Abstract
Background
In cases of EGFR‐tyrosine kinase inhibitor (TKI) failure, re‐biopsy may be useful to understand resistance mechanisms and guide further treatment decisions. However, performing re‐biopsy is challenging because of several hurdles. We assessed the feasibility of re‐biopsy in advanced non‐small cell lung cancer (NSCLC) patients in real‐world clinical practice.
Methods
We retrospectively reviewed the clinical and pathologic data of advanced NSCLC patients who experienced disease progression after previous treatment with EGFR‐TKIs at a single tertiary hospital in Korea between January 2014 and December 2016. Re‐biopsy specimens included small biopsy, surgical tissue, or liquid‐based cytology. EGFR mutation was tested using peptide nucleic acid‐mediated clamping PCR.
Results
Of the 230 NSCLC patients that experienced progression after EGFR‐TKI therapy, 105 (45.7%) underwent re‐biopsy. Re‐biopsy was successfully performed in 94 (89.5%) patients, and 11 patients were diagnosed with no malignancy. The complication rate was 8.6%, including seven cases of pneumothorax. EGFR mutation testing was performed on 75 patients using re‐biopsy specimens. Of the 57 patients who had sensitizing mutations at diagnosis, T790M mutations were found in 19 (33.3%), while 38 (66.7%) had no T790M mutation. Multivariate analysis showed that the re‐biopsy group was younger (P = 0.002) and exhibited a previous response to EGFR‐TKIs (P < 0.001).
Conclusion
Re‐biopsy in advanced NSCLC is feasible in real world clinical practice, particularly in younger patients and those who achieved a previous response to EGFR‐TKIs.
Keywords: EGFR mutation, re‐biopsy, T790M
Introduction
Treatment with EGFR‐tyrosine kinase inhibitors (TKIs) is the standard of care for advanced non‐small cell lung cancer (NSCLC) patients with an EGFR sensitizing mutation; however, acquired resistance is inevitable, occurring within 10–12 months of treatment.1, 2 Various resistance mechanisms have been identified. Understanding such mechanisms is critical to guide further treatment for patients with EGFR‐TKI resistant NSCLC.3 Among the EGFR‐TKI resistance mechanisms, T790M mutations, which substitute threonine (T) with methionine (M) at position 790 of exon 20 of the EGFR gene are the most common, accounting for more than 50%.4 Other resistance mechanisms include amplification of the MET gene, PIK3CA and BRAF mutations, epithelial‐to‐mesenchymal transition, and small cell lung cancer transformation.3, 4, 5 Thus, re‐biopsy of sufficient tissue for molecular analysis is necessary to identify the resistance type and guide further treatment decisions after EGFR‐TKI treatment failure.6
With the development of third generation EGFR‐TKIs, re‐biopsy is even more important in order to identify T790M mutations. Several studies have reported high success rates of re‐biopsy, ranging from 75% to 97%.6, 7, 8, 9, 10, 11 However, re‐biopsy is still challenging in real practice because of several hurdles, including tissue availability, procedural feasibility, and limited accessibility to new anti‐cancer drugs, which differ in different countries. As limited data is available on the feasibility of re‐biopsy and its clinical impact in real‐world clinical practice, the aim of this study was to assess successful re‐biopsy rates and the factors influencing re‐biopsy in Korean real world clinical practice.6, 10, 12, 13 In addition, we evaluated EGFR mutation status and clinical factors associated with an increased frequency of T790M mutations.
Methods
Patients and materials
This retrospective, observational study included all patients diagnosed with NSCLC who experienced disease progression after EGFR‐TKI therapy at the Chonnam National University Hwasun Hospital between January 2014 and December 2016. Disease progression was confirmed by chest computed tomography (CT) according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. Progression was defined not only as progression after initial response or durable (> 6 months) stable disease after EGFR‐TKIs, but also intrinsic resistance to EGFR‐TKIs. Patients who discontinued EGFR‐TKIs before disease progression or whose RECIST responses were not confirmed were excluded. Re‐biopsy procedures included surgery, bronchoscopy, endobronchial ultrasonography (EBUS)‐guided transbronchial needle aspiration (TBNA), percutaneous core needle biopsy (PCNB), excisional biopsy, fine needle aspiration (FNA), lumbar puncture, pericardiocentesis, and thoracentesis.
All data were gathered in accordance with the amended Declaration of Helsinki, following approval from an independent hospital institutional review board (IRB approval no.: CNUHH‐2017‐108). The need for written informed consent was waived because of the retrospective design of the study.
EGFR mutation test
We used the PNA Clamp Mutation Detection Kit (Panagene Inc., Daejeon, Korea) to detect EGFR gene mutations using real‐time PCR from DNA acquired from formalin fixed paraffin‐embedded tumor tissue samples or liquid‐based cytology samples. DNA was isolated using a Gene All Tissue DNA Purification Kit (General Biosystems, Seoul, Korea) according to manufacturer protocol. All reactions were performed in 20 μL volumes using template DNA, primer, PNA probe set, and fluorescence PCR master mix. All reagents were included in the kit. Real‐time PCR reaction of PNA‐mediated clamping PCR was performed using a CFX 96 (BioRad Laboratories Inc., Hercules, CA, USA). PCR cycling conditions were set at a five minute hold at 94°C for 40 cycles, at 94°C for 30 seconds, 70°C for 20 seconds, 63°C for 30 seconds, and 72°C for 30 seconds. The pooled sensitivity and specificity of the PNA clamp methods were 93% and 100%, respectively.14, 15
Statistical analysis
All data were expressed as medians and interquartile ranges or as numbers (%) in the text and tables. Intergroup comparisons were performed using the Mann–Whitney U test for continuous variables, and Pearson's χ2 or Fisher's exact tests for categorical variables. Multivariate analysis was performed to determine the feasibility of re‐biopsy using a binary logistic regression. All analyses were performed using SPSS version 19.0 (IBM Corp., Armonk, NY, USA). A P value < 0.05 indicated statistical significance.
Results
Patients
A total of 230 NSCLC patients who experienced progression after EGFR‐TKI treatment were reviewed (Table 1). The median age was 68.0 years (59.8–74.0) with women comprising 47.4% of the total. Adenocarcinoma (90.4%) was the most common histology. The EGFR‐TKIs used were gefitinib (47.0%), erlotinib (48.7%), and afatinib (4.3%). The median duration of treatment was 247 (82–459) days. The initial biopsy specimen was used to detect EGFR mutation in 211 patients. Of the 230 patients included, 66 (28.7%) had exon 19 deletions, 62 (27.0%) had L858R mutations, and 68 (29.6%) were wild type. In addition, 9 (3.9%) patients had uncommon mutations, such as G719X, S768I, or exon 20 insertion; 2 (0.9%) patients had de novo T790M mutations; and although tests were conducted, the results were not available in 4 (1.7%) cases.
Table 1.
Baseline characteristics of NSCLC patients who experienced EGFR‐TKI failure (n = 230)
| Characteristics | Value |
|---|---|
| Age (years) | 68.0 (59.8–74.0) |
| Gender, male | 109 (47.4%) |
| Smoking | |
| Current smoker | 57 (24.8%) |
| Ex‐smoker | 50 (21.7%) |
| Never smoker | 123 (53.5%) |
| ECOG performance status | |
| 0 | 4 (1.7%) |
| 1 | 167 (72.6%) |
| 2 | 43 (18.7%) |
| 3 | 11 (4.8%) |
| 4 | 5 (2.2%) |
| Histology | |
| ADC | 208 (90.4%) |
| SQC | 15 (6.5%) |
| NSCLC | 5 (2.2%) |
| LCNEC | 1 (0.4%) |
| SCLC + ADC | 1 (0.4%) |
| EGFR‐TKI | |
| Gefitinib | 108 (47.0%) |
| Erlotinib | 112 (48.7%) |
| Afatinib | 10 (4.3%) |
| Treatment line | |
| 1st line | 125 (54.3%) |
| 2nd line | 72 (31.3%) |
| 3rd line | 23 (10.0%) |
| > 4th line | 10 (4.4%) |
| Duration of EGFR‐TKI therapy, days | 247 (82–459) |
| EGFR mutation status at diagnosis (n = 211) | |
| Exon 19 deletion | 66 (28.7%) |
| Exon 21 L858R | 62 (27.0%) |
| Wild type | 68 (29.6%) |
| Uncommon mutations | 9 (3.9%) |
| De novo T790M mutation | 2 (0.9%) |
| Invalid | 4 (1.7%) |
Values are presented as number (%) or median with interquartile range.
ADC, adenocarcinoma; ECOG, Eastern Cooperative Oncology Group; LCNEC, large cell neuroendocrine carcinoma; NSCLC, non‐small cell lung cancer; SCLC, small cell lung cancer; SQC, squamous cell carcinoma; TKI, tyrosine kinase inhibitor.
Characteristics of re‐biopsy
Re‐biopsy was performed on 105 (45.7%) patients, while 125 (53.4%) did not undergo re‐biopsy. Reasons for not performing re‐biopsy included patients receiving salvage systemic chemotherapy without re‐biopsy (n = 69), best supportive care (n = 52), refusal to perform re‐biopsy (n = 2), and patients lost to follow‐up (n = 2). Of the 105 patients who underwent re‐biopsy, 94 (89.5%) were pathologically diagnosed with malignancy from 59 histological and 35 cytological samples (Fig 1). The most common sampling method was intrathoracic, which included video‐assisted thoracoscopic surgery (n = 3), bronchoscopy with EBUS‐TBNA (n = 25), CT‐guided PCNB (n = 17), pericardiocentesis (n = 2), and thoracentesis (n = 30) (Table 2). The primary tumor was more frequently used for the initial biopsy than the re‐biopsy (59.6% vs. 30.5%; P < 0.001). Metastatic lesions were more frequently used for re‐biopsy than initial biopsy (53.3% vs. 13.0%; P < 0.001). Most specimens used for the initial biopsy were tissue samples (84.4%). In contrast, the proportion of cytology sample specimens used was significantly higher in re‐biopsy than initial biopsy (40.0% vs. 15.3%; P < 0.001).
Figure 1.
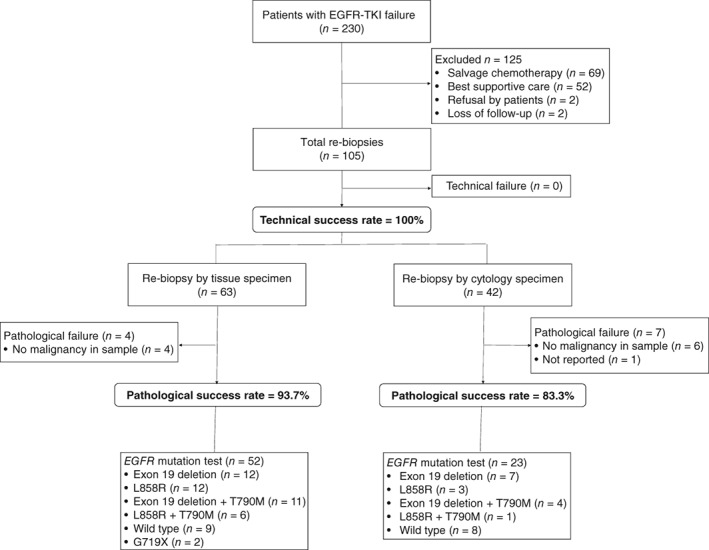
Flow chart of subject enrollment.
Table 2.
Comparison between initial biopsy and re‐biopsy
| Variables | Initial biopsy (n = 230) | Re‐biopsy (n = 105) | P |
|---|---|---|---|
| Site of biopsy | < 0.001 | ||
| Primary tumor | 137 (59.6%) | 32 (30.5%) | |
| Regional lymph node | 63 (27.4%) | 17 (16.2%) | |
| Metastasis | 30 (13.0%) | 56 (53.3%) | |
| Method of biopsy | <0.001 | ||
| Tissue | 195 (84.8%) | 63 (60.0%) | |
| Surgery | 43 (18.7%) | 10 (9.5%) | 0.371 |
| Lung | 35 | 3 | |
| Brain | 4 | 3 | |
| Femur | 3 | 1 | |
| Spine | 1 | 3 | |
| Small biopsy | 152 (66.1%) | 53 (50.5%) | |
| Bronchoscopy + EBUS | 111 | 25 | |
| CT‐guided PCNB | 26 | 17 | |
| SCL biopsy | 13 | 6 | |
| Liver biopsy | 1 | 2 | |
| Adrenal biopsy | 1 | 2 | |
| Gastroscopy | 0 | 1 | |
| Cytology | 35 (15.2%) | 42 (40.0%) | |
| Bronchial brushing cytology | 12 | 0 | |
| Thoracentesis | 20 | 30 | |
| Lumbar puncture | 0 | 6 | |
| FNA from lymph node | 3 | 3 | |
| Pericardiocentesis | 0 | 2 | |
| Paracentesis | 0 | 1 |
Values are presented as number (%).
CT, computed tomography; EBUS, endobronchial ultrasound; FNA, fine needle aspiration; PCNB, percutaneous needle biopsy; SCL, supraclavicular lymph node; VATS, video‐assisted thoracoscopic surgery.
We compared the clinical characteristics of patients who underwent re‐biopsy with those who did not. In univariate analysis, patients who underwent re‐biopsy were younger, had good performance status, more sensitizing mutations, better previous responses (complete or partial response), and a longer treatment duration than those who did not. In multivariate analysis, the frequency of re‐biopsies performed was significantly higher in younger patients (65 vs. 69 years; P = 0.002) and in patients with a previous response to EGFR‐TKIs (P < 0.001) (Table 3).
Table 3.
Comparison between the characteristics of patients who underwent re‐biopsy and those who did not
| Characteristics | No re‐biopsy (n = 125) | Re‐biopsy (n = 105) | P (univariate) | Multivariate | |
|---|---|---|---|---|---|
| HR (95% CI) | P | ||||
| Age, years | 69.0 (63.0–75.0) | 65.0 (54.0–73.0) | 0.001 | 0.950 (0.922–0.980) | 0.001 |
| Gender, male | 53 (42.4%) | 56 (53.3%) | 0.235 | ||
| Smoking history | 0.666 | ||||
| Current smoker | 34 (27.2%) | 23 (21.9%) | |||
| Ex‐smoker | 26 (20.8%) | 24 (22.9%) | |||
| Never smoker | 65 (52.0%) | 58 (55.2%) | |||
| ECOG performance status | 0.004 | 4.830 (0.956–24.391) | 0.057 | ||
| 0–2 | 111 (88.8%) | 103 (98.1%) | |||
| 3–4 | 14 (11.2%) | 2 (1.9%) | |||
| Histology | 0.108 | ||||
| ADC | 110 (88.0%) | 99 (94.3%) | |||
| SQC | 12 (11.4%) | 3 (2.9%) | |||
| NSCLC | 2 (1.9%) | 3 (2.9%) | |||
| EGFR‐TKI | 0.006 | 0.106 | |||
| Gefitinib | 69 (55.2%) | 39 (37.1%) | |||
| Erotinib | 54 (43.2%) | 58 (55.2%) | |||
| Afatinib | 2 (1.6%) | 8 (7.6%) | |||
| EGFR mutation status | 0.001 | 0.265 | |||
| 19 deletion and L858R | 58 (44.9%) | 70 (66.0%) | |||
| Others† | 67 (55.1%) | 35 (34.0%) | |||
| Treatment line | 0.058 | ||||
| 1st line | 60 (48.0%) | 65 (61.9%) | |||
| 2nd line | 39 (31.2%) | 33 (31.3%) | |||
| 3rd line | 18 (14.4%) | 5 (4.8%) | |||
| ≥ 4th line | 8 (6.4%) | 2 (2.0%) | |||
| Best response | < 0.0001 | 7.210 (3.203–16.229) | < 0.0001 | ||
| Complete response | 4 (3.2%) | 4 (3.8%) | |||
| Partial response | 48 (38.4%) | 70 (66.7%) | |||
| Stable disease | 27 (21.6%) | 21 (20.0%) | |||
| Progressive disease | 46 (36.8%) | 10 (9.5%) | |||
| Duration of EGFR‐TKIs, days | 131 (55–353) | 348 (172–508) | < 0.0001 | 0.222 | |
”Others” includes wild‐type, G719X, uncommon mutations, de novo T790M, and invalid cases.
Values are presented as number (%) or median with interquartile range.
ADC, adenocarcinoma; CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; NSCLC, non‐small cell lung cancer; SQC, squamous cell carcinoma; TKI, tyrosine kinase inhibitor.
Eleven cases (7 cytology, 4 tissue) were not pathologically confirmed. The seven cytology specimens consisted of three cases of cerebrospinal fluid and four of pleural fluid. Bronchoscopy was used to obtain the four tissue samples in which malignant cells were not acquired. The pathological success rate was higher in tissue (93.7%) than cytology (83.3%) specimens.
Safety
The complication rate was 8.6% (9/105). Pneumothorax was the most common complication, occurring in seven patients: five who underwent PCNB, and two who underwent bronchoscopy. Of the seven patients, two required tube thoracostomy. Hemoperitoneum occurred in one patient who underwent US‐guided adrenal biopsy and one death occurred as a result of respiratory failure two days after bronchoscopy.
EGFR mutation analysis of re‐biopsied samples
Of 94 patients, 75 (71.4%) were tested for EGFR mutation (Fig 2, Table 4). Nineteen patients were not tested because of the unavailability of effective drugs, such as third generation EGFR‐TKIs (42.1%); the detection of other resistance mechanisms (10.5%); a deteriorated condition (10.5%); enrollment in other clinical trials (15.8%); and a lack of durable response to EGFR‐TKIs (21.1%). When re‐biopsied, 30 (52.6%) of the 57 patients initially detected with sensitizing mutations only harbored their initial mutation, while 19 (33.3%) were positive for T790M mutation. The initial EGFR mutation disappeared in 8 (14.1%) patients. Among the 10 patients with wild type, T790M mutations were newly detected in 2 (20.0%) and sensitizing mutations in 2 (20.0%).
Figure 2.
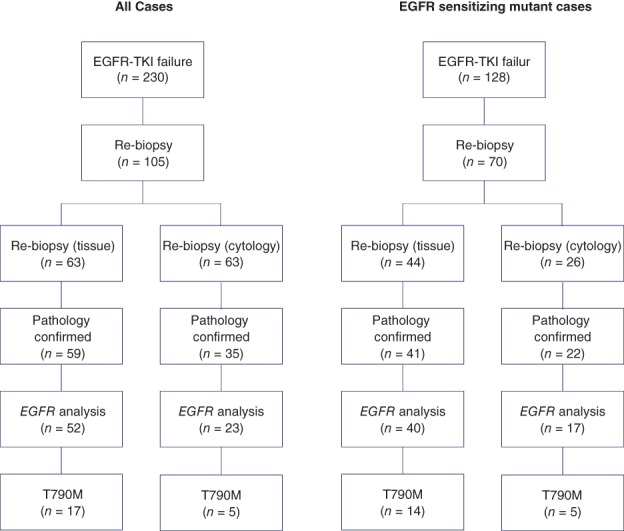
EGFR mutation analysis of re‐biopsied samples.
Table 4.
Changes in EGFR mutation status between initial diagnosis and re‐biopsy specimens (n = 75)
| EGFR mutation status | Value |
|---|---|
| Sensitizing mutation at diagnosis (n = 57) | |
| Sensitizing mutation → only sensitizing mutation | 30 (40.0%) |
| Sensitizing mutation → positive T790M | 19 (25.3%) |
| Sensitizing mutation → wild type | 8 (10.7%) |
| Wild type at diagnosis (n = 10) | |
| Wild type → wild type | 6 (8.0%) |
| Wild type → positive T790M | 2 (2.7%) |
| Wild type → sensitizing mutation | 2 (2.7%) |
| Other types at diagnosis (n = 8) | |
| G719X → 19 deletion and T790M | 1 (1.3%) |
| G719X → G719X | 2 (2.7%) |
| G719X and T790M → 19 deletion | 1 (1.3%) |
| Ins. 3 duplication → wild type | 1 (1.3%) |
| Invalid → wild type | 1 (1.3%) |
| Unknown → L858R | 1 (1.3%) |
| Unknown → wild type | 1 (1.3%) |
Values are presented as number (%).
The prevalence of T790M mutations was not statistically different to sensitive mutations (38.2% in 19 deletion vs. 26.1% in L858R), specimen type (29.4% in cytology vs. 35.0% in tissue samples), pleural effusion (23.1% in pleural effusion vs. 36.4% in other specimens), or cancer burden (27.9% in intrathoracic disease vs. 50.0% in extra‐thoracic metastasis) (Table 5).
Table 5.
Prevalence of T790M mutations according to clinical characteristics
| Characteristics | T790M (%) | P |
|---|---|---|
| EGFR mutation at diagnosis | 0.569 | |
| Exon 19 deletion (n = 34) | 13 (38.2) | |
| L858R mutation (n = 23) | 6 (26.1) | |
| Specimen type of re‐biopsy | 0.845 | |
| Tissue (n = 40) | 14 (35.0) | |
| All cytology (n = 17) | 5 (29.4) | |
| Pleural effusion (n = 13) | 3 (23.1) | |
| Site of re‐biopsy | 0.845 | |
| Intrathoracic lesion (n = 43) | 12 (27.9) | |
| Extrathoracic lesion (n = 14) | 7 (50.0) | |
| EGFR‐TKIs | 0.107 | |
| Gefitinib (n = 21) | 8 (38.1) | |
| Erlotinib (n = 29) | 11(38.0) | |
| Afatinib (n = 7) | 0 (0.0) |
Values are presented as number (%).
Discussion
We conducted a retrospective study on the feasibility of re‐biopsy in 230 NSCLC patients in whom EGFR‐TKI treatment had failed in Korean real‐world clinical practice. Re‐biopsy was performed in 105 (45.7%) patients to decide subsequent treatment or to improve symptoms. With a 100% technical success rate and an 89.5% pathological success rate (93.7% in tissue and 83.3% in cytology samples), re‐biopsy was feasible and EGFR mutation analysis was performed in 71.4% of all patients who had undergone invasive procedures. The complication rate was 8.6% and mortality was 0.95%. T790M mutations were reported in 33.3% of cases, unlike previous reports that detected such mutations in approximately 50% of patients.7, 10, 16, 17, 18 We were unable to determine the specific characteristics related to T790M prevalence.
With the development of new treatments for NSCLC, including third‐generation EGFR‐TKIs and immune checkpoint inhibitors, the importance of re‐biopsy has increased.19, 20, 21, 22, 23 However, re‐biopsy is still challenging in real world clinical practice because of its invasiveness and the different medical environments found in different countries. Recently, several reports reflecting the reality in different countries have been published.8, 22, 24, 25, 26 Most studies reported a high re‐biopsy success rate, ranging from 75% to 97%,6, 7, 9, 11 similar to the results of our study. The re‐biopsy complication rate is reported to be 1.3–5.8%,10, 11 which is similar to that of initial lung biopsy. In this study, the complication rate was 8.6%, with most cases developing pneumothorax that improved after conservative treatment, including a high oxygen supply. One death occurred as a result of respiratory failure two days after bronchoscopic re‐biopsy. In that patient, the right main bronchus was almost completely obstructed by the tumor mass and we believe that the patient progressed to respiratory failure because of the rapidly progressing cancer, not as a result of the procedure. This emphasizes the importance of candidate choice before performing an invasive procedure, such as as re‐biopsy, following EGFR‐TKI failure.
When the biopsy sites and procedures performed in this study were compared, the initial biopsy at diagnosis was more frequently performed on the primary tumor (59.6%), while re‐biopsy was more frequently performed on metastatic lesions (53.3%). We believe this is because there is greater access to the tumor site during the initial biopsy. Most samples for the initial biopsy came from tissue (84.8% vs. 15.2%), while a large proportion of the samples for re‐biopsy came from cytology (60.0% 40.0%). Bronchoscopy and/or EBUS are the main methods used for both initial biopsy and re‐biopsy at our institution. Thoracentesis for pleural effusion is frequently performed during re‐biopsy.
In our study, 11 cases were not pathologically confirmed and the pathological success rate was higher in tissue (93.7%) than cytology (83.3%) specimens. Cerebrospinal and pleural fluid may not yield a sufficient number of cancer cells, thus repeated puncture could increase diagnostic yield. In addition, all tissue specimens that were not pathologically confirmed were obtained by bronchoscopy. However, it may be difficult to determine a site for re‐biopsy by bronchoscopy because of secondary mucosal changes, such as necrosis or fibrosis after previous cancer therapy.
Although re‐biopsy is beneficial for treatment decision‐making it may be impossible to perform in some patients because of difficulty accessing the tumor site or poor performance status; many patients in our study could not undergo re‐biopsy for these reasons. In our study the patients who underwent re‐biopsy were younger and had a better response to previous EGFR‐TKIs. In these cases, it may be that the re‐biopsy was conducted when the patients were performing well, satisfied with previous EGFR targeted therapy, and had high expectations of further treatment.
T790M mutations are the most common acquired resistance mechanism, accounting for 50% to 63% of mutations.4, 7, 10, 16, 17, 18 In this study, using the PNA Clamp method, only 33.9% of patients with sensitizing mutations exhibited T790M mutations. The first explanation for the lower prevalence is our small study sample. Secondly, the PNA Clamp test is not highly sensitive to T790M mutations. Thirdly, T790M prevalence may be low in real world clinical practice, as a result of several confounding factors, including poor sample quality and DNA denaturation. Our results were similar to those of several retrospective studies conducted in Japan, which reported that the prevalence of T790M mutations is 33–34% in real world clinical practice.3, 13
The prevalence of T790M mutations with exon 19 deletions in our study was higher than L858R mutations, although this result was not statistically significant (41.2% vs. 26.1%; P = 0.569). This result is consistent with the results of several previous studies.10, 27 Ke et al. reported a higher prevalence of T790M mutations with exon 19 deletions than L858R mutations. Such patients achieve greater survival periods because third generation EGFR‐TKIs targeting the T790M mutation can be administered.25
First‐line EGFR‐TKI treatment is the standard of care for advanced NSCLC patients with an EGFR sensitizing mutation. However, EGFR‐TKIs could be used as second or third‐line treatments, even in patients with EGFR wild type, based on the results of the INTEREST study.28 We performed re‐biopsies in 10 patients harboring EGFR wild type because the initial detection method may not have been accurate in cases with small biopsy specimens. Tseng et al. reported that a significant portion of the response in EGFR wild‐type patients to erlotinib was related to limitations in detection methods, not only direct sequencing, but also sensitive mutant type‐specific methods.29 In our study, four cases of sensitizing mutation or T790M were newly found by re‐biopsy in patients harboring wild‐type EGFR at initial diagnosis. This result suggested that re‐biopsy may be considered even in EGFR wild type cases, particularly in patients who have shown a good response to EGFR‐TKIs. In addition, we identified several strange changes in EGFR status between the initial biopsy and re‐biopsy, such as alteration from wild‐type to T790M or from G719X/T790M to exon 19 deletions. These discrepancies seem to be related to the insufficient sensitivity of mutation testing or sample processing errors rather than representing new resistance mechanisms.
Recent advances in highly sensitive genotyping technologies have allowed for the development of novel plasma genotyping assays that are capable of noninvasively detecting targetable alterations in circulating tumor cells or plasma cell‐free DNA.30 Plasma genotyping allows oncologists to rapidly obtain information on tumor genotypes while avoiding the inherent risk and discomfort associated with tissue biopsy.30 Liquid biopsy has the advantage of identifying resistance mechanisms with only a blood sample, but despite its rapid development, which led to an explosion in the number of highly sensitive assay platforms available from both commercial and laboratory sources, its sensitivity remains lower than that of re‐biopsy.31 Thus, it is critical to confirm the mechanism of acquired resistance using tumor biopsy as liquid biopsy is limited and can produce false negative results.30, 32
In conclusion, this retrospective study indicates that re‐biopsy is feasible in NSCLC patients who experience EGFR‐TKI treatment failure in Korean real world clinical practice, especially in younger patients and those with a previous response to EGFR targeted therapy. EGFR mutation analysis can be performed in most re‐biopsied patients, which is beneficial to help guide further therapeutic strategies. However, a large‐scale multi‐center study is needed to better understand lung cancer progression.
Disclosure
No authors report any conflict of interest.
Acknowledgment
This work was supported by a clinical research grant from Chonnam National University Hwasun Hospital, 2017.
References
- 1. Maemondo M, Inoue A, Kobayashi K et al Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–8. [DOI] [PubMed] [Google Scholar]
- 2. Zhou C, Wu YL, Chen G et al Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): A multicentre, open‐label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–42. [DOI] [PubMed] [Google Scholar]
- 3. Ko R, Kenmotsu H, Serizawa M et al Frequency of EGFR T790M mutation and multimutational profiles of rebiopsy samples from non‐small cell lung cancer developing acquired resistance to EGFR tyrosine kinase inhibitors in Japanese patients. BMC Cancer 2016; 16: 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu HA, Arcila ME, Rekhtman N et al Analysis of tumor specimens at the time of acquired resistance to EGFR‐TKI therapy in 155 patients with EGFR‐mutant lung cancers. Clin Cancer Res 2013; 19: 2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sequist LV, Waltman BA, Dias‐Santagata D et al Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chouaid C, Dujon C, Do P et al Feasibility and clinical impact of re‐biopsy in advanced non small‐cell lung cancer: A prospective multicenter study in a real‐world setting (GFPC study 12‐01). Lung Cancer 2014; 86: 170–3. [DOI] [PubMed] [Google Scholar]
- 7. Arcila ME, Oxnard GR, Nafa K et al Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid‐based assay. Clin Cancer Res 2011; 17: 1169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hata A, Katakami N, Nanjo S, Okuda C, Kaji R, Imai Y. Rebiopsy of histological samples in pretreated non‐small cell lung cancer: Comparison among rebiopsy procedures. In Vivo 2017; 31: 475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hata A, Katakami N, Yoshioka H et al Rebiopsy of non‐small cell lung cancer patients with acquired resistance to epidermal growth factor receptor‐tyrosine kinase inhibitor: Comparison between T790M mutation‐positive and mutation‐negative populations. Cancer 2013; 119: 4325–32. [DOI] [PubMed] [Google Scholar]
- 10. Nosaki K, Satouchi M, Kurata T et al Re‐biopsy status among non‐small cell lung cancer patients in Japan: A retrospective study. Lung Cancer 2016; 101: 1–8. [DOI] [PubMed] [Google Scholar]
- 11. Simon G, Sharma A, Li X et al Feasibility and efficacy of molecular analysis‐directed individualized therapy in advanced non‐small‐cell lung cancer. J Clin Oncol 2007; 25: 2741–6. [DOI] [PubMed] [Google Scholar]
- 12. Hasegawa T, Sawa T, Futamura Y et al Feasibility of rebiopsy in non‐small cell lung cancer treated with epidermal growth factor receptor‐tyrosine kinase inhibitors. Intern Med 2015; 54: 1977–80. [DOI] [PubMed] [Google Scholar]
- 13. Kawamura T, Kenmotsu H, Taira T et al Rebiopsy for patients with non‐small‐cell lung cancer after epidermal growth factor receptor‐tyrosine kinase inhibitor failure. Cancer Sci 2016; 107: 1001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoon SH, Choi YD, Oh IJ et al Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non‐small cell lung cancer. Cancer Res Treat 2015; 47: 661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song JU, Lee J. Peptide nucleic acid clamping and direct sequencing in the detection of oncogenic alterations in lung cancer: Systematic review and meta‐analysis. Yonsei Med J 2018; 59: 211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuiper JL, Heideman DAM, Thunnissen E et al Incidence of T790M mutation in (sequential) rebiopsies in EGFR‐mutated NSCLC‐patients. Lung Cancer 2014; 85: 19–24. [DOI] [PubMed] [Google Scholar]
- 17. Sun JM, Ahn MJ, Choi YL, Ahn JS, Park K. Clinical implications of T790M mutation in patients with acquired resistance to EGFR tyrosine kinase inhibitors. Lung Cancer 2013; 82: 294–8. [DOI] [PubMed] [Google Scholar]
- 18. Tseng JS, Su KY, Yang TY et al The emergence of T790M mutation in EGFR‐mutant lung adenocarcinoma patients having a history of acquired resistance to EGFR‐TKI: Focus on rebiopsy timing and long‐term existence of T790M. Oncotarget 2016; 7: 48059–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borghaei H, Paz‐Ares L, Horn L et al Nivolumab versus docetaxel in advanced nonsquamous non‐small‐cell lung cancer. N Engl J Med 2015; 373: 1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brahmer J, Reckamp KL, Baas P et al Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med 2015; 373: 123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garon EB, Rizvi NA, Hui R et al Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med 2015; 372: 2018–28. [DOI] [PubMed] [Google Scholar]
- 22. Ishii H, Azuma K, Yamada K et al Accuracy of transbronchial biopsy as a rebiopsy method for patients with relapse of advanced non‐small‐cell lung cancer after systemic chemotherapy. BMJ Open Respir Res 2017; 4: e000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jänne PA, Yang JC, Kim DW et al AZD9291 in EGFR inhibitor‐resistant non‐small‐cell lung cancer. N Engl J Med 2015; 372: 1689–99. [DOI] [PubMed] [Google Scholar]
- 24. Gobbini E, Galetta D, Tiseo M et al Molecular profiling in Italian patients with advanced non‐small‐cell lung cancer: An observational prospective study. Lung Cancer 2017; 111: 30–7. [DOI] [PubMed] [Google Scholar]
- 25. Ke EE, Zhou Q, Zhang QY et al A higher proportion of the EGFR T790M mutation may contribute to the better survival of patients with exon 19 deletions compared with those with L858R. J Thorac Oncol 2017; 12: 1368–75. [DOI] [PubMed] [Google Scholar]
- 26. Tokaca N, Barth S, O'Brien M et al Molecular adequacy of image‐guided rebiopsies for molecular retesting in advanced non‐small cell lung cancer: A single‐center experience. J Thorac Oncol 2017; 13: 63–72. [DOI] [PubMed] [Google Scholar]
- 27. Matsuo N, Azuma K, Sakai K et al Association of EGFR exon 19 deletion and EGFR‐TKI treatment duration with frequency of T790M mutation in EGFR‐mutant lung cancer patients. Sci Rep 2016; 6: 36458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim ES, Hirsh V, Mok T et al Gefitinib versus docetaxel in previously treated non‐small‐cell lung cancer (INTEREST): A randomised phase III trial. Lancet 2008; 372: 1809–18. [DOI] [PubMed] [Google Scholar]
- 29. Tseng JS, Wang CL, Huang MS et al Impact of EGFR mutation detection methods on the efficacy of erlotinib in patients with advanced EGFR‐wild type lung adenocarcinoma. PLoS One 2014; 9: e107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sacher AG, Komatsubara KM, Oxnard GR. Application of plasma genotyping technologies in non‐small cell lung cancer: A practical review. J Thorac Oncol 2017; 12: 1344–56. [DOI] [PubMed] [Google Scholar]
- 31. Thress KS, Brant R, Carr TH et al EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross‐platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015; 90: 509–15. [DOI] [PubMed] [Google Scholar]
- 32. Oxnard GR, Thress KS, Alden RS et al Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non‐small‐cell lung cancer. J Clin Oncol 2016; 34: 3375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]


