Advances in high throughput technologies and big data analytics are rapidly increasing our understanding of the role of various immune cells and factors associated with atopic disease beyond the Th1/Th2 axis. Many children and adults with atopy can suffer from co-morbid conditions like having food allergy and chronic asthma, which can increase the risk of anaphylaxis and fatal reactions to a food allergen. Further understanding of the contributions and roles of the different immune cell subsets would be of great value in understanding the food allergy-asthma co-morbid clinical phenotype. Therefore, we performed a pilot study using immune monitoring of blood samples from 4 cohorts: non-food allergic asthmatics, food allergic asthmatics, food allergic non-asthmatics, and healthy controls.
In recent years, we have made modest inroads in identifying some of the immune cells, cytokines, pathways, and interactions that play a role in mediating allergic diseases and allergic tolerance. Among immune cells, the important role of T regulatory cells in promoting allergic tolerance has been shown by a number of studies1, 2. Other immune cells that have been shown to play a role in allergic diseases are basophils, eosinophils, mast cells, B cells, Th9, Th17, Th22, gamma delta T (GDT), and type 2 innate lymphoid cells. Some of the regulatory cytokines known to be involved in allergic diseases are IL-4, IL-5, IL-9, and IL-13 (secreted by immune cells) and IL-25, IL-31, IL-33, and TSLP (secreted by tissue cells), IFN-γ, IL-9, IL-22, IL-10, TGFβ, etc. There are a number of excellent reviews on the mechanisms of allergic disease 3-5. However, there is still much that we need to understand regarding allergic pathways and their regulatory factors. To study these and other cells in the samples from our cohorts and attempt to immunophenotype allergic conditions, such as allergy and asthma, we used a novel high dimensional immunophenotyping technology based on mass spectrometry (i.e. Cytometry Time of Flight or CyTOF).
We determined differences between immune cell subsets from previously frozen peripheral blood mononuclear cells to avoid batch effects6 and obtained from a convenience sample (i.e. those with enough samples stored) of 54 participants from 4 study cohorts: non-food allergic asthmatics (n=16), food allergic asthmatics (n=18), food allergic non-asthmatics (n=6), and healthy controls (n=14) using CyTOF. See Table e1 for participant demographics. Samples were obtained from participants from three studies. Healthy and only-asthmatic participant samples were obtained from 2 well-described cohorts, respectively, 7 and foodallergic-only and food allergic-with-asthma participant samples were obtained from 2 well-defined cohorts of participants with food allergy, respectively8. All participants consented via a Stanford IRB approved protocol. Food allergy and asthma status were identified via standardized double-blind placebo-controlled food challenge (DBPCFC) and NHLBI criteria (https://www.nhlbi.nih.gov/health-pro/guidelines/current/asthma-guidelines), respectively. Blood samples of food allergic participants were drawn at the same day of the DBPCFC before the DBPCFC. Food allergy participants had no environmental allergies. Healthy controls were negative for any allergy diagnosis or allergy testing (IgE or skin). The non-food allergic asthmatics had no food allergies. No participants had autoimmune disease, cancer, or active infections. Frozen samples of human peripheral blood mononuclear cells were thawed according to standard, reproducible methods, resulting in over 80% viability upon defrosting (http://iti.stanford.edu/himc/blood-processing.html). CyTOF mass cytometry with standardized antibodies (see Table e2) was then performed according to previously publishedprotocols.7, 9
The frequency and percent of peripheral blood monocytes, B cells, GDT cells, CD3+ cells, CD8+ cells, CD4+ T cells, naïve T cells, memory T cells, effector memory T cells, central memory T cells, CD4+ IFN-γ + Th 1, CD4+ IL-4+ Th2, Th17, and Tregs were determined by hand gating techniques (FloJo software). Additionally, since ratios of cells to each other could be important in the pathology of the disease, 10 ratios of cell expression were derived based on combinations of Th1, Th2, Th17, GDT cells, and Treg. Multivariate linear regression models were fit to the percentage of specific cell expression as a function of the cohorts controlling for age, gender, and race. All tests were adjusted for multiple comparisons by controlling the false discovery rate to be ≤ 0.05 using the Benjamini and Hochberg approach. Analyses were conducted using R software version 3.2.2.10
The majority of participants were Caucasian (80%) and male (61%), with a median age of 13 years old. There were no significant differences between cohorts for age or sex. However distributional differences existed for race and ethnicity, which could impact the results and future studies are needed with larger sample sizes.
Most of the T cell subsets had low levels of expression (below 10%) with smaller variations in values compared to other cell types (Figure 1). In general, asthmatics had little variation within each cell type, while the healthy controls had the largest range of values. Treg, central and effector memory T cells, memory cells, monocytes, naïve cells, Th17 cells, and ratios of Th1 to Treg, Th17 to GDT, Th17 to Treg, and Treg to GDT had statistically significant expression differences across the 4 allergy cohorts.
Figure 1.
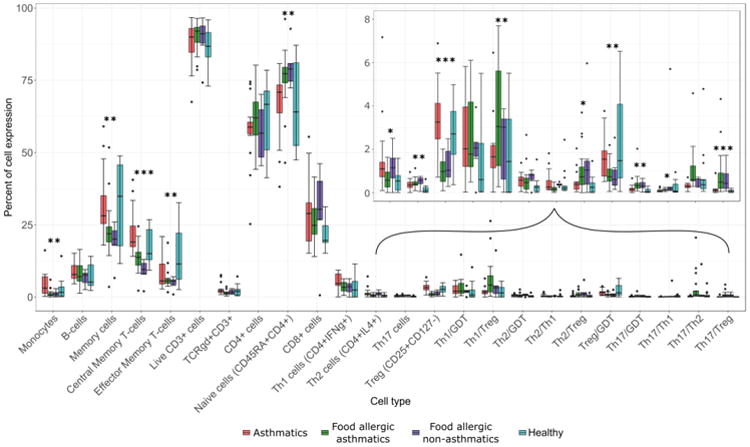
Percent of cell expression by cell type and cohort, with an inset for cells with low expression. astnmatics non-asinmaucs.
*** p < 0.01, ** p < 0.05, * p < 0.10 for adjusted p-values from the multivariable linear regression model (controlling for age, gender, and race) are reported.
GDT, Gamma delta T cells; Treg, T regulatory cells.
Some cell types were significantly different between asthmatics vs. food allergic non-asthmatics, and asthmatics vs. food allergic asthmatics. Asthmatics had significantly higher expressions of monocytes compared with food allergic asthmatics and food allergic non-asthmatics. Asthmatics had almost 15% higher central memory T-cell expression compared with food allergic non-asthmatics. Healthy participants also had significantly higher levels of expression of central memory T-cells compared with food allergic non-asthmatics. Food allergic non-asthmatics and food allergic asthmatics had very similar expression for each cell type.
Adverse events between the cohorts of food allergic asthmatics vs. food allergic non-asthmatics were compared since, for these, adverse events (CTCAE v4.03)11 were collected during oral immunotherapy.12 The 24 subjects experienced a total of 480 adverse events associated with food allergen. The majority of adverse events were gastrointestinal (59%), followed by respiratory (17%) and skin (10%). Twenty-five percent of adverse events were classified as moderate grade, and none were considered severe. None of the cell types were associated with gastrointestinal or respiratory adverse events. However, higher values of the ratio of Th17 to Th1 were significantly associated with a lower likelihood of having moderate skin reactions (OR: 0.69, 95% CI: 0.53, 0.89).
In this pilot study, we used a high dimensional immunophenotyping approach to evaluate cellular heterogeneity associated with asthmatics, food allergy asthmatics, food allergic non-asthmatics, vs. healthy controls. One of the major differences observed was the lower percentages of memory T cells and Treg cells in food allergic participants, which has been observed by other studies13. Using CyTOF, we were able to determine statistically significant differences in expression in Tregs, effector and central memory T cells among others, as well as ratios of Treg/GDT, Th17/GDT, and Th17/Treg. We also found associations between immune cell profiles and adverse events. Higher Th17/Th1 ratios were associated with lower odds of skin reactions. However, these differences should be interpreted with caution and further studies with larger sample sizes, different ethnicities, and other allergic conditions such as atopic dermatitis, are needed in the future. Testing additional T cell subsets such as Th9 and Th22 are of great interest. A few participants had environmental allergies in the asthma group and further studies are needed to determine the extent to which concomitant environmental allergies affect asthma phenotypes. These preliminary results report possible associative signals that should be explored in larger studies. Further investigation could aid in the identification of biomarkers, new pathways, and cell types that are dysregulated in food allergic diseases.
Supplementary Material
Table e1. Demographics by Allergy Cohort
Acknowledgments
We would like to thank Holden Maecker of the Human Immune Monitoring Center, Olivia Raeber and Andrew Long of the Sean N Parker Center for Allergy and Asthma Research at Stanford University, Manisha Desai of the Quantitative Sciences Unit, Stanford University School of Medicine, Lucile Packard Children's Hospital, and participants and their families.
Funding Source: EAT (End Allergies Together) Foundation, Sean N. Parker Center for Allergy and Asthma Research at Stanford University, NIH R01 ES020926, and Child Health Research Institute, Stanford University.
Abbreviations/Acronyms
- CyTOF
Cytometry by Time-Of-Flight
- Treg
Regulatory T cells
- GD T cells
Gamma delta T cells
- DBPCFC
Double-blind placebo-controlled food challenge
Footnotes
Conflict Of Interest: None
Trial Registration: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berker M, Frank LJ, Gessner AL, et al. Allergies - A T cells perspective in the era beyond the TH1/TH2 paradigm. Clin Immunol. 2017;174:73–83. doi: 10.1016/j.clim.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Prince BT, Devonshire AL, Erickson KA, et al. Regulatory T-cell populations in children are affected by age and food allergy diagnosis. J Allergy Clin Immunol. 2017;140:1194–1196 e1116. doi: 10.1016/j.jaci.2017.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palomares O, Akdis M, Martin-Fontecha M, Akdis CA. Mechanisms of immune regulation in allergic diseases: the role of regulatory T and B cells. Immunol Rev. 2017;278:219–236. doi: 10.1111/imr.12555. [DOI] [PubMed] [Google Scholar]
- 4.Rothenberg ME, Saito H, Peebles RS., Jr Advances in mechanisms of allergic disease in 2016. J Allergy Clin Immunol. 2017;140:1622–1631. doi: 10.1016/j.jaci.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Chinthrajah RS, Hernandez JD, Boyd SD, Galli SJ, Nadeau KC. Molecular and cellular mechanisms of food allergy and food tolerance. J Allergy Clin Immunol. 2016;137:984–997. doi: 10.1016/j.jaci.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burel JG, Qian Y, Lindestam Arlehamn C, et al. An Integrated Workflow To Assess Technical and Biological Variability of Cell Population Frequencies in Human Peripheral Blood by Flow Cytometry. J Immunol. 2017;198:1748–1758. doi: 10.4049/jimmunol.1601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prunicki M, Stell L, Dinakarpandian D, et al. Exposure to NO2, CO, and PM2.5 is linked to regional DNA methylation differences in asthma. Clin Epigenetics. 2018;10:2. doi: 10.1186/s13148-017-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andorf S, Purington N, Block WM, et al. Anti-IgE treatment with oral immunotherapy in multifood allergic participants: a double-blind, randomised, controlled trial. Lancet Gastroenterol Hepatol. 2018;3:85–94. doi: 10.1016/S2468-1253(17)30392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R Core Team. R Foundation for Statistical Computing; 2015. [Accessed 3, 2017]. R: A language and environment for statistical computing. https://www.r-project.org/ [Google Scholar]
- 11.US Department of Health and Human Services. Common Terminology Criteria for Adverse Events (CTCAE), v4.03. [Accessed 6, 2017];2010 https://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03_2010-06-14_QuickReference_8.5×11.pdf.
- 12.Sampson HA, Aceves S, Bock SA, et al. Food allergy: a practice parameter update-2014. J Allergy Clin Immunol. 2014;134:1016–1025 e1043. doi: 10.1016/j.jaci.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Syed A, Garcia MA, Lyu SC, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3) J Allergy Clin Immunol. 2014;133:500–510. doi: 10.1016/j.jaci.2013.12.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table e1. Demographics by Allergy Cohort


