Abstract
Myeloid-derived suppressor cells (MDSCs) are highly immunosuppressive myeloid cells that show increased expression in cancer patients; however, the molecular mechanisms underlying their generation and function are unclear. Whereas granulocytic-MDSCs correlate with poor overall survival in breast cancer (BC), the presence and relevance of monocytic (Mo)-MDSCs are unknown. Here, we report for the first time increased chemokine and chemokine receptor production by Mo-MDSCs in BC patients. A clear population of Mo-MDSCs with the typical cell surface phenotype (human leukocyte antigen-antigen D related [HLA-DR]low/− CD11b+ CD33+ CD14+) increased significantly during disease progression. In addition, the chemokine receptor expression level on Mo-MDSCs in patients with invasive BC was the highest. Furthermore, different chemokine receptor expression patterns were noted in Mo-MDSCs between healthy controls (HC) and BC patients. Additionally, CD4 T cells proliferations were significantly decreased in the invasive BC groups compared with the HC group. However, the ductal carcinoma in situ (DCIS) group had no significantly compared with the HC group. Our data suggest that monitoring chemokine and chemokine receptor production by Mo-MDSCs may represent a novel and simple biomarker for assessing disease progression in BC patients.
Keywords: Myeloid-derived suppressor cells, Chemokines, Chemokine receptors, Breast neoplasms
INTRODUCTION
Breast cancer (BC) are the most common human cancers and the leading cause of cancer-related deaths worldwide. Cancer tumorigenesis involves numerous pathological factors and suppression of the immune response (1,2). Tumors avoid the immune system by inhibiting the development of immune responses for tumor progression (3,4).
In patients with cancer, tumors inhibit the differentiation of common myeloid progenitor cells and enhance the accumulation of immature myeloid cells (IMCs). A block in IMC differentiation during acute or chronic infection and enhancement of the tumor environment expand this population of cells, known as myeloid-derived suppressor cells (MDSCs).
Therefore, the characterization of these immunosuppressive cells has important implications for cancer diagnosis and therapy (5). MDSC-mediated immunity is also believed to be involved in this mechanism, but research on this topic is limited (6,7).
Monocytic (Mo)-MDSCs and granulocytic/polymorphonuclear (PMN)-MDSCs are 2 main MDSC subtypes that inhibit immune responses via different mechanisms (8). Although several studies of MDSC subtypes have been conducted in mice, their human counterparts have no universal marker, and their functions and pathophysiological relevance in human oncology are less well defined. Similar to murine MDSCs, MDSCs are referred to as human leukocyte antigen-antigen D related (HLA-DR) −CD33+ cells in pancreatic cancer patients and different combinations of markers including HLA-DR, CD33, CD11b, and CD14 have been used to investigate human MDSCs subtype (9). The balance of MDSC subtypes should be affected by cancer type.
In human, the ratios of PMN-MDSCs and Mo-MDSCs are very variable in different tumor environment and the factors regulating their proportion are not entirely known (10,11).
Recently, much focus has been put on the PMN-MDSCs that are frequently enriched in cancer patients (12,13).
Furthermore, while local induction of MDSCs has been extensively investigated and involves tumor derived factors such as IL-10, transforming growth factor (TGF) β, vascular endothelial growth factor (VEGF), and granulocyte-macrophage colony-stimulating factor (GM-CSF) the origin and mechanism of generation of circulating Mo-MDSCs is, as of yet, largely unknown (14,15).
Although the differential expression of chemokines and chemokine receptors plays a key role in determining the tissue infiltration of different leukocyte subsets, the mechanism by which MDSCs are recruited to the tumor microenvironment remains unclear (16,17).
Recent research has determined that several chemokines and their cognate receptors are responsible for the migration of MDSCs into the tumor microenvironment (18). Additionally, some evidence suggests that chemokine receptors are differentially expressed by Mo-MDSCs and PMN-MDSCs, which aids in their differential recruitment by C-C chemokine receptor type 2 (CCR2) and C-X-C chemokine receptor type 1 (CXCR1), type 2 (CXCR2), and type 4 (CXCR4) (17,19,20,21,22,23,24).
Therefore, we analyzed the presence and function of circulating and tumor-infiltrating Mo-MDSCs and chemokine receptors with the aim to investigate the clinical relevance of Mo-MDSCs in BC patients.
MATERIALS AND METHODS
Peripheral blood samples
This study was approved by the Institutional Review Board of Konkuk University Medical Center (KUH-1160090). Patients were excluded if they had histories of 1) chemotherapy, 2) radiotherapy, 3) immunosuppressive agent use, 4) bone marrow transplantation, and 5) any present symptom or sign of infection. Patients were assigned to the ductal carcinoma in situ (DCIS) non-invasive BC group or invasive BC group according to the presence of tumor, nodes, metastasis (TNM) stage; the latter was determined based on preoperative computed tomography (CT) and bone scans, intraoperative surgical and pathological findings, and postoperative pathological findings. The stage of BC was confirmed by perioperative evaluations and pathological findings. Staging followed the American Joint Committee on Cancer (AJCC) scheme using TNM classifications. The T stage describes the size of the primary colorectal cancer tumor and whether it has invaded nearby tissue (depth). The N stage describes the regional lymph nodes that are involved (regional lymph node metastasis). The M stage describes distant metastasis.
Peripheral blood mononuclear cell (PBMC) isolation
Heparin-anticoagulated peripheral blood samples were obtained from each patient with cancer, and PBMCs were isolated by density gradient centrifugation using Biocoll gradient solution (Biochrom, Berlin, Germany). PBMCs were collected in conical tubes and washed in PBS (Gibco, Gaithersburg, MD, USA). Before staining, PBMCs were washed with FACS buffer (PBS, 1% bovine serum albumin [BSA] and 0.01% NaN3).
FACS of MDSCs
After PBMC isolation, cells were stained with the following antibodies to detect MDSCs: PE anti-human CD11b (clone ICRF44; BioLegend, San Diego, CA, USA), peridinin chlorophyll protein anti-human HLA-DR (clone L243; BioLegend), allophycocyanin (APC) anti-human CD33 (clone WM53; BioLegend), and APC-cy7 anti-human CD14 (clone M5E2; BioLegend). To detect chemokine receptors, the antibodies used were FITC anti-human CCR2 (clone K036C2; BioLegend), CXCR1 (clone 8F1/CXCR1; BioLegend), PE-cy7 anti-human CXCR2 (clone 5E8/CXCR2; BioLegend), and CXCR4 (clone 12G5; BioLegend). For MDSC detection, bone marrow cells were stained with FITC anti-mouse Ly-6G (clone 1A8; Miltenyi Biotec, Gladbach, Germany), APC anti-mouse Ly-6C (clone 1G7.G10; Miltenyi Biotec), and APC-cy7 anti-mouse/human CD11b (clone M1/70; BioLegend).
Staining was performed for 30 min at room temperature and cells were washed with FACS buffer. After washing, samples were collected on a BD FACSAria (Becton Dickinson, Franklin Lakes, NJ, USA). All statistical analyses were performed with FlowJo software (Tree Star Inc., Ashland, OR, USA).
Isolation of Mo-MDSCs for the proliferation assay
PBMCs were isolated using density-gradient centrifugation over a Ficoll-Hypaque gradient (GE Healthcare, Piscataway, NJ, USA) to collect PBMCs. PBMCs were washed with PBS (137 mM NaCl, 2.7 M KCl, 10 mM Na2HPO4 and 2 mM KH2PO4, pH 7.4) and re-suspended in flow cytometry (FACS) buffer (0.1% BSA in PBS). The cells were stained with PE anti-human CD11b (clone ICRF44, Biolegend), peridinin chlorophyll protein (PerCP) anti-human HLA-DR (clone L243, Biolegend), APC anti-human CD33 (clone WM53, Biolegend) and APC-cy7 anti-human CD14 (clone M5E2, Biolegend) for 30 min to detect Mo-MDSCs. After washing, samples were isolated on BD FACSAria (Becton Dickinson) according to the manufacturer's protocol (Becton Dickson, Brea, CA, USA).
T cell suppression assay
The 0, 1,000 and 5,000 Mo-MDSCs were co-cultured with 10,000 naive CD4+ T cells from healthy blood donors at indicated stimulator:responder ratios in Roswell Park Memorial Institute medium 1640 (RPMI 1640) supplemented with penicillin/streptomycin (Thermo Scientific, Rockford, IL, USA), 10 ng/ml GM-CSF in all cultures and controls and CD3+/CD28+ T cell activating dynal beads according to the manufacturer's instructions (Gibco Life Technologies, AS, Oslo, Norway) for a total of 48 h. The 1 μCi/ml [methyl-3H] thymidine was added for the last 18 h and incorporation was measured in a Microbeta Counter (PerkinElmer, Boston, MA, USA). The background signal from monocytes was subtracted before calculating the relative proliferation of CD4+ T lymphocytes.
Statistical analysis
Data were analyzed using IBM SPSS Statistics 22.0 software (IBM Inc., Armonk, NY, USA) and GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA). Data are presented as the mean±standard deviation or the mean±standard error of the mean. Unpaired Student's t-test or 1-way analysis of variance was used to determine significant differences between groups. The p-values <0.05 were considered to be significant.
RESULTS
In total, 69 subjects were eligible for this study, which was conducted from 2015 to 2016. Informed consent was obtained from all subjects, who were divided into three groups: 1) 29 healthy controls (HC) with a mean age of 46.90±15.99; 2) 21 DCIS patients with a mean age of 48.57±9.93, and 3) 19 invasive BC patients with a mean age of 46.84±13.37 (Table 1). Table 1 are showed the clinical characteristics of all subjects in the study. We were used standard laboratory methods. And we were gained the blood from patients before surgery. There are no significantly difference in age between HCs, DCIS and invasive BC patients. This study confirmed the existence of 2 circulating subtypes of MDSCs using a flow cytometry analysis of the peripheral blood of patients with BC. To detect the percentage of MDSCs, PBMCs were stained with CD11b, CD33, HLA-DR, and CD14 monoclonal antibodies. Mo-MDSCs were defined as CD11b+CD33+HLA-DR−CD14+, and PMN-MDSCs as CD11b+CD33+HLA-DR−CD14−.
Table 1. Baseline characteristics.
| Characteristics | Group | |||
|---|---|---|---|---|
| HC (n=29) | DCIS (n=21) | Invasive BC (n=19) | ||
| Age (yr) | 46.90±15.99 | 48.57±9.93 | 46.84±13.37 | |
| Final stage | ||||
| 0 | 9 (42.9) | |||
| I | 12 (57.1) | |||
| II | 15 (78.9) | |||
| III | 4 (21.1) | |||
| Estrogen receptor | ||||
| Positive | 15 (71.4) | 17 (89.5) | ||
| Negative | 4 (19.0) | 2 (10.5) | ||
| Unknown | 2 (9.6) | |||
Values are presented as mean±standard deviation or number (%).
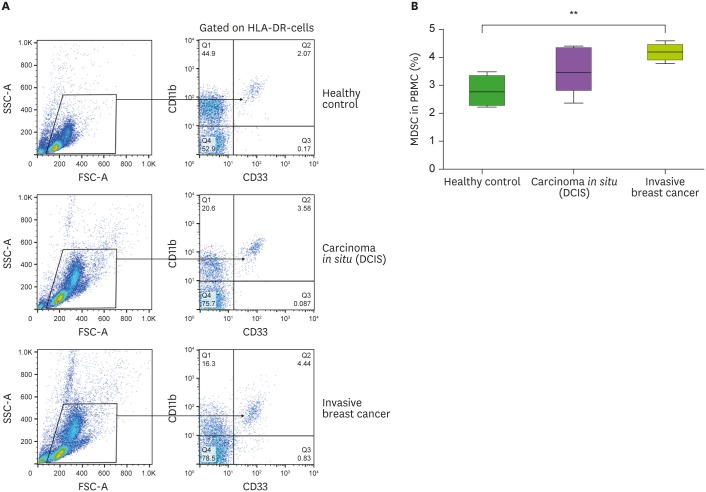
The frequency of circulating MDSCs was high in patients with invasive BC (2.78±0.54 in the HC group vs. 3.52±0.82 in the DCIS group vs. 4.14±0.29 in the invasive BC group, p<0.01; Fig. 1).
Figure 1. (A) Representative dot plots of the population of HLA-DRlow/− CD11b+ CD33+ MDSCs. MDSCs are presented by the percentages of CD11b+ CD33+ of gated HLA-DRlow/− cells. (B) Expression of circulating HLA-DRlow/− CD11b+ CD33+ MDSC in 3 groups.
**p<0.01.
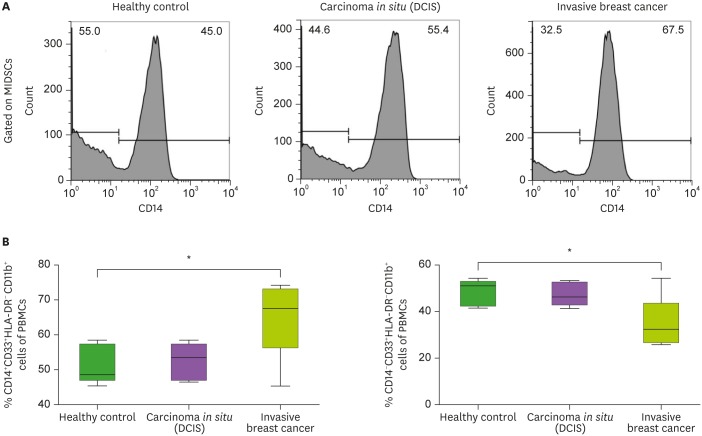
In addition, the frequency of Mo-MDSCs showed a pattern similar to that of MDSCs. On the contrary, the frequency of PMN-MDSC significantly decreased in patients with invasive BC (Fig. 2). However, the circulating of MDSCs, and Mo- and PMN-MDSCs did not differ between the DCIS and HC groups (Mo-MDSCs; 51.36±5.63 in the HC group vs. 52.27±5.19 in the DCIS group vs. 65.32±11.58 in the invasive BC group, p<0.05; Fig. 2).
Figure 2. (A) Representative histogram of flow cytometry analysis of Mo-MDSCs and PMN-MDSCs. (B) The frequency of Mo-MDSCs (CD14+ HLA-DRlow/−CD33+ CD11b+) and PMN-MDSCs (CD14− HLA-DRlow/− CD33+ CD11b+) in MDSCs.
*p<0.05.
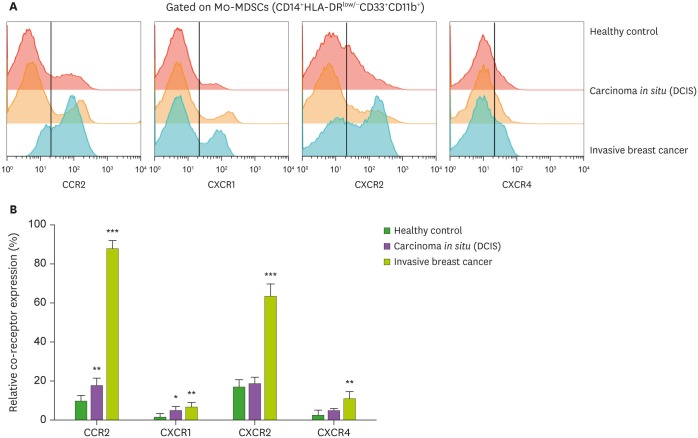
To confirm the frequency of chemokine receptors in cancer, I analyzed the expression of CCR2, CXCR1, CXCR2, and CXCR4 in each cancer. The amount of all chemokine receptors in Mo-MDSCs was significantly higher in the invasive BC groups than in the HC group (Fig. 3). However, the CXCR2/CXCR4 receptors showed no significant increases between the DCIS and HC groups (Fig. 3).
Figure 3. (A) Representative histogram of flow cytometry analysis of chemokines/chemokine receptors. (B) The frequency of chemokines/chemokine receptors among 3 groups.
*p<0.05, **p<0.01, ***p<0.001.
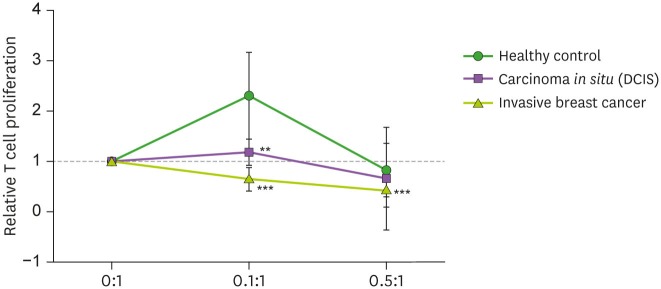
Additionally, CD4 T cells proliferations were significantly decreased in the invasive BC groups, compared to the HC group. However, CD4 T cell proliferation was not significantly different between the DCIS group and the HC group (Fig. 4).
Figure 4. In vitro CD4 T-cell proliferation assay with Mo-MDSCs from patients. T cell proliferation was assessed by thymidine incorporation among 3 groups; HC (n=10), DCIS (n=10), and invasive BC (n=10).
**p<0.01, ***p<0.001.
DISCUSSION
In this study, we analyzed the peripheral blood MDSCs populations from patients with DCIS as well as with invasive BC in relation to HC.
This study revealed that demonstrates that expression of 2 main subsets in MDSCs, Mo-MDSCs and granulocytic-MDSCs. Moreover, several chemokines and chemokine receptors significantly correlate as Mo-MDSCs.
In this study, we investigated the distribution of 2 MDSC subsets between BC and HC in peripheral blood by staining for CD11b, CD33, HLA-DR, and CD14. Importantly, presence of Mo-MDSCs Association correlated with more severe disease, as patients with high frequency of Mo-MDSCs presented with more severe stage and metastases.
Zhang et al. (25), demonstrated that the increase in circulating inflammatory myeloid cells in peripheral blood affects tumor progression via local immune suppression and the stimulation of tumor neovasculogenesis (26).
Also, in tumor tissue, MDSCs are believed to play an important role in inhibiting immune responses by T cells and natural killer cells, and elevated MDSC infiltration is correlated with a poor prognosis and resistance to therapy (27,28,29,30). Yu et al. (31) reported that CD33+ myeloid cells displayed the characteristic phenotype of MDSCs in tumor tissue based on immunohistochemical staining. They obtained CD33+ myeloid cells from primary cancer tissue. In this study, MDSCs were detected by immunohistochemical staining by using CD33 antibody. In a microscope of 40× magnification, CD33+ cells were also more abundant invasive BC tissue compared with normal tissue (data not shown).
Additionally, Zhao et al. (32) reported that the RNA level of MDSCs of mice, there has been reported that CCR2 and CXCR4 are highly expressed in Mo-MDSCs and CXCR1 and CXCR2 are highly expressed in PMN-MDSCs. Therefore, we focused on the CCR2, CXCR1, CXCR2 and CXCR4 in the present study. Unlike previous studies in mice, CXCR2 and CXCR4 were significantly increased in patients with invasive BC than in HC and DCIS. Interestingly, CXCR2 and CXCR4 have been demonstrated to stimulate MDSCs generation and migration to tumor sites (20,32). Our data further support these findings, as Mo-MDSCs from invasive BC patients expressed CCR2, CXCR1, CXCR2, and CXCR4. This finding is consistent with previous studies. Indeed, T cell proliferation was suppressed by Mo-MDSCs from patients invasive BC than in HC and DCIS. It may also be of interest to note that the higher ratio used for the T cell suppression assay may have limited relevance for the HC as the CXCR2 and CXCR4 ratio is lower in HC compared to invasive BC patients.
One possible scenario for this observation is that factors secreted from tumor cells stimulate the overexpansion of chemokines in MDSCs, and recruited Mo-MDSCs promote tumor cell survival and invasion. Therefore, a disruption of this vicious cycle may hold great promise for enhancing treatment efficacy for cancer. Further studies are required in order to elucidate the impact that invasive BC have on the Mo-MDSC population.
Reports also indicate that a co-inhibition of CXCR2 and CXCR4 is more effective in reducing gastric cancer metastasis (33,34). It also shows that the expression of CXCR2 and CXCR4 each other to promote the metastasis of gastric cancer (35).
In conclusion, our results also indicate that an increased level of CXCR2 and CXCR4 of Mo-MDSCs with invasive BC.
Therefore, CXCR2 and CXCR4 may serve as a prognostic indicator in patients with invasive BC.
Based on our findings, we suggest that monitoring the chemokines and chemokine receptors of Mo-MDSC levels in BC patients may represent a novel and simple biomarker for assessing disease progression.
ACKNOWLEDGEMENTS
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (NRF-2016R1A5A2012284, 2017R1D1A1B03028011)
Abbreviations
- APC
allophycocyanin
- BC
breast cancer
- CCR2
chemokine receptor type 2
- CXCR
C-X-C chemokine receptor
- DCIS
ductal carcinoma in situ
- HC
healthy controls
- HLA-DR
human leukocyte antigen-antigen D related
- MDSC
myeloid-derived suppressor cell
- Mo
monocytic
- PBMC
peripheral blood mononuclear cell
- PMN
polymorphonuclear
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
Authors Contributions: Conceptualization: Lee SH; Data curation: Namgung JH, Kim SH, Lee SH; Formal analysis: Seo EH, Namgung JH, Oh CS, Lee SH; Investigation: Seo EH, Namgung JH, Oh CS, Lee SH; Writing - original draft: Seo EH, Namgung JH, Oh CS, Lee SH.
References
- 1.Kusmartsev S, Gabrilovich DI. Role of immature myeloid cells in mechanisms of immune evasion in cancer. Cancer Immunol Immunother. 2006;55:237–245. doi: 10.1007/s00262-005-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara HM, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35(Suppl):S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Marcus A, Gowen BG, Thompson TW, Iannello A, Ardolino M, Deng W, Wang L, Shifrin N, Raulet DH. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. 2014;122:91–128. doi: 10.1016/B978-0-12-800267-4.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato Y, Shimizu K, Shinga J, Hidaka M, Kawano F, Kakimi K, Yamasaki S, Asakura M, Fujii SI. Characterization of the myeloid-derived suppressor cell subset regulated by NK cells in malignant lymphoma. OncoImmunology. 2015;4:e995541. doi: 10.1080/2162402X.2014.995541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wesolowski R, Duggan MC, Stiff A, Markowitz J, Trikha P, Levine KM, Schoenfield L, Abdel-Rasoul M, Layman R, Ramaswamy B, et al. Circulating myeloid-derived suppressor cells increase in patients undergoing neo-adjuvant chemotherapy for breast cancer. Cancer Immunol Immunother. 2017;66:1437–1447. doi: 10.1007/s00262-017-2038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toor SM, Syed Khaja AS, El Salhat H, Faour I, Kanbar J, Quadri AA, Albashir M, Elkord E. Myeloid cells in circulation and tumor microenvironment of breast cancer patients. Cancer Immunol Immunother. 2017;66:753–764. doi: 10.1007/s00262-017-1977-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaled YS, Ammori BJ, Elkord E. Increased levels of granulocytic myeloid-derived suppressor cells in peripheral blood and tumour tissue of pancreatic cancer patients. J Immunol Res. 2014;2014:879897. doi: 10.1155/2014/879897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albeituni SH, Ding C, Yan J. Hampering immune suppressors: therapeutic targeting of myeloid-derived suppressor cells in cancer. Cancer J. 2013;19:490–501. doi: 10.1097/PPO.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandau S, Trellakis S, Bruderek K, Schmaltz D, Steller G, Elian M, Suttmann H, Schenck M, Welling J, Zabel P, et al. Myeloid-derived suppressor cells in the peripheral blood of cancer patients contain a subset of immature neutrophils with impaired migratory properties. J Leukoc Biol. 2011;89:311–317. doi: 10.1189/jlb.0310162. [DOI] [PubMed] [Google Scholar]
- 12.Wynn TA. Myeloid-cell differentiation redefined in cancer. Nat Immunol. 2013;14:197–199. doi: 10.1038/ni.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zilio S, Serafini P. Neutrophils and granulocytic MDSC: the Janus God of Cancer Immunotherapy. Vaccines (Basel) 2016;4:31. doi: 10.3390/vaccines4030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Wu T, Shao S, Shi B, Zhao Y. Phenotype, development, and biological function of myeloid-derived suppressor cells. OncoImmunology. 2015;5:e1004983. doi: 10.1080/2162402X.2015.1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, Cerwenka A. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J Immunol. 2012;189:5602–5611. doi: 10.4049/jimmunol.1201018. [DOI] [PubMed] [Google Scholar]
- 17.Sevko A, Umansky V. Myeloid-derived suppressor cells interact with tumors in terms of myelopoiesis, tumorigenesis and immunosuppression: thick as thieves. J Cancer. 2013;4:3–11. doi: 10.7150/jca.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer C, Sevko A, Ramacher M, Bazhin AV, Falk CS, Osen W, Borrello I, Kato M, Schadendorf D, Baniyash M, et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc Natl Acad Sci U S A. 2011;108:17111–17116. doi: 10.1073/pnas.1108121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chun E, Lavoie S, Michaud M, Gallini CA, Kim J, Soucy G, Odze R, Glickman JN, Garrett WS. CCL2 promotes colorectal carcinogenesis by enhancing polymorphonuclear myeloid-derived suppressor cell population and function. Cell Reports. 2015;12:244–257. doi: 10.1016/j.celrep.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stromnes IM, Greenberg PD, Hingorani SR. Molecular pathways: myeloid complicity in cancer. Clin Cancer Res. 2014;20:5157–5170. doi: 10.1158/1078-0432.CCR-13-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebrun A, Lo Re S, Chantry M, Izquierdo Carerra X, Uwambayinema F, Ricci D, Devosse R, Ibouraadaten S, Brombin L, Palmai-Pallag M, et al. CCR2+ monocytic myeloid-derived suppressor cells (M-MDSCs) inhibit collagen degradation and promote lung fibrosis by producing transforming growth factor-β1. J Pathol. 2017;243:320–330. doi: 10.1002/path.4956. [DOI] [PubMed] [Google Scholar]
- 23.Alfaro C, Teijeira A, Oñate C, Pérez G, Sanmamed MF, Andueza MP, Alignani D, Labiano S, Azpilikueta A, Rodriguez-Paulete A, et al. Tumor-produced interleukin-8 attracts human myeloid-derived suppressor cells and elicits extrusion of neutrophil extracellular traps (NETs) Clin Cancer Res. 2016;22:3924–3936. doi: 10.1158/1078-0432.CCR-15-2463. [DOI] [PubMed] [Google Scholar]
- 24.Qu P, Wang LZ, Lin PC. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. 2016;380:253–256. doi: 10.1016/j.canlet.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Wang Z, Wu L, Zhang M, Li W, Ding J, Zhu J, Wei H, Zhao K. Circulating and tumor-infiltrating myeloid-derived suppressor cells in patients with colorectal carcinoma. PLoS One. 2013;8:e57114. doi: 10.1371/journal.pone.0057114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. 2013;91:493–502. doi: 10.1038/icb.2013.29. [DOI] [PubMed] [Google Scholar]
- 27.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, Mackall CL. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med. 2014;6:237ra67. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol. 2012;22:275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connor MA, Fu WW, Green KA, Green WR. Subpopulations of M-MDSCs from mice infected by an immunodeficiency-causing retrovirus and their differential suppression of T- vs B-cell responses. Virology. 2015;485:263–273. doi: 10.1016/j.virol.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finke J, Ko J, Rini B, Rayman P, Ireland J, Cohen P. MDSC as a mechanism of tumor escape from sunitinib mediated anti-angiogenic therapy. Int Immunopharmacol. 2011;11:856–861. doi: 10.1016/j.intimp.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Du W, Yan F, Wang Y, Li H, Cao S, Yu W, Shen C, Liu J, Ren X. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. 2013;190:3783–3797. doi: 10.4049/jimmunol.1201449. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Xu Y, Xu J, Wu D, Zhao B, Yin Z, Wang X. Subsets of myeloid-derived suppressor cells in hepatocellular carcinoma express chemokines and chemokine receptors differentially. Int Immunopharmacol. 2015;26:314–321. doi: 10.1016/j.intimp.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Satomura H, Sasaki K, Nakajima M, Yamaguchi S, Onodera S, Otsuka K, Takahashi M, Muroi H, Shida Y, Ogata H, et al. Can expression of CXCL12 and CXCR4 be used to predict survival of gastric cancer patients? Anticancer Res. 2014;34:4051–4057. [PubMed] [Google Scholar]
- 34.Ying J, Xu Q, Zhang G, Liu B, Zhu L. The expression of CXCL12 and CXCR4 in gastric cancer and their correlation to lymph node metastasis. Med Oncol. 2012;29:1716–1722. doi: 10.1007/s12032-011-9990-0. [DOI] [PubMed] [Google Scholar]
- 35.Xiang Z, Zhou ZJ, Xia GK, Zhang XH, Wei ZW, Zhu JT, Yu J, Chen W, He Y, Schwarz RE, et al. A positive crosstalk between CXCR4 and CXCR2 promotes gastric cancer metastasis. Oncogene. 2017;36:5122–5133. doi: 10.1038/onc.2017.108. [DOI] [PubMed] [Google Scholar]