Abstract
To examine the functional role of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) and PPARγ in skin cancer, stable cell lines were created in the A431 human squamous cell carcinoma cell line. Expression of PPAR target genes was greatly enhanced in response to ligand activation of PPARβ/δ or PPARγ in A431 cells expressing these receptors. PPARβ/δ expression blocked the cell cycle at the G2/M phase, and this effect was increased by ligand activation. Ligand activation of PPARβ/δ markedly inhibited clonogenicity as compared to vehicle-treated controls. Similarly, ligand activation of PPARγ in A431 cells expressing PPARγ resulted in reduced clonogenicity. Expression of either PPARβ/δ or PPARγ markedly reduced tumor volume in ectopic xenografts, while ligand activation of these receptors had little further influence on tumor volume. Collectively, these studies demonstrate that stable expression and activation of PPARβ/δ or PPARγ in A431 cells led to reduced tumorigenicity. Importantly, PPAR expression or ligand activation had major impacts on clonogenicity and/or tumor volume. Thus, PPARβ/δ or PPARγ could be therapeutically targeted for the treatment of squamous cell carcinomas.
Keywords: skin, carcinoma, PPARβ/δ, PPARγ, proliferation, cell cycle
1. Introduction
Targeting peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) or PPARγ has potential for the prevention and treatment of skin cancer because these transcription factors are nodal in nature and target multiple signaling pathways (reviewed in (Peters et al. 2015; Peters et al. 2012)). The first observation to suggest that PPARβ/δ could prevent chemically-induced skin cancer was the enhanced tumorigenicity found in Pparβ/δ-null mice as compared to controls (Kim et al. 2004). This was later supported by numerous studies showing that ligand activation of PPARβ/δ inhibits chemically-induced non-melanoma skin cancer through mechanisms that include induction of terminal differentiation, inhibition of mitosis, and promoting oncogene-induced senescence by modulating extracellular signal-regulated kinase (ERK) and protein kinase B (AKT) activities, and repressing endoplasmic reticulum stress (Bility et al. 2008; Bility et al. 2010; Zhu et al. 2011; Zhu et al. 2014a; Zhu et al. 2014b; Zhu et al. 2012). These mechanisms were verified using both in vivo and in vitro mouse models. Interestingly, the role of PPARβ/δ in other forms of cancer is less clear due to conflicting reports in the literature and may be due to differences in the models examined (reviewed in (Muller 2017; Peters et al. 2015; Peters et al. 2012)). Thus, clinical studies examining the role of PPARβ/δ ligands in cancer prevention and therapy are lacking due to disparate results in rodent cancer and cell culture studies showing both anti- and pro-carcinogenic effects. This illustrates the need for more experimentation using alternative approaches to help resolve the role of PPARβ/δ in carcinogenesis.
Interestingly, it was also initially suggested that ligand activation of PPARγ promoted colon cancer, but subsequent studies showed that activation of PPARγ inhibited colon cancer, as well as other types of cancer (reviewed in (Peters et al. 2015; Peters et al. 2012)). This has led to clinical trials examining the efficacy of PPARγ ligands as cancer chemopreventive or chemotherapeutic agents in humans (reviewed in (Peters et al. 2012)). With respect to non-melanoma skin cancer, it was originally shown that mice with reduced expression and activity of PPARγ exhibited enhanced sensitivity to chemically-induced skin cancer as compared to controls (Nicol et al. 2004); a phenotype similar to that observed of Pparβ/δ-null mice (Kim et al. 2004). Dietary or topical administration of two different PPARγ ligands (rosiglitazone or troglitazone) did not inhibit chemically-induced or ultraviolet (UV)-induced skin cancer, although dietary administration of troglitazone inhibited basal keratinocyte proliferation (He et al. 2005). A retrospective study in humans suggested that therapeutic use of the PPARγ ligand rosiglitazone may reduce the risk of non-melanoma skin cancer (Tseng 2015). While clinical trials examining the effect of PPARγ ligands for cancer chemoprevention or chemotherapy are ongoing, similar to PPARβ/δ, there remain studies suggesting that PPARγ ligands may promote some, but not all, cancers (reviewed in (Peters et al. 2012)). This also illustrates the need for more experimentation using alternative approaches to help resolve the role of PPARγ in carcinogenesis.
One major variable that has led to much of the disparities with respect to the roles of PPARβ/δ and PPARγ in human cancer centers on relative expression of these proteins in tumor cells compared to normal control tissue. For example, it was originally reported that expression of PPARγ or PPARβ/δ were elevated in epithelial tumor cells as compared to non-transformed tissue (DuBois et al. 1998; He et al. 1999), but subsequent, more quantitative studies showed that relative expression of both PPARγ or PPARβ/δ is actually lower in epithelial tumors as compared to non-transformed tissue (Foreman et al. 2011; Modica et al. 2010; Thul et al. 2017; Thul and Lindskog 2018; Uhlen et al. 2015; Uhlen et al. 2017). To better understand the effect of higher expression of PPARβ/δ and PPARγ in epithelial cancer cells, stable cell lines over-expressing these nuclear receptors were created and used to investigate the effect of ligand activation of these receptors on target gene expression, cell cycle regulation, anchorage-dependent cell growth, and in vivo tumorigenesis.
2. Materials and methods
2.1. Materials and cell culture
The PPARβ/δ ligand GW0742 synthesized and provided by GlaxoSmithKline (Research Triangle Park, NC) and was dissolved in dimethylsulfoxide (DMSO). The PPARγ ligand rosiglitazone maleate was purchased from Santa Cruz Biotechnologies (Santa Cruz, CA) and was dissolved in DMSO. Primers for quantitative real-time polymerase chain reaction (qPCR) were purchased from Integrated DNA Technologies (IDT, Coralville, IA). A431 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). The identity of the A431 cell lines was confirmed by Research Animal Diagnostic Laboratories (Columbia, MO). Cell lines were cultured in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin solution at 37°C and 5% CO2. Athymic NCr-nu/nu mice were purchased from the National Cancer Institute (NCI, Frederick, MD).
2.2. Establishment of stable A431 cell lines over-expressing PPARβ/δ or PPARγ
Stable human A431 squamous cell carcinoma cell lines over-expressing PPARβ/δ or PPARγ were generated using the Migr1 bicistronic retrovirus vector, which has been described previously (Borland et al. 2011; Pear et al. 1998). The Migr1 vector has a mouse stem cell virus promoter to drive expression of the cloned PPAR and an internal ribosome entry site (IRES) directly upstream of the gene encoding enhanced green fluorescent protein (eGFP). This methodology was previously described and yields high eGFP fluorescence to facilitate identification and sorting of cells that have stably integrated the Migr1 vectors encoding either PPARβ/δ or PPARγ (Borland et al. 2011; Borland et al. 2017; Foreman et al. 2011; Yao et al. 2017; Yao et al. 2015a; Yao et al. 2015b; Yao et al. 2014; Zhu et al. 2014a). Parental control A431 cells, A431-Migr1 vector control (A431-Migr1), A431-Migr1-hPPARβ/δ, or A431-Migr1-hPPARγ cells were used for the different experiments.
2.3. Characterization of the PPAR gain-of-function A431 cell lines
Quantitative real-time PCR (qPCR) and Western blot analysis was performed to confirm that the PPARs were over-expressed at the mRNA and protein levels as previously described (Borland et al. 2011). The primary antibodies used were anti-human PPARβ/δ (ab21209, Abcam, Cambridge, MA), anti-human PPARγ (2430, Cell Signaling Technology, Danvers, MA) or anti-ACTIN (Rockland, Gilbertsville, PA). To determine the ability of ligand activation to alter gene expression, the different cell lines were cultured with or without high affinity PPAR agonists as previously described (Borland et al. 2011). Ligand activation of PPARβ/δ was examined in cells cultured in medium with vehicle (0.02% DMSO) or the PPARβ/δ ligand GW0742 (0.01 – 10.0 μM) for 8 h. Ligand activation of PPARγ was examined in cells cultured in medium with vehicle (0.02% DMSO) or the PPARγ ligand rosiglitazone (0.01 – 10.0 μM) for 24 h. qPCR was used to quantify mRNA expression of PPARβ/δ, PPARγ, and the generic PPAR target gene angiopoietin-like protein 4 (ANGPTL4) as previously described (Borland et al. 2011).
2.4. Flow cytometry analysis of cell cycle progression
A431 cells from each cell line were seeded onto 6-well tissue culture dishes at a concentration of 250,000 cells per well and cultured in DMEM (with 10% FBS and 1% penicillin/streptomycin) for 24 h in the absence of any ligand. Flow cytometric analysis was then performed on this cohort of cells as described below. A second cohort of cells, also 24 h post-plating, were cultured with control (0.02% DMSO), GW0742 (0.01 – 10.0 μM) or rosiglitazone (0.01 – 10.0 μM) for an additional 24 h. After these treatments, culture medium was removed and the cells were trypsinized, pelleted and fixed in ice cold 70% ethanol. Prior to analysis, cells were stained with propidium iodide (PI) solution containing 1 μg PI/μL and 0.125% RNase A (Sigma Aldrich, St. Louis, MO). Approximately 10,000 cells/sample were analyzed using an EPICS-XL-MCL flow cytometer (Beckman Coulter, Miami Lakes, FL) fitted with a single 15-mW argon ion laser providing excitation at 488 nm. The percentage of cells at each phase of the cell cycle was determined with MultiCycle® analysis software. Values were calculated from a minimum of three independent samples per treatment.
2.5. Effect of PPARβ/δ and PPARγ on modulation of cell proliferation
The different A431 cell lines were plated in 12-well plates at a density of 25,000 cells/well 24 h before cell counting at time 0. Cell proliferation was determined using a Z1 Coulter particle counter (Beckman Coulter, Hialeah, FL). After this seeding period, cells were maintained in DMEM treated with control (0.02% DMSO), GW0742 (0.01 – 10.0 μM), or rosiglitazone (0.01 – 10.0 μM). Cells were counted every 24 h through 72 h post-ligand treatment. Triplicate samples for each treatment were used for each time point, and each replicate was counted three times.
2.6. Effect of PPARβ/δ and PPARγ on anchorage-dependent clonogenicity
The different A431 cell lines were plated in 60-mm culture dishes at a density of 1,200 cells per dish. After allowing the cells to adhere for 6 h, cell culture medium was replaced with medium containing either vehicle (0.02% DMSO), GW0742 (0.01 – 10.0 μM), or rosiglitazone (0.01 – 10.0 μM). After 14 d in culture, cell colonies were fixed and stained with a 6% (v/v) glutaraldehyde/0.5% (w/v) crystal violet solution. Colonies were counted with a stereomicroscope. Plating efficiency and surviving fractions were calculated as previously described from three independent samples per treatment group (Franken et al. 2006).
2.7. Ectopic xenografts
All animal studies were approved by The Pennsylvania State University Animal Care and Use Committee. Athymic nude mice were injected subcutaneously with A431 cells to produce ectopic xenografts as previously described (Yao et al. 2017; Yao et al. 2015b; Yao et al. 2014). Briefly, 6-week-old female immune-deficient athymic nude (nu/nu) mice were injected subcutaneously with two million (2 × 106) cells per hind flank. The A431-Migr1 (Migr1) cells were injected in the left rear flank and the A431-hPPARβ/δ (hPPARβ/δ) cells were injected in the right rear flank. Alternatively, the A431-Migr1 cells (Migr1) were injected in the left rear flank and the A431-hPPARγ cells (hPPARγ) were injected in the right rear flank. Groups of mice (N = 5) were then treated with or without GW0742 (2.5 mg/kg/d) or rosiglitazone (20 mg/kg/d) for up to 15 days. The PPAR ligands were provided 3 times per week by dosing with a pellet made with Bacon-flavored Transgenic Dough Diet (Bioserv, Inc., Flemington, New Jersey) mixed with either vehicle control (0.02% DMSO), GW0742 or rosiglitazone. Tumor volumes were measured 3 times a week.
2.8. Statistical analysis
Data were analyzed for statistical significance using one-way analysis of variance (ANOVA) and the Bonferroni’s multiple comparison tests, or Student’s T-test as described in the figure legends. All data are presented as the mean ± standard error of the mean (SEM) using Prism 5.0 (GraphPad Software Inc., La Jolla, CA).
3. Results
3.1. Enhanced receptor activity in A431 cells expressing PPARβ/δ or PPARγ
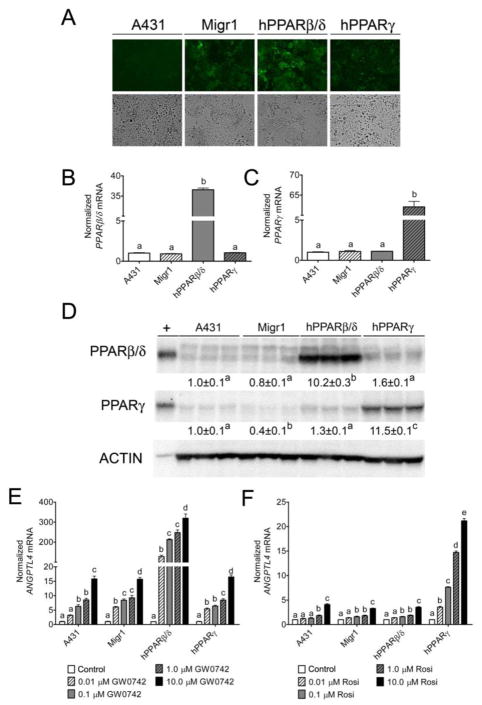
A431-Migr1, A431-Migr1-hPPARβ/δ, and A431-Migr1-hPPARγ cells exhibited eGFP fluorescence that was not observed in control A431 cells (Fig. 1A). No macroscopic changes in cell morphology were observed in any of these cell lines as compared to the parental A431 cells (Fig. 1A). Stable integration of the Migr1-hPPARβ/δ or Migr1-hPPARγ vectors caused increased PPARB or PPARG mRNA (Fig. 1B and 1C) and protein (Fig. 1D) in respective cell lines as compared to controls. ANGPTL4 mRNA was measured to determine whether the increase in expression of PPARs led to functional changes in their ability to modulate ligand-dependent transcription as expression of this gene can be increased by both PPARβ/δ or PPARγ (Mandard et al. 2004). A dose-dependent increase in expression of ANGPTL4 mRNA was observed in parental A431 cells and A431-Migr1 control cells in response to 0.01 μM to 10 μM GW0742 (Fig. 1E). Markedly higher increases in ligand induced expression of ANGPTL4 mRNA was observed in A431-Migr1-hPPARβ/δ cells in response to 0.01 μM to 10 μM GW0742 as compared to both parental A431 cells and A431-Migr1 vector control cells (Fig. 1E). Similarly, a dose-dependent increase in expression of ANGPTL4 mRNA was observed in parental A431 cells and A431-Migr1 control cells in response to 0.01 μM to 10 μM rosiglitazone (Fig. 1F). Additionally, higher increases in ligand-induced expression of ANGPTL4 mRNA were observed in A431-Migr1-hPPARγ cells in response to 0.01 μM to 10 μM rosiglitazone as compared to both control A431 cells and A431-Migr1 control cells (Fig. 1F). It is also worth noting that no difference in rosiglitazone-induced expression of ANGPTL4 mRNA was observed between the A431 cell line, the Migr1 control cell line, or the A431-Migr1-hPPARβ/δ cell line (Fig. 1F). Combined, these data establish that over-expression of PPARβ/δ or PPARγ in A431 cells can cause enhanced ligand-induced receptor activity and provides a useful model for examining the functional roles of these receptors in a squamous cell carcinoma model.
Fig. 1.
Characterization of a human squamous cell carcinoma cell line (A431) over-expressing human PPARβ/δ or PPARγ. (A) Representative photomicrographs of A431 cells, A431-Migr1 control cells (Migr1), A431-Migr1-hPPARβ/δ cells (hPPARβ/δ), and A431-Migr1-hPPARγ cells (hPPARγ) examined by fluorescent microscopy (upper panels) or light microscopy (lower panels). qPCR analysis for mRNA expression of (B) PPARβ/δ or (C) PPARγ in the A431 cell lines, normalized to GAPDH mRNA. (D) Western blot analysis of PPARβ/δ or PPARγ in the A431 cell lines, normalized to ACTIN expression. +positive control: cell lysate from COS-1 cells transfected with hPPARβ/δ or hPPARγ expression vector. qPCR analysis of ANGPTL4 mRNA in response to (E) the PPARβ/δ ligand GW0742 for 8 h or (F) the PPARγ ligand rosiglitazone (Rosi) for 24 h, normalized to the GAPDH mRNA. Data represents triplicate independent sample means ± S.E.M.. Values with different letters are significantly different (p ≤ 0.05).
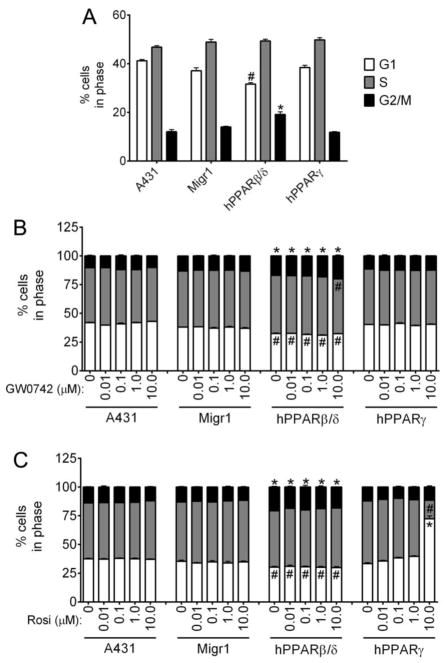
3.2. Effect of over-expressing PPARβ/δ or PPARγ on A431 cell cycle kinetics and cell proliferation
A431-Migr1-hPPARβ/δ cells exhibited a decrease in the percentage of cells at the G1 phase and an increase in the percentage of cells at the G2/M phase of the cell cycle as compared to control A431 and control A431-Migr1 cells (Fig. 2A). Over-expression of PPARγ in A431 cells did not alter the percentage of cells in any phase of the cell cycle as compared to controls (Fig. 2A). Ligand activation of PPARβ/δ did not influence the distribution of control A431, A431-Migr1, or A431-Migr1-hPPARγ cells in any phase of the cell cycle (Fig. 2B). While the same decrease in the percentage of cells at the G1 phase and increase in the percentage of cells at the G2/M phase of the cell cycle was observed in A431-Migr1-hPPARβ/δ cells compared to control, ligand activation of PPARβ/δ with GW0742 (0.01 – 1.0 μM) did not further influence this distribution (Fig. 2B). However, ligand activation of PPARβ/δ with 10 μM GW0742 further decreased the percentage of cells in the S phase in A431-Migr1-hPPARβ/δ cells compared to the other three cell lines (Fig. 2B). Ligand activation of PPARγ did not influence the distribution of control A431, A431-Migr1, or A431-Migr1-hPPARβ/δ cells in any phase of the cell cycle, although A431-Migr1-hPPARβ/δ cells exhibited the same decrease in the percentage of cells at the G1 phase and increase in the percentage of cells at the G2/M phase of the cell cycle was observed in A431-Migr1-hPPARβ/δ cells compared to controls (Fig. 2B).
Fig. 2.
Effect of ligand activation and/or over-expression of PPARβ/δ and PPARγ on cell cycle progression. Cell cycle progression was examined in A431, A431-Migr1 vector control cells (Migr1), A431-Migr1-hPPARβ/δ cells (hPPARβ/δ), and A431-Migr1-hPPARγ cells (hPPARγ) by flow cytometry. (A) Effect of PPARβ/δ or PPARγ over-expression on cell cycle progression. Effect of ligand activation of (B) PPARβ/δ or (C) PPARγ on cell cycle progression. Data represents triplicate independent sample means ± S.E.M.. *Significantly higher compared to A431 control (p ≤ 0.05). #Significantly lower compared to A431 control (p ≤ 0.05).
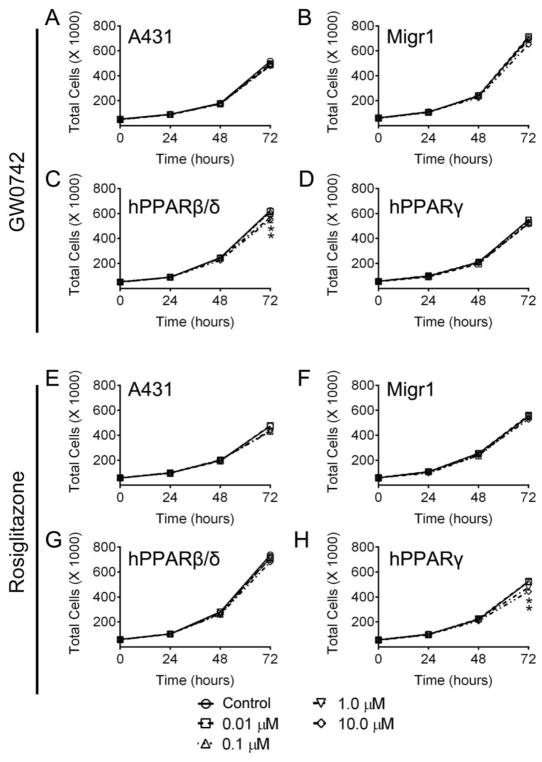
Ligand activation of PPARγ with rosiglitazone (0.01 – 1.0 μM) did not change the distribution of cells in any phase of the cell cycle in A431-Migr1-hPPARγ cells compared to controls (Fig. 2C). However, ligand activation of PPARγ with 10 μM rosiglitazone caused a marked decrease in the percentage of cells in the S phase and an increase in the percentage of cells in the G1 phase of the cell cycle as compared to controls (Fig. 2C). Ligand activation of PPARβ/δ did not influence cell proliferation in control A431 cells, control A431-Migr1 or A431-Migr1-hPPARγ cells compared to controls (Fig. 3A–D). By contrast, ligand activation of PPARβ/δ with 1.0 or 10 μM GW0742 modestly inhibited proliferation of A431-Migr1-hPPARβ/δ cells after 72 h compared to controls (9% ± 2% decrease and 12% ± 4% decrease, respectively, Fig. 3C). Ligand activation of PPARγ did not influence cell proliferation of A431, A431-Migr1, or A431-Migr1-hPPARβ/δ cells (Fig. 3E–H). Ligand activation of PPARγ with 1.0 or 10 μM rosiglitazone inhibited cell proliferation of A431-Migr1-hPPARγ cells after 72 h compared to controls (9% ± 3% decrease and 15% ± 1% decrease, respectively, Fig. 3H).
Fig. 3.
Effect of PPARβ/δ and PPARγ over-expression and/or ligand activation on cell proliferation. Proliferation of A431, A431-Migr1 vector control cells (Migr1), A431-Migr1-hPPARβ/δ cells (hPPARβ/δ), or A431-Migr1-hPPARγ cells (hPPARγ) was examined over a 72 h period by Coulter Counting. Cells were treated with indicated concentration of the PPARβ/δ ligand GW0742 (A–D) or the PPARγ ligand rosiglitazone (E–H), at time 0. Data represents triplicate independent sample means ± S.E.M.. *Significantly different than cell line-specific control (p ≤ 0.05).
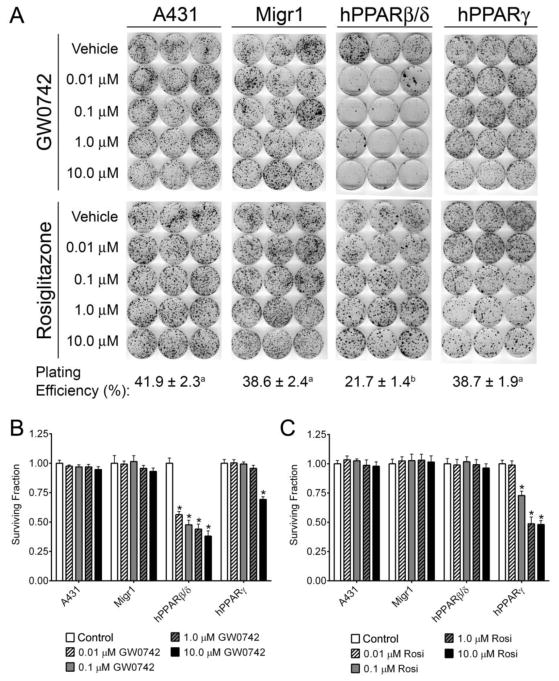
3.3. Effect of ligand activation and PPAR gain-of-function on anchorage-dependent clonogenicity of A431 cells
Anchorage-dependent clonogenicity was not influenced in either A431 or A431-Migr1 cells by expression of PPARβ/δ and was also unchanged by ligand activation (Fig. 4A). By contrast expression of PPARβ/δ and ligand activation of PPARβ/δ in A431-Migr1-hPPARβ/δ cells markedly inhibited anchorage-dependent clonogenicity compared to controls (Fig. 4A, B). The plating efficiency [(number of colonies/total cells plated) X 100] was significantly lower in A431-Migr1-hPPARβ/δ cells compared to controls and A431-Migr1-hPPARγ cells in the groups treated with GW0742 (Fig. 4A), but was unaffected in any of the cell types treated with rosiglitazone (data not shown). Expression of PPARβ/δ and ligand activation of PPARβ/δ in A431-Migr1-hPPARγ cells with 0.01 – 1.0 μM GW0742 did not influence anchorage-dependent clonogenicity compared to controls, while inhibition of anchorage-dependent clonogenicity was observed in A431-Migr1-hPPARγ cells in response to ligand activation of PPARβ/δ with 10 μM GW0742 (Fig. 4A). Anchorage-dependent clonogenicity was not influenced in either A431 or A431-Migr1 by expression of PPARγ, nor by ligand activation of PPARγ in A431, A431-Migr1, or A431-Migr1-hPPARβ/δ cells compared to controls (Fig. 4B). However, ligand activation of PPARγ inhibited anchorage-dependent clonogenicity in A431-Migr1-hPPARγ cells with 0.1 – 1.0 μM rosiglitazone as compared to controls (Fig. 4B).
Fig. 4.
Effect of PPARβ/δ and PPARγ over-expression and/or ligand activation on anchorage-dependent clonogenicity. (A) Clonogenicity was examined in A431, A431-Migr1 control cells (Migr1), A431-Migr1-hPPARβ/δ cells (hPPARβ/δ), or A431-Migr1-hPPARγ cells (hPPARγ), after ligand activation of PPARβ/δ with GW0742 or PPARγ with rosiglitazone. Values for plating efficiency with different letters are significantly different (p ≤ 0.05). Quantification of the surviving fraction following ligand activation of PPARβ/δ with GW0742 (B) or PPARγ with rosiglitazone (C). *Significantly different than cell line-specific vehicle control (p ≤ 0.05).
3.4. Effect of ligand activation and PPAR gain-of-function on tumorigenicity of A431 cells
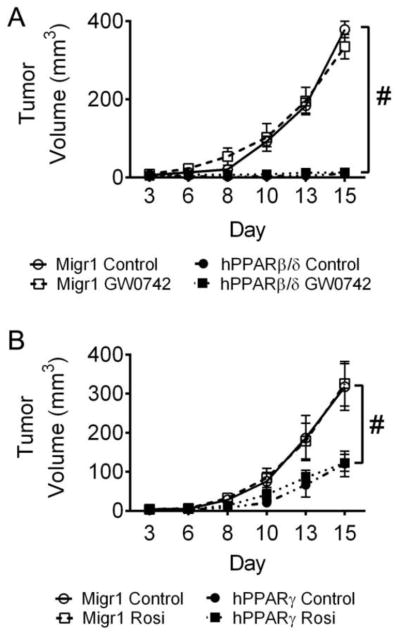
Over-expression of PPARβ/δ markedly inhibited the average volume of tumors derived from A431-Migr1 cells as compared to controls (Fig. 5A). Ligand activation of PPARβ/δ did not influence the average volume of tumors derived from A431-Migr1 cells, nor in tumors derived from A431-Migr1-hPPARβ/δ cells (Fig. 5A). However, the average volume of tumors derived from A431-Migr1-hPPARβ/δ cells was very low and similar to that observed with over-expression of PPARβ/δ (Fig. 5A). Similarly, over-expression of PPARγ inhibited the average volume of tumors derived from A431-Migr1 cells as compared to controls, while ligand activation of PPARγ did not influence the average volume of tumors derived from A431-Migr1 cells, nor in tumors derived from A431-Migr1-hPPARγ cells (Fig. 5B). The average volume of tumors derived from A431-Migr1-hPPARγ cells was low and similar to that observed with over-expression of PPARγ, but this effect was not as profound as that observed with over-expression of PPARβ/δ (Fig. 5A and B).
Fig. 5.
Over-expression of PPARβ/δ or PPARγ suppresses ectopic tumors from squamous cell carcinoma cell line A431. Two million cells were subcutaneously injected into the hind quarter leg of athymic NCR-nu/nu male mice (N=5). Mice were orally administered control, GW0742 (5 mg/kg), or rosiglitazone (Rosi; 20 mg/kg) three days per week beginning on Day 3. Data represents the mean ± SEM. (A) Average tumor volumes from A431-MigR1 (MigR1) or A431-hPPARβ/δ (hPPARβ/δ) cells with or without ligand activation of PPARβ/δ with GW0742. (B) Average tumor volumes from A431-MigR1 (MigR1) or A431-hPPARγ (hPPARγ) cells with or without ligand activation of PPARγ with rosiglitazone. #Significantly different than control (p ≤ 0.05).
4. Discussion
Two, new models to study the role of PPARβ/δ and PPARγ in squamous cell carcinomas were developed in these studies. Multiple approaches were used but collectively, the evidence clearly supports the notion that both PPARβ/δ and PPARγ can be therapeutically targeted for the treatment of squamous cell carcinoma. The fact that relatively higher expression of both PPARβ/δ and PPARγ in A431 cells inhibited cell cycle and proliferation, is consistent with the observed reduction in anchorage-dependent clonogenicity and the striking inhibition of tumor volume in ectopic xenografts. It is of interest to note that the relative expression levels of both PPARβ/δ and PPARγ is low in parental A431 cells but that when expression is markedly increased, effects associated with inhibition of cell cycle and proliferation, and measures of tumorigenesis are observed. While it is clear that ligand activation causes an enhanced effect in the cells expressing the PPARs, there are likely endogenous, high affinity ligands present that prevent further diminished effects of the endpoints examined as have been observed in previous studies as well (Borland et al. 2011; Borland et al. 2017; Yao et al. 2017; Yao et al. 2015b; Yao et al. 2014).
The mechanism(s) by which PPARβ/δ or PPARγ inhibit tumorigenicity of A431 squamous cell carcinomas is likely mediated at least in part by modulation of the cell cycle. For example, activation of PPARβ/δ can cause a block in the G2/M phase in human or mouse keratinocytes, in particular those with a mutant Hras gene (Burdick et al. 2007; Zhu et al. 2012). The latter is mediated via shuttling of PPARβ/δ into the nucleus with other proteins that in turn modulates E2F signaling (Zhu et al. 2012). Alternatively, there is also evidence that PPARβ/δ and PPARγ ligands can induce terminal differentiation, which is known to cause inhibition of the cell cycle consistent with that observed in these studies (Demerjian et al. 2006; Mao-Qiang et al. 2004; Schmuth et al. 2004; Westergaard et al. 2001; Yan et al. 2015). While ligand activation of PPARβ/δ and PPARγ caused modulation of cell cycle and proliferation, only expression of PPARβ/δ and PPARγ was required for inhibition of tumorigenicity. Thus, it will be of interest to determine if endogenous ligands underlie this effect. This is of interest to note because as noted above there is evidence that endogenous ligands for PPARβ/δ and PPARγ do exist and mediate the non-exogenous ligand effects (Borland et al. 2011; Borland et al. 2017; Yao et al. 2017; Yao et al. 2015b; Yao et al. 2014), as indicated by the marked changes in PPARβ/δ target gene expression as a results of knocking our the receptor in both mouse keratinocytes and human myeloid cells (Adhikary et al. 2011; Khozoie et al. 2012). Further work will be required to identify these endogenous ligands and characterize their role in tumorigenesis. Moreover, we neglected to perform histopathological analysis on the tumors obtained from these studies, which may have provided further insight into the potential mechanisms underlying the striking differences in tumor volumes observed in the stable cell lines. This will also be of interest to examine in future studies. Regardless, results from these studies illustrate that targeting PPARβ/δ and PPARγ may be useful for the treatment of squamous cell carcinoma.
Highlights.
Expression of peroxisome PPARβ/δ blocks G2/M in A431 cancer cells
Expression of PPARβ/δ or PPARγ inhibits xenografts from A431 cells
PPARβ/δ and PPARγ are potential nodal targets for cancer therapy
Acknowledgments
The authors gratefully acknowledge Dr. Gary H. Perdew at The Pennsylvania State University for the use of a fluorescent microscope, and the Center for Quantitative Cell Analysis at the Huck Institutes of Life Sciences of The Pennsylvania State University for their technical support in sorting the fluorescent cells. This work supported in part by the National Institutes of Health (CA140369, CA124533; JMP), the United States Department of Agriculture (Project 4607; JMP), the Bloomsburg University (BU) Research & Scholarship Grant (MGB), and a BU Margin of Excellence Grant (MGB).
Abbreviations
- ADRP
adipocyte differentiation-related protein
- ANGPTL4
angiopoietin-like protein 4
- DMEM
Dulbecco’s minimal essential medium
- DMSO
dimethylsulfoxide
- eGFP
enhanced green fluorescent protein
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IRES
internal ribosome entry site
- PPAR
peroxisome proliferator-activated receptor
- qPCR
quantitative real-time polymerase chain reaction
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adhikary T, et al. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) PLoS One. 2011;6:e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bility MT, et al. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits chemically-induced skin tumorigenesis. Carcinogenesis. 2008;29:2406–2414. doi: 10.1093/carcin/bgn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bility MT, et al. Ligand activation of peroxisome proliferator-activated receptor-β/δ and inhibition of cyclooxygenase-2 enhances inhibition of skin tumorigenesis. Toxicol Sci. 2010;113:27–36. doi: 10.1093/toxsci/kfp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland MG, et al. Stable over-expression of PPARβ/δ and PPARγ to examine receptor signaling in human HaCaT keratinocytes. Cell Signal. 2011;23:2039–2050. doi: 10.1016/j.cellsig.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland MG, et al. Editor’s Highlight: PPARβ/δ and PPARγ Inhibit Melanoma Tumorigenicity by Modulating Inflammation and Apoptosis. Toxicol Sci. 2017;159:436–448. doi: 10.1093/toxsci/kfx147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick AD, et al. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits cell growth of human N/TERT-1 keratinocytes. Cell Signal. 2007;19:1163–1171. doi: 10.1016/j.cellsig.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerjian M, et al. Topical treatment with thiazolidinediones, activators of peroxisome proliferator-activated receptor-γ, normalizes epidermal homeostasis in a murine hyperproliferative disease model. Exp Dermatol. 2006;15:154–160. doi: 10.1111/j.1600-0625.2006.00402.x. [DOI] [PubMed] [Google Scholar]
- DuBois RN, et al. The nuclear eicosanoid receptor, PPARγ, is aberrantly expressed in colonic cancers. Carcinogenesis. 1998;19:49–53. doi: 10.1093/carcin/19.1.49. [DOI] [PubMed] [Google Scholar]
- Foreman JE, et al. Functional characterization of peroxisome proliferator-activated receptor-β/δ expression in colon cancer. Mol Carcinog. 2011;50:884–900. doi: 10.1002/mc.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken NA, et al. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- He G, et al. The effect of PPARγ ligands on UV- or chemically-induced carcinogenesis in mouse skin. Mol Carcinog. 2005;43:198–206. doi: 10.1002/mc.20111. [DOI] [PubMed] [Google Scholar]
- He TC, et al. PPARδ is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozoie C, et al. Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4. BMC Genomics. 2012;13:665. doi: 10.1186/1471-2164-13-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, et al. Peroxisome proliferator-activated receptor β (δ)-dependent regulation of ubiquitin C expression contributes to attenuation of skin carcinogenesis. J Biol Chem. 2004;279:23719–23727. doi: 10.1074/jbc.M312063200. [DOI] [PubMed] [Google Scholar]
- Mandard S, et al. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J Biol Chem. 2004;279:34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- Mao-Qiang M, et al. Peroxisome-Proliferator-Activated Receptor (PPAR)-γ Activation Stimulates Keratinocyte Differentiation. J Invest Dermatol. 2004;123:305–312. doi: 10.1111/j.0022-202X.2004.23235.x. [DOI] [PubMed] [Google Scholar]
- Modica S, et al. The intestinal nuclear receptor signature with epithelial localization patterns and expression modulation in tumors. Gastroenterology. 2010;138:636–648. 648 e631–612. doi: 10.1053/j.gastro.2009.09.060. [DOI] [PubMed] [Google Scholar]
- Muller R. PPARβ/δ in human cancer. Biochimie. 2017;136:90–99. doi: 10.1016/j.biochi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- Nicol CJ, et al. PPARγ influences susceptibility to DMBA-induced mammary, ovarian and skin carcinogenesis. Carcinogenesis. 2004;25:1747–1755. doi: 10.1093/carcin/bgh160. [DOI] [PubMed] [Google Scholar]
- Pear WS, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Peters JM, et al. Establishing the Role of PPARβ/δ in Carcinogenesis. Trends Endocrinol Metab. 2015;26:595–607. doi: 10.1016/j.tem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, et al. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12:181–195. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuth M, et al. Peroxisome proliferator-activated receptor (PPAR)-β/δ stimulates differentiation and lipid accumulation in keratinocytes. J Invest Dermatol. 2004;122:971–983. doi: 10.1111/j.0022-202X.2004.22412.x. [DOI] [PubMed] [Google Scholar]
- Thul PJ, et al. A subcellular map of the human proteome. Science. 2017:356. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018;27:233–244. doi: 10.1002/pro.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH. Rosiglitazone may reduce non-melanoma skin cancer risk in Taiwanese. BMC Cancer. 2015;15:41. doi: 10.1186/s12885-015-1057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Uhlen M, et al. A pathology atlas of the human cancer transcriptome. Science. 2017:357. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- Westergaard M, et al. Modulation of keratinocyte gene expression and differentiation by PPAR-selective ligands and tetradecylthioacetic acid. J Invest Dermatol. 2001;116:702–712. doi: 10.1046/j.1523-1747.2001.01329.x. [DOI] [PubMed] [Google Scholar]
- Yan Y, et al. Various peroxisome proliferator-activated receptor (PPAR)-γ agonists differently induce differentiation of cultured human keratinocytes. Exp Dermatol. 2015;24:62–65. doi: 10.1111/exd.12571. [DOI] [PubMed] [Google Scholar]
- Yao PL, et al. Peroxisome proliferator-activated receptor-β/δ inhibits human neuroblastoma cell tumorigenesis by inducing p53- and SOX2-mediated cell differentiation. Mol Carcinog. 2017;56:1472–1483. doi: 10.1002/mc.22607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PL, et al. Peroxisome Proliferator-activated Receptor-D (PPARD) Coordinates Mouse Spermatogenesis by Modulating Extracellular Signal-regulated Kinase (ERK)-dependent Signaling. J Biol Chem. 2015a;290:23416–23431. doi: 10.1074/jbc.M115.664508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PL, et al. Inhibition of testicular embryonal carcinoma cell tumorigenicity by peroxisome proliferator-activated receptor-β/δ and retinoic acid receptor-dependent mechanisms. Oncotarget. 2015b;6:36319–36337. doi: 10.18632/oncotarget.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PL, et al. Activation of peroxisome proliferator-activated receptor-β/δ (PPAR-β/δ) inhibits human breast cancer cell line tumorigenicity. Mol Cancer Ther. 2014;13:1008–1017. doi: 10.1158/1535-7163.MCT-13-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, et al. Chemoprevention of chemically induced skin tumorigenesis by ligand activation of peroxisome proliferator-activated receptor-β/δ and inhibition of cyclooxygenase 2. Mol Cancer Ther. 2011;9:3267–3277. doi: 10.1158/1535-7163.MCT-10-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, et al. PPARβ/δ promotes HRAS-induced senescence and tumor suppression by potentiating p-ERK and repressing p-AKT signaling. Oncogene. 2014a;33:5348–5359. doi: 10.1038/onc.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, et al. The nuclear receptor peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) promotes oncogene-induced cellular senescence through repression of endoplasmic reticulum stress. J Biol Chem. 2014b;289:20102–20119. doi: 10.1074/jbc.M114.551069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, et al. Peroxisome proliferator-activated receptor β/δ cross talks with E2F and attenuates mitosis in HRAS-expressing cells. Mol Cell Biol. 2012;32:2065–2082. doi: 10.1128/MCB.00092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]