Abstract
Inflorescences in the Asteraceae plant family, flower heads, or capitula, mimic single flowers but are highly compressed structures composed of multiple flowers. This transference of a flower-like appearance into an inflorescence level is considered as the key innovation for the rapid tribal radiation of Asteraceae. Recent molecular data indicate that Asteraceae flower heads resemble single flowers not only morphologically but also at molecular level. We summarize this data giving examples of how rewiring of conserved floral regulators have led to evolution of morphological innovations in Asteraceae. Functional diversification of the highly conserved flower meristem identity regulator LEAFY has shown a major role in the evolution of the capitulum architecture. Furthermore, gene duplication and subsequent sub- and neofunctionalization of SEPALLATA- and CYCLOIDEA-like genes in Asteraceae have been shown to contribute to meristem determinacy, as well as flower type differentiation—key traits that specify this large family. Future challenge is to integrate genomic, as well as evolutionary developmental studies in a wider selection of Asteraceae species to understand the detailed gene regulatory networks behind the elaborate inflorescence architecture, and to promote our understanding of how changes in regulatory mechanisms shape development.
Introduction
Asteraceae, commonly referred as the sunflower or daisy family, has a prominent place in our daily life as it includes many economically important food crops (sunflower, lettuce, artichoke, endive, and safflower), herbal and medicinal species (Calendula, Artemisia, Echinacea), as well as popular ornamentals (gerbera, chrysanthemum, aster, dahlia, zinnia, and marigold). Also some noxious weeds, such as dandelion or thistle are members of the family. Asteraceae represents the largest family of flowering plants with around 25,000 species that are widespread into nearly all terrestrial habitats except of the Antarctica1,2. From evolutionary perspective, Asteraceae is a young family. Relatively recent discoveries of fossils have proposed scenarios that the split of Asteraceae from its ancestor occurred in Patagonia, southern South America some time before the Eocene 56–34 million years ago, and it spread to Africa before the two continents became geographically isolated3,4. The family then rapidly diversified and colonized the earth. This explosive tribal radiation has been associated with complex history of whole-genome duplication events, and especially with a paleopolyploidization event shared with Asteraceae and its sister family Calycearaceae, as well as a second round of genome duplications among the core Asteraceae tribes5,6. At the genetic level, functional analyses are necessary to explore how the ancestral polyploidization has contributed to the evolution of the extensive diversity of developmental traits, secondary metabolites, as well as life histories discovered in Asteraceae.
The key morphological innovation that has been associated with the evolutionary success of Asteraceae is the unique head-like inflorescence, capitulum that is the distinguishing feature of the whole family. Capitulum is a pseudanthium, a false flower that superficially mimics a single flower but is a highly aggregated structure comprised of multiple flowers with specialized functions. Asteraceae is largely pollinated by insects (rarely by birds), and several studies have indicated that the unique organization of the capitulum has selective advantage to pollinator attraction and plant vigor7–9. Moreover, the showy capitulum with all the diversity in shapes and colors generated by breeders is surely a key attraction to consumers contributing to the economic significance of this family in the ornamental industry. In this review, we aim to summarize the recent research in Asteraceae models to understand genetic regulation of capitulum development, as well as its organization and evolution. Altogether, the data demonstrate novel gene functions for both conserved floral regulators as well as duplicated genes that through sub- and neofunctionalization have gained specific roles in regulation of the elaborate inflorescence architecture.
Organization of Asteraceae heads
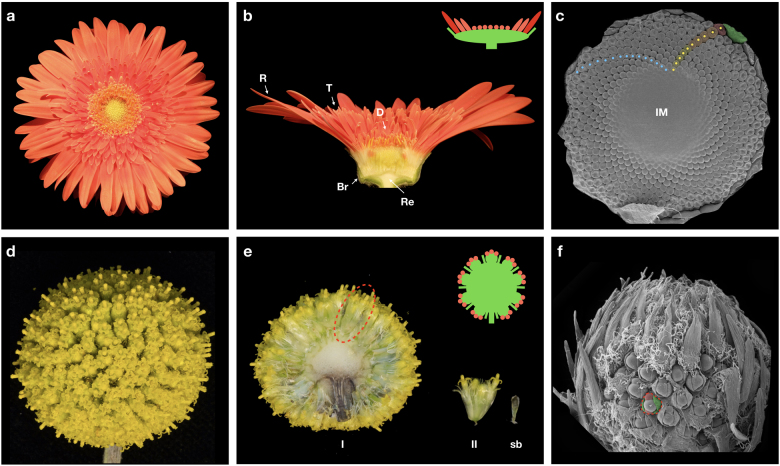
Asteraceae flower heads combine multiple flowers attached on a single receptacle (Fig. 1). After induction of flowering, the early ontogeny of the head is characterized by rapid expansion of the inflorescence meristem (receptacle), while later the meristematic area gradually shrinks and is fully consumed by emerging flower primordia10,11. In simple heterogamous, or radiate flower heads, the margin of the capitulum is occupied by showy ray flowers and the center with less conspicuous disc flowers. In sunflower (Helianthus), the marginal ray flowers are sterile and they develop extended ligules while the central disc flowers are perfect, and produce pollen. In gerbera (Gerbera) the ray flowers are female, and it may also develop intermediate trans flowers that resemble rays but are just smaller in size (Fig. 1a, b). In contrast, the homogamous heads are formed of only single-flower types. For example, the heads of lettuce (Lactuca) are composed of only ligulate ray flowers while the discoid heads in thistles, representing several genera in Asteraceae, develop solely disc flowers. The discoid capitula have been considered as an ancestral condition in Asteraceae1,12 where the emergence of ray flowers has occurred multiple times independently. The entire head is surrounded by involucral bracts that behave like sepals in single flowers, giving protection to the developing head at early developmental stages (Fig. 1b).
Fig. 1. Organization of flower heads in Asteraceae.
a Top view of a gerbera head. b Cross section of a gerbera head showing marginal ray (R), intermediate trans (T), and central disc (D) flowers that are all attached to a single, expanded receptacle (Re). The entire structure is surrounded by involucral bracts (Br) giving an impression of a single flower. c Scanning electron microscopy image focusing on early developmental stage of a growing head. The inflorescence meristem (IM) produces flower primordia in clockwise (blue dots) and counterclockwise (yellow dots) spirals. d Top view of a syncephalium of Craspedia globosa. e Cross section of a Craspedia primary head (I) shows multiple secondary heads (II, red circle) attached to a single receptacle, and developing a subtending bract (sb). f Scanning electron microscopy image of a Craspedia syncephalium at early developmental stage. A single-secondary head is highlighted (red circle) showing secondary inflorescence meristem producing bract (shaded in green) and flower primordia (shaded in red)
Asteraceae includes also species that show a higher level of complexity in their inflorescences. As examples of simple capitula, sunflower and gerbera heads appear as solitary structures on top of a floral stem. However, several capitula may also form synflorescences being arranged in distinct branched systems, such as racemes (e.g., Artemisia pycnocephala), cymes (e.g., Senecio vulgaris), or corymbs (e.g., Achillea millefolium)13. Another hierarchical level of complexity is generated when flower heads are aggregated onto a single receptacle, and form higher order structures, ‘heads within heads’, also known as capitulescence or syncephalium (e.g., Craspedia, Echinops, Lagascea)14–16 (Fig. 1d–f).
A characteristic feature shared by distinct head-like inflorescences is the spatial arrangement of individual flowers on the expanded receptacle (Fig. 1c). They appear in a phyllotactic pattern that follow strict clockwise and counterclockwise spirals (contact parastichies), the numbers of which always follow the two consecutive numbers in the mathematical Fibonacci series: 1, 1, 2, 3, 5, 8, 13, 21, 34, etc. where each number is a sum of the two previous ones. The spirals can usually reach high numbers; for example flowers on gerbera and sunflower heads are typically arranged in 34/55 and 55/89 spirals, respectively. The initiation of individual flowers in a capitulum has been thought to occur in an acropetal manner, from the margins toward the center of the capitulum. This is indeed the case in homogamous heads with only single-flower types, however, morphological studies have indicated that in heterogamous heads the early ontogeny of marginal ray flower primordia is distinct10,13. In fact, in extreme cases, ray flower development may proceed basipetally toward the margins of the head (instead of the center), or in milder cases ray primordia show a developmental arrest compared to the development of an adjacent disc or trans flower primordia10,13,17–19. This differential early ontogeny has turned out to be crucial for understanding how distinct flower types have evolved (see below).
Flower meristem identity gene functions give light for understanding the evolution of the capitulum architecture
Inflorescences are typically branched structures that bear flowers. As known from the conventional model species, the major difference between the two basic types of inflorescences, racemes in Arabidopsis or Antirrhinum, and cymes in Solanaceae species such as petunia or tomato, is in their determinacy20. The elongating, indeterminate racemes continuously produce flowers in the flanks of the inflorescence meristem (IM) while in cymes the meristem always terminates in a flower, but forms a new axillary IM that continues the growth. Two highly conserved genes LEAFY (LFY) and UNUSUAL FLORAL ORGANS (UFO), encode the key regulators that define flower meristem (FM) identity. However, these genes function in an opposite manner in racemes and cymes20,21. In Arabidopsis, LFY expression is specifically localizing to FMs22, and through interaction with UFO23 and SEPALLATA3 (SEP3)24, LFY activates flower organ identity genes, and floral developmental program. In cymes, instead, the FM identity is defined by the UFO homologs, DOUBLE TOP (DOT) in petunia25 or ANANTHA (AN) in tomato26.
The unique, and apparently successful, organization of the capitulum has inspired botanists to investigate its evolutionary origin. Capitulum is often interpreted as a compressed raceme or cyme27. However, Claβen-Bockhoff and Bull-Hereñu28 proposed that it has evolved from a single, determinate meristem that through subdivision gave rise to multi-flowered head. Claβen-Bockhoff and Bull-Hereñu present of concept of a floral unit meristem (FUM) that resembles a single flower meristem (FM) by its histological organization and development, being flat and enlarged structure with inhibited internodes. The distinction of FUM is that it produces flower primordia in an acropetal manner while a FM develops floral organ primordia. An alternative scenario was presented based on morphological studies on the basal relatives of Asteraceae (Menyanthaceae, Goodeniaceae, and Calyceraceae) where the main axis of the inflorescence shows a racemous, and the basal lateral branches cymous branching pattern29,30. According to Pozner et al.30 Asteraceae heads evolved from these mixed inflorescences through the loss of the marginal, cymose branches (called cymose units) and a terminal flower still found in the compressed inflorescences (cephalioids) of Calyceraceae, a sister family to Asteraceae. The marginal ray flowers are thus considered to originate from the cymose units, experiencing further functional differentiation.
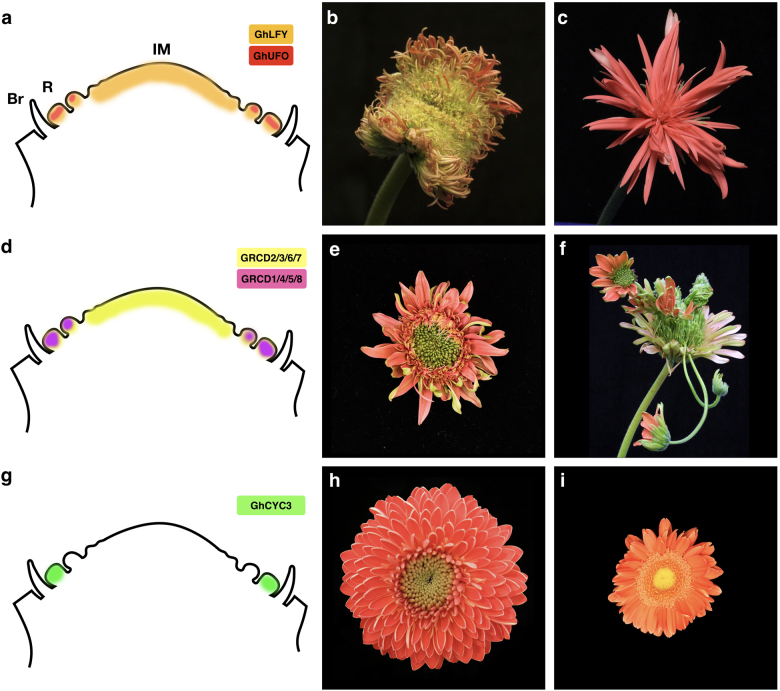
In order to test these distinct hypotheses of the evolutionary origin of flower heads in Asteraceae, Zhao et al.10 conducted functional studies of FM gene orthologs in gerbera. As an assumption, loss of FM identity would occur similarly in both flower types if they originated from a single meristem, or differentially, if they originated form distinct branching system. Most interestingly, and in contrast to conventional model species, the gerbera GhLFY showed uniform expression across the entire, naked IM (Fig. 2a). This pattern defines the capitulum as a determinate structure with resemblance of single FM. Suppression of GhLFY expression in transgenic RNAi lines led to loss of flower organ identity, as expected (Fig. 2b). However, the lines also revealed a specific role for GhLFY in regulation of ray flower development. The ray primordia were converted into branched units resembling the marginal cymose units as found in Calyceraceae. Moreover, GhLFY expression domain was found to be associated with the early ontogeny (developmental delay) of ray flowers emerging at the axils of involucral bracts. The gerbera GhUFO instead showed FM-specific expression, and a conserved role in regulating the FM identity (Fig. 2a). Ectopic expression of GhUFO was sufficient to convert the capitulum into a single flower associated with a dramatic change from spiral to whorled phyllotaxis, and development of numerous organ primordia instead of flowers10 (Fig. 2c).
Fig. 2. Schematic representation of expression domains, and examples of representative transgenic phenotypes for key regulatory genes affecting head development in gerbera.
a The expression of flower meristem identity gene GhLFY is detected both in undifferentiated inflorescence meristem (IM) as well as in emerging flower primordia while GhUFO expression in localizing to flower primordia10. b Transgenic phenotype of a GhLFY RNAi line showing loss of organ identity. c Ectopic expression of GhUFO converts the meristem into a single flower that develops only multiple flower organs instead of florets. d The expression domains of the duplicated SEP-like GRCD genes. e A mild phenotype of a transgenic GRCD4/5 double RNAi line shows defects in petal development. f Downregulation of multiple GRCD genes lead to floral reversion where carpels or ovaries of individual flowers are replaced by developing new heads. g CYC2 clade gene GhCYC3 is specifically expressed in ray flower primordia. h A crested gerbera cultivar develops only ray flowers as a result of ectopic expression of GhCYC3 throughout the capitulum. i Transgenic gerbera GhCYC5 RNAi lines shows significantly reduced petal length in ray and trans flowers
Altogether, the molecular studies in gerbera suggest that none of the previous hypotheses of patterning and evolution of the capitulum architecture are sufficient by themselves. In fact, the current data gives support for both hypotheses. The differential role GhLFY in controlling flower type identity suggests that floral dimorphism is connected with their distinct origin from separate branching systems10,30. However, it cannot be excluded that the centre of the capitulum, harboring the disc flowers, may indeed have originated by subdivision of a single meristem as proposed by Claβen-Bockhoff and Bull-Hereñu28. As one possibility, this could have occurred as a result of temporal changes in GhUFO expression needed to define FM identity. Most importantly, the data also emphasizes that the capitulum can be seen as an analog of a single flower not only morphologically but also at molecular level. Comparative evo-devo studies should be extended to the basal relatives of Asteraceae as well as to syncephalious species to clarify the current hypotheses, and to better understand the molecular networks behind this vast morphological diversity.
Gerbera SEPALLATA-like gene functions in regulation of IM determinacy
Based on the absence of a clearly defined, single terminal flower, earlier botanical studies have classified flower heads as indeterminate inflorescences13,31. However, functional studies of the gerbera GhLFY indicate that the expanding IM is in fact determinate, and eventually fully packed with flower primordia10,32. In transgenic lines with suppressed GhLFY expression, the IM never got fully consumed with emerging primordia that were randomly initiating in the center of the capitulum10. Furthermore, functional studies have indicated that also the E class SEPALLATA-like MADS box genes, GERBERA REGULATOR of CAPITULUM DEVELOPMENT 2/7 (GRCD2/7), are required for the determinacy of the IM11,33. The number of the flowers produced in the inflorescences of gerbera antisense-GRCD2 lines were nearly doubled compared to the wild type, and this was attributed to the indefinite growth of IM that remained undifferentiated until senescence33. Similar phenotypes were observed in transgenic gerbera lines when GRCD7, a close paralog of GRCD2, was downregulated11. Both GRCD2 and GRCD7 belong to the SEP1/2/4 clade (also known as LOFSEP clade) of E function genes11,34,35. Like GhLFY, both GRCD2 and GRCD7 show ubiquitous expression in the undifferentiated IM throughout the early ontogeny of flower heads11 (Fig. 2d).
LFY gene is present as a single copy gene in most of the land plant species21. It is an example of a gene in Asteraceae that has evolved a novel function to regulate early ontogeny of ray flowers as well as determinacy of the capitulum10 (Fig. 2a). In contrast, the SEP-like GRCD genes are represented by a medium size gene family, composed of altogether eight members in gerbera11,35. In contrast to high level of redundancy among the four Arabidopsis SEP genes, affecting the identity of all organ whorls36,37, the gerbera GRCD genes show subfunctionalization. At the level of individual flowers, GRCD genes have evolved whorl-specific functions, with distinct gene family members contributing to the identity of stamens, carpels as well as petals, respectively11,33,38,39 (Fig. 2d, e). Furthermore, similarly as SEP orthologs in Arabidopsis, petunia or tomato37,40,41, GRCD genes show conserved roles in regulating the maintenance of flower meristem identity. In extreme cases, loss of GRCD expression led to floral reversion replacing carpels or ovaries by a new capitulum11 (Fig. 2f). What is specific to Asteraceae is that the duplicated SEP-like genes have been recruited to regulate IM development, an example of potential neofunctionalization. In addition to GRCD2 and GRCD7, two other genes, GRCD6 of the SEP1/2/4 clade, and GRCD3 of the closely related AGL6 clade were identified as candidates to regulate IM patterning in gerbera11 (Fig. 2d). However, their detailed functions are not yet defined. The determinate fate of IM in flower heads could be, at least partially, explained by the temporal and spatial changes in expression and functions of these floral meristem/organ identity genes contributing to their novel roles in regulating Asteraceae inflorescence architecture10,11.
Understanding of patterning of the Asteraceae IM still awaits answers for many open questions. How is the determinacy of the IM established? What are the genetic factors up- and downstream of GRCD2/7 and GhLFY? A determinate meristem is programmed to terminate its activity in a precise manner, at correct time and location. This is exemplified in the Arabidopsis flower meristem, where termination is executed through the activity of AGAMOUS (AG) in its central domain leading to suppression WUSCHEL (WUS) expression, a key gene to promote stem cell activity42–44. Aerial meristems in Arabidopsis are histologically following a zonal arrangement with separate central, peripheral and rib zones45. Stem cells of the meristem are maintained within the central zone, through a well-known signaling cascade involving a negative feedback loop between WUS and CLAVATA3 (CLV3) peptide46–48. In contrast, the IM in Asteraceae flower heads is organized in a distinct’mantle-core’ configuration, consisting an enlarged meristematic mantle in addition to a quiescent core underneath45,49. As necessary factors for maintaining the determinate fate of the IM in heads, GRCD2/7 and GhLFY must have master roles in regulating the genetic program that determines first the shrinkage of the meristematic volume, and later its consumption and termination. So far, we still lack the genetic evidence to explain how stem cell homeostasis is regulated in the mantle-core configurated meristem.
CYCLOIDEA2 clade genes define the identity of the marginal ray flowers
The presence of distinct flower types is a characteristic feature of Asteraceae, and so far mostly studied trait at genetic level. Typically, the flower types differ in their symmetry—marginal ray (and trans) flowers being bilaterally symmetrical (zygomorphic) and central disc flowers radially symmetrical (actinomorphic) (Fig. 1a, b). At the level of single flowers, the symmetry regulation by CYCLOIDEA/TEOSINTE BRANCHED1-like TCP domain transcription factors is well-established across core eudicots50,51. During the last decade, dissection of functions of the CYC/TB1-like genes in Gerbera, Helianthus, Senecio, and Chrysanthemum indicate their independent recruitment in defining flower type identity in Asteraceae52–59. This function can specifically be addressed to the CYC2 subclade gene family members that have experienced frequent duplications in Asteraceae53,54,56,59. The CYC2 clade homologues are predominantly expressed in the periphery of the capitulum, in emerging ray flower primordia while low, or no expression is detected in the central disc primordia17,53,55,56,58,59 (Fig. 2g). The functional data indicate that ray flower identity is controlled by different CYC2 paralogs in distinct Asteraceae lineages54,56,58 giving molecular support that ray identity evolved multiple times independently in the family60. Interestingly, duplicated CYC/TB1-like genes have also been identified in Dipsacaceae61 and Myrtaceae62 where pseudanthium-type inflorescences with distinct flower types originated independently of Asteraceae.
Functional studies have shown variability in phenotypes of transgenic plants and/or in mutant lines indicating that, although CYC2 homologues clearly affect flower type differentiation, they may function in a species-specific manner (Table 1). Conversion of disc flowers into ray-like, with elongated ligules and disrupted stamen development, was discovered in transgenic gerbera by ectopic activation of GhCYC2, GhCYC3 and GhCYC4, respectively52,57. In the so-called crested or double-flowered cultivars of gerbera (Fig. 2h) and Chrysanthemum morifolium, as well as in double-flowered (dbl) and chrysanthemoides (Chry) mutants of sunflower, all flowers are of ray identity. All these phenotypes have been shown to arise by ectopic activation of CYC2 clade genes throughout the capitulum55,57,59,63. Interestingly, in Senecio and chrysanthemum, ectopic expression of ray-specific CYC genes did not affect disc flower development53,59. Furthermore, also the ligule length of ray and/or trans flowers is differentially affected by genetic transformation in distinct species52,53,59 (Table 1) indicating that the given genes may function in distinct regulatory networks. So far, gene silencing has been challenging, and has not led to complete absence of ray flowers suggesting genetic redundancy among the gene family members, or involvement of yet unknown factors (Fig. 2i). The development of trumpet shaped, actinomorphic ray flowers has been associated with reduced CYC gene expression in tubular-rayed (tub) sunflower mutant55,64, and in genus Anacyclus (tribe Anthemidae)17 but also with ectopic expression as the Senecio RAY253. Further functional studies, e.g. with CRISPR/Cas9 approaches, are necessary to study whether complete suppression of CYC2 activity is sufficient to fully abolish ray flower initiation or whether it only affects organ differentiation.
Table 1.
Summary of phenotypes in transgenic lines, cultivars and mutants in Asteraceae family affected by CYC2 clade TCP genes
| Species | Modification/mutant | Observed phenotype | |||
|---|---|---|---|---|---|
| Ray | Trans | Disc | |||
| Gerbera hybrida | Ectopic expression of GhCYC2, GhCYC3 or GhCYC452,57 | Reduced ligule length | No effect | Ray-like with elongated ligules and disrupted stamen development | |
| Cosuppression of GhCYC252 | No effect | Reduced ligule length, occasional splitting of ligules | No effect | ||
| Crested cultivar CH02.663: upregulation of GhCYC3 expression in disc flowers57 | Ray flower identity only | ||||
| Senecio vulgaris | Ectopic expression of RAY153 | Reduced ligule length, complete absence of ray flowers | — | No effect | |
| Ectopic expression of RAY253 | Tubular ray flowers by promoting elongation of all petal lobes | — | No effect | ||
| Suppressed expression of RAY358 | Reduced ligule length | — | No effect | ||
| Chrysanthemum lavandulifolium | Ectopic expression of CmCYC2c59 | Increased ligule length | Decreased ligule length | No effect | |
| Chrysanthemum morifolium | ‘Double-ray flowered’ heads: upregulation of CmCYC2c expression in disc flowers59 | Ray flower identity only | |||
| Anacyclus clavatus | Trumpet-type: low expression of AcCYC2b, AcCYC2c and AcCYC2d in the ray flowers17 | Tubular ray flowers | — | No effect | |
| Helianthus annuus | Double-flowered (dbl) | Ectopic expression of HaCYC2c by an insertion in the promoter region of HaCYC2c55,63,79 | No effect | — | Ray-like |
| Chrysanthemoides (Chry) | No effect | — | Ray-like, reduction in stamen length, absence of ovules | ||
| Tubular-rayed (tub) | Inactivation HaCYC2c by an insertion of transposable element in the coding region of HaCYC2c6,55,64 | Tubular ray flowers | — | No effect | |
| Tubular ray flower (turf) | Tubular ray flowers, fertile stamens and ovules | — | No effect | ||
The role of CYC-like proteins in regulating flower symmetry was originally discovered in bilaterally symmetrical flowers of Antirrhinum majus65,66, as also their interaction with MYB-domain regulators RADIALIS (RAD) and DIVARICATA (DIV)67. In Antirrhinum, CYC defines the identity of the dorsal domain of the flowers by promoting the enlargement of the dorsal petals and disrupting the development of the dorsal stamen65,66. CYC is positively regulating RAD68 whereas DIV, that defines the identity of ventral petals, is repressed by RAD from the dorsal domain67. Recent studies in Senecio suggest that similar regulatory network exists in Asteraceae but involves diversified expression domains of these key regulatory genes as well as modifications in their interactions58. Functional studies indicate that the Senecio RAY3 promotes elongation of the large ventral ligule of ray flowers while SvDIV1B apparently functions as a negative regulator of elongation by inhibiting RAY3 and SvRAD. Garcês et al.58 further suggest complex regulatory relationships between the CYC2 genes, and identified two other CYC2 clade genes, RAY1 and RAY2, as potential target genes of RAY3. However, detailed understanding of the network still awaits further functional studies.
The pairwise protein–protein interaction studies conducted with gerbera and sunflower indicated that CYC/TB1-like proteins function in complexes that are likely to direct their functional specificity56. Interestingly, the capacity of CYC/TB1-like proteins to form homodimers varied between gerbera and sunflower while heterodimer formation was more similar between the two species that represent distant tribes in Asteraceae. Positive autoregulatory feedback loops of CYC2 clade genes themselves as well as cross-regulation among each other has been identified in Gesneriaceae (Lamiales) as a mechanism for maintaining CYC2 gene expression in the dorsal domain of the developing flowers69. Cross-regulation has already been suggested for the RAY genes in Senecio58 as well as for CmCYC2 genes in C. morifolium59. Further studies are still required to identify potential upstream as well as interacting regulators of CYC2 clade genes. The mechanisms of how CYC2 clade genes acquire their specificity to regulate early flower type determination as well as later stages of organ differentiation are fully unknown. Transcriptome analyses in gerbera and chrysanthemum have revealed candidate transcription factor families, including MADS-box proteins, but also members of NAC, MYB, AP2/ERF, WUS, CYC2, and other TCP families, among differentially expressed genes between the ray and disc flowers70,71, however, their connections with CYC2 proteins have not been established.
Future prospects
The advances in next-generation sequencing already have, and still will dramatically change biological research. For Asteraceae, the reference genome sequences for lettuce (Lactuca sativa)72 and sunflower73, as well as draft sequences for horseweed (Conyza canadensis)74, globe artichoke (Cynara cardunculus)75, and sweet wormwood (Artemisia annua)76 have been published. Genome information has established the evolutionary history of the family. Sunflower, lettuce, and artichoke have experienced a whole-genome triplication (WGT-1, 38-50 Mya) event in addition to the ancestral WGT-γ (gamma triplication, 122–164 Mya) common for eudicots72,73. Moreover, younger, lineage specific duplications (WGD-2, 29 Mya) have occurred in sunflower73, and also in the gerbera lineage5. As indicated for gene regulatory networks of flowering time in sunflower73, these polyploidization events have resulted in more complex regulatory networks, involving additional paralogs in Asteraceae compared to the Arabidopsis model. Also in lettuce, enrichment of some transcription factors and DNA-binding proteins in the triplicated regions of the genome, as well as sequence divergence was discovered72. This is also apparent for the specific developmental regulators reviewed here, highlighting the value of Asteraceae as a model to understand evolution of gene functions through gene duplication and subsequent diversification.
Detailed understanding of floral and inflorescence diversity both at evolutionary and developmental context is of utmost importance as they form the basis for plant reproduction, yield and human sustenance. Evolutionary developmental (evo-devo) approaches combine plant systematics (phylogenetics), developmental genetics and genomics in an unprecedented way76. In a context of a robust phylogenetic framework for angiosperms, large sequence datasets are produced not only from single-model species, but from several species that represent the phenotypic variation across ‘model clades’77. Asteraceae clearly represents yet not very extensive studied, but promising and emerging model clade in this respect. Recently, genome skimming, or whole-genome shotgun sequencing, as a low-coverage and cost-effective method was used for mining of low-abundant CYC-like genes from 24 Goodeniaceae species, basal relatives of Asteraceae78. The possibility for simultaneous comparisons of multiple taxa in association of trait gains, losses, or modifications will facilitate our understanding of how changes in regulatory mechanisms shape development78. Combined with functional studies, for example in well-established models, this will also pave the way for new applications in breeding of novel traits in important agricultural and ornamental crops.
Acknowledgements
We thank all colleagues and collaborators who have contributed to Asteraceae research, and specifically to our research in the Gerbera Lab, University of Helsinki, Finland. This work has been funded by the Academy of Finland grants for P.E. (1139092; 1310318), Jenny and Antti Wihuri Foundation for Y.Z. and Doctoral Programme in Plant Sciences for Y.Z. and T.Z.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Funk, V. A., Susanna, A., Stuessy, T. F. & Bayer, R. J. Systematics, evolution, and biogeography of Compositae. (International Association for Plant Taxonomy, Vienna, 2009).
- 2.Jeffrey, C. Compositae: Introduction with key to tribes. in Flowering plants: Eudicots; Asterales (eds. Kadereit, J. J. W. & Jeffrey, C.) 61–87 (Springer-Verlag, Berlin, 2007).
- 3.Barreda VD, et al. Eocene Patagonia fossils of the daisy family. Science. 2010;329:1621. doi: 10.1126/science.1193108. [DOI] [PubMed] [Google Scholar]
- 4.Barreda VD, et al. An extinct Eocene taxon of the daisy family (Asteraceae): evolutionary, ecological and biogeographical implications. Ann. Bot. 2012;109:127–134. doi: 10.1093/aob/mcr240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker MS, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker MS, et al. Most Compositae (Asteraceae) are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am. J. Bot. 2016;103:1203–1211. doi: 10.3732/ajb.1600113. [DOI] [PubMed] [Google Scholar]
- 7.Stuessy TF, Spooner DM, Evans KA. Adaptive significance of ray corollas in Helianthus grosseserratus (Compositae) Am. Midl. Nat. 1986;115:191–197. doi: 10.2307/2425849. [DOI] [Google Scholar]
- 8.Sun M, Ganders FR. Outcrossing rates and allozyme variation in rayed and rayless morphs of Bidens pilosa. Heredity. 1990;64:139–143. doi: 10.1038/hdy.1990.18. [DOI] [Google Scholar]
- 9.Andersson S. Pollinator and nonpollinator selection on ray morphology in Leucanthemum vulgare (Oxeye daisy, Asteraceae) Am. J. Bot. 2008;95:1072–1078. doi: 10.3732/ajb.0800087. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, et al. Evolutionary co-option of floral meristem identity genes for patterning of the flower-like Asteraceae inflorescence. Plant Physiol. 2016;172:284–296. doi: 10.1104/pp.16.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, et al. Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 2017;216:939–954. doi: 10.1111/nph.14707. [DOI] [PubMed] [Google Scholar]
- 12.Bremer, K. Asteraceae: Cladistics and Classification. (Timber Press, Portland, OR, 1994).
- 13.Harris EM. Inflorescence and floral ontogeny in Asteraceae: a synthesis of historical and current concepts. Bot. Rev. 1995;61:93–278. doi: 10.1007/BF02887192. [DOI] [Google Scholar]
- 14.Claβen-Bockhoff, R. Functional units beyond the level of the capitulum and cypsela in Compositae. in Compositae: biology & utilization (eds. Cagliari, R. D. S. & Hind, D. J. N.) 129–160 (Royal Botanic Gardens, Kew, 1996).
- 15.Leins P, Gemmeke V. Infloreszenz- und Blutenentwicklung bei der Kugeldistel Echinops exaltatus (Asteraceae) Plant Syst. Evol. 1979;132:189–204. doi: 10.1007/BF00990465. [DOI] [Google Scholar]
- 16.Harris EM. Developmental evidence for the derivation of syncephalia in Lagascea (Heliantheae; Asteraceae) Am. J. Bot. 1994;81:1139–1148. doi: 10.1002/j.1537-2197.1994.tb15608.x. [DOI] [Google Scholar]
- 17.Bello MA, Cubas P, Álvarez I, Sanjuanbenito G, Fuertes-Aguilar J. Evolution and expression patterns of CYC/TB1 genes in Anacyclus: phylogenetic insights for floral symmetry genes in Asteraceae. Front. Plant Sci. 2017;8:1–12. doi: 10.3389/fpls.2017.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris EM, Tucker SC, Urbatsch LE. Floral initiation and early development in Erigeron philadelphicus (Asteraceae) Am. J. Bot. 1991;78:108–121. doi: 10.1002/j.1537-2197.1991.tb12577.x. [DOI] [Google Scholar]
- 19.Broholm, S. K., Teeri, T. H. & Elomaa, P. Molecular control of inflorescence development in Asteraceae. in Advances in Botanical Research, The molecular genetics of floral transition and flower development (eds. Jacquot, J. -P., Gadal, P. & Fornara, F.) 72, 297–333 (Academic Press, Elsevier, Oxford, 2014).
- 20.McKim S, Hay A. Patterning and evolution of floral structures - marking time. Curr. Opin. Genet. Dev. 2010;20:448–453. doi: 10.1016/j.gde.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F. LEAFY blossoms. Trends Plant. Sci. 2010;15:346–352. doi: 10.1016/j.tplants.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Weigel D, Alvarez J, Smyth D, Yanofsky M, Meyerowitz E. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-N. [DOI] [PubMed] [Google Scholar]
- 23.Chae E, Tan QKG, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–1245. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Xi W, Shen L, Tan C, Yu H. Regulation of floral patterning by flowering time genes. Dev. Cell. 2009;16:711–722. doi: 10.1016/j.devcel.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Souer E, et al. Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of Petunia. Plant Cell. 2008;20:2033–2048. doi: 10.1105/tpc.108.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lippman ZB, et al. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008;6:2424–2435. doi: 10.1371/journal.pbio.0060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cronquist A. The Compositae revisited. Brittonia. 1977;29:137–153. doi: 10.2307/2805847. [DOI] [Google Scholar]
- 28.Claßen-Bockhoff R, Bull-Hereñu K. Towards an ontogenetic understanding of inflorescence diversity. Ann. Bot. 2013;112:1523–1542. doi: 10.1093/aob/mct009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endress PK. Disentangling confusions in inflorescence morphology: patterns and diversity of reproductive shoot ramification in angiosperms. J. Syst. Evol. 2010;48:225–239. doi: 10.1111/j.1759-6831.2010.00087.x. [DOI] [Google Scholar]
- 30.Pozner R, Zanotti C, Johnson LA. Evolutionary origin of the Asteraceae capitulum: insights from Calyceraceae. Am. J. Bot. 2012;99:1–13. doi: 10.3732/ajb.1100256. [DOI] [PubMed] [Google Scholar]
- 31.Weberling, F. Morphology of flowers and inflorescences. (Cambridge University Press, Cambridge,1992).
- 32.Teeri TH, et al. Reproductive meristem fates. J. Exp. Bot. 2006;57:3445–3455. doi: 10.1093/jxb/erl181. [DOI] [PubMed] [Google Scholar]
- 33.Uimari A, et al. Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc. Natl Acad. Sci. USA. 2004;101:15817–15822. doi: 10.1073/pnas.0406844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahn LM, et al. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH. Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol. 2010;10:129. doi: 10.1186/1471-2229-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 37.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 38.Kotilainen M, et al. GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell. 2000;12:1893–1902. doi: 10.1105/tpc.12.10.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teeri TH, Elomaa P, Kotilainen M, Albert VA. Mining plant diversity: Gerbera as a model system for plant developmental and biosynthetic research. Bioessays. 2006;28:756–767. doi: 10.1002/bies.20439. [DOI] [PubMed] [Google Scholar]
- 40.Ferrario S, Immink RGH, Shchennikova A, Busscher-Lange J, Angenent GC. The MADS box gene FBP2 is required for SEPALLATA function in Petunia. Plant Cell. 2003;15:914–925. doi: 10.1105/tpc.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao J. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002;130:605–617. doi: 10.1104/pp.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/S0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 43.Sun B, Xu Y, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, et al. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell. 2011;23:3654–3670. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwiatkowska D. Flowering and apical meristem growth dynamics. J. Exp. Bot. 2008;59:187–201. doi: 10.1093/jxb/erm290. [DOI] [PubMed] [Google Scholar]
- 46.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 47.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 48.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 49.Marc JAN, Palmer JH. Changes in mitotic acitivity and cell size in the apical meristem of Helianthus annuus L. during the transition to flowering. Am. J. Bot. 1982;69:768–775. doi: 10.1002/j.1537-2197.1982.tb13317.x. [DOI] [Google Scholar]
- 50.Hileman LC. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130348. doi: 10.1098/rstb.2013.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hileman LC. Bilateral flower symmetry - how, when and why? Curr. Opin. Plant. Biol. 2014;17:146–152. doi: 10.1016/j.pbi.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Broholm SK, et al. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl Acad. Sci. USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim M, et al. Regulatory genes control a key morphological and ecological trait transferred between species. Science. 2008;322:1116–1119. doi: 10.1126/science.1164371. [DOI] [PubMed] [Google Scholar]
- 54.Chapman MA, Leebens-Mack JH, Burke JM. Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol. Biol. Evol. 2008;25:1260–1273. doi: 10.1093/molbev/msn001. [DOI] [PubMed] [Google Scholar]
- 55.Chapman MA, et al. Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet. 2012;8:e1002628. doi: 10.1371/journal.pgen.1002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tähtiharju S, et al. Evolution and diversification of the CYC/TB1 gene family in Asteraceae - a comparative study in gerbera (Mutisieae) and sunflower (Heliantheae) Mol. Biol. Evol. 2012;29:1155–1166. doi: 10.1093/molbev/msr283. [DOI] [PubMed] [Google Scholar]
- 57.Juntheikki-Palovaara I, et al. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae) Plant J. 2014;79:783–796. doi: 10.1111/tpj.12583. [DOI] [PubMed] [Google Scholar]
- 58.Garcês HMP, Spencer VMR, Kim M. Control of floret symmetry by RAY3, SvDIV1B, and SvRAD in the capitulum of Senecio vulgaris. Plant Physiol. 2016;171:2055–2068. doi: 10.1104/pp.16.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang D, et al. Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front. Plant Sci. 2016;7:1–15. doi: 10.3389/fpls.2016.01633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol. Phylogenet. Evol. 2008;47:757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Carlson SE, Howarth DG, Donoghue MJ. Diversification of CYCLOIDEA-like genes in Dipsacaceae (Dipsacales): implications for the evolution of capitulum inflorescences. BMC Evol. Biol. 2011;11:1–13. doi: 10.1186/1471-2148-11-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Claßen-Bockhoff R, Ruonala R, Bull-Hereñu K, Marchant N, Albert VA. The unique pseudanthium of Actinodium (Myrtaceae) - morphological reinvestigation and possible regulation by CYCLOIDEA-like genes. EvoDevo. 2013;4:8. doi: 10.1186/2041-9139-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fambrini M, Salvini M, Basile A, Pugliesi C. Transposon-dependent induction of Vincent van Gogh’s sunflowers: exceptions revealed. Genesis. 2014;52:315–327. doi: 10.1002/dvg.22743. [DOI] [PubMed] [Google Scholar]
- 64.Fambrini M, Salvini M, Pugliesi C. A transposon-mediate inactivation of a CYCLOIDEA-like gene originates polysymmetric and androgynous ray flowers in Helianthus annuus. Genetica. 2011;139:1521–1529. doi: 10.1007/s10709-012-9652-y. [DOI] [PubMed] [Google Scholar]
- 65.Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 66.Luo D, et al. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/S0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- 67.Corley SB, Carpenter R, Copsey L, Coen E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in. Proc. Natl Acad. Sci. USA. 2005;102:5068–5073. doi: 10.1073/pnas.0501340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costa MMR, Fox S, Hanna AI, Baxter C, Coen E. Evolution of regulatory interactions controlling floral asymmetry. Development. 2005;132:5093–5101. doi: 10.1242/dev.02085. [DOI] [PubMed] [Google Scholar]
- 69.Yang X, et al. Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell. 2012;24:1834–1847. doi: 10.1105/tpc.112.099457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laitinen RAE, Broholm S, Albert VA, Teeri TH, Elomaa P. Patterns of MADS-box gene expression mark flower-type development in Gerbera hybrida (Asteraceae) BMC Plant Biol. 2006;6:11. doi: 10.1186/1471-2229-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu H, et al. Whole-transcriptome analysis of differentially expressed genes in the vegetative buds, floral buds and buds of Chrysanthemum morifolium. PLoS ONE. 2015;10:1–24. doi: 10.1371/journal.pone.0128009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reyes-Chin-Wo S, et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017;8:14953. doi: 10.1038/ncomms14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Badouin H, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 74.Peng Y, et al. De novo genome assembly of the economically important weed horseweed using integrated data from multiple sequencing platforms. Plant Physiol. 2014;166:1241–1254. doi: 10.1104/pp.114.247668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scaglione D, et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016;6:1–17. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen, Q. et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant (2018), in press. 10.1016/j.molp.2018.03.015 [DOI] [PubMed]
- 77.Chanderbali AS, Berger BA, Howarth DG, Soltis PS, Soltis DE. Evolving ideas on the origin and evolution of flowers: new perspectives in the genomic era. Genetics. 2016;202:1255–1265. doi: 10.1534/genetics.115.182964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berger BA, et al. The unexpected depths of genome-skimming data: a case study examining Goodeniaceae floral symmetry genes. Appl. Plant Sci. 2017;5:1700042. doi: 10.3732/apps.1700042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berti F, Fambrini M, Turi M, Bertini D, Pugliesi C. Mutations of corolla symmetry affect carpel and stamen development in Helianthus annuus. Can. J. Bot. 2005;83:1065–1072. doi: 10.1139/b05-047. [DOI] [Google Scholar]