Abstract
Contrast enhanced MRI is commonly used in imaging and treatment planning of prostate cancer. However, no tumor targeting contrast agent is commercially available for accurate detection and characterization prostate cancer with MRI. Extradomain B fibronectin (EDB-FN), an oncoprotein present in aggressive tumors, is a promising molecular target for detection and stratification of high-risk prostate cancer. In this work, we have identified four small peptides (GVK, IGK, SGV, and ZD2) specific to EDB-FN for tumor targeting. In-silico simulations of the binding patterns and affinities of peptides to the EDB protein fragment revealed different binding site to different peptide in the ligand-receptor interactions. Tumor specificity and organ distribution of the peptides were assessed using fluorescence imaging in male mice bearing PC-3 human prostate cancer xenografts. Targeted contrast agents were synthesized by conjugating tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) to the peptides in the solid phase, followed by complexation with GdCl3. The contrast agents were characterized by MALDI-TOF mass spectrometry and relaxivity measurements. All four peptide Gd-DOTA conjugates resulted in robust tumor contrast enhancement in MR imaging of the PC3 mouse prostate cancer model. The peptide Gd-DOTA conjugates specific to EDB-FN are promising targeted small molecular macrocyclic contrast agents for MR molecular imaging of prostate cancer.
Keywords: EDB fibronectin, MRI, targeted contrast agent, peptide, prostate cancer
Graphical abstract
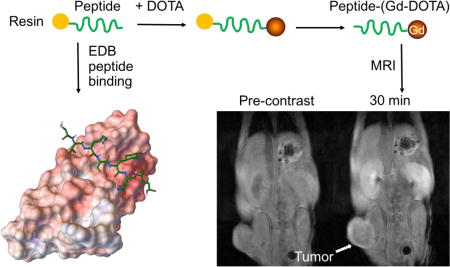
Introduction
Prostate cancer (PCa) is the most common cancer in men in the United States, with a lifetime incidence rate of 1 case per 7 men and a mortality of 1 out of 38 patients.[1] Current clinical practice for prostate cancer screening requires digital rectal examination and prostate specific antigen (PSA) measurement. If necessary, MRI and needle biopsy sampling are performed. However, these methods are inadequate either in sensitivity or specificity, often resulting in overdiagnosis and overtreatment with limited improvements to patient survival.[2–3] While no cancer-specific contrast agents are commercially available, there has been significant progress in developing novel technologies for diagnostic imaging of PCa. For example, multiparametric MRI, which combines morphological and functional imaging, dynamic contrast enhancement (DCE), and magnetic resonance spectroscopic imaging (MRSI) sequences, provides a non-invasive way to characterize PCa.[4–9] Additionally, PET probes targeting prostate-specific membrane antigen (PSMA) have entered clinical trials for detection of PCa.[10–12] Unfortunately, these strategies have had only limited success in locating and differentiating malignant PCa from low risk indolent lesions.[13]
We have identified extradomain B fibronectin (EDB-FN) as a molecular target for detection and differential diagnosis of high-risk PCa. EDB-FN, an oncofetal isoform of fibronectin (FN), is highly expressed in various aggressive cancers, including PCa, but is absent from normal tissues.[14–16] Strong expression of oncofetal FN, including EDB-FN, is correlated with a high rate of metastasis and poor overall survival in patients with prostate, breast, ovarian, head and neck, and other cancers.[15, 17–27] Clinical evidence and our preliminary data have demonstrated high expression of EDB-FN in high-risk PCa and low expression in benign lesions, including benign prostatic hyperplasia.[26–30] As revealed in one study, EDB-FN expression in prostate carcinoma at the mRNA level was 3.5-fold higher than in benign prostatic hyperplasia, and it had a distinct distribution pattern in the PCa stroma.[27] Thus, EDB-FN is a promising target for detection and differential localization of high-risk prostate tumors with molecular imaging.
In previous work, we synthesized and tested an EDB-FN specific contrast agent ZD2-Gd(HP-DO3A) for detection and differential diagnosis of high and low risk prostate tumors with MRI.[31] The targeted contrast agent was able to produce differential signal enhancement in mouse models of prostate tumors with different expression levels, which correlated with tumor aggressiveness. It shows the promise for non-invasive detection and risk-stratification of aggressive prostate cancer in MRI. Nevertheless, further studies may be needed to optimize the targeted contrast agent specific to EDB-FN for clinical translation.
In this work, we have identified three new small peptides specific to EDB-FN with phage display and synthesized four peptide Gd-DOTA conjugates as targeted contrast agents for MR molecular imaging of high-risk PCa. The binding pattern and affinity of ZD2 and three other peptides to the EDB-FN protein was simulated with the AutoDock software package. Specific binding of the peptides to aggressive PCa was further investigated in a mouse tumor model with fluorescence imaging. Macrocyclic chelate Gd-DOTA was used for the new targeted MRI contrast agents because of its high in vivo chelation stability.[32] The efficacy of the targeted contrast agents for MR molecular imaging was assessed in mice with aggressive PC3 prostate tumor xenografts. All targeted contrast agents exhibited stronger tumor enhancement than a clinical contrast agent Gd(HP-DO3A).
2 EXPERIMENTAL SECTION
2.1 Materials
The ligand 2,2',2''-(10-(1-carboxy-4-((4-isothiocyanatobenzyl)amino)-4-oxobutyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (p-NCS-Bz-DOTA-GA) was purchased from CheMatech, France. All the other reagents for chemical synthesis were bought from Sigma Aldrich unless stated otherwise. PC3 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in RPMI medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD, USA) and 1% penicillin and streptomycin (Thermo Fisher) at 37°C in 5% CO2. To construct GFP-expressing cell lines, cells were transfected with lentivirus as previously reported.[33]
2.2 Animals
BALB/c nude mice were purchased from Charles River Laboratories (Ashland, OH, USA) and housed in the Animal Resource Center Core Facility at Case Western Reserve University. All animal experiments were performed in accordance with the animal protocol approved by the CWRU Institutional Animal Care and Use Committee. Athymic nude mice (male) were subcutaneously injected with a 100 µL cell suspension (4×107 cells/mL) in Matrigel (Corning Bioscience, Corning, NY) to initiate tumor growth. Mice with tumors 5–8 mm in diameter were used for imaging studies.
2.3 Phage Display
The Ph.D C7C library (New England Biolabs, Beverly, MA) was used to screen for EDB-specific cyclic nonapeptides. Candidate peptides were selected by panning for four rounds.[30] In each round, purified EDB fragment (100 µg/mL) was immobilized by overnight coating on non-treated 96-well plates (Corning Costar, Tewksbury, MA, USA) 4°C. BSA (0.5%) was used to block non-specific binding (1 h, room temperature) followed by incubating with phages for 1 h at room temperature. Extensive washing with PBST (0.1%, 0.3%, 0.5% BSA, respectively, three times) was performed to remove non-binding phages before eluting the bound phages with 0.1 M glycine-HCl (pH 2.2) and neutralizing with Tris-HCl (pH 9.1). The eluted phages were titered and amplified with E. coli (ER2758), according to the user’s manual. Amplified phages in the medium were purified by ultrafiltration and PEG/NaCl precipitation. At the end of round 4, properly diluted phages were cultured on LB/IPTG/Xgal plates and DNA from 29 random blue plaques was sequenced using supplied primers (New England Biolabs) along with the phage library. Peptide sequences were acquired after translating the corresponding DNA sequences.
2.4 Molecular Docking
The molecular binding models were simulated with AutoDock Vina[34] and visualized with Python Molecular Viewer software.[35] Before running the docking command, the receptor (EDB fragment) and all the peptides were prepared as pdbqt files. For the receptor, the nuclear magnetic resonance spectroscopy (NMR) confirmed 3D crystal structure[36] was obtained from RSCB Protein Data Bank (PDB ID: 2fnb) as pdb file. The receptor then was opened in AutoDock Tools. Its coordinates center was set to x = −1.306, y = 1.677, z = 2.236. The size of the grid was set to 60 × 66 × 60. After this, all the H2O molecules were deleted and polar hydrogens were added to the receptor. At the same time, the non-polar hydrogens were merged. After this, Kollman Charge was added to the receptor. At last, the receptor was saved as pdbqt file.
For the linear peptides, their 3D conformations were converted from the 2D structures via ChemDraw 15.0 3D software. Their conformations were optimized by minimizing the energies with the force field method and then saved as mol2 files. Then the peptides were opened in AutoDock Tools. Polar hydrogens and charges were added in the same manner as described for the receptor. The rotatable bonds were selected and active torsions were set to 9. At last, the ligands were saved as pdbqt files. The AutoDock Vina was run in cmd.exe in Window10 with a configuration script which contains the coordination of the center, size of the grid, number of modes and location of the receptor, ligand and output file. The output files were obtained as txt and pdbqt files containing all the modes.
2.5 Fluorescence probe synthesis
The peptide was first synthesized in solid phase using Fmoc chemistry, followed by conjugation of a short PEG spacer (Fmoc-12-amino-4,7,10-trioxadode-canoic acid). Synthesis of the fluorescent probes, peptide-Cy5.5, was achieved by conjugating peptide-PEG on resin to Cy5.5 NHS ester (Lumiprobe, Hallandale Beach, FL, USA). The cleavage cocktail composed of TFA:H2O:triisobutylsilane (96.5:2.5:1) was used to remove the peptide Cy5.5 conjugates from the resin. The product was precipitated in cold ether and freeze dried. The conjugates were purified by preparative HPLC on an Agilent 1100 HPLC system equipped with a semi-preparative C18 column and characterized by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry on a Voyager DE-STR spectrometer (PerSeptive BioSystems) in linear mode with R 2,5-dihydroxybenzoic acid as a matrix. Concentration of peptide-Cy5.5 was quantified from the absorbance at 450 nm and a fluorphore extinction coefficient of ε = 209,000 L·mol−1·cm−1 as provided by the manufacturer.
2.6 Fluorescence Imaging
The peptide-Cy5.5 conjugates in 0.1 mL PBS (10 nmol) was administered by tail vein injection for each mouse. At 1.5 h post injection, the mice were sacrificed to image the tumors and the organs. The binding of peptide-Cy5.5 to the PC3 tumor and biodistribution were assessed in vivo before sacrifice and ex vivo using Maestro FLEX In Vivo Imaging System (Caliper Life Sciences, Hopkinton, MA) with a red filter set (spectral range of 630−910 nm, 1000 ms exposure time).
2.7 Synthesis of MRI contrast agent
Peptides were synthesized using solid phase chemistry. A short PEG spacer (Fmoc-12-amino-4,7,10-trioxadode-canoic acid) was then conjugated, followed by p-NCS-Bz-DOTA-GA in the presence of N,N-diisopropylethylamine in DMSO for 24 h in dark at room temperature. A cocktail of TFA:H2O:triisobutylsilane (96.5:2.5:1) was applied to harvest the precursor peptide-DOTA from the resin. After the precursors were precipitated in cold ether, centrifuged and freeze-dried. The peptide DOTA conjugates were dissolved in water and complexed with GdCl3 at room temperature overnight to obtain the targeted contrast agents. The peptide-(Gd-DOTA) conjugates were purified by HPLC on an Agilent 1100 HPLC system equipped with a semi-preparative C18 column. The gradient of HPLC was 100% water for 10 min and 0–20% acetonitrile in water for another 20 min and 50–100% acetonitrile in water for 5 min. MALDI-TOF mass spectra were acquired on a Voyager DE-STR spectrometer in linear mode with R 2,5-dihydroxybenzoic acid as a matrix. The lyophilized contrast agents were reconstituted in saline before use.
2.7 In vivo MRI
All MRI experiments were performed on an Aspect M3 compact MRI scanner (1.0 Telsa, Aspect Imaging, Israel). Imaging experiments were performed when the tumor size reached 5–8 mm in diameter. Each contrast agent was administered intravenously at a dose of 0.1 mmol/kg Gd3+ after acquiring pre-contrast images. ProHance® (gadoteridol) was used as a control in the imaging experiments. Post-contrast images were obtained at 10 min intervals up to 30 min. Axial slices of the mouse at the tumor location were acquired using a T1-weighted spin echo sequence with the following parameters: TR = 500 ms, TE = 8.144 ms, slice thickness = 1 mm, inter-slice gap = 0.1mm, field of view (FOV) = 3 cm × 3 cm. The total imaging time is 216 seconds. Contrast-to-noise ratio (CNR) at each time point was calculated by measuring the ratios of signal intensities in manually drawn regions of interest (ROIs) in tumor and muscle, and then normalizing to image noise. A 3D FLASH gradient echo sequence was used for whole-body coronal imaging. Imaging parameters were TR = 17.362 ms, TE = 6 ms, flip angle = 15°, slice thickness = 1.5 mm, FOV = 3.5 cm × 80 cm. The total imaging time is 226 seconds. Each of the 16 slices was aquired immediately after the acquisitions with the T1-weighted spin echo sequence.
3. RESULTS AND DISCUSSION
3.1 Receptor–ligand interaction studies
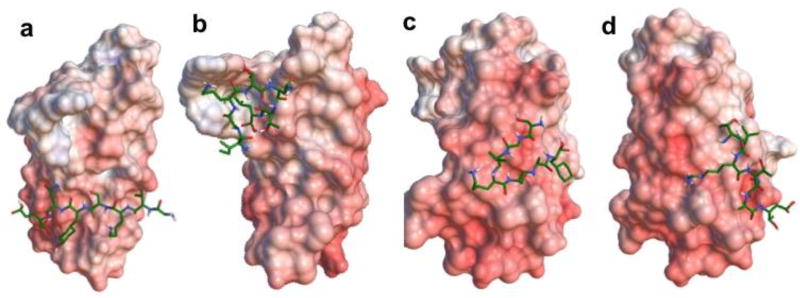
Beside ZD2 (TVRTSAD) peptide reported previously,[30] three other sequences namely GVKSYNE (GVK), IGKTNTL (IGK), SGVKSAF (SGV) were also identified to bind to EDB-FN from the phage display. To verify the potential for ligand-receptor interactions, in-silico simulation of the binding patterns and affinities of the linear peptides to EDB-FN was investigated with Autodock Vina[34]. The molecular docking generally visualized all the necessary interactions between the peptides and EDB fragment, which may be the binding sites for the ligands. These binding interactions include electrostatic interactions, aromatic-aromatic interactions, lipophilic-lipophilic interactions, hydrogen bond interactions and hydrophobic interactions. Even though the data from simulation is only predictive, it provides a prediction of how the peptide might bind to the receptor. Nine interaction modes for each peptide were generated and the site with the lowest binding energy and zero distance was selected. The best mode of each peptide is shown in Figure 1. The EDB fragment is highly acidic with negatively charged residues evenly distributed on the surface, as shown in red. The predicted affinity and distance from the best mode, isoelectric point (PI), grand average of hydropathicity (GAH)[37] of the peptides are listed in Table 1. A binding affinity above 5 kcal/mol was predicted for each of the peptides.
Figure 1.
Electrostatic surface of EDB fragment and 3D stick molecular models of linear peptides GVK (a), IGK (b), SGV (c), and ZD2 (d) fitted to the protein. For the surface of EDB, blue indicates positive charged residues, red represents negative areas and white are neutral regions. Active residues for docking calculations are numbered. For the peptide stick models, green indicates the carbon atoms, blue indicates the nitrogen atoms, white indicates the hydrogen atoms and red indicates the oxygen atoms.
Table 1.
Parameters related to peptide-EDB binding.
| Peptide | Affinity (kcal/mol) | Distance from best model |
Grand average of hydropathicity |
Isoelectric point |
|
|---|---|---|---|---|---|
|
| |||||
| RMSD/lb | RMSD/ub | ||||
| GVK | −7.3 ± 0.18 | 0.000 | 0.000 | − 1.31 | 6.00 |
| IGK | −4.96 ± 0.21 | 0.000 | 0.000 | − 0.13 | 8.75 |
| SGV | −5.91 ± 0.20 | 0.000 | 0.000 | 0.41 | 8.47 |
| ZD2 | −5.68 ± 0.15 | 0.000 | 0.000 | − 0.60 | 5.50 |
3.2 Tumor specific binding of the peptides
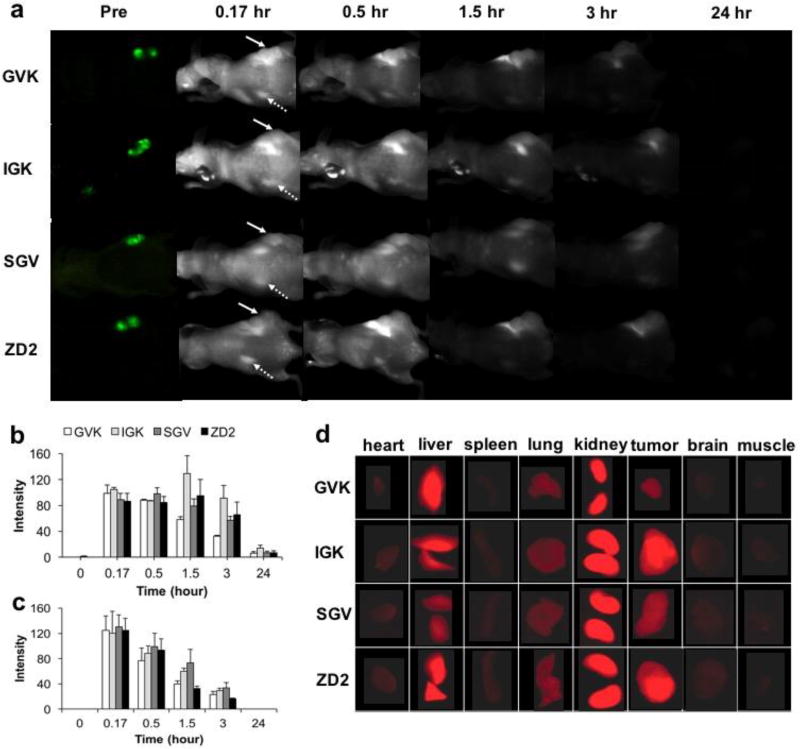
The tumor specific targeting profile and organ distribution of the peptides were investigated with in vivo and ex vivo fluorescence imaging in green fluorescence protein (GFP) labeled PC3 tumor bearing mice. The peptides were labeled with Cy5.5, a far-red emitting dye, via a short PEG spacer for fluorescence detection of the peptide. Figure 2a shows whole body fluorescence images for in vivo tumor targeting of the peptides. Tumors were located by imaging of green fluorescence prior to injection of fluorescently labeled peptides. Substantial Cy5.5 fluorescence signal was seen in the tumor of all 4 groups at 15 min after injection and lasted for 3 hrs. At 24 hrs post injection, trace amounts of fluorescence still could be observed (Figures 2a and b) in the tumors. By comparison, the fluorescence intensity in the kidneys (Figures 2a and c) of all groups was increased significantly at 15 min after injection but decreased faster than that in the tumors. At 3 h after injection, the fluorescence intensity decreased to 1/3 of its level at 15 min (Figure 2c) and no fluorescence could be observed at 24 hr.
Figure 2.
In vivo fluorescence imaging of PC3-GFP xenograft mice injected with various peptide-Cy5.5 at 10 nmol. Images were taken before and at 0.17, 0.5, 1.5, 3 and 24 hrs after injection (a); in vivo fluorescence intensity of tumors (b) and kidneys (c) under arrows over the period of 24 hrs. Tumor are indicated by full arrows, while kidneys are indicated by dotted arrows; ex vivo fluorescence images of organs at 1.5 hr post injection(d) various peptide-Cy5.5 at 10 nmol. (n = 5).
The distribution of Cy5.5 labeled peptides in organs was evaluated by ex vivo fluorescence imaging at 1.5 hr after injection. As shown in Figure 2d, the kidney exhibited the highest fluorescence intensity, followed by liver, tumor lung and heart. Spleen, brain and muscle showed only trace amount of fluorescence. In the tumor, a difference in intensity was observed among the peptides at 90 min after administration. Specifically, greater fluorescence signals of IGK and ZD2 were seen in tumors compared to SGV, while GVK was the weakest (Figures 2d). In normal tissues, no EDB-FN is present[14–16], so the fluorescence intensity was weak in the brain, muscle, spleen, heart and lung, consistent with our previous finding.[30, 31] Significant fluorescence was observed in the kidneys and liver because the small water soluble peptides with molecular weights around 1.6 kD mainly cleared via the kidneys, and then by the liver. This is logical since the molecular weight cutoff for glomerular filtration is arround 30–50 kDa,[38] far beyond these peptides with 7 amino acid residues. The short half-life and fast clearance of small peptide from circulation has been discussed extensively in literature.[39]
3.3 Synthesis of the targeted contrast agents
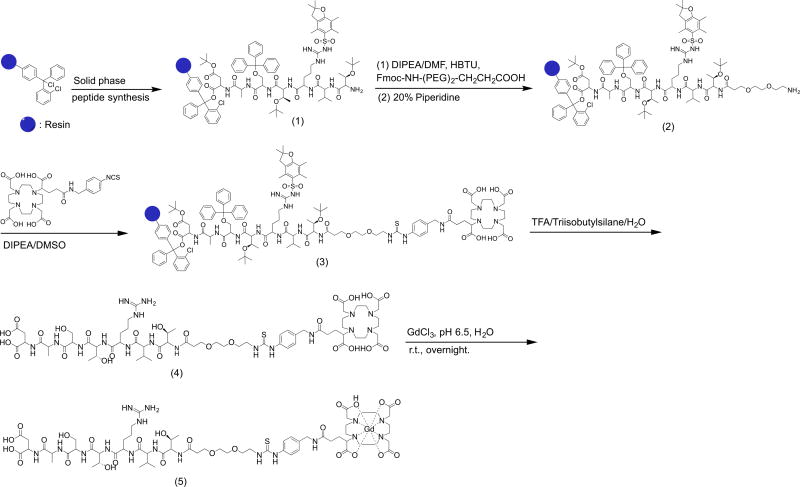
The synthesis of the MRI contrast agent peptide-(Gd-DOTA) is demonstrated in Figure 3 with ZD2 as an example peptide. The peptides were first synthesized in the solid phase using standard Fmoc chemistry. The overall yield of compound 1 was greater than 90%. It should be noted that the addition of a spacer between the peptide and DOTA with isothiocyanate is necessary for conjugation in solid phase when TFA is used for cleaving the products from the resin. Directly linking the peptide to DOTA via isothiocyanate was unsuccessful because it underwent a cyclization with isothiocyanate and subsequent removal of the last amino acid.[40] The addition of a small PEG spacer at the N-terminus of the peptides prevented the side reaction during cleavage of the product from the resin with TFA.[41] The final yield of compound 4 was 80% after purification.
Figure 3.
A typical synthetic procedure of the targeted contrast agents with ZD2-(Gd-DOTA) as an example.
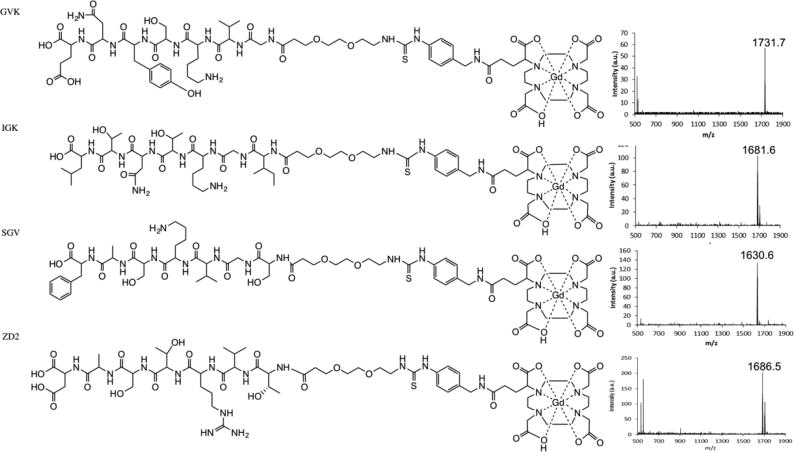
The peptide targeted MRI contrast agents were finally synthesized by complexation of the ligands with gadolinium (III) ions. The structures of the 4 peptide-based GBCAs are shown in Figure 4. The products were purified by preparative HPLC and characterized by MALDI-TOF mass spectrometry. The MALDI-TOF measured m/z of GVK, IGK, SGV and ZD2 targeted contrast agents were 1731.7, 1681.6, 1630.6 and 1686.5, respectively. The calculated m/z values for the agents were 1731.6, 1681.7, 1630.6 and 1686.5, correspondingly. Although the peptide-(Gd-DOTA) had an increased molecular weight around 1.6 kD as compared to the corresponding peptides, they all had good water solubility. The solid phase reaction conditions were advantageous as they allowed for easy removal of all impurities from the final product. Overall, this synthetic approach provided a facile and efficient way to synthesize the peptide-(Gd-DOTA) with good overall yield.
Figure 4.
Chemical structure of each contrast agent and m/z measurements of the contrast agents, GVK, IGK, SGV and ZD2.
3.4 Relaxivity
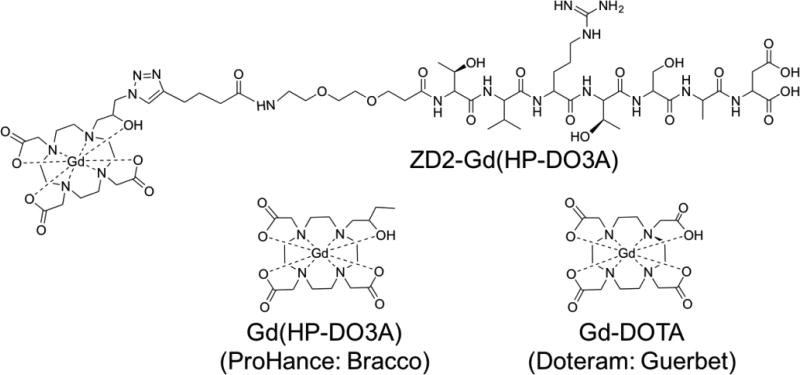
Relaxivity is an important parameter for an MR contrast agent. It reflects how the relaxation rate of the surrounding water proton changes as a function of concentration, and it is directly associated with the agent’s contrast enhancing capability during imaging. The relaxivity of the 4 contrast agents was tested at 37°C in water and compared to Gd(HP-DO3A), Gd-DOTA and ZD2-Gd(HP-DO3A) (Figure 5). As shown in Table 2, The r1 relaxivity for the GVK, IGK, SGV and ZD2 Gd-DOTA conjugates was 4.3, 4.6, 4.7 and 4.1 mM−1sec−1 at 1.5T, respectively, significantly higher than that of Gd-DOTA or Gd(HP-DO3A), which were 2.9 and 3.2 mM−1sec−1. The r2 relaxivities for GVK, IGK, SGV and ZD2 targeted contrast agents were 5.0, 5.2, 5.6 and 4.8 mM−1sec−1 at 1.5T, respectively, which were also significantly higher than that of Gd-DOTA or Gd(HP-DO3A) (both 3.2 mM−1sec−1). ZD2-Gd(HP-DO3A) exhibited high r1 and r2 relaxivities of 5.4 and 6.1 mM−1sec−1, respectively. Compared to Gd(HP-DO3A), Gd-DOTA, the size increase of the targeted contrast agents due to the peptides and PEG spacer could result in an increase of τR,[42, 43] thus leading to higher relaxivities.
Figure 5.
Chemical structure of ZD2-Gd(HP-DO3A), Gd(HP-DO3A) and Gd-DOTA.
Table 2.
Relaxivities of the contrast agents in 1.5 T in water, 37 °C.
| Agents | r1 (mM−1sec−1) | r2 (mM−1sec−1) |
|---|---|---|
| Gd(HP-DO3A) | 2.9 | 3.2 |
| Gd-DOTA | 2.9 | 3.2 |
| GVK-(Gd-DOTA) | 4.3 | 5.0 |
| IGK-(Gd-DOTA) | 4.6 | 5.2 |
| SGV-(Gd-DOTA) | 4.7 | 5.6 |
| ZD2-(Gd-DOTA) | 4.1 | 4.8 |
| ZD2-Gd(HP-DO3A) | 5.4 | 6.1 |
3.5 Magnetic Resonance Imaging
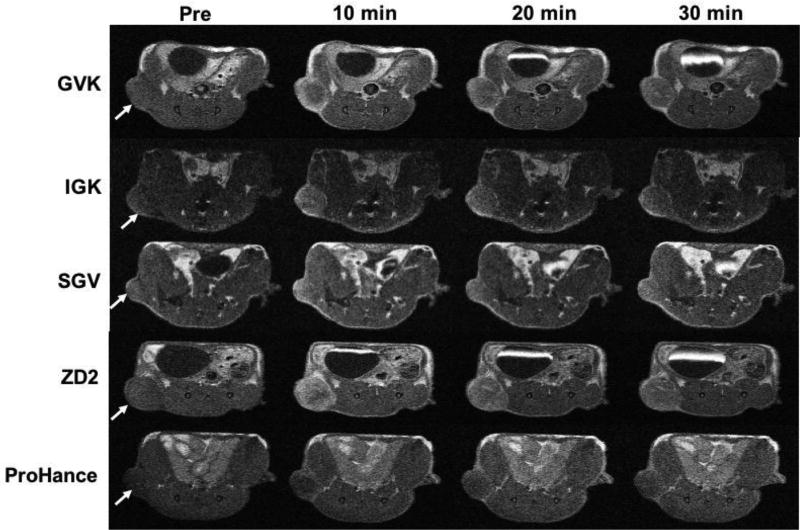
The MRI contrast enhancement of each of the agents was tested in PC3 tumor-bearing mice with different MRI sequences. Figure 6 shows the high-resolution 2D axial MR images of tumor tissues obtained a T1-weighted spin-echo sequence before (pre) and at 10, 20 and 30 min after i.v. injection at a dose of 0.1 mmol/kg. Strong tumor signal enhancement was observed at 10 minutes and remained for at least 30 min post injection for all the targeted contrast agents. The results were consistent with observation from the fluorescence imaging. The non-specific clinical agent Gd(HP-DO3A) only resulted in modest contrast enhancement in the PC3 tumors. The robust signal enhancement is the tumor remained in the tumors for at least 30 minutes for the targeted contrast agents when background signal were significantly decreased. Prolonged tumor enhancement indicated the binding of the targeted contrast agents to the tumor tissues. Background noise reduction at later time points would provide better tumor delineation. The findings demonstrated the effectiveness of the contrast agents specific to EDB-FN for MR molecular imaging of aggressiveness PCa.
Figure 6.
T1-weighted axial MRI images of PC3 tumor-bearing mice acquired pre-contrast (pre) and at 10, 20 and 30 min after i.v. injection of each contrast agent at 0.1 mmol/kg. The tumors are indicated by arrows (n = 5).
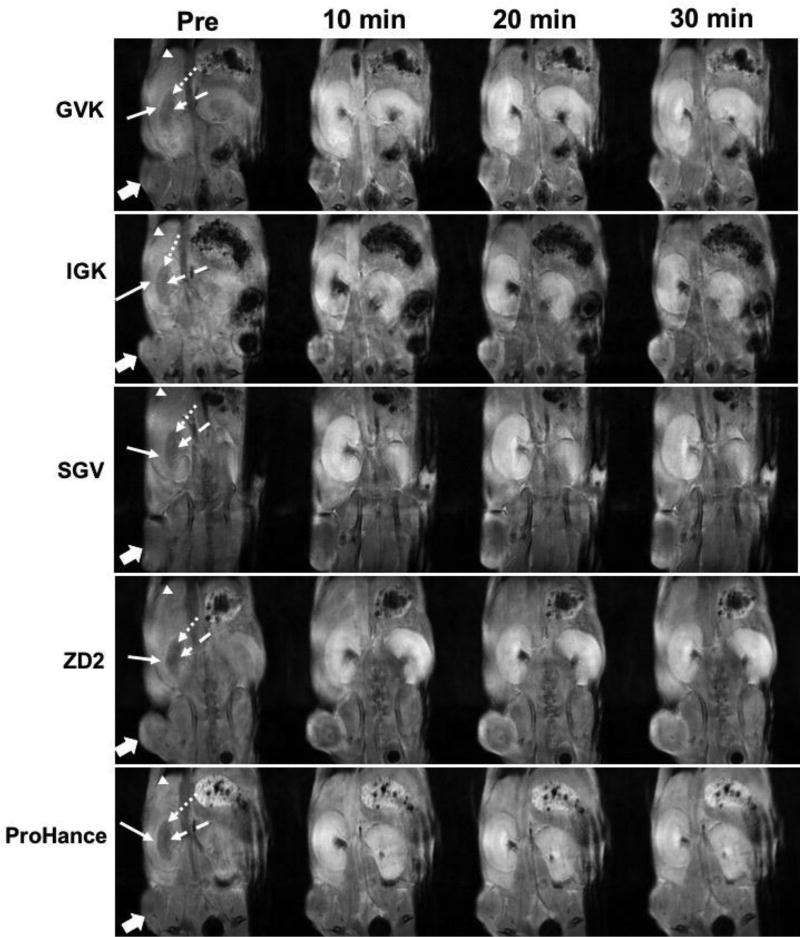
Whole-body MR images enhanced with the targeted contrast agents were obtained with a T1-weighted 3D FLASH sequence to assess signal enhancement in the tumors and the major organs, including the liver and kidneys. Figure 7 shows the 2D coronal images enhanced by the targeted contrast agents. All four peptide targeted contrast agents produced significant tumor enhancement as compared to the non-specific control agent Gd(HP-DO3A), consistent with the results obtained with the spin-echo sequence. Significant signal enhancement was observed in the cortex and outer medulla of the kidneys at 10 and 20 min after injection for all the 5 agents including ProHance.[44] The signal was increased after 20 minutes in the inner medulla, where the urine and other metabolites, including the contrast agents, concentrate and pass through the collecting duct.[45] In the liver, only slight signal increases were observed for all the contrast agents, including Gd(HP-DO3A), suggesting that the liver was not the main pathway for clearance. Considering the same kidney signal intensity and rhythm between the peptide based GBCAs and Gd(HP-DO3A), it can be inferred that the peptide based contrast agents underwent a similar renal excretion pattern as Gd(HP-DO3A). These findings were consistent with observation from the fluorescence imaging as well as our previous report on ZD2-Gd(HP-DO3A).[31]
Figure 7.
T1-weighted 3D FLASH coronal MR images of PC3 tumor-bearing mice acquired pre-contrast (Pre) and at 10, 20 and 30 min after i.v. injection of GVK (a), IGK(b), SGV(c), ZD2(d) Gd-DOTA conjugates and Gd(HP-DO3A) (e) at 0.1 mmol/kg respectively. In the kidney cortex (full arrow) and outer medulla (round dot arrow), significant signal enhancement was found at 10, 20 and 30 min following injection of all peptide agents. In the inner medulla (dash arrow), high signal enhancement was found at 20 and 30 min as compared to at 10 min for the agents. In the liver, slight increase of signal (arrow head) at 10, 20 and 30 min after injection of the agents; In the tumor (block full arrow), all targeted agents produced significantly high tumor contrast enhancement as compared to the contrast agent Gd(HP-DO3A) (n = 5).
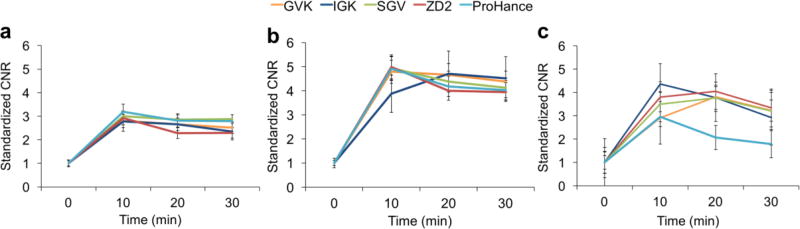
Quantitative analysis of contrast-to-noise ratios (CNR) in the tumor, liver and kidneys in T1-weighted 3D FLASH images further demonstrated the tumor enhancement of the targeted contrast agents. As shown in Figure 8a, the CNR in the liver increased about three fold above baseline at 10 min after injection of all agents, and then gradually decreased to an average CNR of 2.6 at 30 min. There was no significant difference between the peptide-based contrast agents and ProHance. By contrast, the CNR in the kidneys was much higher than in the liver, in a range of 3.9–5.0, as shown in Figure 8b. The GVK, IGK, SGV, ZD2 and Prohance groups had no significant difference in kidney CNR enhancement over the imaging period of 30 min. As shown in Figure 8c, the CNR in PC3 tumors increased 2.9–4.4 fold at 10 min after injection and peaked at 20 min for the GVK, SGV and ZD2 targeted agents. The IGK and ProHance, however, showed a peak CNR at 10 min and then gradually decreased. Overall, ProHance resulted in lower CNR in PC3 tumors, especially at the later time points (20 and 30 min) after the injection, compared to the peptide targeted contrast agents.
Figure 8.
Contrast to noise ratio of liver(a), kidney (b) and tumor(c). (n = 5) Error bars represent ± standard deviation.
MRI provides high-resolution images of soft tissues and has been routinely used in cancer detection, diagnosis, therapeutic efficacy evaluation, and image-guided interventions. However, the potential of molecular MRI for accurate detection and delineation of human cancers, including prostate cancer, has not been fully utilized because of non-specificity of the existing contrast agents. The development of cancer-specific MRI contrast agents is hampered by the low sensitivity of MRI for molecular imaging. Although significant efforts have been devoted to design nanosized targeted MRI contrast agents to improve the sensitivity of molecular MRI, the slow excretion of the nanosized contrast agents has impeded their clinical development because of the safety concerns about their long-term accumulation. For the first time, we have demonstrated the effectiveness of MR molecular imaging of cancer with clinical translatable small molecular contrast agents in animal tumor models.[30,31,33,46,47] These agents are designed to target abundant oncoproteins in tumor extracellular matrix, including fibrin-fibronectin clots and EDB-FN. Because of the abundance of the molecular targets, a sufficient amount of the targeted contrast agents can bind to the tumors to generate robust signal enhancement for effective MR molecular imaging. The small size of the peptide targeted contrast agents allows rapid and complete clearance as the clinical contrast agents, which will alleviate the safety concerns for clinical translation.
We have shown in this study that the three new peptides are equally effective for MR molecular imaging of prostate cancer as the previous reported ZD2 peptide, although they bind to different sites of EDB protein fragment. The peptide targeted contrast agents are based on a macrocyclic contrast agent, Gd-DOTA, which is approved for its high chelation stability and minimal tissue retention, especially in the brain[48–50]. The robust and prolonged signal enhancement of the targeted contrast agents in the aggressive PCa model suggests that they are promising for effective molecular MRI of prostate cancer. Nevertheless, further investigations are needed to determine their pharmacokinetics, tissue retention and safety for clinical translation. Based on our previous publications about the correlation of EDB-FN expression with tumor aggressiveness,[30,31] these agents have a potential to non-invasively assess tumor aggressiveness with MRI. They can be readily incorporated into the existing protocols of multiparametric MRI for clinical management of prostate cancer.
4. CONCLUSION
In this work, we synthesized four new peptide Gd-DOTA conjugates specific to EDB-FN as targeted contrast agents for MR molecular imaging of prostate cancer. Computational simulation indicates that the peptides bind to different sites of the EDB fragment. The specific tumor binding of the peptides is demonstrated in mice bearing PC3 human prostate cancer xenografts. The peptide targeted contrast agents have a good water solubility and relatively high relaxivities. All of the targeted contrast agents produce robust and prolonged signal enhancement in the tumor as compared to a clinical contrast agent. Because of the high in vivo stability and good safety profile of Gd-DOTA, it is expected that the peptide targeted Gd-DOTA conjugates would have a good safety profile as the clinical agent. Besides MR molecular imaging, the EDB-FN specific peptides have a potential to deliver other imaging probes and therapeutics for cancer imaging and therapy. The targeted contrast agents have a potential to provide accurate detection and diagnosis of aggressive prostate cancer with molecular MRI.
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Health grants (R01 EB00489 and R01 CA211762) and (R43 CA199826). Zheng-Rong Lu is the M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Bill-Axelson A, Holmberg L, Garmo H, Rider JR, Taari K, Busch C, Nordling S, Haggman M, Andersson SO, Spangberg A, Andren O, Palmgren J, Steineck G, Adami HO, Johansson JE. Radical prostatectomy or watchful waiting in early prostate cancer. N. Engl. J. Med. 2014;370(10):932–42. doi: 10.1056/NEJMoa1311593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter HB, Albertsen PC, Barry MJ, Etzioni R, Freedland SJ, Greene KL, Holmberg L, Kantoff P, Konety BR, Murad MH, Penson DF, Zietman AL. Early detection of prostate cancer: AUA Guideline. J. Urol. 2013;190(2):419–26. doi: 10.1016/j.juro.2013.04.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durmus T, Reichelt U, Huppertz A, Hamm B, Beyersdorff D, Franiel T. MRI-guided biopsy of the prostate: correlation between the cancer detection rate and the number of previous negative TRUS biopsies. Diagn. Interv. Radiol. 2013;19(5):411–7. doi: 10.5152/dir.2013.13055. [DOI] [PubMed] [Google Scholar]
- 5.van de Ven WJ, Barentsz JO. Prostate cancer: MRI/US-guided biopsy-a viable alternative to TRUS-guidance. Nat. Rev. Urol. 2013;10(10):559–60. doi: 10.1038/nrurol.2013.179. [DOI] [PubMed] [Google Scholar]
- 6.Turkbey B, Mena E, Aras O, Garvey B, Grant K, Choyke PL. Functional and molecular imaging: applications for diagnosis and staging of localised prostate cancer. Clin. Oncol. (R. Coll. Radiol.) 2013;25(8):451–60. doi: 10.1016/j.clon.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puech P, Rouviere O, Renard-Penna R, Villers A, Devos P, Colombel M, Bitker MO, Leroy X, Mege-Lechevallier F, Comperat E, Ouzzane A, Lemaitre L. Prostate cancer diagnosis: multiparametric MR-targeted biopsy with cognitive and transrectal US-MR fusion guidance versus systematic biopsy--prospective multicenter study. Radiology. 2013;268(2):461–9. doi: 10.1148/radiol.13121501. [DOI] [PubMed] [Google Scholar]
- 8.Roobol MJ, Carlsson SV. Risk stratification in prostate cancer screening. Nat. Rev. Urol. 2013;10(1):38–48. doi: 10.1038/nrurol.2012.225. [DOI] [PubMed] [Google Scholar]
- 9.Afshar-Oromieh A, Haberkorn U, Hadaschik B, Habl G, Eder M, Eisenhut M, Schlemmer HP, Roethke MC. PET/MRI with a (68)Ga-PSMA ligand for the detection of prostate cancer. Eur. J. Nucl. Med. Mol. Imaging. 2013;40(10):1629–30. doi: 10.1007/s00259-013-2489-5. [DOI] [PubMed] [Google Scholar]
- 10.Giesel FL, Sterzing F, Schlemmer HP, Holland-Letz T, Mier W, Rius M, Afshar-Oromieh A, Kopka K, Debus J, Haberkorn U, Kratochwil C. Intra-individual comparison of (68)Ga-PSMA-11-PET/CT and multi-parametric MR for imaging of primary prostate cancer. Eur. J. Nucl. Med. Mol. Imaging. 2016;43(8):1400–6. doi: 10.1007/s00259-016-3346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorin MA, Pomper MG, Rowe SP. PSMA-targeted imaging of prostate cancer: the best is yet to come. BJU Int. 2016;117(5):715–6. doi: 10.1111/bju.13435. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee SR, Pullambhatla M, Byun Y, Nimmagadda S, Green G, Fox JJ, Horti A, Mease RC, Pomper MG. 68Ga-labeled inhibitors of prostate-specific membrane antigen (PSMA) for imaging prostate cancer. J. Med. Chem. 2010;53(14):5333–41. doi: 10.1021/jm100623e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegde JV, Mulkern RV, Panych LP, Fennessy FM, Fedorov A, Maier SE, Tempany CM. Multiparametric MRI of prostate cancer: an update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J. Magn. Reson. Imaging. 2013;37(5):1035–54. doi: 10.1002/jmri.23860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64(21):7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 15.Inufusa H, Nakamura M, Adachi T, Nakatani Y, Shindo K, Yasutomi M, Matsuura H. Localization of oncofetal and normal fibronectin in colorectal cancer. Correlation with histologic grade, liver metastasis, and prognosis. Cancer. 1995;75(12):2802–8. doi: 10.1002/1097-0142(19950615)75:12<2802::aid-cncr2820751204>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 16.Sauer S, Erba PA, Petrini M, Menrad A, Giovannoni L, Grana C, Hirsch B, Zardi L, Paganelli G, Mariani G, Neri D, Durkop H, Menssen HD. Expression of the oncofetal ED-B-containing fibronectin isoform in hematologic tumors enables ED-B-targeted 131I-L19SIP radioimmunotherapy in Hodgkin lymphoma patients. Blood. 2009;113(10):2265–74. doi: 10.1182/blood-2008-06-160416. [DOI] [PubMed] [Google Scholar]
- 17.Freire-de-Lima L, Gelfenbeyn K, Ding Y, Mandel U, Clausen H, Handa K, Hakomori SI. Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process. Proc. Natl. Acad. Sci. USA. 2011;108(43):17690–5. doi: 10.1073/pnas.1115191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaspar M, Zardi L, Neri D. Fibronectin as target for tumor therapy. Int. J. Cancer. 2006;118(6):1331–9. doi: 10.1002/ijc.21677. [DOI] [PubMed] [Google Scholar]
- 19.Menzin AW, Loret de Mola JR, Bilker WB, Wheeler JE, Rubin SC, Feinberg RF. Identification of oncofetal fibronectin in patients with advanced epithelial ovarian cancer: detection in ascitic fluid and localization to primary sites and metastatic implants. Cancer. 1998;82(1):152–8. doi: 10.1002/(sici)1097-0142(19980101)82:1<152::aid-cncr19>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Loridon-Rosa B, Vielh P, Matsuura H, Clausen H, Cuadrado C, Burtin P. Distribution of oncofetal fibronectin in human mammary tumors: immunofluorescence study on histological sections. Cancer Res. 1990;50(5):1608–12. [PubMed] [Google Scholar]
- 21.Santimaria M, Moscatelli G, Viale GL, Giovannoni L, Neri G, Viti F, Leprini A, Borsi L, Castellani P, Zardi L, Neri D, Riva P. Immunoscintigraphic detection of the ED-B domain of fibronectin, a marker of angiogenesis, in patients with cancer. Clin. Cancer Res. 2003;9(2):571–9. [PubMed] [Google Scholar]
- 22.Hesse E, Musholt PB, Potter E, Petrich T, Wehmeier M, von Wasielewski R, Lichtinghagen R, Musholt TJ. Oncofoetal fibronectin--a tumour-specific marker in detecting minimal residual disease in differentiated thyroid carcinoma. Br. J. Cancer. 2005;93(5):565–70. doi: 10.1038/sj.bjc.6602741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons AJ, Bateman AC, Spedding A, Primrose JN, Mandel U. Oncofetal fibronectin and oral squamous cell carcinoma. Br. J. Oral Maxillofac. Surg. 2001;39(6):471–7. doi: 10.1054/bjom.2001.0702. [DOI] [PubMed] [Google Scholar]
- 24.Richter P, Junker K, Franz M, Berndt A, Geyer C, Gajda M, Kosmehl H, Wunderlich H. IIICS de novo glycosylated fibronectin as a marker for invasiveness in urothelial carcinoma of the urinary bladder (UBC) J. Cancer Res. Clin. Oncol. 2008;134(10):1059–65. doi: 10.1007/s00432-008-0390-6. [DOI] [PubMed] [Google Scholar]
- 25.Mhawech P, Dulguerov P, Assaly M, Ares C, Allal AS. EB-D fibronectin expression in squamous cell carcinoma of the head and neck. Oral Oncol. 2005;41(1):82–8. doi: 10.1016/j.oraloncology.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Jankovic MM, Kosanovic MM. Fibronectin pattern in benign hyperplasia and cancer of the prostate. Dis. Markers. 2008;25(1):49–58. doi: 10.1155/2008/308420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albrecht M, Renneberg H, Wennemuth G, Moschler O, Janssen M, Aumuller G, Konrad L. Fibronectin in human prostatic cells in vivo and in vitro: expression, distribution, and pathological significance. Histochem. Cell Biol. 1999;112(1):51–61. doi: 10.1007/s004180050391. [DOI] [PubMed] [Google Scholar]
- 28.Sonmez H, Suer S, Karaarslan I, Baloglu H, Kokoglu E. Tissue fibronectin levels of human prostatic cancer, as a tumor marker. Cancer Biochem. Biophys. 1995;15(2):107–10. [PubMed] [Google Scholar]
- 29.Suer S, Sonmez H, Karaaslan I, Baloglu H, Kokoglu E. Tissue sialic acid and fibronectin levels in human prostatic cancer. Cancer Lett. 1996;99(2):135–7. doi: 10.1016/0304-3835(95)04084-6. [DOI] [PubMed] [Google Scholar]
- 30.Han Z, Zhou Z, Shi X, Wang J, Wu X, Sun D, Chen Y, Zhu H, Magi-Galluzzi C, Lu Z-R. EDB Fibronectin Specific Peptide for Prostate Cancer Targeting. Bioconjug. Chem. 2015;26(5):830–38. doi: 10.1021/acs.bioconjchem.5b00178. [DOI] [PubMed] [Google Scholar]
- 31.Han Z, Li Y, Roelle S, Zhou Z, Liu Y, Sabatelle R, DeSanto A, Yu X, Zhu H, Magi-Galluzzi C, Lu Z-R. A Targeted Contrast Agent Specific to an Oncoprotein in Tumor Microenvironment with the Potential for Detection and Risk Stratification of Prostate Cancer with MRI. Bioconjugate. Chem. 2017;28(4):1031–40. doi: 10.1021/acs.bioconjchem.6b00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morcos SK. Extracellular gadolinium contrast agents: differences in stability. Eur. J. Radiol. 2008;66(2):175–9. doi: 10.1016/j.ejrad.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z, Wu X, Kresak A, Griswold M, Lu ZR. Peptide targeted tripod macrocyclic Gd(III) chelates for cancer molecular MRI. Biomaterials. 2013;34(31):7683–93. doi: 10.1016/j.biomaterials.2013.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31(2):455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanner MF. Python: A Programming Language for Software Integration and Development. J. Mol. Graphics Mod. 1999;17(1):57–61. [PubMed] [Google Scholar]
- 36.Fattorusso R, Pellecchia M, Viti F, Neri P, Neri D, Wuthrich K. NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis. Structure. 1999;7(4):381–390. doi: 10.1016/s0969-2126(99)80051-3. [DOI] [PubMed] [Google Scholar]
- 37.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157(1):105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.Ruggiero A, Villa CH, Bander E, Rey DA, Bergkvist M, Batt CA, Manova-Todorova K, Deen WM, Scheinberg DA, McDevitt MR. Paradoxical glomerular filtration of carbon nanotubes. Proc. Natl. Acad. Sci. 2010;107(27):12369–12374. doi: 10.1073/pnas.0913667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di L. Strategic Approaches to Optimizing Peptide ADME Properties. AAPS J. 2015;17(1):134–143. doi: 10.1208/s12248-014-9687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jullian M, Hernandez A, Maurras A, Puget K, Amblard M, Martinez J, Subra G. N-terminus FITC labeling of peptides on solid support: the truth behind te spacer. Tetrahedron Lett. 2009;50(3):260–3. [Google Scholar]
- 41.Toth E, van Uffelen I, Helm L, Merbach AE, Ladd D, Briley- Saebo KK, Kellar KE. Gadolinium-based linear polymer with temperature-independent proton relaxivities: a unique interplay between the water exchange and rotational contributions. Magn. Reson. Chem. 1998;36(S1):S125–34. [Google Scholar]
- 42.Doble DMJ, Botta M, Wang J, Aime S, Barge A, Raymond KN. Optimization of the Relaxivity of MRI Contrast Agents: Effect of Poly(ethylene glycol) Chains on the Water-Exchange Rates of GdIIIComplexes. J. Am. Chem. Soc. 2011;123(43):10758–9. doi: 10.1021/ja011085m. [DOI] [PubMed] [Google Scholar]
- 43.Raymond KN, Pierre VC. Next Generation, High Relaxivity Gadolinium MRI Agents. Bioconjugate Chem. 2005;16(1):3–8. doi: 10.1021/bc049817y. [DOI] [PubMed] [Google Scholar]
- 44.Xie L, Bennett KM, Liu C, Johnson AG, Zhang JL, Lee VS. MRI tools for assessment of microstructure and nephron function of the kidney. Am. J. Physiol. Renal Physiol. 2016;311(6):F1109–24. doi: 10.1152/ajprenal.00134.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pannabecker PL. Comparative physiology and architecture associated with the mammalian urine concentrating mechanism: role of inner medullary water and urea transport pathways in the rodent medulla. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;304(7):R488–503. doi: 10.1152/ajpregu.00456.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Z, Lu Z-R. Targeting Fibronectin for Cancer Imaging and Therapy. J. Mater. Chem. B. 2017;5(4):639–654. doi: 10.1039/C6TB02008A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Lu Z-R. Gadolinium-based contrast agents for MR cancer imaging. Wiley Interdiscip Rev. Nanomed. Nanobiotechnol. 2013;5(1):1–18. doi: 10.1002/wnan.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robert P, Violas X, Grand S, Lehericy S, Idée J-M, Ballet S, Corot C. Linear gadolinium-based contrast agents are associated with brain gadolinium retention in healthy rats. Invest. Radiol. 2016;51(2):73–82. doi: 10.1097/RLI.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aime S, Caravan P. Biodistribution of gadolinium-based contrast agents, including gadolinium deposition. J Magn Reson Imaging. 2009;30(6):1259–1267. doi: 10.1002/jmri.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramalho J, Semelka RC, Ramalho M, Nunes RH, AlObaidy M, Castillo M. Gadolinium-based contrast agent accumulation and toxicity: an update. Am. J. Neuroradiol. 2016;37(7):1192–1198. doi: 10.3174/ajnr.A4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.