Abstract
Lung cancer is the leading cause of cancer‐associated death, and non‐small cell lung cancer (NSCLC) accounts for 85% of all lung cancer cases. Many drugs have been used to treat NSCLC in order to improve patient prognosis. Platinum‐based chemotherapy is the first‐line treatment for locally advanced or metastatic patients. For patients with activating EGFR mutations, tyrosine kinase inhibitors are the best treatment choice. NSCLC initially exhibits an excellent response to treatment; however, acquired resistance has been observed in many patients, leading to ineffective treatment. Clinical resistance is an impediment in the treatment of patients with advanced NSCLC. Many sequencing technologies have shown that long non‐coding RNA (lncRNA) is expressed differently between drug‐resistant and drug‐sensitive lung cancer cells. We review the literature on lncRNA in drug resistance of NSCLC. The aim of this review is to gain insight into the molecular mechanisms of drug resistance, mainly focusing on the role of lncRNA in NSCLC.
Keywords: Cisplatin, drug resistance, EGFR‐TKI, lncRNA, lung cancer
Introduction
Lung cancer is the leading cause of cancer death worldwide as a result of high incidence and mortality.1 Lung cancer is traditionally classified as small cell lung cancer (SCLC) and non‐small lung cancer (NSCLC).2, 3, 4 NSCLC accounts for 85% of lung cancer cases.5 NSCLC patient prognosis is poor as nearly half present with metastatic disease at diagnosis.6 Platinum‐based combination chemotherapy is currently recommended as a standard treatment for patients with advanced NSCLC.
Cisplatin (DDP) remains the most widely employed first‐line chemotherapeutic agent for the treatment of lung cancer.7 EGFR‐directed therapies have now emerged as the best option for NSCLC patients with an EGFR mutation in exons 19 or 21.8 However, individuals respond to drug therapy differently and the efficacy of drug treatment is often impaired after the emergence of drug resistance. Clinical resistance is considered an impediment to the treatment of patients with advanced NSCLC, and can be affected by many factors.9 In addition to patients failing to receive standardized treatment on time, factors such as the abnormal regulation of molecular levels, including DNA damage repair, regulation of signaling pathways, and epithelial‐to‐mesenchymal transition (EMT), can contribute to drug resistance. Investigating the mechanisms of drug resistance and identifying strategies to overcome such resistance are thus important clinical goals.
Long non‐coding RNAs (lncRNAs) and non‐small cell lung cancer (NSCLC)
The human transcription comprises a large number of protein‐coding messenger RNAs (mRNAs), and a large set of non‐protein coding RNAs (ncRNAs).10 With the help of advanced techniques, scientists have determined that 87.3% of the human genome is actively transcribed, although < 3% of the total human sequence encodes proteins.11 These discoveries revealed widespread expression of ncRNA. NcRNA can further be divided into two classes: small ncRNAs with a length of < 200 bps and long ncRNAs (lncRNA) with a length > 200 bps.12
LncRNAs are non‐protein coding transcripts and play an important regulatory role in cancer development, metastasis, and prognosis. Various studies have suggested that the dysregulation of lncRNA is associated with lung cancer. Compared to normal lung tissue, linc01433 is significantly overexpressed and promotes migration and invasion in NSCLC.13 LINC00094 is highly expressed in lung cancer tissues and could be a molecular target for therapy for smoking‐related lung cancer.14 In recent years, emerging evidence has confirmed the role of lncRNAs in drug resistance.15, 16
The many lncRNAs associated with lung cancer are listed in Table 1. However, the mechanism by which changes in lncRNA levels affect the expression of gene products that may contribute to drug resistance remains largely unknown. Drug resistance poses a great challenge to clinical treatment; therefore, the relationship between lncRNAs and drug resistance has attracted much attention. Herein, we review the literature on lncRNA in drug resistance of NSCLC. We focus on the roles of lncRNA in DDP resistant NSCLC and EGFR‐tyrosine kinase inhibitor (TKI) resistant NSCLC.
Table 1.
NSCLC related lncRNAs
| LncRNA | Key factors | Functions | Reference |
|---|---|---|---|
| linc01433 | Promotes migration and invasion | 13 | |
| LINC00094 | Highly expressed in lung cancer tissues | 14 | |
| Trp53corl | cdkn1a | DDP resistance | 26 |
| DDSR1 | BRCA1, hnRNPUL1 | DDP resistance | 27 |
| HOTAIR | p21, EZH2 | DDP resistance, poor prognosis, advanced stage, shorter disease‐free survival | 28 |
| TRPM2‐AS | P66shc | DDP resistance | 29 |
| ROR | DDP resistance | 30 | |
| H19 | FAS, BAX, BAK | DDP resistance, suppresses apoptosis, promotes cell growth | 31 |
| MEG3 | DDP resistance, suppresses cell apoptosis, induces apoptosis | 32, 33, 34, 35 | |
| SNHG12 | MAPK1, MAP2K1 | DDP resistance | 37 |
| NEAT1 | CTR1 | DDP resistance | 38 |
| AK126698 | NKD | DDP resistance | 40, 41, 42 |
| RP11‐15H7.2 | CITED2 | DDP resistance | 43 |
| BC087858 | FOXC1 | EGFR‐TKI resistance | 59, 60 |
| MALAT1 | ZEB1, ZEB2, slug, E‐cadherin | EGFR‐TKI resistance, poor prognosis, shorter overall survival, metastasis survival | 61, 62, 63, 64 |
| MIR31HG | EGFR‐TKI resistance | 70 | |
| UCA1 | E‐cadherin, vimentin, snail, N‐cadherin | EGFR‐TKI and DDP resistance | 67, 72 |
| GAS5 | IGF‐1R | EGFR‐TKI resistance, induces apoptosis | 73 |
DDP, cisplatin; NSCLC, non‐small cell lung cancer; TKI, tyrosine kinase inhibitor.
LncRNAs and cisplatin (DDP) resistance
Combination chemotherapy based on platinum is a standard adjunctive treatment strategy for advanced NSCLC following surgical resection. DDP is the most commonly used platinum drug that inhibits DNA replication and destroys cell membrane structure. However, DDP resistance is the primary cause of chemotherapy failure. Therefore, investigation of the molecular mechanisms underlying DDP resistance in NSCLC is of great significance for improving patient outcomes. Recent studies have reported that lncRNA plays a vital role in NSCLC and that some lncRNAs are associated with DDP resistance.
LncRNAs promote DDP resistance by repairing DNA damage
Platinum‐based chemotherapy plays a key role in the treatment of NSCLC by damaging DNA and inducing tumor cell death. An increasing number of studies have proven that sensitivity to chemotherapy varies from person to person, because large interindividual variability exists in the capacity for DNA damage response and repair.17, 18 Previous studies have indicated that NSCLC cells with different functional status of the various DNA repair pathways display different responses to DDP. A growing body of evidence indicates that enhanced DNA damage repair ability is involved in conferring DDP resistance.19, 20, 21, 22 DNA damage response and repair capacity is closely related to sensitivity to chemotherapy (Fig 1). Directly targeting the DNA pathway involved in DDP resistance is an effective strategy to overcome such resistance in NSCLC.23, 24, 25, 26, 27
Figure 1.
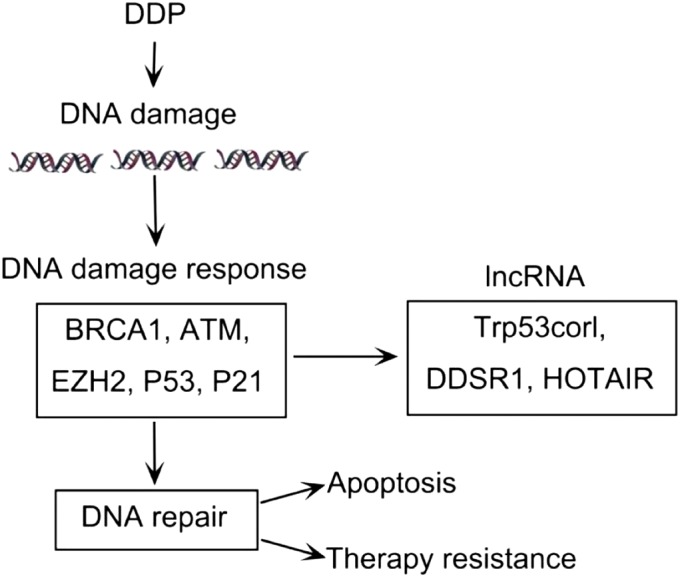
Long non‐coding RNAs (LncRNAs) promote cisplatin (DDP) resistance by repairing DNA damage. DDP induces DNA damage, which activates the DNA damage response. The damage response system initiates a series of related regulating factors to repair damage. The DNA damage response consists of two results: the damaged reactants interact with other regulating factors and eventually repair the damage, leading to resistance; or, if the damage cannot be repaired, the tumor cells become apoptotic.
With the evolution of DNA sequencing and bioinformatics, lncRNAs are emerging as a novel research field as they interact with DNA, RNA, and proteins. Recently, numerous lncRNAs were shown to correlate with drug resistance in lung cancer in a p53‐dependent manner. LncRNA‐p21 (Trp53corl) affects the expression of hundreds of gene targets that are normally repressed by p53. Moreover, lncRNA‐p21 knockout mice have enforced the G1/S checkpoint, which causes increased proliferation by decreasing Cdkn1a expression.28 LncRNA DDSR1, induced by the ataxia telangiectasia mutated (ATM)‐NF‐κB pathway, can increase the capacity for DNA repair by homologous recombination through interaction with BRCA1 and hnRNPUL1. However, whether DDSR1 promotes DDP resistance via the DNA repair pathway requires further verification.29 P21, a cyclin‐dependent kinase inhibitor, has the ability to inhibit cell proliferation and is induced by p53 upon DNA damage or p53 overexpression. Low p21 expression has been found to promote resistance to DDP. EZH2 is reported to participate in the modifications of DNA binding proteins, subsequently regulating global gene expression via cooperation with lncRNA HOTAIR. In conclusion, lncRNA HOTAIR contributes to DDP resistance via the downregulation of p21 expression and interaction with EZH2, which leads to chromosome modifications.30
LncRNAs play critical roles in DNA repair. Differentially expressed lncRNA modulates repair capacity in NSCLC, thus presenting a novel strategy to overcome resistance. Although many studies have been carried out to identify the molecular mechanisms of drug resistance, current research regarding the roles of lncRNA regulation in the DNA repair pathway is still limited, thus further studies are urgently required.
LncRNAs promote DDP resistance by regulating the cell signaling pathway
In addition to regulating the ability of DNA repair to promote DDP resistance, lncRNAs can also cause resistance through many other pathways. Many studies have shown that lncRNAs can lead to drug resistance by regulating cellular signaling pathways. P66shc, a pro‐oxidant protein in mitochondria, could negatively regulate lifespan. Downregulated lncRNA TRPM2‐AS inhibits DDP resistance via activation of the p53‐p66shc pathway in NSCLC.31 Silencing lncRNA ROR improves sensitivity to DDP in NSCLC by regulating the PI3K/AKT/mTOR signaling pathway.32 LncRNA H19 is related to apoptosis proteins FAS, BAX, and BAK, and high H19 levels are negatively associated with patient response to DDP‐based chemotherapy.33 Therefore, H19 may be an oncogenic factor in DDP resistance of NSCLC.
MEG3, an imprinted lncRNA within the DLK1‐MEG3 locus located at human chromosome 14q32, is reported to regulate DDP sensitivity through many mechanisms.34 Liu et al. found that MEG3 overexpression increased DDP sensitivity both in vitro and in vivo by hindering cell proliferation and promoting apoptosis.35 Xia et al. reported that MEG3 downregulation enhances the DDP resistance of lung cancer cells by activating the Wnt/β‐catenin signaling pathway.36 Wang et al. proved that MEG3 enhances DDP sensitivity in NSCLC by regulating the miR‐21‐5p/SOX7 axis.37 Therefore, MEG3 may be a potential target for reversing DDP resistance in lung cancer. MEG3 acts as a competing endogenous RNA by sponging specific micro RNAs (miRNAs). Previous studies have confirmed that lncRNAs can act as miRNA sponges, reducing the regulatory effects of miRNA.38 LncRNA SNHG12 also acts as a competing endogenous RNA to regulate MAPK1 and MAP2K1 by sponging miR‐181a in NSCLC. SNHG12 knockdown enhances DDP sensitivity in NSCLC in vivo.39 Upregulation of lncRNA NEAT1 could function as a competing endogenous lncRNA in lung cancer, mediating CTR1 by sponging miRNA‐98‐5p to enhance DDP sensitivity.40 All of these findings present new strategies to overcome chemoresistance in NSCLC.
The development of molecular biotechnologies makes it possible to detect molecular differences between different subsets of cells. Hou et al. identified 1702 lncRNAs that were differentially expressed between DDP‐sensitive and DDP‐resistant patients. Compared with DDP‐resistant patients, lncRNA AC006050.3‐003 was significantly downregulated in DDP‐sensitive patients, proving that AC006050.3‐003 may be a biomaker for DDP resistance.41 Another study confirmed that there are 1380 lncRNAs differentially expressed between A549/DDP and A549 parental cells. A gene co‐expression network identified that lncRNAs, including BX648420, ENST00000366408, and AK126698, potentially play a key role in DDP resistance.42 AK126698, discovered by direct sequencing in 2003, is a 3820bp lncRNA in the cerebellum. The Wnt/β‐catenin canonical signaling pathway was previously believed to play a central role in determining cell fate and is controlled by many regulators.43 The naked cuticle (NKD) family, as one of the regulators of signaling pathways, includes Drosophila NKD and its two vertebrate orthologs, NKD1 and NKD2, is reported to negatively regulate the Wnt/β‐catenin canonical signaling pathway inhibited by binding to the Dvl protein.44 AK126698 may play an important role in NSCLC DDP resistance through the Wnt pathway. However, the exact mechanism by which AK126698 regulates the Wnt pathway requires further elucidation.
Research has also suggested that lncRNAs may lead to lung cancer resistance to drugs by regulating adjacently located genes. LncRNA‐RP11‐15H7.2, a 1580bp intergenic lncRNA, is found to be located near CITED2, which is a transcriptional modulator involved in the resistance of cancer cells to DDP.45 Therefore, LncRNA‐RP11‐15H7.2 may influence chemoresistance by regulating neighboring genes.
LncRNAs have become a hot research topic in recent years and many experiments have been conducted to prove their relationship with DDP resistance; however, the exact mechanisms have not yet been identified. There are significant differences in microarray expression of lncRNAs between DDP‐resistant and DDP‐sensitive cell lines. Therefore, lncRNAs offer an opportunity to develop potential predictors for chemotherapeutic targets in NSCLC.
LncRNAs and EGFR‐tyrosine kinase inhibitor (TKI) resistance
EGFR has been identified as an oncogenic driver. Blockade of EGFR with specific TKIs is the first‐line treatment for advanced NSCLC.46, 47 The development of EGFR‐TKIs (gefitinib, erlotinib, and afatinib) is a milestone for the treatment of NSCLC harboring EGFR‐activating mutations; however, to date, drug resistance still greatly limits the usefulness of anti‐EGFR agents.48
LncRNAs promote EGFR‐TKI resistance by regulating the epithelial‐to‐mesenchymal transition (EMT) process
Sensitive mutation in the EGFR gene is associated with a dramatic clinical response to EGFR‐TKIs in NSCLC.49, 50 Despite an initial response to EGFR‐TKI treatment, most patients eventually acquire drug resistance and experience disease progression.51 A secondary T790M mutation,52 MET amplification,53 and overexpression of hepatocyte growth factor (HGF)54 are well‐studied mechanisms underlying acquired resistance to EGFR‐TKIs. EMT also plays a part in determining the sensitivity to EGFR‐TKIs.55 EMT is a process in which the expression of epithelial markers, such as E‐cadherin and gamma catenin, is decreased, and mesenchymal markers, such as vimentin and fibronectin, is increased.56
Increasing evidence shows that the mesenchymal phenotype is more resistant to EGFR‐TKIs than the epithelial phenotype in vitro. For example, lung cancer cell lines undergoing EMT, with expression of vimentin and/or fibronectin, were insensitive to EGFR‐TKIs.57 Moreover, the restoration of epithelial markers, such as E‐cadherin and gamma catenin, enhances the sensitivity of cancer cells (Fig 2).58, 59, 60 At the same time, many lncRNAs are reported to promote EGFR‐TKI resistance by regulating the EMT process.
Figure 2.
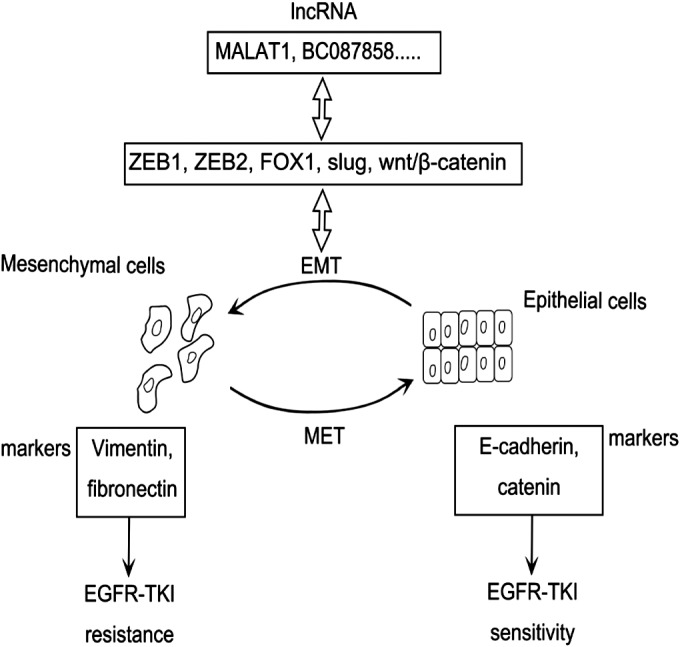
Long non‐coding RNAs (LncRNAs) promote EGFR‐tyrosine kinase inhibitor (TKI) resistance by regulating the epithelial‐to‐mesenchymal transition (EMT) process. LncRNAs interact with related factors to promote EMT. Cells with vimentin and/or fibronectin expression are more resistant to EGFR‐TKIs. By contrast, cells expressing E‐cadherin and gamma catenin are less tolerant of EGFR‐TKIs. MET, mesenchymal to epithelial transition.
As a member of the FOX transcription factor family, FOXC1 is important for cancer development. LncRNA‐BC087858 is an intergenic lncRNA located near the FOXC1 gene. FOXC1 promotes EMT by inhibiting E‐cadherin expression and inducing cell migration and invasion,61, 62 suggesting that BC087858 may be associated with EGFR‐TKI resistance through EMT. As the first identified lncRNA in lung cancer, MALAT1 (also known as NEAT2) is downregulated in gefitinib‐resistant cells. This highly conserved lncRNA has been well studied in recent years. Its potential roles in the regulation of EMT‐associated gene transcription, such as ZEB1, ZEB2, slug, and E‐cadherin, have been reported.63 MALAT1 is linked to EMT associated transcription factors. ZEB1 is reported to mediate acquired EGFR‐TKI resistance in NSCLC. Moreover, MALAT1 promotes EMT by activating the Wnt signaling pathway.64, 65, 66 The mechanism by which MALAT1 regulates EMT to affect sensitivity to EGFR‐TKIs is unclear. Therefore, future studies are needed to elucidate the possible mechanism by which lncRNAs may promote resistance to EGFR‐TKIs.
lncRNAs promote EGFR‐TKI resistance by regulating the cell signaling pathway
Previous studies have proven that activation of the PI3K/AKT and MEK/ERK cell signaling pathways, as well as EMT, is associated with EGFR‐TKI resistance in NSCLC.67, 68 Blocking the PI3K/AKT and MEK/ERK pathways could restore sensitivity to gefitinib‐resistant NSCLC cell lines.69 This evidence indicates that lncRNAs regulate the cell signaling pathway that leads to EGFR‐TKI resistance.
PI3K/AKT is an important downstream mediator of the EGFR signaling cascade.70 Dysregulation of the PI3K/AKT signaling pathway is associated with reduced rates of apoptosis and the phenotype of multidrug resistance.71 Wang et al. revealed that overexpression of MIR31HG lncRNA contributes to gefitinib resistance in NSCLC cell lines, affecting cell proliferation, apoptosis, and the cell cycle by activating the EGFR/PI3K/AKT pathway.72
LncRNA UCA1 was first identified in bladder cancer cells and is involved in the invasion and progression of bladder cancer.73 In lung cancer, overexpression of UCA1 induces non‐T790M acquired resistance to EGFR‐TKIs by activating the AKT/mTOR pathway.74 Knockdown of UCA1 enhances E‐cadherin expression, but attenuates vimentin, snail, and N‐cadherin expression.69 UCA1 is upregulated in lung cancer and induces chemoresistance. These factors suggest that lncRNA UCA1 regulates resistance to EGFR‐TKIs, not only by activating the AKT/mTOR pathway but also by activating EMT.74 The downregulation of lncRNA GAS5 is not only related to tumorigenesis and progression, but also to EGFR‐TKI resistance. Although IGF‐1R has been identified as a key downstream mediator of GAS5, specific signaling pathways still need to be elucidated.75
Cheng et al. found a total of 22 578 differentially expressed lncRNAs between EGFR‐TKI‐sensitive and EGFR‐TKI‐resistant human lung cancer cells by microarray. Based on analysis using the Kyoto Encyclopedia of Genes and Genomes database, the enriched pathways of these lncRNAs are associated with cell proliferation and apoptosis.76 Previous studies have reported that EGFR‐TKI resistance is connected to cell proliferation and apoptosis.69, 70, 77 LncRNAs may, therefore, present novel candidate biomarkers for future therapeutic strategies involving EGFR‐TKIs.
Other drugs related to lncRNAs
Besides DDP and TKIs, many other drugs are used to treat NSCLC. As time goes on, these drugs eventually become insensitive to cancer cells. Studies have shown that resistance is associated with lncRNAs. LncRNA KCNQ1OT1 expression is much higher in lung adenocarcinoma patients sensitive to paclitaxel than in those not sensitive to paclitaxel. Knockdown of KCNQ1OT1 depresses chemoresistance to paclitaxel in lung adenocarcinoma patients.78 In SCLC, lncRNA TUG1 is involved in chemoresistance by regulating LIMK2b via EZH2.79
Conclusion
Previous studies have suggested that lncRNA dysregulation in NSCLC is associated with lymph node metastasis, advanced stage, metastasis development, and poor patient prognosis. Anti‐tumor drug resistance in various carcinomas, including colon,80 bladder,81 ovarian,82 and gastric83 cancers, and NSCLC, is associated with lncRNAs. Although advances have been achieved in diagnosis and treatment, NSCLC is still one of the most common malignancies, with five‐year survival rates < 15%.84 A combination of DDP‐based chemotherapy and EGFR‐TKIs is commonly used as a treatment regimen for advanced NSCLC patients. Drug resistance remains one of the most important predictors associated with patient prognosis. Therefore, further elucidation of the molecular mechanisms of drug resistance is required to improve outcomes for NSCLC patients.
As previously mentioned, DDP can lead to DNA damage and induce tumor cell death, and different patients exhibit different drug sensitivities and abilities to repair DNA damage. Various studies have suggested that lncRNA dysregulation is involved in DDP resistance by regulating the nearby genes, modulating repairing factors, and affecting signaling pathways. Not surprisingly, targeting lncRNAs may be a potential strategy for reversing NSCLC resistance to DDP‐based chemotherapy. EMT contributes to drug resistance, rather than metastasis.85 LncRNAs are involved in EGFR‐TKI resistance by regulating the EMT phenotype. The molecular mechanisms by which lncRNAs regulate EGFR‐TKI resistance are still unknown. Furthermore, many signaling pathways could be targets of certain lncRNAs, regulating EGFR‐TKI resistance.78, 79
In conclusion, research of lncRNAs and drug resistance has attracted much attention in recent years. Numerous lncRNAs have been proven to be involved in drug resistance. Further studies focusing on lncRNA function are required to improve the response to therapeutic drugs. The relationship between lncRNAs and drug resistance may serve as a new prognostic biomarker for lung cancer.
Disclosure
No authors report any conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81602182), the Natural Science Foundation of Shandong Province (ZR2016HP42 and ZR2017MH062), and the Science and Technology for People's Livelihood Project of Qingdao (17‐3‐3‐33‐nsh).
Contributor Information
Jia Liu, Email: dadaliujia@qdu.edu.cn.
Zhuang Yu, Email: yuzhuang2002@163.com.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Zhang ZY, Fu SL, Xu SQ et al By downregulating Ku80, hsa‐miR‐526b suppresses non‐small cell lung cancer. Oncotarget 2015; 6: 1462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang HH, Pang M, Dong W et al miR‐511 induces the apoptosis of radioresistant lung adenocarcinoma cells by triggering BAX. Oncol Rep 2014; 31: 1473–9. [DOI] [PubMed] [Google Scholar]
- 4. Gao F, Chang J, Wang H et al Potential diagnostic value of miR‐155 in serum from lung adenocarcinoma patients. Oncol Rep 2014; 31: 351–7. [DOI] [PubMed] [Google Scholar]
- 5. Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet 2000; 355: 479–85. [DOI] [PubMed] [Google Scholar]
- 6. Patel AN, Simone CB II, Jabbour SK. Risk factors and management of oligometastatic non‐small cell lung cancer. Ther Adv Respir Dis 2016; 10: 338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer 2011; 71: 3–10. [DOI] [PubMed] [Google Scholar]
- 8. Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin 2011; 61: 91–112. [DOI] [PubMed] [Google Scholar]
- 9. Cobo M, Isla D, Massuti B et al Customizing cisplatin based on quantitative excision repair cross‐complementing 1 mRNA expression: A phase III trial in non‐small‐cell lung cancer. J Clin Oncol 2007; 25: 2747–54. [DOI] [PubMed] [Google Scholar]
- 10. Ørom UA, Derrien T, Beringer M et al Long noncoding RNAs with enhancer‐like function in human cells. Cell 2010; 143: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Djebali S, Davis CA, Merkel A et al Landscape of transcription in human cells. Nature 2012; 489: 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81: 145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qian B, Wang X, Mao C et al Long non‐coding RNA linc01433 promotes migration and invasion in non‐small cell lung cancer. Thorac Cancer 2018; 9: 589–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S, Sun X, Miao S, Liu J, Jiao W. Differential protein‐coding gene and long noncoding RNA expression in smoking‐related lung squamous cell carcinoma. Thorac Cancer 2017; 8: 672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu X, Li Z. Long non‐coding RNA HOTAIR: A novel oncogene (Review). Mol Med Rep 2015; 12: 5611–8. [DOI] [PubMed] [Google Scholar]
- 16. Gutschner T, Hämmerle M, Eissmann M et al The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013; 73: 1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bentzen SM, Overgaard J. Patient‐to‐patient variability in the expression of radiation‐induced normal tissue injury. Semin Radiat Oncol 1994; 4: 68–80. [DOI] [PubMed] [Google Scholar]
- 18. Burdett S, Pignon JP, Tierney J et al Adjuvant chemotherapy for resected early‐stage non‐small cell lung cancer. Cochrane Database Syst Rev 2015; CD011430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voutsadakis IA. The chemosensitivity of testicular germ cell tumors. Cell Oncol (Dordr) 2014; 37: 79–94. [DOI] [PubMed] [Google Scholar]
- 20. Galluzzi L, Senovilla L, Vitale I et al Molecular mechanisms of cisplatin resistance. Oncogene 2012; 31: 1869–83. [DOI] [PubMed] [Google Scholar]
- 21. Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 2003; 22: 7265–79. [DOI] [PubMed] [Google Scholar]
- 22. Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 2008; 18: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamazoe M, Sonoda E, Hochegger H, Takeda S. Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair (Amst) 2004; 3: 1175–85. [DOI] [PubMed] [Google Scholar]
- 24. Chen J, Dexheimer TS, Ai Y et al Selective and cell‐active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non‐small cell lung cancer cells. Chem Biol 2011; 18: 1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chirnomas D, Taniguchi T, de la Vega M et al Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol Cancer Ther 2006; 5: 952–61. [DOI] [PubMed] [Google Scholar]
- 26. Duan W, Gao L, Aguila B et al Fanconi anemia repair pathway dysfunction, a potential therapeutic target in lung cancer. Front Oncol 2014; 4: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arora S, Kothandapani A, Tillison K, Kalman‐Maltese V, Patrick SM. Downregulation of XPF‐ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair (Amst) 2010; 9: 745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dimitrova N, Zamudio JR, Jong RM et al LincRNA‐p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014; 54: 777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma V, Khurana S, Kubben N et al A BRCA1‐interacting lncRNA regulates homologous recombination. EMBO Rep 2015; 16: 1520–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Z, Sun M, Lu K et al The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS ONE 2013; 8: e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma LY, Xie XW, Ma L et al Downregulated long non‐coding RNA TRPM2‐AS inhibits cisplatin resistance of non‐small cell lung cancer cells via activation of p53‐p66shc pathway. Eur Rev Med Pharmacol Sci 2017; 21: 2626–34. [PubMed] [Google Scholar]
- 32. Shi H, Pu J, Zhou XL, Ning YY, Bai C. Silencing long non‐coding RNA ROR improves sensitivity of non‐small‐cell lung cancer to cisplatin resistance by inhibiting PI3K/Akt/mTOR signaling pathway. Tumour Biol 2017; 39: 1010428317697568. [DOI] [PubMed] [Google Scholar]
- 33. Wang Q, Cheng N, Li X et al. Correlation of long non‐coding RNA H19 expression with cisplatin‐resistance and clinical outcome in lung adenocarcinoma. Oncotarget 2017; 8: 2558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyoshi N, Wagatsuma H, Wakana S et al Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 2000; 5: 211–20. [DOI] [PubMed] [Google Scholar]
- 35. Liu J, Wan L, Lu K et al The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS ONE 2015; 10: e0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xia Y, He Z, Liu B et al Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/beta‐catenin signaling pathway. Mol Med Rep 2015; 12: 4530–7. [DOI] [PubMed] [Google Scholar]
- 37. Wang P, Chen D, Ma H, Li Y. LncRNA MEG3 enhances cisplatin sensitivity in non‐small cell lung cancer by regulating miR‐21‐5p/SOX7 axis. Onco Targets Ther 2017; 10: 5137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng K, Wang H, Guo X, Xia J. The cross talk between long, non‐coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai) 2016; 48: 111–6. [DOI] [PubMed] [Google Scholar]
- 39. Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR‐181a in non‐small cell lung cancer. Oncotarget 2017; 8: 84086–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang P, Wu X, Wang X, Huang W, Feng Q. NEAT1 upregulates EGCG‐induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016; 7: 43337–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hou Z, Xu C, Xie H et al Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS ONE 2014; 9: e108133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non‐small‐cell lung cancer cell. PLoS ONE 2013; 8: e65309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li F, Chong ZZ, Maiese K. Winding through the WNT pathway during cellular development and demise. Histol Histopathol 2006; 21: 103–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu T, Li C, Cao Z et al Myristoylated Naked2 antagonizes Wnt‐beta‐catenin activity by degrading Dishevelled‐1 at the plasma membrane. J Biol Chem 2010; 285: 13561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu ZZ, Sun NK, Chao CC. Knockdown of CITED2 using short‐hairpin RNA sensitizes cancer cells to cisplatin through stabilization of p53 and enhancement of p53‐dependent apoptosis. J Cell Physiol 2011; 226: 2415–28. [DOI] [PubMed] [Google Scholar]
- 46. Tsao MS, Sakurada A, Cutz JC et al Erlotinib in lung cancer ‐ molecular and clinical predictors of outcome. N Engl J Med 2005; 353: 133–44 (Published erratum appears in N Engl J Med 2006; 355: 1746). [DOI] [PubMed] [Google Scholar]
- 47. Shaw AT, Kim DW, Nakagawa K et al Crizotinib versus chemotherapy in advanced ALK‐positive lung cancer. N Engl J Med 2013; 368: 2385–94 (Published erratum appears in N Engl J Med 2015; 373: 1582). [DOI] [PubMed] [Google Scholar]
- 48. Lin Y, Wang X, Jin H. EGFR‐TKI resistance in NSCLC patients: Mechanisms and strategies. Am J Cancer Res 2014; 4: 411–35. [PMC free article] [PubMed] [Google Scholar]
- 49. Uramoto H, Sugio K, Oyama T et al Epidermal growth factor receptor mutations are associated with gefitinib sensitivity in non‐small cell lung cancer in Japanese. Lung Cancer 2006; 51: 71–7. [DOI] [PubMed] [Google Scholar]
- 50. Uramoto H, Mitsudomi T. Which biomarker predicts benefit from EGFR‐TKI treatment for patients with lung cancer? Br J Cancer 2007; 96: 857–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Uramoto H, Sugio K, Oyama T, Sugaya M, Hanagiri T, Yasumoto K. Resistance to gefitinib. Int J Clin Oncol 2006; 11: 487–91. [DOI] [PubMed] [Google Scholar]
- 52. Gazdar AF. Activating and resistance mutations of EGFR in non‐small‐cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009; 28 (Suppl 1): S24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Engelman JA, Zejnullahu K, Mitsudomi T et al MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–43. [DOI] [PubMed] [Google Scholar]
- 54. Yano S, Wang W, Li Q et al Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor‐activating mutations. Cancer Res 2008; 68: 9479–87. [DOI] [PubMed] [Google Scholar]
- 55. Suda K, Tomizawa K, Fujii M et al Epithelial to mesenchymal transition in an epidermal growth factor receptor‐mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol 2011; 6: 1152–61. [DOI] [PubMed] [Google Scholar]
- 56. Thiery JP. Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–54. [DOI] [PubMed] [Google Scholar]
- 57. Thomson S, Buck E, Petti F et al Epithelial to mesenchymal transition is a determinant of sensitivity of non‐small‐cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res 2005; 65: 9455–62. [DOI] [PubMed] [Google Scholar]
- 58. Witta SE, Gemmill RM, Hirsch FR et al Restoring E‐cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 2006; 66: 944–50. [DOI] [PubMed] [Google Scholar]
- 59. Miyanaga A, Gemma A, Ando M et al E‐cadherin expression and epidermal growth factor receptor mutation status predict outcome in non‐small cell lung cancer patients treated with gefitinib. Oncol Rep 2008; 19: 377–83. [PubMed] [Google Scholar]
- 60. Yauch RL, Januario T, Eberhard DA et al Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res 2005; 11: 8686–98. [DOI] [PubMed] [Google Scholar]
- 61. Xia L, Huang W, Tian D et al Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology 2013; 57: 610–24. [DOI] [PubMed] [Google Scholar]
- 62. Wang J, Ray PS, Sim MS et al FOXC1 regulates the functions of human basal‐like breast cancer cells by activating NF‐kappaB signaling. Oncogene 2012; 31: 4798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ji P, Diederichs S, Wang W et al MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene 2003; 22: 8031–41. [DOI] [PubMed] [Google Scholar]
- 64. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT‐1 contributes to bladder cancer cell migration by inducing epithelial‐to‐mesenchymal transition. Mol Biosyst 2012; 8: 2289–94. [DOI] [PubMed] [Google Scholar]
- 65. Sequist LV, Waltman BA, Dias‐Santagata D et al Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Samatov TR, Tonevitsky AG, Schumacher U. Epithelial‐mesenchymal transition: Focus on metastatic cascade, alternative splicing, non‐coding RNAs and modulating compounds. Mol Cancer 2013; 12: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hampton KK, Craven RJ. Pathways driving the endocytosis of mutant and wild‐type EGFR in cancer. Oncoscience 2014; 1: 504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rolfo C, Giovannetti E, Hong DS et al Novel therapeutic strategies for patients with NSCLC that do not respond to treatment with EGFR inhibitors. Cancer Treat Rev 2014; 40: 990–1004. [DOI] [PubMed] [Google Scholar]
- 69. Li H, Schmid‐Bindert G, Wang D et al Blocking the PI3K/AKT and MEK/ERK signaling pathways can overcome gefitinib‐resistance in non‐small cell lung cancer cell lines. Adv Med Sci 2011; 56: 275–84. [DOI] [PubMed] [Google Scholar]
- 70. Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib‐sensitizing EGFR mutations in lung cancer activate anti‐apoptotic pathways. Science 2004; 305: 1163–7. [DOI] [PubMed] [Google Scholar]
- 71. Qiao M, Sheng S, Pardee AB. Metastasis and AKT activation. Cell Cycle 2008; 7: 2991–6. [DOI] [PubMed] [Google Scholar]
- 72. Wang B, Jiang H, Wang L et al Increased MIR31HG lncRNA expression increases gefitinib resistance in non‐small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol Lett 2017; 13: 3494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non‐protein‐coding RNA up‐regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett 2008; 582: 1919–27. [DOI] [PubMed] [Google Scholar]
- 74. Cheng N, Cai W, Ren S et al Long non‐coding RNA UCA1 induces non‐T790M acquired resistance to EGFR‐TKIs by activating the AKT/mTOR pathway in EGFR‐mutant non‐small cell lung cancer. Oncotarget 2015; 6: 23582–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dong S, Qu X, Li W et al The long non‐coding RNA, GAS5, enhances gefitinib‐induced cell death in innate EGFR tyrosine kinase inhibitor‐resistant lung adenocarcinoma cells with wide‐type EGFR via downregulation of the IGF‐1R expression. J Hematol Oncol 2015; 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cheng N, Li X, Zhao C et al Microarray expression profile of long non‐coding RNAs in EGFR‐TKIs resistance of human non‐small cell lung cancer. Oncol Rep 2015; 33: 833–9. [DOI] [PubMed] [Google Scholar]
- 77. Ng KP, Hillmer AM, Chuah CT et al A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 2012; 18: 521–8. [DOI] [PubMed] [Google Scholar]
- 78. Ren K, Xu R, Huang J, Zhao J, Shi W. Knockdown of long non‐coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol 2017; 80: 243–50. [DOI] [PubMed] [Google Scholar]
- 79. Niu Y, Ma F, Huang W et al Long non‐coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 2017; 16: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M. LncRNA BCAR4 promotes colon cancer progression via activating Wnt/beta‐catenin signaling. Oncotarget 2017; 8: 92815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xie D, Zhang H, Hu X, Shang C. Knockdown of long non‐coding RNA taurine up‐regulated 1 inhibited doxorubicin resistance of bladder urothelial carcinoma via Wnt/beta‐catenin pathway. Oncotarget 2017; 8: 88689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang L, Hu Y, Xiang X, Qu K, Teng Y. Identification of long non‐coding RNA signature for paclitaxel‐resistant patients with advanced ovarian cancer. Oncotarget 2017; 8: 64191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang L, Chunyan Q, Zhou Y et al BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci 2017; 21: 4064–70. [PubMed] [Google Scholar]
- 84. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60: 277–300 (Published erratum appears in CA Cancer J Clin 2011; 61: 133–4.).20610543 [Google Scholar]
- 85. Fischer KR, Durrans A, Lee S et al Epithelial‐to‐mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015; 527: 472–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


