Abstract
Background
Tumor‐associated immune factors are heterogeneous and play an important role in determining outcome in cancer patients. In this study, the expression levels of immune factors in tumor tissue‐conditioned media from lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) were analyzed.
Methods
LUAD and LUSC tissue specimens were collected immediately after surgery for antibody array analysis and real‐time quantitative PCR.
Results
Higher levels of chemokines MCP1/CCL2 (21.11‐fold increase) and MIP‐1β/CCL4 (19.33‐fold increase) were identified in LUAD than in LUSC. Western blot and quantitative real‐time PCR analyses showed higher co‐expression of CCL2 and CCL4 in LUAD tissues compared to LUSC (P < 0.0001). Immunofluorescent co‐staining showed a high percentage of CCL2+/CD68+ and CCL4+/CD68+ tumor‐associated macrophages in LUAD compared to LUSC tissues, which might be responsible for the higher expression of CCL2 and CCL4 in LUAD samples. Kaplan–Meier curves showed that CCL2 overexpression in patients with LUSC was associated with beneficial overall survival (OS; P = 0.048) and progression‐free survival (PFS; P = 0.012); however, LUAD patients with higher CCL2 expression had unfavorable OS (P = 6.7e−08) and PFS (P = 0.00098). Similarly, CCL4 overexpression predicted favorable PFS (P = 0.021) in patients with LUSC, but patients with high CCL4 levels in LUAD had shorter OS (P = 0.013).
Conclusion
Our study revealed that CCL2 and CCL4 expression levels could serve as potential prognostic biomarkers and therapeutic targets for NSCLC patients.
Keywords: Biomarker, CCL2, CCL4, non‐small cell lung cancer, tumor microenvironment
Introduction
Non‐small cell lung cancer (NSCLC) is a highly aggressive malignant neoplasm with an unfavorable prognosis. Lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) are the main pathological types of NSCLC. However, patients with LUAD have poorer prognosis than those with LUSC, indicating that some key compositions in the LUAD microenvironment may play a regulatory role during cancer development and metastasis formation.
Lung squamous cell carcinoma, more common in men than in women, is closely correlated with a history of tobacco smoking.1 LUSC is an epidermoid carcinoma, and begins in the tissue that lines the air passages in the lungs. Most LUSCs are located centrally, usually in the larger bronchi that join the trachea to the lung.2 Advanced LUSC always undergoes central necrosis and cavitation because of a lack of blood supply.3
Lung adenocarcinoma is currently the most common type of NSCLC in lifelong non‐smokers and women, but in recent years incidence has increased in smokers.4 LUAD accounts for approximately 40% of NSCLC cases, and causes ~400 000 cases of cancer‐associated mortality annually worldwide.5 This cancer usually develops peripherally in the lungs, as opposed to LUSC. LUAD contains certain distinct architectural, cytological, or molecular malignant tissue features, and tends to metastasize at an early stage.6 The prognosis of patients with LUAD is poorer than LUSC patients because of the different pathogenesis, cancer cell characteristics, or tumor microenvironment.7 In particular, non‐cellular components in the tumor microenvironment, such as soluble proteins, molecules, and the extracellular matrix, have emerged as an important regulator of cancer development.8 Therefore, investigating novel biomarkers for diagnosis and prognosis would be beneficial to improve the survival rates of NSCLC patients.
The NSCLC tumor microenvironment is a pivotal factor in tumorigenesis, particularly in tumor progression, and the pathogenesis of cancer is largely dependent on its interactions with microenvironmental components.9 In particular, the signaling molecules in the microenvironment can affect the growth and metastasis of cancer cells.10 VEGF is a potent mediator of angiogenesis that enhances endothelial cell survival, induces vasodilatation, and regulates pericyte coverage in the NSCLC tumor microenvironment.11 A recent study showed that high IL‐2 expression in NSCLC cells is associated with recurrence or metastasis.12 TNFβ expression in tumor‐infiltrating lymphocytes or in cancer cells is reported to significantly reduce postoperative survival, especially in patients with LUSC.13 Following the recent impressive benefit to cancer patients observed by targeting the immune system in cancer therapy, biomarker research focus has turned to the tumor immune microenvironment. Different subsets of tumor infiltrating immune cells interact with cancer cells in a complex and dynamic ecosystem, mediating immune surveillance and the destruction of cancer cells, as well as pro‐tumoral inflammation.14 In fact, extensive research has shown that the immune contexture has an impact on cancer patient outcomes.15, 16 However, the key tumor‐associated immune factors in NSCLC are not yet known.
In the present study, we show that chemokines CCL2 and CCL4 are overexpressed in the LUAD microenvironment compared to LUSC. Moreover, high expression of CCL2 or CCL4 is associated with unfavorable overall survival (OS) and progression‐free survival (PFS) in patients with LUAD. By contrast, high levels of CCL2 or CCL4 predict good prognosis in LUSC patients. These factors may have significant clinical implications for the treatment and prognosis of NSCLC.
Methods
Patients and tissue samples
All NSCLC tissue specimens were obtained from the Sun Yat‐sen University Cancer Center (Guangzhou, China). All recruited patients provided written informed consent, and the Committee for Ethical Review of Research at Sun Yat‐sen University Cancer Center approved the study protocol.
Three LUAD and three LUSC fresh tissue specimens for antibody array analysis were collected immediately after surgery (cohort 1). In this cohort, no recruited patients received preoperative chemotherapy or radiotherapy. Twenty‐eight primary LUSC and 18 LUAD tissues were obtained immediately after surgical resection between March and September 2013 for quantitative real‐time (qRT)‐PCR (cohort 2).
The enrollment criteria were as follows: (i) definitive LUAD and LUSC diagnosis by pathology based on World Health Organization criteria; (ii) exclusive treatment with chemotherapy or radiotherapy before tumor excision; (iii) no simultaneous use of local treatment modalities (i.e. radiofrequency ablation, microwave ablation); and (iv) complete surgical resection.
Preparation of tissue culture‐conditioned medium
Fresh tissues were collected immediately into sterile tubes containing 5 mL Dulbecco's modified Eagle medium (DMEM, without fetal bovine serum [FBS]) after surgery. To avoid blood contamination, all tissues were rinsed five times with phosphate buffer solution (PBS) and cut into 5–10 mm3 fragments with sterile scalpels, and then rinsed again three times with PBS. Tissues were then incubated with 1 mL DMEM (without FBS) in a six‐well plate for six hours at 37°C in a humidified chamber containing 5% CO2. The supernatant was then collected into a centrifuge tube and centrifuged at 2000 rpm for 10 minutes at 4°C, and filtered with a 0.22 μm filter to remove the potential contamination of bacteria and cell debris. Subsequently, the tissue culture‐conditioned media were analyzed by antibody array (Fig 1).
Figure 1.
Diagram for analyzing the immune factors in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) microenvironments.
Inflammation antibody array analysis
Three LUSC and three LUAD tissue culture supernatants were mixed equally in volume, respectively. Two mixed conditioned media (1 mL) were added to antibody arrays against 40 unique immune factors (Catalog #AAH‐INF‐G3‐4; RayBiotech Inc., Norcross, GA, USA) and processed according to the manufacturer's protocol.17 The inflammation antibody array map is shown in Figure 2a. Antibodies to specified proteins, and positive and negative controls were printed in duplicate. Positive controls (POS1, POS2, POS3) were treated with equal amounts of biotinylated immunoglobulin G (IgG) printed directly onto the array, and negative control spots with a protein‐containing buffer. The images were captured using a GenePix 4000B Microarray Scanner (Molecular Devices, Sunnyvale, CA, USA), and fluorescence intensity data were obtained by RayBio Analysis Tool software that automatically measured the local background around each spot. Normalized values were calculated by subtracting the background and normalizing to the positive control signals.
Figure 2.
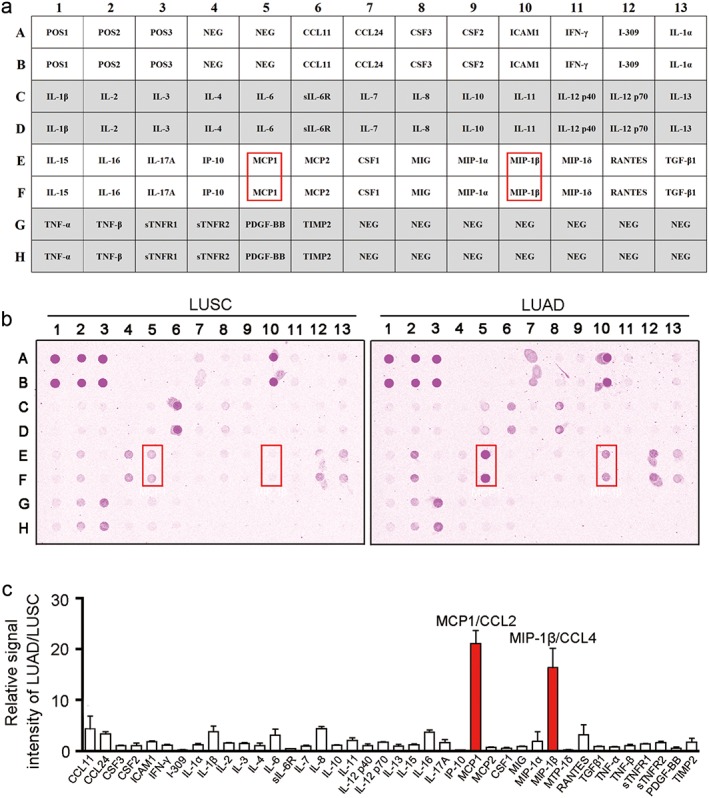
Antibody array identification of immune factors in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). (a) Antibody map of the inflammation antibody array used in panel (b), adapted from the Human Inflammation Antibody Array protocol (RayBiotech, Catalog #AAH‐INF‐G3). (b) Conditioned media from LUSC (left panel) and LUAD (right panel) were analyzed using an antibody array associated with inflammation factors. The overexpressed proteins in LUAD are indicated by red boxes. (c) Signal intensity of arrays in panel (b) were analyzed using densitometry, and the relative fold changes (LUAD vs. LUSC) in individual proteins were calculated after normalizing to the positive controls on each array (mean ± standard deviation, n = 2).
Western blotting
Fresh tissue homogenates were lysed in ice‐cold radioimmunoprecipitation assay buffer (Cell Signaling Technology, Danvers, MA, USA), with 1 mg/ml phenylmethylsulfonyl fluoride and 1% protease inhibitor cocktail (Roche, Basel, Switzerland) to obtain the whole cell lysate. Protein aliquots (30 μg per sample) were resolved on 15% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride Western blotting membranes (Roche, Basel, Switzerland) using the Bio‐Rad Blotting System (Hercules, CA, USA). After washing, the membranes were blocked with 5% blocking solution (nonfat dry milk dissolved in Tris‐buffered saline/Tween 20 [TBST]) at room temperature for one hour, and incubated with the primary antibodies CCL2 (Catalog #MAB279), CCL4 (Catalog #MAB271; R&D Systems, Minneapolis, MN, USA); and β‐Actin (Catalog #3700, Cell Signaling Technology) overnight at 4°C. After washing three times with 1 × TBST for eight minutes, the membranes were incubated for two hours at room temperature with 1:1000 diluted horseradish peroxidase (HRP)‐conjugated secondary antibody goat anti‐rabbit IgG‐HRP (Catalog #7054, Cell Signaling Technology) or goat anti‐mouse IgG‐HRP (Catalog #sc‐2005, Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washing three times with 1 × TBST for eight minutes, the protein bands were detected using Western Lightening Chemiluminescence Reagent (Life Technologies, Gaithersburg, MD, USA).
RNA extraction and quantitative real‐time PCR
Total RNA was extracted from cells or tissues using TRIzol (Roche) and was reverse transcribed using reverse transcriptase (Takara, Dalian, China) according to the manufacturer's instructions. qRT‐PCR was performed to detect levels of the corresponding CCL2, CCL4, and ACTB using SYBR Green SuperMix (Roche) and the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). ACTB was used as an internal control. The gene‐specific primers were as follows: CCL2: 5′‐TTCCCCTAGCTTTCCCCAGA‐3′ (forward) and 5′‐TCCCAGGGGTAGAACTGTGG‐3′ (reverse); CCL4 5′‐TGCTAGTAGCTGCCTTCTGC‐3′ (forward) and 5′‐TTCACTGGGATCAGCACAGAC‐3′ (reverse); ACTB: 5′‐CATGTACGTTGCTATCCAGGC‐3′ (forward) and 5′‐CTCCTTAATGTCACGCACGAT‐3′ (reverse). The relative expression level (defined as fold change) of the target gene (2−ΔΔCt) was normalized to the endogenous ACTB reference (ΔCt).
Immunofluorescent staining
In brief, slides with frozen sections were rinsed with PBS. Nonspecific binding was blocked with 5% normal goat serum for 30 minutes at room temperature. The slides were incubated with monoclonal mouse anti‐CD68 (ab955, 1:200 dilution) and rabbit anti‐CCL2 (ab9669, 1:200 dilution) or CCL4 (ab25129, 1:300 dilution) at 4°C overnight in a humidified chamber (Abcam, Cambridge, UK). After washing with PBS, the slides were then incubated with Alexa Fluor 594 (rabbit) or 488 (mouse) conjugated secondary antibodies (Invitrogen, Carlsbad, CA, USA). Finally, all slides were mounted with anti‐fade reagent with 4′,6‐diamidino‐2‐phenylindole (s36938, Life Technologies). Images were captured by OLYMPUS FV1000 fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA). An independent student's t‐test was used to assess the statistical significance of CCL2 and CCL4 expression between LUSC and LUAD. Correlation analyses of CCL2 and CCL4 in LUSC and LUAD were extracted from GEPIA (http://gepia.cancer-pku.cn/) by Pearson's test.18 The prognostic value was calculated by Kaplan–Meier analysis and log‐rank test. All survival curves were obtained from Kaplan–Meier Plotter (http://kmplot.com/analysis/).13 Co‐expressed genes and Gene Ontology analyses were based on COEXPEDIA (http://www.coexpedia.org/).19 P < 0.05 was considered statistically significant.
Results
Antibody array identification of immune factors in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD)
Non‐small cell lung cancer is the leading cause of cancer‐related death, and despite strong therapeutic efforts, mortality remains high. Therefore, there is an urgent need for novel prognostic biomarkers to improve the prediction of NSCLC patient outcomes, aid clinical decision‐making, and improve survival. The tumor microenvironment is a complex network including secreted macromolecules that regulate cancer progression.20 Unlike LUSC, the carcinogenesis, pathological type, and survival of LUAD are distinctive, which indicates that key factors play an important role in accelerating LUAD progression. Immune‐related factors in the tumor microenvironment are important participators of the extracellular milieu in regulating cancer progression, and are associated with patient survival.
To analyze the native immune‐associated factors deposited in tumor microenvironments, we used a short time tissue culture method that enabled us to further compare the protein expression levels of factors between LUSC and LUAD using antibody array (Fig 1).21 The antibody array used to identify the immune cytokines in tissue culture‐conditioned supernatants could detect 40 human inflammatory factors in a test (Fig 2a). The fluorescent signals of antibody array testing for LUSC and LUAD are shown in Figure 2b. Signal intensity analysis showed overexpression of chemokines MCP1/CCL2 (21.11‐fold increase) and MIP‐1β/CCL4 (19.33‐fold increase) in LUAD compared to LUSC (Fig 2c, Table 1). Low expression of I‐309/CCL2 (0.21‐fold decrease), IP‐10/CCL10 (0.20‐fold decrease), and MIP‐1δ/CCL15 (0.20‐fold decrease) in LUAD compared to LUSC were also observed (Table 1). The results of antibody array suggest that LUSC and LUAD have different immune microenvironments.
Table 1.
Antibody array relative signal intensity
| Spots no. | Array | Description | LUAD/LUSC |
|---|---|---|---|
| A6,B6 | CCL11 | CC motif chemokine ligand 11 | 1.71 |
| A7,B7 | CCL24 | CC motif chemokine ligand 24 | 3.33 |
| A8,B8 | CSF3 | Colony stimulating factor 3/G‐CSF | 1.04 |
| A9,B9 | CSF2 | Colony stimulating factor 2/GM‐CSF | 1.05 |
| A10,B10 | ICAM1 | Intercellular adhesion molecule 1 | 1.84 |
| A11,B11 | IFN‐γ | Interferon gamma | 1.16 |
| A12,B12 | I‐309 | CC motif chemokine ligand 1/CCL1 | 0.21 |
| A13,B13 | IL‐1α | Interleukin 1 alpha | 1.22 |
| C1,D1 | IL‐1β | Interleukin 1 beta | 3.65 |
| C2,D2 | IL‐2 | Interleukin 2 | 1.56 |
| C3,D3 | IL‐3 | Interleukin 3 | 1.51 |
| C4,D4 | IL‐4 | Interleukin 4 | 0.90 |
| C5,D5 | IL‐6 | Interleukin 6 | 2.99 |
| C6,D6 | sIL‐6R | Soluble interleukin 6 receptor | 0.44 |
| C7,D7 | IL‐7 | Interleukin 7 | 0.99 |
| C8,D8 | IL‐8 | Interleukin 8 | 4.40 |
| C9,D9 | IL‐10 | Interleukin 10 | 1.14 |
| C10,D10 | IL‐11 | Interleukin 11 | 1.93 |
| C11,D11 | IL‐12 p40 | IL‐12 subunit p40/IL‐12B | 1.04 |
| C12,D12 | IL‐12 p70 | IL‐12 subunit p35/IL‐12A, heterodimer | 1.77 |
| C13,D13 | IL‐13 | Interleukin 13 | 0.89 |
| E1,F1 | IL‐15 | Interleukin 15 | 1.20 |
| E2,F2 | IL‐16 | Interleukin 16 | 3.73 |
| E3,F3 | IL‐17A | Interleukin 17A | 1.68 |
| E4,F4 | IP‐10 | CXC motif chemokine ligand 10/CXCL10 | 0.20 |
| E5,F5 | MCP1 | CC motif chemokine ligand 2/CCL2 | 21.11 |
| E6,F6 | MCP2 | CC motif chemokine ligand 8/CCL8 | 0.73 |
| E7,F7 | CSF1 | Colony stimulating factor 1/M‐CSF | 0.54 |
| E8,F8 | MIG | CXC motif chemokine ligand 9/CXCL9 | 0.89 |
| E9,F9 | MIP‐1α | CC motif chemokine ligand 3/CCL3 | 1.36 |
| E10,F10 | MIP‐1β | CC motif chemokine ligand 4/CCL4 | 19.33 |
| E11,F11 | MIP‐1δ | CC motif chemokine ligand 15/CCL15 | 0.20 |
| E12,F12 | RANTES | CC motif chemokine ligand 5/CCL5 | 2.50 |
| E13,F13 | TGF‐β1 | Transforming growth factor beta 1 | 0.93 |
| G1,H1 | TNF‐α | Tumor necrosis factor alpha | 0.79 |
| G2,H2 | TNF‐β | Tumor necrosis factor beta | 1.07 |
| G3,H3 | sTNFR1 | Soluble tumor necrosis factor receptor 1 | 1.44 |
| G4,H4 | sTNFR2 | Soluble tumor necrosis factor receptor 2 | 1.60 |
| G5,H5 | PDGF‐BB | Platelet derived growth factor subunit B | 0.55 |
| G6,H6 | TIMP2 | Tissue inhibitor of metalloproteinase 2 | 1.58 |
Any ≥ 5‐fold increase or ≤ 0.20‐fold decrease in signal intensity is shown in bold. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Higher levels of CCL2 and CCL4 in LUAD than in LUSC
To verify the CCL2 and CCL4 expression levels in LUSC and LUAD, four LUSC and four LUAD tissue samples were tested by Western blotting. CCL2 was obviously overexpressed in 4/4 LUAD tissues compared to LUSC tissues (Fig 3a). In addition, high expression of CCL4 in 3/4 LUAD tissues was also observed, while CCL4 expression was significantly low in all LUSC tissues (Fig 3a). Using qRT‐PCR, we compared CCL2 and CCL4 expression in RNA levels in patients with LUSC (n = 29) and LUAD (n = 18), respectively. Consistent with the Western blot results, both CCL2 and CCL4 were highly expressed in LUAD compared to LUSC at RNA levels (P < 0.001) (Fig 3b). Co‐staining of CCL2 or CCL4 with macrophage marker CD68 in normal lung, LUSC, and LUAD tissues showed that CCL2 and CCL4 could be secreted by alveolar macrophages and tumor‐associated macrophages (TAMs) (Fig 3c). Moreover, higher percentages of CCL2+/CD68+ and CCL4+/CD68+ cells in LUAD compared to LUSC tissues were observed, which might be responsible for the high expression of CCL2 and CCL4 in LUAD samples (Fig 3d). Interestingly, the Pearson correlation tests based on data from The Cancer Genome Atlas (TCGA) database (http://gepia.cancer-pku.cn/) showed a positive correlation between CCL2 or CCL4 expression in LUSC (P = 5.1e−14) and LUAD (P = 7.3e−12) tissues (Fig 3e). These findings suggest that the abundant CCL2 and CCL4 in the LUAD microenvironment may play an interrelated role in cancer progression and account for the poor prognosis of LUAD patients.
Figure 3.
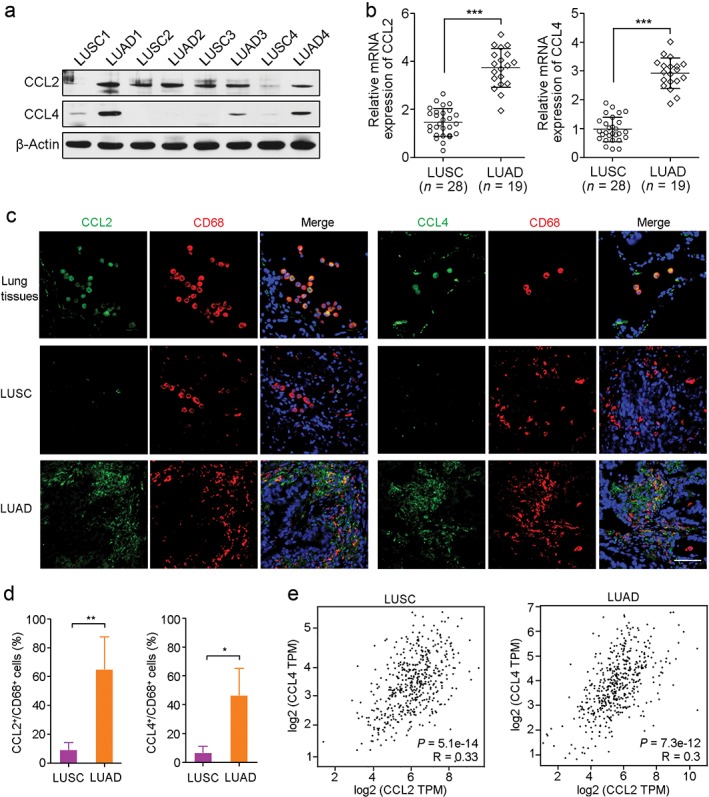
CCL2 and CCL4 overexpression in lung adenocarcinoma (LUAD). (a) Western blot analysis showed that CCL2 and CCL4 were upregulated in LUAD but not in lung squamous cell carcinoma (LUSC). β‐Actin was used as a loading control. (b) Quantitative real‐time‐PCR showed high expression of CCL2 and CCL4 at RNA level in LUAD (n = 19) compared to LUSC (n = 28). ***P < 0.001. (c) Co‐staining of CCL2 or CCL4 (green) with macrophage marker CD68 (red) in normal lung, LUSC, and LUAD tissues. Scale bar, 100 μm. Cell nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (blue). (d) The percentages of CCL2+/CD68+ and CCL4+/CD68+ cells per visual field in LUAD and LUSC tissues were analyzed. *P < 0.05. **P < 0.01. (e) Correlation analyses of CCL2 and CCL4 expression between LUSC and LUAD. mRNA, messenger RNA.
Association between CCL2 expression and non‐small cell lung cancer (NSCLC) patient survival
The association between CCL2 expression level and survival of NSCLC patients was then determined via Kaplan–Meier analysis based on data from the TCGA database (http://kmplot.com/analysis/). Survival curves showed that high CCL2 expression in LUSC patients was significantly associated with favorable OS (P = 0.048) and PFS (P = 0.012) (Fig 4a,b). However, in LUAD patients, high levels of CCL2 predicted unfavorable OS (P = 6.8e−08) and PFS (P = 0.00098) (Fig 4c,d). The opposing prognostic value of CCL2 in LUSC and LUAD suggests that CCL2 plays key but different roles in NSCLC progression.
Figure 4.
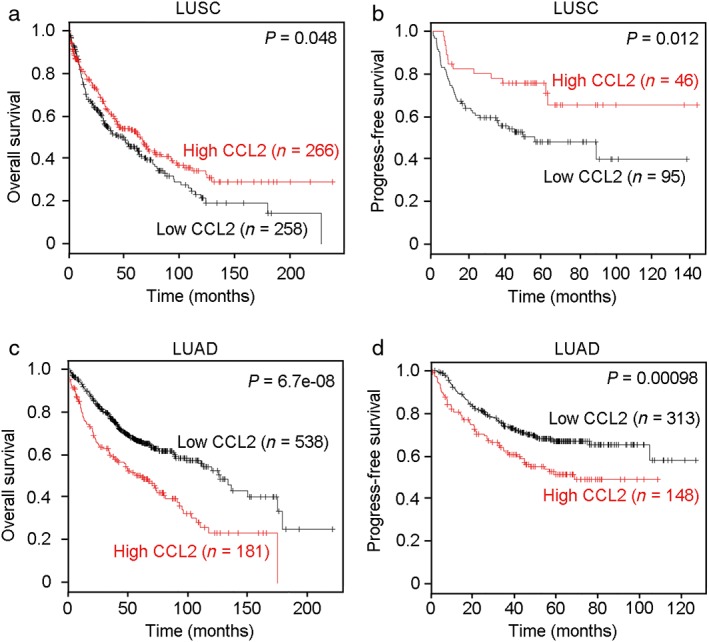
Prognostic value of CCL2 in the lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) patient cohorts. A high level of CCL2 was associated with good (a) overall survival (OS) (P = 0.048) and (b) progression‐free survival (PFS) (P = 0.012) in patients with LUSC. However, high expression of CCL2 in LUAD indicated poor (c) OS (P = 6.8e−08) and (d) PFS (P = 0.00098).
CCL4 expression predicts survival in NSCLC patients
The prognostic role of CCL4 in LUSC and LUAD was further analyzed using Kaplan–Meier Plotter (http://kmplot.com/analysis/). CCL4 expression was not correlated with OS in LUSC patients (P = 0.22) (Fig 5a). High levels of CCL4 could predict good PFS in LUSC patients (P = 0.021) (Fig 5b). However, in LUAD, high expression of CCL4 was significantly associated with poorer OS (P = 0.013) (Fig 5c). Moreover, there was no significant correlation between CCL4 expression and PFS in LUAD patients (P = 0.077) (Fig 5d). Our results suggest that CCL2 and CCL4 expression could serve as potential prognostic biomarkers for NSCLC patients.
Figure 5.
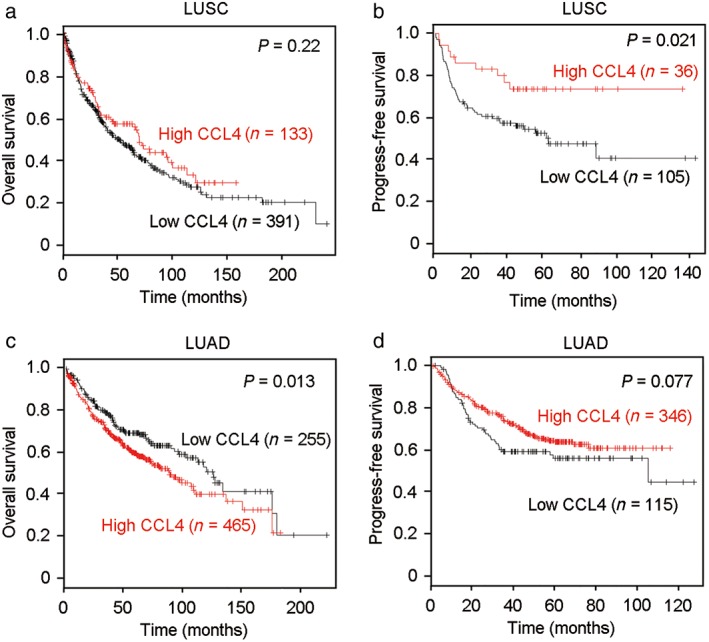
CCL4 expression was associated with survival in patients with lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). (a) There was no correlation between CCL4 expression and overall survival (OS) in patients with LUSC (P = 0.22). (b) High levels of CCL4 were associated with good progression‐free survival (PFS) (P = 0.021) in LUSC patients. However, in LUAD patients, high CCL4 expression indicated (c) poor OS (P = 0.013), (d) but not PFS (P = 0.077).
Discussion
In the tumor microenvironment, cancer cells grow within an intricate network of epithelial and endothelial cells, cytokines and chemokines, and infiltrating immune cells. Different types of infiltrating immunocytes and immune factors have different effects on tumor progression, which can vary according to cancer type.22 In this study, we compared the immune factors in LUSC and LUAD by antibody array and observed higher expression of CCL2 and CCL4 in LUAD than in LUSC, which was significantly associated with unfavorable survival in LUAD patients. Our findings indicate that both CCL2 and CCL4 may play key and contrary roles in the development of LUAD and LUSC.
CCL2, also known as monocyte chemoattractant protein‐1, is a small cytokine, belonging to the CC chemokine family. CCL2 could recruit monocytes, memory T cells, and dendritic cells to the sites of inflammation produced either by tissue injury or infection, as well as the tumor microenvironment.23 CCL2 is produced by a variety of tumors and plays an important role in cancer progression, particularly in promoting cancer cell migration and invasion.24 Our study showed that CCL2 was highly expressed in the LUAD microenvironment; however, the reason for this is unclear. In Lewis lung carcinoma, TNFα released by tumor cell‐activated macrophages is critical for increased CCL2 production by tumor cells.25 Wang et al. reported that IL‐6 increased the number of migrated macrophages to tumor cells after irradiation by upregulating CCL2 and CCL5 in A549 and H157 cells.26 Moreover, type I interferons can also induce CCL2 expression to initiate the recruitment and activation of leukocytes.27 Collectively, CCL2 expression in lung cancer cells is tightly regulated by the complex tumor microenvironment.
CCL4 is well known as a macrophage inflammatory protein 1 beta, and belongs to the inflammatory CC chemokine subfamily. It can be secreted by activated leukocytes, lymphocytes, and endothelial and muscle cells in response to inflammation. It is a chemoattractant for natural killer cells, monocytes, and a variety of other immune cells involved in immune responses.28 Except for roles in inflammation and immune‐regulation, CCL4 is implicated in carcinogenesis by facilitating instability in the tumor environment.29 Moreover, CCL4 could also promote tumor growth through regulation of antitumor immunity.30 In a mouse model of peritoneal dissemination of a colon tumor, intraperitoneal injection of hemagglutinating virus of Japan cationic liposomes containing the MIP‐1β gene resulted in local expression of MIP‐1β and local accumulation of CD4+ T lymphocytes and significantly increased survival of the cancer cell‐injected mice, suggesting that CCL4 might serve as a potential therapeutic target against peritoneal disseminated cancer.31 The properties of tumor microenvironments have a strong impact on cancer progression, response to therapy, and patient prognosis.32
In addition, a study showed that the messenger RNA levels of CCL2 and CCL4 in NSCLC tissues were apparently higher than those in normal contrast tissues.33 Cao et al. also showed higher CCL2 and CCL4 expression in NSCLC tissues than in non‐tumor tissues.34 Moreover, Zhang et al. reported that the presence of CCL2‐positive cancer cells was high in LUAD relative to LUSC and other histological types.35 A recent study demonstrated that Lewis lung carcinoma cells harbor the activating Kras G12C mutation, which upregulates CCL2 production linking cancer‐associated inflammation.36 A previous study reported that knockdown of CCL2 and CCL4 in lung cancer cells could suppress macrophage invasion under co‐culture conditions.37 This evidence suggests that lung cancer cell‐derived CCL2 and CCL4 can recruit macrophages into the tumor microenvironment, promoting cancer progression. In the present study, using antibody array, Western blot, and qRT analyses, we found higher RNA and protein levels of CCL2 and CCL4 in the LUAD than in LUSC microenvironment. Dynamic interaction between cancer cells and the microenvironment is critical for tumorigenesis, and cancer immunosurveillance plays an important role in tumor evolution.38 Therefore, these results indicate the different compositions of tumor immune microenvironments between LUSC and LUAD.
The composition of the tumor microenvironment can play a regulatory role during cancer development and metastasis formation, and affects patient survival. However, correlations between CCL2 or CCL4 expression and survival of patients with LUSC or LUAD remain largely unknown. Based on data from the TCGA database, we found that high CCL2 or CCL4 expression was significantly associated with survival in LUSC patients but predicted poor outcomes in LUAD patients. These findings suggest that CCL2 and CCL4 may play opposite roles in LUSC and LUAD progression. In addition, TAMs can be recruited by CCL2 and CCL4 into the tumor microenvironment. There are two main phenotypes of TAMs: M1 (anti‐tumor) and M2 (pro‐tumor). The higher percentage of M2 TAMs in LUAD than LUSC tissues suggests that TAMs may tend to differentiate into the M2 subtype in the LUAD microenvironment, while most TAMs differentiate into the M1 subtype in LUSC. This differentiation may be responsible for the opposing roles of CCL2 or CCL4 for predicting prognosis in patients with LUAD and LUSC.39
CCL2 preferentially binds to the CC chemokine receptor type 2 (CCR2). Elevated CCL2 and CCR2 expression has been observed in a variety of malignancies and is associated with adverse prognosis in patients with breast cancer,40 nasopharyngeal carcinoma,41 and colorectal cancer.42 Moreover, high CCL2 and CCR2 expression are also remarkably correlated with shortened survival time and increased risk of recurrence in renal cell carcinoma.43 However, a previous study reported that higher levels of CCL2 in ovarian cancer cell lines were associated with increased chemosensitivity and decreased invasion in vitro, and increased ovarian tumoral expression of CCL2 was associated with improved chemotherapy response and survival outcomes.44 In addition, the CCL4 axis can contribute to breast cancer metastasis to bone by mediating the interaction between cancer cells and fibroblasts in bone cavity by bonding to CC chemokine receptor type 5.45 Inversely, increased CCL4 expression shows a significant association with prolonged OS in patients with esophageal squamous cell carcinoma.46 This evidence suggests that the effect of CCL2 and CCL4 on cancer progression is inconclusive, with different results in different types of cancers. Further research is required to determine the functional mechanisms.
In summary, our data demonstrate that CCL2 and CCL4 are highly expressed in the LUAD microenvironment compared to LUSC. CCL2 and CCL4 predict adverse prognosis in LUAD patients but favorable survival in LUSC patients. These findings highlight the importance of chemokines in immune surveillance and survival of NSCLC patients. Although CCL2 and CCL4 may serve as potential predictors and therapeutic targets of survival for NSCLC patients, further research is needed to investigate their reciprocal roles in LUSC and LUAD.
Disclosure
No authors report any conflict of interest.
Acknowledgment
This project was supported by grants from the National Natural Science Foundation of China (81772554 and 81672357).
References
- 1. Zhong J, Zheng Q, Gao E et al Influence of body mass index on the therapeutic efficacy of gemcitabine plus cisplatin and overall survival in lung squamous cell carcinoma. Thorac Cancer 2018; 9: 291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sikaria D, Tu YN, Fisler DA, Mauro JA, Blanck G. Identification of specific feed‐forward apoptosis mechanisms and associated higher survival rates for low grade glioma and lung squamous cell carcinoma. J Cancer Res Clin Oncol 2018; 144: 459–68. [DOI] [PubMed] [Google Scholar]
- 3. Zhou X, Liu Q, Wada Y, Liao L, Liu J. CDKN1A‐interacting zinc finger protein 1 is a novel biomarker for lung squamous cell carcinoma. Oncol Lett 2018; 15: 183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yousem SA. Pulmonary intestinal‐type adenocarcinoma does not show enteric differentiation by immunohistochemical study. Mod Pathol 2005; 18: 816–21. [DOI] [PubMed] [Google Scholar]
- 5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 7–30. [DOI] [PubMed] [Google Scholar]
- 6. Kenfield SA, Wei EK, Stampfer MJ, Rosner BA, Colditz GA. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control 2008; 17: 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanazawa H, Ebina M, Ino‐Oka N et al Transition from squamous cell carcinoma to adenocarcinoma in adenosquamous carcinoma of the lung. Am J Pathol 2000; 156: 1289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep 2014; 15: 1243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Witz IP. The tumor microenvironment: The making of a paradigm. Cancer Microenviron 2009; 2 (Suppl 1): 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korneev KV, Atretkhany KN, Drutskaya MS, Grivennikov SI, Kuprash DV, Nedospasov SA. TLR‐signaling and proinflammatory cytokines as drivers of tumorigenesis. Cytokine 2017; 89: 127–35. [DOI] [PubMed] [Google Scholar]
- 11. Kowanetz M, Ferrara N. Vascular endothelial growth factor signaling pathways: Therapeutic perspective. Clin Cancer Res 2006; 12: 5018–22. [DOI] [PubMed] [Google Scholar]
- 12. Yan X, Jiao SC, Zhang GQ, Guan Y, Wang JL. Tumor‐associated immune factors are associated with recurrence and metastasis in non‐small cell lung cancer. Cancer Gene Ther 2017; 24: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sterlacci W, Wolf D, Savic S et al High transforming growth factor β expression represents an important prognostic parameter for surgically resected non‐small cell lung cancer. Hum Pathol 2012; 43: 339–49. [DOI] [PubMed] [Google Scholar]
- 14. Fridman WH, Pagès F, Sautès‐Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer 2012; 12: 298–306. [DOI] [PubMed] [Google Scholar]
- 15. Lovitch SB, Rodig SJ. The role of surgical pathology in guiding cancer immunotherapy. Annu Rev Pathol 2016; 11: 313–41. [DOI] [PubMed] [Google Scholar]
- 16. Yuan J, Hegde PS, Clynes R et al Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer 2016; 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Al‐Aidaroos AQ, Yuen HF, Guo K et al Metastasis‐associated PRL‐3 induces EGFR activation and addiction in cancer cells. (Published erratum appears in J Clin Invest 2013;123:4540. J Clin Invest 2013; 123: 3459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45(W1): W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang S, Kim CY, Hwang S et al COEXPEDIA: Exploring biomedical hypotheses via co‐expressions associated with medical subject headings (MeSH). Nucleic Acids Res 2017; 45 (D1): D389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hynes RO. The extracellular matrix: Not just pretty fibrils. Science 2009; 326: 1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Zhu YH, Li Y, Guan XY. Identification of chemokine CXCL10 in tumor microenvironment by antibody array as a prognostic marker in hepatocellular carcinoma. Neoplasma 2017; 64: 778–86. [DOI] [PubMed] [Google Scholar]
- 22. Wang RF. Regulatory T cells and innate immune regulation in tumor immunity. (Published erratum appears in Springer Semin Immunopathol 2006;28:77. Springer Semin Immunopathol 2006; 28: 17–23. [DOI] [PubMed] [Google Scholar]
- 23. Xu LL, Warren MK, Rose WL, Gong W, Wang JM. Human recombinant monocyte chemotactic protein and other C‐C chemokines bind and induce directional migration of dendritic cells in vitro. J Leukoc Biol 1996; 60: 365–71. [DOI] [PubMed] [Google Scholar]
- 24. Zhang J, Chang L, Jin H et al Benzopyrene promotes lung cancer A549 cell migration and invasion through up‐regulating cytokine IL8 and chemokines CCL2 and CCL3 expression. (Published erratum appears in (Published erratum appears in Exp Biol Med 2016;241:1620. Exp Biol Med 2016; 241: 1516–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoshimura T, Liu M, Chen X, Li L, Wang JM. Crosstalk between tumor cells and macrophages in stroma renders tumor cells as the primary source of MCP‐1/CCL2 in Lewis lung carcinoma. Front Immunol 2015; 6: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X, Yang X, Tsai Y et al IL‐6 mediates macrophage infiltration after irradiation via up‐regulation of CCL2/CCL5 in non‐small cell lung cancer. Radiat Res 2017; 187: 50–9. [DOI] [PubMed] [Google Scholar]
- 27. Lehmann MH, Torres‐Domínguez LE, Price PJ, Brandmüller C, Kirschning CJ, Sutter G. CCL2 expression is mediated by type I IFN receptor and recruits NK and T cells to the lung during MVA infection. J Leukoc Biol 2016; 99: 1057–64. [DOI] [PubMed] [Google Scholar]
- 28. Bystry RS, Aluvihare V, Welch KA, Kallikourdis M, Betz AG. B cells and professional APCs recruit regulatory T cells via CCL4. Nat Immunol 2001; 2: 1126–32. [DOI] [PubMed] [Google Scholar]
- 29. Sadeghi M, Lahdou I, Oweira H et al Serum levels of chemokines CCL4 and CCL5 in cirrhotic patients indicate the presence of hepatocellular carcinoma. Br J Cancer 2015; 113: 756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lien MY, Lin CW, Tsai HC et al Impact of CCL4 gene polymorphisms and environmental factors on oral cancer development and clinical characteristics. Oncotarget 2017; 8: 31424–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miyata T, Yamamoto S, Sakamoto K, Morishita R, Kaneda Y. Novel immunotherapy for peritoneal dissemination of murine colon cancer with macrophage inflammatory protein‐1beta mediated by a tumor‐specific vector, HVJ cationic liposomes. Cancer Gene Ther 2001; 8: 852–60. [DOI] [PubMed] [Google Scholar]
- 32. Klein B, Schuppe HC, Bergmann M, Hedger MP, Loveland BE, Loveland KL. An in vitro model demonstrates the potential of neoplastic human germ cells to influence the tumour microenvironment. Andrology 2017; 5: 763–70. [DOI] [PubMed] [Google Scholar]
- 33. Zou W, Hu TH. Effect of chemokine interleukin‐8, monocyte chemoattractant protein‐1 and macrophage inflammatory protein‐1 on the angiogenesis of non‐small cell lung cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2007; 32: 665–70 In Chinese. [PubMed] [Google Scholar]
- 34. Cao Y, Huang H, Wang Z, Zhang G. The inflammatory CXC chemokines, GROαhigh, IP‐10low, and MIGlow, in tumor microenvironment can be used as new indicators for non‐small cell lung cancer progression. Immunol Invest 2017; 46: 361–74. [DOI] [PubMed] [Google Scholar]
- 35. Zhang XW, Qin X, Qin CY, Yin YL, Chen Y, Zhu HL. Expression of monocyte chemoattractant protein‐1 and CC chemokine receptor 2 in non‐small cell lung cancer and its significance. Cancer Immunol Immunother 2013; 62: 563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agalioti T, Giannou AD, Krontira AC et al Mutant KRAS promotes malignant pleural effusion formation. Nat Commun 2017; 8: 15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen S, Jiao J, Jiang D et al T‐box transcription factor Brachyury in lung cancer cells inhibits macrophage infiltration by suppressing CCL2 and CCL4 chemokines. Tumour Biol 2015; 36: 5881–90. [DOI] [PubMed] [Google Scholar]
- 38. Wiggins JM, Opoku‐Acheampong AB, Baumfalk DR, Siemann DW, Behnke BJ. Exercise and the tumor microenvironment: Potential therapeutic implications. Exerc Sport Sci Rev 2018; 46: 56–64. [DOI] [PubMed] [Google Scholar]
- 39. Almatroodi SA, McDonald CF, Darby IA, Pouniotis DS. Characterization of M1/M2 tumour‐associated macrophages (TAMs) and Th1/Th2 cytokine profiles in patients with NSCLC. Cancer Microenviron 2016; 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P, Cheng N. CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein‐ and p42/44 mitogen‐activated protein kinase (MAPK)‐dependent mechanisms. J Biol Chem 2012; 287: 36593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J, Lv X, Chen J et al CCL2‐CCR2 axis promotes metastasis of nasopharyngeal carcinoma by activating ERK1/2‐MMP2/9 pathway. Oncotarget 2016;7:15632–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao L, Lim SY, Gordon‐Weeks AN et al Recruitment of a myeloid cell subset (CD11b/Gr1 mid) via CCL2/CCR2 promotes the development of colorectal cancer liver metastasis. Hepatology 2013; 57: 829–39. [DOI] [PubMed] [Google Scholar]
- 43. Wang Z, Xie H, Zhou L et al CCL2/CCR2 axis is associated with postoperative survival and recurrence of patients with non‐metastatic clear‐cell renal cell carcinoma. Oncotarget 2016; 7: 51525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fader AN, Rasool N, Vaziri SA et al CCL2 expression in primary ovarian carcinoma is correlated with chemotherapy response and survival outcomes. Anticancer Res 2010; 30: 4791–8. [PubMed] [Google Scholar]
- 45. Sasaki S, Baba T, Nishimura T et al Essential roles of the interaction between cancer cell‐derived chemokine, CCL4, and intra‐bone CCR5‐expressing fibroblasts in breast cancer bone metastasis. Cancer Lett 2016; 378: 23–32. [DOI] [PubMed] [Google Scholar]
- 46. Liu JY, Li F, Wang LP et al CTL‐ vs Treg lymphocyte‐attracting chemokines, CCL4 and CCL20, are strong reciprocal predictive markers for survival of patients with oesophageal squamous cell carcinoma. Br J Cancer 2015; 113: 747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]


