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). First-strand cDNA synthesis (160 ng RNA) was performed using Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Braunschweig, Germany) with oligo-dT primers according to the manufacturer’s instructions. Each PCR reaction was performed with 1/20th of the cDNA mix and gene-specific primers (N-1471 5′-GGCCCAGCAGAGCAAGTGATG-3′, NmiRTS-rev 5′-GATGGAGGGTAGGCGGATGTTG-3′, F-6747 5′-CAAGTCGGGAGCAGGAGGTATC-3′, FmiRTS-rev 5′-TTAATTAATGGAAGACTAGTGATTTTGTTG-3′, H-8862 5′-CTTCCAGGGTTGAACATGCTGTG-3′, H-9202+ 5′-TTAATTCTGATGTCTATTTCACACT-3′, L-15240 5′-GCAATTGTGGGAGACGCAGTTAGT-3′, LSmiRTS-rev 5′-GCAAATAATGCCTAACCACCTAGG-3′) to detect the respective MV mRNA transcripts. Additionally, β-actin primers (β-actin Ex6 for 5′-TCATTGCTCCTCCTGAGCGCA-3′, β-actin Ex6 rev 5′-CTAGAAGCATTTGCGGTGGAC-3′) were used to detect the amount of β-actin in each sample as an input control. PCR reactions were conducted using Q5 polymerase (New England Biolabs) with the following conditions (25 cycles): 10 s at 98°C, 30 s at 60°C (F and H gene primers) or 67°C (N and L gene primers), 15 s at 72°C.
MicroRNA Quantification
Reverse transcription of 300 ng total brain RNA (Human Adult Normal Tissue Total RNA - Brain, Biocat, Heidelberg, Germany) was performed using the miScript II RT Kit (QIAGEN) according to the manufacturer’s instructions. Relative quantification of microRNA expression levels was performed from 15 ng of cDNA using the miScript SYBR Green PCR Kit (QIAGEN) and miScript Primer Assays (QIAGEN) on a Light Cycler 480 (Roche, Mannheim, Germany) according to the manufacturer’s instructions. The expression level of miR-7-5p in total brain RNA was quantified by qPCR using a microRNA standard curve as described previously.14 Expression levels of miR-124-3p and miR-125b-5p were calculated relative to miR-7-5p.
RACE Experiments
Vero cells were seeded in 6-well plates at a density of 2.5 × 105 cells/well. After 24 hr, cells were transfected with microRNA mimics as described above. Fifteen hours post-transfection, cells were washed with 1 mL PBS (Life Technologies) and infected with MV-EGFP-HmiRTS7, MV-EGFP-HmiRTS7rc, MV-EGFP-HmiRTS122, MV-EGFP-HmiRTS122rc, or MV-EGFP-HmiRTS124 at an MOI of 0.03. Thirty-five hours p.i., medium was aspirated, cells were washed with 1 mL PBS, and total RNA was isolated using the RNeasy Mini Kit (QIAGEN). Poly(A) tail synthesis was performed using E. coli Poly(A) Polymerase (New England Biolabs) according to the manufacturer’s instructions.
First-strand cDNA synthesis was performed using Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) with RACE-T anchor primer (5′-AAGGCTCCGTCGGCATCGATCGCGCGACTCGAGGCGGAGGTTTTTTTTTTTTTTTTTV). Two reactions of a nested PCR with RACE primers (RACE-O 5′-AAGGCTCCGTCGGCATCG, RACE-I 5′-GCATCGATCGCGCGACTC, H-8633 5′-ACTATCCCGCCAATGAAGAACCTA, H-8862 5′-CTTCCAGGGTTGAACATGCTGTG) were performed to detect the respective MV mRNA transcripts. Indicated PCR products (see Figure 7B) were excised, and gel purification was performed using QIAquick Gel Extraction Kit (QIAGEN). For MV-EGFP-HmiRTS122, MV-EGFP-HmiRTS122rc, and MV-EGFP-HmiRTS124, the procedure was performed analogously. Individual PCR fragments were subsequently cloned into the pJet1.2 vector (Thermo Fisher Scientific) and analyzed by Sanger sequencing (GATC Biotech, Konstanz, Germany).
Author Contributions
Conceptualization, M.F.L., M.-A.B., S.B., G.U.; Investigation, M.F.L., M.-A.B., S.C.A., L.H., R.B., B.H., J.A.; Writing – Original Draft, M.F.L., M.-A.B.; Writing – Review & Editing, G.U., C.E.E., S.B., S.C.A., C.S., H.M.S., D.J., C.v.K., M.F.L., M.-A.B.; Supervision, M.F.L., S.B., G.U., C.S., D.J., C.v.K.; Funding Acquisition, G.U., D.J., C.v.K.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We thank John Bell (Ottawa Hospital Research Institute, Ottawa, ON, Canada) and Roberto Cattaneo (Mayo Clinic, Rochester, MN, USA) for their continuous support. This work was supported by Deutsche Krebshilfe (German Cancer Aid), Max Eder Program No. 110702 (to G.U.), DKFZ Heinrich F.C. Behr fellowships (to M.-A.B. and H.M.S.), a Mildred-Scheel fellowship (by Deutsche Krebshilfe, to S.C.A.), a Helmholtz-Association/DKFZ PhD fellowship, and a physician-scientist fellowship of the Medical Faculty, Heidelberg University (both to M.F.L.). This work was done in Heidelberg, Germany.
Footnotes
Supplemental Information includes one figure and can be found with this article online at https://doi.org/10.1016/j.omto.2018.04.002.
Supplemental Information
References
- 1.Kelly E., Russell S.J. History of oncolytic viruses: genesis to genetic engineering. Mol. Ther. 2007;15:651–659. doi: 10.1038/sj.mt.6300108. [DOI] [PubMed] [Google Scholar]
- 2.Vacchelli E., Eggermont A., Sautès-Fridman C., Galon J., Zitvogel L., Kroemer G., Galluzzi L. Trial watch: oncolytic viruses for cancer therapy. OncoImmunology. 2013;2:e24612. doi: 10.4161/onci.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pol J., Bloy N., Obrist F., Eggermont A., Galon J., Cremer I., Erbs P., Limacher J.M., Preville X., Zitvogel L. Trial Watch: oncolytic viruses for cancer therapy. OncoImmunology. 2014;3:e28694. doi: 10.4161/onci.28694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pol J., Buqué A., Aranda F., Bloy N., Cremer I., Eggermont A., Erbs P., Fucikova J., Galon J., Limacher J.M. Trial watch—oncolytic viruses and cancer therapy. OncoImmunology. 2015;5:e1117740. doi: 10.1080/2162402X.2015.1117740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pol J., Kroemer G., Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. OncoImmunology. 2015;5:e1115641. doi: 10.1080/2162402X.2015.1115641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ottolino-Perry K., Diallo J.S., Lichty B.D., Bell J.C., McCart J.A. Intelligent design: combination therapy with oncolytic viruses. Mol. Ther. 2010;18:251–263. doi: 10.1038/mt.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miest T.S., Cattaneo R. New viruses for cancer therapy: meeting clinical needs. Nat. Rev. Microbiol. 2014;12:23–34. doi: 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichty B.D., Breitbach C.J., Stojdl D.F., Bell J.C. Going viral with cancer immunotherapy. Nat. Rev. Cancer. 2014;14:559–567. doi: 10.1038/nrc3770. [DOI] [PubMed] [Google Scholar]
- 9.Leber M.F., Bossow S., Leonard V.H., Zaoui K., Grossardt C., Frenzke M., Miest T., Sawall S., Cattaneo R., von Kalle C., Ungerechts G. MicroRNA-sensitive oncolytic measles viruses for cancer-specific vector tropism. Mol. Ther. 2011;19:1097–1106. doi: 10.1038/mt.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edge R.E., Falls T.J., Brown C.W., Lichty B.D., Atkins H., Bell J.C. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol. Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- 11.Kelly E.J., Hadac E.M., Greiner S., Russell S.J. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 12.Ylösmäki E., Hakkarainen T., Hemminki A., Visakorpi T., Andino R., Saksela K. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J. Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly E.J., Russell S.J. MicroRNAs and the regulation of vector tropism. Mol. Ther. 2009;17:409–416. doi: 10.1038/mt.2008.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baertsch M.A., Leber M.F., Bossow S., Singh M., Engeland C.E., Albert J., Grossardt C., Jäger D., von Kalle C., Ungerechts G. MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Ther. 2014;21:373–380. doi: 10.1038/cgt.2014.40. [DOI] [PubMed] [Google Scholar]
- 15.Russell S.J., Peng K.W. Measles virus for cancer therapy. Curr. Top. Microbiol. Immunol. 2009;330:213–241. doi: 10.1007/978-3-540-70617-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cattaneo R., Rebmann G., Schmid A., Baczko K., ter Meulen V., Billeter M.A. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 1987;6:681–688. doi: 10.1002/j.1460-2075.1987.tb04808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plumet S., Duprex W.P., Gerlier D. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 2005;79:6900–6908. doi: 10.1128/JVI.79.11.6900-6908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao N.Y., Hu H.Y., Yan Z., Xu Y., Hu H., Menzel C., Li N., Chen W., Khaitovich P. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics. 2010;11:409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whelan S.P., Barr J.N., Wertz G.W. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 20.Rima B.K., Duprex W.P. The measles virus replication cycle. Curr. Top. Microbiol. Immunol. 2009;329:77–102. doi: 10.1007/978-3-540-70523-9_5. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banzhaf-Strathmann J., Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014;12:30. doi: 10.1186/1478-811X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tili E., Michaille J.J., Luo Z., Volinia S., Rassenti L.Z., Kipps T.J., Croce C.M. The down-regulation of miR-125b in chronic lymphocytic leukemias leads to metabolic adaptation of cells to a transformed state. Blood. 2012;120:2631–2638. doi: 10.1182/blood-2012-03-415737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., Lee J., Fine H., Chiocca E.A., Lawler S., Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz A.J., Russell S.J. MicroRNAs and oncolytic viruses. Curr. Opin. Virol. 2015;13:40–48. doi: 10.1016/j.coviro.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Visani M., de Biase D., Marucci G., Cerasoli S., Nigrisoli E., Bacchi Reggiani M.L., Albani F., Baruzzi A., Pession A., PERNO study group Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol. Oncol. 2014;8:417–430. doi: 10.1016/j.molonc.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster R.J., Giles K.M., Price K.J., Zhang P.M., Mattick J.S., Leedman P.J. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J. Biol. Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 28.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 29.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 30.Balbo P.B., Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ungerechts G., Springfeld C., Frenzke M.E., Lampe J., Johnston P.B., Parker W.B., Sorscher E.J., Cattaneo R. Lymphoma chemovirotherapy: CD20-targeted and convertase-armed measles virus can synergize with fludarabine. Cancer Res. 2007;67:10939–10947. doi: 10.1158/0008-5472.CAN-07-1252. [DOI] [PubMed] [Google Scholar]
- 32.Radecke F., Spielhofer P., Schneider H., Kaelin K., Huber M., Dötsch C., Christiansen G., Billeter M.A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin A., Staeheli P., Schneider U. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J. Virol. 2006;80:5708–5715. doi: 10.1128/JVI.02389-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.