Activation of innate immunity is essential for host cells to restrict the spread of invading viruses and other pathogens. MAVS plays a critical role in innate immune response to RNA viral infection. In this study, we demonstrated that TRIM21 targets MAVS to positively regulate innate immunity. Notably, TRIM21 targets and catalyzes K27-linked polyubiquitination of MAVS and then promotes the recruitment of TBK1 to MAVS, leading to upregulation of innate immunity. Our study outlines a novel mechanism by which the IFN signaling pathway blocks RNA virus to escape immune elimination.
KEYWORDS: IL-28A, IL-29, MAVS, TRIM21, innate immunity, interferon, virus
ABSTRACT
Human innate immunity responds to viral infection by activating the production of interferons (IFNs) and proinflammatory cytokines. The mitochondrial adaptor molecule MAVS plays a critical role in innate immune response to viral infection. In this study, we show that TRIM21 (tripartite motif-containing protein 21) interacts with MAVS to positively regulate innate immunity. Under viral infection, TRIM21 is upregulated through the IFN/JAK/STAT signaling pathway. Knockdown of TRIM21 dramatically impairs innate immune response to viral infection. Moreover, TRIM21 interacts with MAVS and catalyzes its K27-linked polyubiquitination, thereby promoting the recruitment of TBK1 to MAVS. Specifically, the PRY-SPRY domain of TRIM21 is the key domain for its interaction with MAVS, while the RING domain of TRIM21 facilitates the polyubiquitination chains of MAVS. In addition, the MAVS-mediated innate immune response is enhanced by both the PRY-SPRY and RING domains of TRIM21. Mutation analyses of all the lysine residues of MAVS further revealed that Lys325 of MAVS is catalyzed by TRIM21 for the K27-linked polyubiquitination. Overall, this study reveals a novel mechanism by which TRIM21 promotes the K27-linked polyubiquitination of MAVS to positively regulate innate immune response, thereby inhibiting viral infection.
IMPORTANCE Activation of innate immunity is essential for host cells to restrict the spread of invading viruses and other pathogens. MAVS plays a critical role in innate immune response to RNA viral infection. In this study, we demonstrated that TRIM21 targets MAVS to positively regulate innate immunity. Notably, TRIM21 targets and catalyzes K27-linked polyubiquitination of MAVS and then promotes the recruitment of TBK1 to MAVS, leading to upregulation of innate immunity. Our study outlines a novel mechanism by which the IFN signaling pathway blocks RNA virus to escape immune elimination.
INTRODUCTION
To restrict the invasion of microbial pathogens, the human innate immune system is the first line of defense that recognizes pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs) (1). Major PRRs include cytosolic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), membrane-bound Toll-like receptors (TLRs), and nucleotide-binding domain- and leucine-rich repeat-containing molecules (NLRs) (2, 3), as well as some DNA sensors, such as cyclic GMP-AMP synthase (cGAS), DDX41, and IFI16 (4–8). RLRs, a family of cytoplasmic RNA helicases, comprise three major members: RIG-I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), all of which contain the DEXD/H helicase domain (9). The recognition of viral RNA by the C terminus of RIG-I and MDA5 induces a conformational change of the CARD domains located at the N terminus, which then interact with the CARD domain of the mitochondrial adaptor protein MAVS (also known as IPS1, VISA, or CARDIF) (10–13). As an adaptor of PRRs, MAVS recruits and activates the classical TAK-binding kinase 1 (TBK1) and IKK-related kinase. These kinases further induce the activation of two transcription factors, IRF3 and NF-κB, which then translocate into the nucleus to initiate the production of interferons (IFNs) and certain proinflammatory cytokines (14). Once IFNs are induced through the RLR pathway, they act in an autocrine or paracrine fashion to induce hundreds of IFN-stimulated genes (ISGs) that eliminate the invasion of microbial pathogens (15).
Insufficient IFN production can cause chronic infection, while excessive IFN production may induce autoimmune or inflammatory diseases. To maintain an appropriate level of IFN production, cells have developed some mechanisms to modulate the IFN signaling pathway. MAVS is a common adaptor protein that links RLRs to their downstream signaling molecules, which is regulated by viral or host factors. For example, hepatitis C virus (HCV) NS3/4A protein cleaves MAVS to block the MAVS-mediated signaling pathway, thereby allowing HCV to evade the innate immune system and establish persistent infection (16–18). NLRX1 interacts with PCBP2 and facilitates the K48-linked polyubiquitination degradation of MAVS by recruiting the E3 ligase AIP4 (19, 20). Moreover, Ndfip1 interacts with the E3 ligase Smurf1 to mediate the ubiquitination and degradation of MAVS (21). The mitochondrial E3 ligase MARCH5 also promotes the degradation of MAVS in a ubiquitination-dependent manner (22).
The tripartite motif-containing (TRIM) superfamily contains 82 members that have highly conserved E3 ligases that take part in a broad range of biological processes, such as cell proliferation, differentiation, development, and apoptosis (23). TRIM4, TRIM5, TRIM6, TRIM8, TRIM14, TRIM15, TRIM23, TRIM25, and TRIM56 can positively regulate antiviral responses in the IFN signaling pathway (24–32). In contrast, TRIM27, TRIM30, and TRIM38 are negative regulators of the IFN signaling pathway by promoting the K48-linked polyubiquitination of their targeted proteins (33–35). Furthermore, TRIM31 facilitates the K63-linked polyubiquitination and aggregation of MAVS to induce the expression of IFNs against viral infection (36). TRIM44 stabilizes MAVS by removing the K48-linked polyubiquitination of the MAVS (37).
TRIM21 (tripartite motif-containing protein 21), also known as Ro52 or SS-A, belongs to the TRIM superfamily and is commonly expressed in a great variety of human cells (38). Similar to other members of the TRIM superfamily, TRIM21 contains a RING motif in the N-terminal domain, a B-box motif, a coiled-coil domain, and a PRY-SPRY region in the C-terminal domain (39). The RING motif is a zinc-binding domain that has E3 ligase-like activity, whereas the B-box and coiled-coil domains are required for protein-protein interaction and the conformation of homo- or heterodimerization. Furthermore, the PRY-SPRY domain can bind to proteins with antiviral activities (38, 40). TRIM21 was originally identified as an autoantigen in autoimmune diseases, such as rheumatoid arthritis, systemic lupus erythematosus (SLE), and Sjogren's syndrome (39). Recent studies have suggested that TRIM21 may play an essential role in the tumor suppressor or drug-induced apoptosis of cancer cells (41, 42). However, contradictory results, that TRIM21 may target IRF3 after RNA viral infection, have been reported (43, 44). Therefore, the exact mechanisms of TRIM21 against viral infection remain unclear.
In this study, we aimed to reveal the antiviral activity of TRIM21 and its interplay with MAVS during infection by HCV, Newcastle disease virus (NDV), Sendai virus (SeV), and vesicular stomatitis virus (VSV). Our findings suggested that TRIM21 acts as a positive regulator of virus-triggered innate immune response. Upon viral infection, TRIM21 is induced and interacts with MAVS to promote K27-linked polyubiquitination at Lys325 of MAVS. Furthermore, the PRY-SPRY domain of TRIM21 plays a key role in its interaction with MAVS, while the RING domain of TRIM21 is critical for facilitating the polyubiquitination chains of MAVS. Notably, the K27-linked polyubiquitination of MAVS mediated by TRIM21 results in the recruitment of TBK1 to MAVS, which positively regulates innate immune response. Overall, our findings revealed novel activities of TRIM21 in the innate immune response, thereby shedding light on the positive regulation of the innate immune system against viral infection.
RESULTS
Viral infection induces the expression of TRIM21.
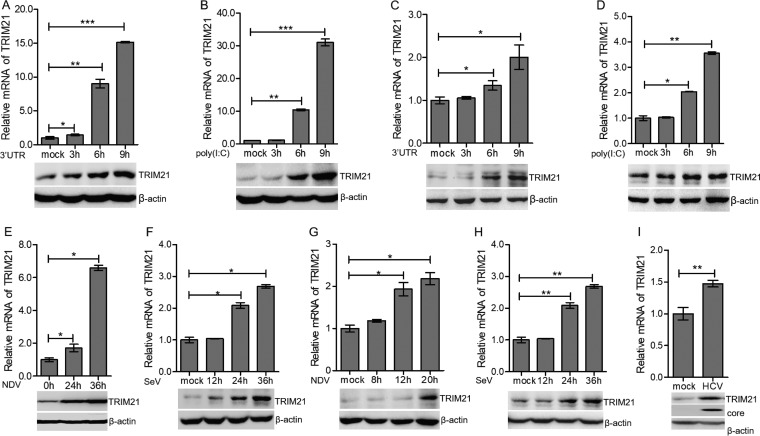
To evaluate the activity of TRIM21 during viral infection, we first measured the expression level of TRIM21 in human hepatocytes stimulated by nucleic acid mimics, the 3′ untranslated region of HCV RNA (HCV 3′ UTR) or long poly(I·C), which were recognized by RIG-I or MDA5, respectively (45–48). After stimulation with the HCV 3′ UTR or poly(I·C), the mRNA and protein of TRIM21 were upregulated in HLCZ01 cells, which support the entire life cycle of HCV and hepatitis B virus (HBV) (Fig. 1A and B) (49). To rule out the possibility of cell specificity, we performed the above-described experiments in Huh7 cells. TRIM21 was also induced by the HCV 3′ UTR or poly(I·C) in the Huh7 cells (Fig. 1C and D). To further evaluate the expression level of TRM21 in the context of viral infection, we infected HLCZ01 and Huh7 cells with NDV or SeV. Both NDV and SeV induced the expression of TRIM21 mRNA and protein in the HLCZ01 (Fig. 1E and F) and Huh7 (Fig. 1G and H) cells. Of note, HCV is a positive-sense single-stranded RNA virus belonging to the family Flaviviridae. HCV can be recognized by RIG-I or MDA5 and further activate IFN signaling (45, 47). TRIM21 expression was induced by HCV infection (Fig. 1I). These data suggested that RNA viruses can induce the expression of TRIM21, indicating a potential role of TRIM21 in innate immune response to viral infection.
FIG 1.
Viral infection induces the expression of TRIM21. (A and B) HLCZ01 cells were transfected with 200 ng of HCV 3′ UTR or poly(I·C) for the indicated times. TRIM21 mRNA (top) and protein (bottom) were analyzed by real-time PCR and immunoblotting, respectively. (C and D) Huh7 cells were transfected with 200 ng of HCV 3′ UTR or poly(I·C) for the indicated times. TRIM21 mRNA (top) and protein (bottom) were analyzed by real-time PCR and immunoblotting, respectively. (E to H) HLCZ01 (E and F) or Huh7 (G and H) cells were infected with NDV (E and G) or SeV (F and H) for the indicated times. TRIM21 mRNA was determined by real-time PCR and normalized with GAPDH. TRIM21 protein was analyzed by immunoblotting. (I) HLCZ01 cells were infected with HCV (multiplicity of infection [MOI] = 0.2) for 72 h. TRIM21 mRNA (top) was analyzed by real-time PCR and normalized with GAPDH. TRIM21 protein and HCV core protein were detected by immunoblotting. β-Actin was used as the loading control. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
The induction of TRIM21 depends on the JAK/STAT signaling pathway.
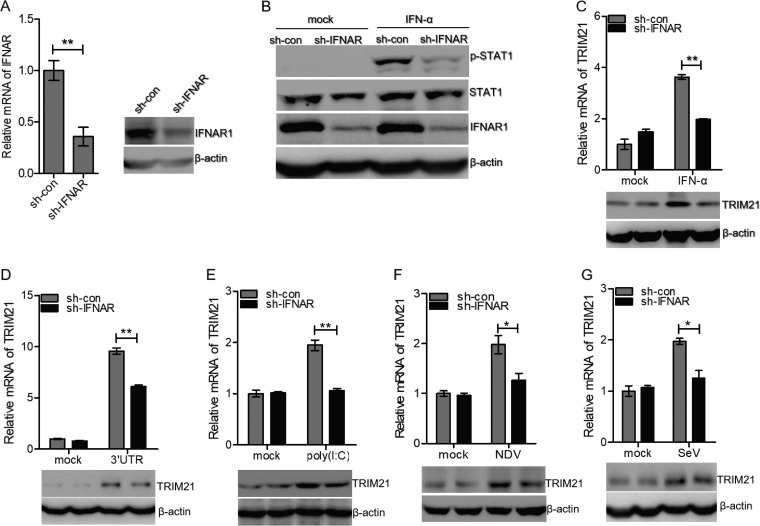
Viral infection induces the production of type I and type III IFNs. IFNs recognize their receptors to activate the JAK/STAT pathway and promote the expression of ISGs, leading to the elimination of invading pathogens (14, 15). To investigate whether the induction of TRIM21 by RNA viruses depends on the JAK/STAT pathway, we constructed stable IFNAR1-silenced cells using HLCZ01 cell lines. IFNAR1 was knocked down effectively in HLCZ01 cells (Fig. 2A). To ensure the efficiency of IFNAR1 knockdown, we detected the phosphorylation of STAT1 with IFN-α treatment. Phosphorylation of STAT1 was attenuated in IFNAR1-silenced cells compared to control cells following IFN-α treatment (Fig. 2B). Knockdown of IFNAR1 significantly decreased the level of TRIM21 in HLCZ01 cells, which was treated by IFN-α (Fig. 2C). After stimulation with the HCV 3′ UTR or poly(I·C), the levels of TRIM21 mRNA and protein were reduced in IFNAR1-silenced cells compared to control cells (Fig. 2D and E). Furthermore, the induction of TRIM21 by NDV or SeV was impaired when we silenced IFNAR1 in HLCZ01 cells (Fig. 2F and G). These data demonstrated that the induction of TRIM21 is dependent on the IFN/JAK/STAT signaling pathway.
FIG 2.
Induction of TRIM21 depends on the JAK/STAT signaling pathway. (A) HLCZ01 cells were stably transfected with either scrambled shRNA (sh-con) or IFNAR shRNA (sh-IFNAR). (Left) IFNAR1 mRNA was analyzed by real-time PCR and normalized with GAPDH. (Right) IFNAR1 protein was analyzed by immunoblotting. (B) Immunoblot analysis of the indicated proteins in HLCZ01-sh-con and HLCZ01-sh-IFNAR1 cell lines treated with IFN-α (500 U/ml) for 30 min. (C) HLCZ01 cells stably transfected with scrambled shRNA or IFNAR shRNA were treated with IFN-α (100 U/ml) for 6 h. TRIM21 mRNA was determined by real-time PCR and normalized with GAPDH. (D and E) TRIM21 protein was analyzed by immunoblotting. HLCZ01 cells stably transfected with scrambled shRNA or IFNAR shRNA were transfected with HCV 3′ UTR (D) or poly(I·C) (E) for 6 h. TRIM21 mRNA and protein were analyzed by real-time PCR and immunoblotting, respectively. (F and G) HLCZ01 cells stably transfected with scrambled shRNA or IFNAR shRNA were infected with NDV (F) or SeV (G) for 16 h. TRIM21 mRNA was determined by real-time PCR and normalized with GAPDH. TRIM21 protein was detected by immunoblotting. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01 versus control.
TRIM21 positively regulates innate immune response to RNA nucleic acid mimics.
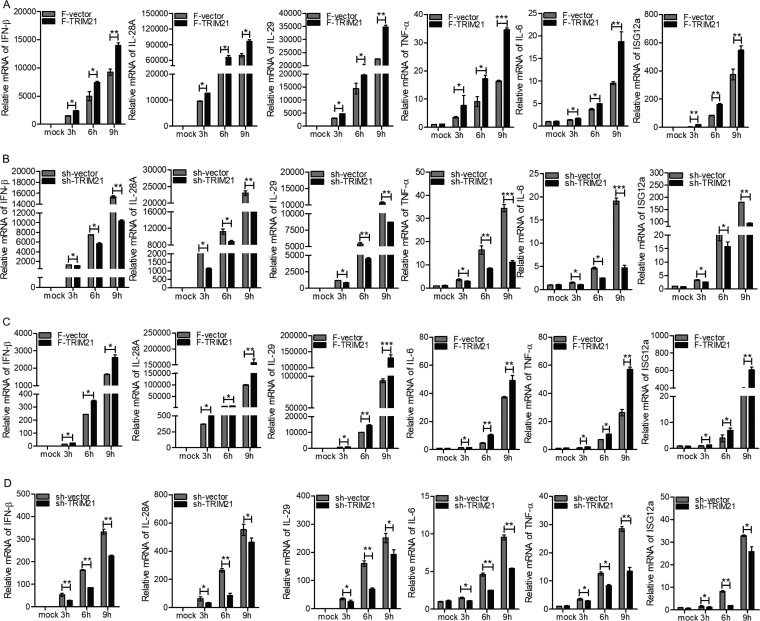
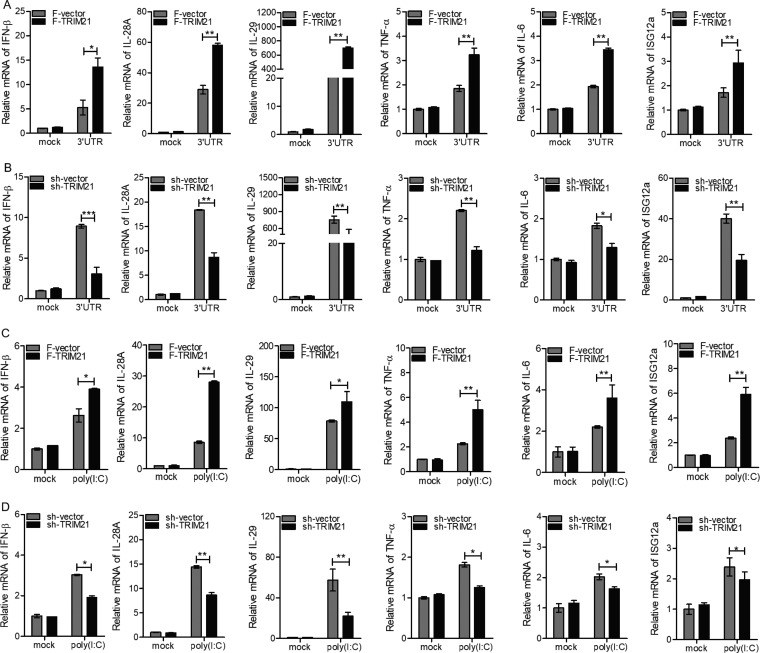
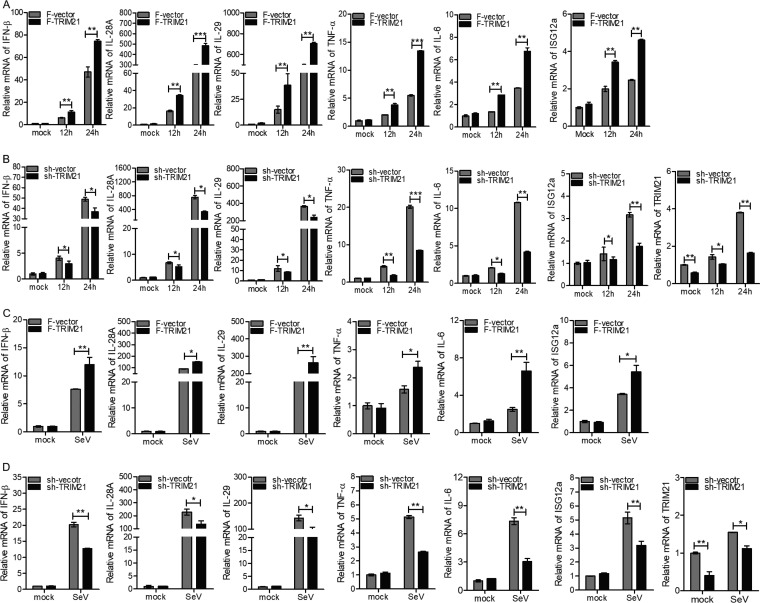
Based on the above-mentioned finding that TRIM21 was induced by viral infection and its production required the IFN/JAK/STAT pathway, we speculated that TRIM21 may play an important role in innate immune response to viral infection. To assess our hypothesis, we measured the expression levels of a series of genes participating in host antiviral defense via ectopic expression or knockdown of TRIM21 in human hepatocytes. Upon stimulation with the HCV 3′ UTR, the ectopic expression of TRIM21 in HLCZ01 cells significantly enhanced the expression of type I IFN (IFN-β), type III IFNs (interleukin 28A [IL-28A] and IL-29), tumor necrosis factor alpha (TNF-α), and IL-6 in a time-dependent manner (Fig. 3A). The representative ISGs, including ISG12a, were upregulated when TRIM21 was overexpressed in HLCZ01 cells (Fig. 3A). Importantly, our previous studies revealed the antiviral activity of ISG12a (50, 51). In fact, silencing of TRIM21 in HLCZ01 cells remarkably impaired the expression of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a (Fig. 3B). Next, we measured the expression levels of innate immune genes stimulated by poly(I·C) in HLCZ01 cells. Ectopic expression or silencing of TRIM21 enhanced or attenuated the expression of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a, (Fig. 3C and D). Moreover, we performed the above-described experiments in Huh7 cells, and similar results were observed (Fig. 4A to D). These data suggested that TRIM21 positively regulates innate immune response to RNA nucleic acid mimics.
FIG 3.
TRIM21 positively regulates innate immune response to RNA nucleic acid mimics in HLCZ01 cells. (A to D) HLCZ01 cells were transfected with p3X-Flag-CMV-vector or p3X-Flag-TRIM21 plasmid (A and C) or pSilencer-vector or pSilencer-TRIM21 plasmid (B and D) for 48 h and then transfected with 200 ng of HCV 3′ UTR (A and B) or 200 ng of poly(I·C) (C and D) for the indicated times. The levels of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a mRNAs were analyzed by real-time PCR and normalized with GAPDH. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control. F, Flag.
FIG 4.
TRIM21 positively regulates innate immune response to RNA nucleic acid mimics in Huh7 cells. (A to D) Huh7 cells were transfected with p3X-Flag-CMV-vector or p3X-Flag-TRIM21 (A and C) or pSilencer-vector or pSilencer-TRIM21 (B and D) for 48 h and then transfected with 200 ng of HCV 3′ UTR (A and B) or 200 ng of poly(I·C) (C and D) for the indicated times. The levels of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a mRNAs were examined by real-time PCR and normalized with GAPDH. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
TRIM21 positively regulates innate immune response to RNA viral infection.
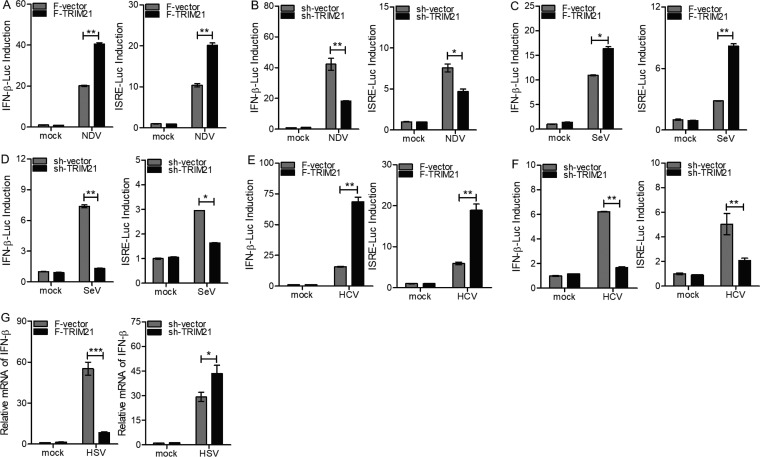
To determine the role of TRIM21 in the context of viral infection, we performed the following experiments using NDV or SeV. First, the ectopic expression or knockdown of TRIM21 increased or decreased the levels of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a upon infection with NDV (Fig. 5A and B). TRIM21 was knocked down efficiently in HLCZ01 cells with or without NDV infection (Fig. 5B). Second, the induction of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a by SeV was enhanced or attenuated when TRIM21 was overexpressed or silenced, respectively (Fig. 5C and D). Moreover, we evaluated the effect of TRIM21 on the regulation of IFN signaling in Huh7 cells transfected with IFN-β–Luc and ISRE-Luc reporters and infected with NDV or SeV. Both IFN-β–Luc and ISRE-Luc were activated by NDV or SeV. Moreover, such activation was increased or attenuated by ectopic expression or knockdown of TRIM21, respectively (Fig. 6A to D).
FIG 5.
TRIM21 positively regulates innate immune response to RNA viral infection in HLCZ01 cells. (A to D) HLCZ01 cells were transfected by p3X-Flag-CMV-vector or p3X-Flag-TRIM21 (A and C) or pSilencer-vector or pSilencer-TRIM21 (B and D) for 24 h and then infected with NDV (A and B) or SeV (C and D) for the indicated times. The levels of IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a mRNAs were tested by real-time PCR and normalized with GAPDH. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
FIG 6.
TRIM21 positively regulates innate immune response to RNA viral infection in Huh7 cells. (A to D) Luciferase activity of lysates in Huh7 cells cotransfected with IFN-β–Luc or ISRE-Luc and p3X-Flag-CMV-vector or p3X-Flag-TRIM21 (A and C) or pSilencer-vector or pSilencer-TRIM21 (B and D) for 12 h and then infected with NDV (A and B) or SeV (C and D) for 12 h. The results are presented relative to the luciferase activity in control cells. (E and F) Luciferase activity of lysates in Huh7.5.1(MAVS-C508R) cells infected with HCV (MOI = 2) for 48 h, followed by cotransfection with IFN-β–Luc or ISRE-Luc and p3X-Flag-CMV vector or p3X-Flag-TRIM21 (E) or pSilencer-vector or pSilencer-TRIM21 (F) for 24 h. (G) Huh7 cells were transfected with p3X-Flag-CMV vector or p3X-Flag-TRIM21 or with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with HSV for 24 h. The level of IFN-β mRNA was tested by real-time PCR and normalized with GAPDH. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
HCV NS3/4A protein cleaves MAVS to abrogate host antiviral innate immunity (16–18). It was reported that MDA5 senses HCV RNA to trigger innate immune response after introduction of a C508R mutant of MAVS (MAVSR) into Huh7.5.1 cells (45). Therefore, to evaluate the effect of TRIM21 on the IFN signaling pathway induced by HCV, Huh7.5.1(MAVS-C508R) cells were transfected with IFN-β–Luc or ISRE-Luc reporters. The activity of IFN-β promoter and ISRE-Luc induced by HCV infection was enhanced or attenuated when TRIM21 was overexpressed or silenced, respectively (Fig. 6E and F). The ectopic expression or knockdown of TRIM21 in Huh7 cells decreased or increased the level of IFN-β upon herpes simplex virus (HSV) infection (Fig. 6G). These data suggested that TRIM21 promotes innate immune response to RNA viral infection.
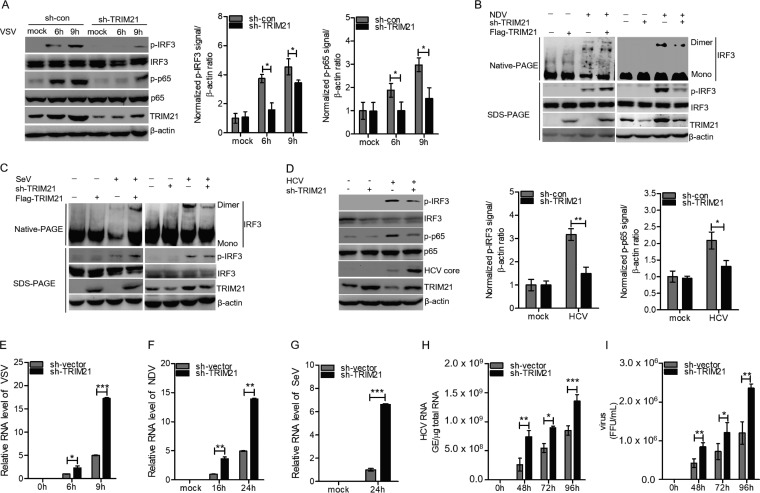
TRIM21 promotes the activation of IRF3 and NF-κB signaling and suppresses viral infection.
It is known that the induction of IFNs by viral infection mainly depends on IRF3. Activation of IRF3 requires phosphorylation by TBK1 and IKK-ε (52). The phosphorylated IRF3 undergoes homodimerization and translocates to the nucleus, leading to the transcription of IFN genes (52). The induction TNF-α and IL-6 by viral infection mainly requires the transcription factor NF-κB. Activation of NF-κB needs the IKK-related kinase complexes (53). To investigate the effect of TRIM21 on the activation of IRF3 and NF-κB, we silenced TRIM21 in HLCZ01 cells, which were then infected with VSV. Knockdown of TRIM21 remarkably attenuated the phosphorylation of IRF3 and p65 in HLCZ01 cells (Fig. 7A). In HLCZ01 cells infected with NDV (Fig. 7B) or SeV (Fig. 7C), the phosphorylation and dimerization of IRF3 could be enhanced or attenuated by the ectopic expression or knockdown of TRIM21, respectively. Activation of IRF3 and p65 was also detected in HCV-infected Huh7.5.1(MAVS-C508R) cells, and TRIM21 knockdown impaired the phosphorylation of IRF3 and p65 (Fig. 7D). Moreover, TRIM21 knockdown increased the level of HCV core in HCV-infected cells (Fig. 7D). These results supported the idea that TRIM21 promotes the activation of IRF3 and NF-κB signaling during viral infection.
FIG 7.
TRIM21 promotes the activation of IRF3 and NF-κB signaling and suppresses viral infection. (A) Immunoblot analysis of the indicated proteins in HLCZ01-shcon and HLCZ01-shTRIM21 cells infected with VSV for the indicated times. (B and C) HLCZ01 cells were transfected with p3X Flag-CMV-TRIM21 or pSilencer-TRIM21 for 36 h and then infected with NDV (B) or SeV (C) for 16 h. The dimer and monomer of IRF3 were detected by native PAGE. Phosphorylated IRF3 was examined by immunoblotting. (D) Huh7.5.1(MAVS-C508R) cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with HCV (MOI = 2) for 48 h. Phosphorylated IRF3, phosphorylated P65, and HCV core protein were detected by immunoblotting. (E to G) HLCZ01 cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with VSV (E), NDV (F), or SeV (G) for the indicated times. RNA of VSV, NDV, and SeV was analyzed by real-time PCR and normalized with GAPDH. (H) Huh7.5.1(MAVS-C508R) cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with HCV (MOI = 2) for the indicated times. HCV RNA was analyzed by real-time PCR. (I) Huh7.5.1(MAVS-C508R) cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with HCV (MOI = 2) for the indicated times. Extracellular infectious virus particles were detected by FFU assay. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
Considering the significance of TRIM21 in the expression of IRF3- and NF-κB-dependent innate antiviral genes, we further assessed the antiviral activities of TRIM21. We found that knockdown of TRIM21 increases the levels of VSV, NDV, and SeV RNAs at the indicated time points (Fig. 7E to G). Similarly, knockdown of TRIM21 in HCV-infected Huh7.5.1(MAVS-C508R) cells remarkably enhanced viral replication and the production of infectious viruses compared to the control group (Fig. 7H to I). These data demonstrated that TRIM21 plays a positive role in the activation of IRF3 and NF-κB signaling triggered by viral infection, thereby inhibiting viral infection.
TRIM21 targets and interacts with MAVS.
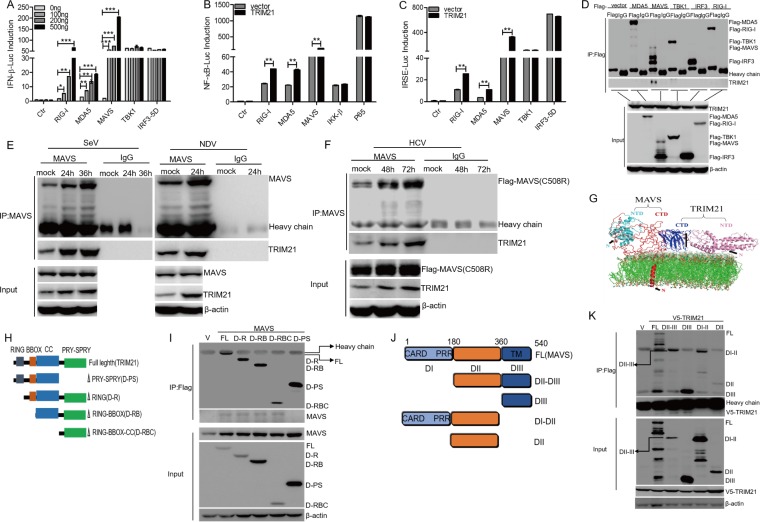
Upon RNA virus infection, RIG-I and MDA5 recognize PAMPs and recruit MAVS, thereby activating IRF3 and NF-κB, leading to the expression of type I and type III IFNs or proinflammatory cytokines for antiviral activity. To determine the molecular mechanism of TRIM21 in enhancing the IFN signaling pathway, we cotransfected pcDNA3.1a-TRIM21 with IFN-β–Luc, NF-κB–Luc, or ISRE-Luc in HEK293T cells. Overexpression of TRIM21 enhanced the activation of the IFN-β promoter by RIG-I, MDA5, or MAVS, whereas the activation of the IFN-β promoter triggered by TBK1 or IRF3-5D (S385D, S386D, S396D, S398D, S402D, S404D, and S405D) was unaffected (Fig. 8A). Similarly, NF-κB activity induced by RIG-I, MAD5, or MAVS was substantially improved by the overexpression of TRIM21, whereas the NF-κB activity mediated by IKK-β or P65 was not altered in the presence of TRIM21 (Fig. 8B). Moreover, the ISRE activity induced by TBK1 or IRF3-5D was barely changed by TRIM21, but overexpression of TRIM21 dramatically enhanced the activation of ISRE by RIG-I, MDA5, or MAVS (Fig. 8C). Taken together, our findings suggested that TRIM21 promotes the IFN signaling pathway upstream of TBK1 and IKK-β.
FIG 8.
TRIM21 targets and interacts with MAVS. (A to C) Luciferase activity of lysates in HEK293T cells transfected for 24 h with IFN-β–Luc (A) or ISRE-Luc (C) plus Flag-RIG-I, Flag-MDA5, Flag-MAVS, Flag-TBK1, or Flag-IRF3-5D, or with NF-κB–Luc (B) plus Flag-RIG-I, Flag-MDA5, Flag-MAVS, Flag-IKKβ, or Flag-P65, along with pcDNA3.1a-TRIM21 or an empty vector. The results are presented relative to the luciferase activity in control cells. (D) HEK 293T cells were transfected with plasmids as indicated for 24 h. Cellular lysates were immunoprecipitated with anti-Flag or IgG. Immunoprecipitates were analyzed by Western blotting (WB) with anti-Flag or anti-TRIM21. (E) HLCZ01 cells were infected with SeV or NDV for the indicated times. Immunoprecipitation and WB analysis were performed with antibodies against the indicated proteins. (F) Huh7.5.1(MAVS-C508R) cells were infected with HCV (MOI = 2) for the indicated times. Immunoprecipitation and WB analysis were performed with antibodies against the indicated proteins. (G) Bioinformatics analysis was conducted to predict the pivotal domains responsible for the interaction of TRIM21 and MAVS. CTD, C-terminal domain; NTD, N-terminal domain. (H) Schematic illustration of TRIM21 truncations. CC, coiled-coil domain. (I) HEK 293T cells were cotransfected with the plasmids encoding MAVS and the indicated domains of TRIM21 for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. (J) Schematic illustration of TRIM21 truncations. (K) HEK 293T cells were cotransfected with the plasmids encoding V5-TRIM2 and the indicated domains of MAVS for 48 h. Co-IP and immunoblotting were performed with the indicated antibodies. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
To determine the key factors in the IFN signaling pathway that could be targeted by TRIM21, we transfected Flag-tagged signaling components into HEK293T cells. Our coimmunoprecipitation (co-IP) experiments revealed that TRIM21 interacts with MAVS but not with RIG-I, MDA5, TBK1, or IRF3 (Fig. 8D). Furthermore, a strong association of endogenous TRIM21 and MAVS was confirmed in HLCZ01 cells, while SeV or NDV infection enhanced TRIM21-MAVS interaction (Fig. 8E). The interaction between MAVS and TRIM21 was also enhanced in HCV-infected Huh7.5.1(MAVS-C508R) cells (Fig. 8F). All the data suggested that TRIM21 targets and interacts with MAVS and that the interaction between TRIM21 and MAVS is enhanced upon viral infection.
Characterization of the interaction between TRIM21 and MAVS is important for exploring the activities of TRIM21 in host cells. Potential interaction interfaces between TRIM21 and MAVS were next estimated by using in silico approaches, which suggested that the PRY-SPRY domain of TRIM21 is most likely involved in the interaction with the DIII domain of MAVS (Fig. 8G). To further confirm the prediction, we constructed a series of truncations of TRIM21 and cotransfected them with the full-length MAVS in HEK293T cells (Fig. 8H). We found that the PRY-SPRY domain of TRIM21 interacts with MAVS (Fig. 8I). Also, the truncation experiments with MAVS revealed that the DIII domain of MAVS is the key domain for the MAVS-TRIM21 interaction (Fig. 8J and K). Our data supported the idea that TRIM21 targets MAVS and that the PRY-SPRY domain of TRIM21 is sufficient for its interaction with the DIII domain of MAVS.
TRIM21 promotes K27-linked ubiquitination of MAVS.
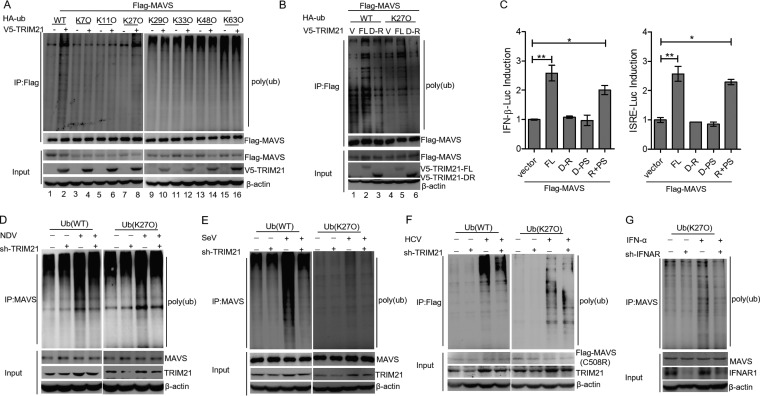
TRIM21 is a RING finger domain-containing E3 ligase that belongs to the TRIM superfamily (39). To investigate whether the E3 ligase activity of TRIM21 is involved in the regulation of MAVS, we transfected Flag-tagged MAVS together with the hemagglutinin (HA)-tagged ubiquitin (HA-ub) wild type (WT) or ubiquitin mutants in HEK293T cells in the presence or absence of TRIM21. Our data showed that TRIM21 promotes the ubiquitination of MAVS (Fig. 9A, lanes 1 and 2). Importantly, TRIM21 catalyzed the formation of the K27-linked polyubiquitin chains on MAVS (Fig. 9A, lanes 7 and 8).
FIG 9.
TRIM21 promotes K27-linked ubiquitination of MAVS. (A) TRIM21 promotes the K27-linked ubiquitination of MAVS. HEK293T cells were cotransfected with the indicated plasmids for 42 h and treated with MG132 (25 μM; Sigma) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (B) The RING domain of TRIM21 is essential for the K27-linked polyubiquitination of MAVS. HEK293T cells were cotransfected with the indicated plasmids for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (C) Luciferase activity of lysates in HEK 293T cells transfected for 24 h with IFN-β–Luc or ISRE-Luc plus Flag-MAVS, along with V5-TRIM21-FL, V5-TRIM21-DR, V5-TRIM21-DPS, or an empty vector. (D and E) HLCZ01 cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with NDV (D) or SeV (E) for 16 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (F) Huh7.5.1(MAVS-C508R) cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with HCV (MOI = 2) for 72 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (G) HLCZ01-sh-con and HLCZ01-sh-IFNAR1 cell lines were transfected with pHA-ub (K27) plasmid for 36 h and then treated with IFN-α (200U/ml) for 6 h. Ubiquitination assays and immunoblotting were performed with the indicated antibodies. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01 versus control.
Previous studies demonstrated that the RING domain of TRIM21 is essential for its E3 ligase activity (8, 39). To determine the role of the RING domain in the ubiquitination of MAVS, HEK293T cells were cotransfected with V5-TRIM21 or truncated TRIM21 that lacked the RING domain (V5-TRIM21-D-R), and HA-ub (WT) or HA-ub (K27O) with Flag-MAVS. Ubiquitination analyses revealed that the deletion of the RING domain in TRIM21 rescued MAVS ubiquitination (Fig. 9B, lanes 1 to 3) and also obstructed the K27-linked ubiquitination of MAVS (Fig. 9B, lanes 4 to 6). These data suggested that the ubiquitination of MAVS induced by TRIM21 requires the RING domain of TRIM21.
Essential regions within TRIM21 were identified, but the functions of these regions in viral restriction remain to be elucidated. We performed luciferase assays to demonstrate that the full-length TRIM21 augmented the MAVS-induced activation of the IFN-β promoter or ISRE activity, whereas the MAVS-triggered activation of the IFN-β promoter or ISRE activity was unaffected by TRIM21-D-R or TRIM21 lacking the PRY-SPRY domain (TRIM21-D-PS). However, these effects were reversed by the combination of TRIM21-D-R and TRIM21-D-PS (Fig. 9C), indicating that TRIM21 promotes MAVS-mediated IFN production through both the RING and PRY-SPRY domains. To confirm TRIM21-induced ubiquitination of endogenous MAVS, we measured the polyubiquitination of MAVS in NDV- or SeV-infected HLCZ01 cells. The ubiquitination of MAVS induced by NDV or SeV was rescued by silencing TRIM21, as well as the K27-linked ubiquitination of MAVS (Fig. 9D and E). Moreover, the K27-linked ubiquitination of MAVS was also decreased by TRIM21 knockdown in HCV-infected Huh7.5.1 cells (MAVS-C508R) (Fig. 9F). Because the induction of TRIM21 depended on the IFN/JAK/STAT signaling pathway (Fig. 2B and C), the induction of K27-linked ubiquitination of MAVS by IFN-α was impaired in IFNAR1-silenced cells compared to the control group (Fig. 9G). Overall, TRIM21 catalyzes the K27-linked polyubiquitination of MAVS for the positive regulation of the IFN signaling pathway.
TRIM21 promotes the recruitment of TBK1 to MAVS.
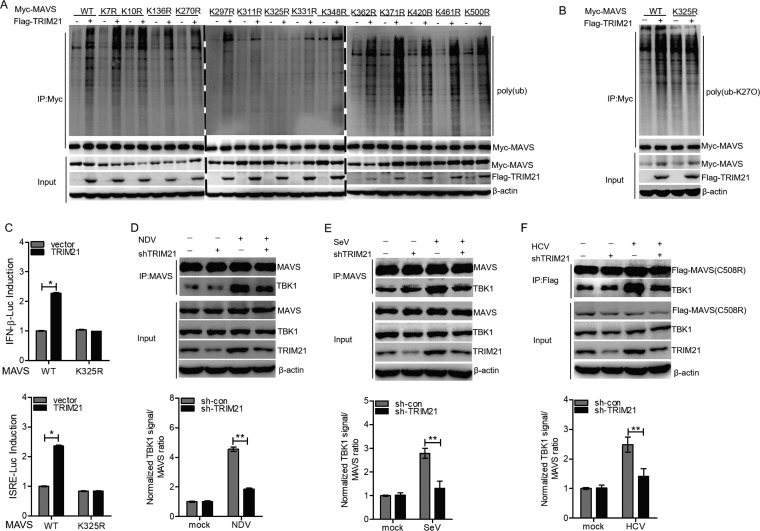
Since MAVS contains 14 lysine residues, we aimed to identify the lysine residues for TRIM21-mediated polyubiquitination by transfecting HA-ub with Myc-tagged wild-type MAVS [Myc-MAVS (WT)] or 14 Myc-tagged MAVS mutants [Myc-MAVS (K7R), (K10R), (K136R), (K270R), (K279R), (K311R), (K325R), (K331R), (K348R), (K362R), (K371R), (K420R), (K461R), and (K500R)] into HEK293T cells. Our co-IP and ubiquitination experiments revealed that TRIM21 does not trigger the ubiquitination of MAVS with the K325R mutation, suggesting K325 within MAVS is the key lysine residue for TRIM21-induced polyubiquitination of MAVS (Fig. 10A). TRIM21 promoted the K27-linked polyubiquitination of MAVS (WT) but had no effect on the K27-linked polyubiquitination of mutant MAVS (K325R) (Fig. 10B). These data suggested that TRIM21 promotes the K27-linked polyubiquitination of MAVS on Lys325. In comparison to wild-type MAVS, TRIM21 could not enhance the activity of the IFN-β promoter or ISRE-Luc in the presence of the MAVS K325R mutant (Fig. 10C). All the data strongly supported the idea that TRIM21 enhances the IFN signaling pathway by catalyzing the K27-linked polyubiquitination of MAVS on K325.
FIG 10.
TRIM21 promotes the K27-linked polyubiquitination of MAVS on Lys325 and the recruitment of TBK1 to MAVS. (A) HEK293T cells were cotransfected with the indicated plasmids for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (B) HEK293T cells were cotransfected with the indicated plasmids for 42 h and treated with MG132 (25 μM) for an additional 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (C) Luciferase activity of lysates in HEK 293T cells transfected for 24 h with IFN-β–Luc or ISRE-Luc plus Myc-MAVS (WT) or Myc-MAVS (K325R), along with Flag-TRIM21 or an empty vector. (D and E) TRIM21 promoted interaction between MAVS and TBK1. HLCZ01 cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with NDV (D) or SeV (E) for 16 h. IP and immunoblotting were performed with the indicated antibodies. (F) Huh7.5.1(MAVS-C508R) cells were transfected with pSilencer-vector or pSilencer-TRIM21 for 24 h and then infected with HCV (MOI = 2) for 72 h. IP and immunoblotting assays were performed with the indicated antibodies. The results are presented as means ± standard deviations. *, P < 0.05; **, P < 0.01 versus control.
The K27-linked polyubiquitination of components of the IFN signaling pathway plays an important role in antiviral responses. For example, the K27-linked polyubiquitination of cGAS or STING is essential for mediating antiviral signaling (54, 55). Moreover, USP13 removes the K27-linked polyubiquitination chains of STING and impairs the recruitment of TBK1 to STING, which negatively regulates IFN production during DNA virus infection (56). Therefore, we speculated that the TRIM21-mediated K27-linked polyubiquitination of MAVS may promote its interaction with TBK1. The interaction between MAVS and TBK1 was impaired in TRIM21-silenced cells with NDV or SeV infection (Fig. 10D and E). Furthermore, the MAVS-TBK1 interaction was also attenuated in HCV-infected Huh7.5.1(MAVS-C508R) cells when we silenced TRIM21 in the cells (Fig. 10F). All the data demonstrated that TRIM21 promotes the K27-linked polyubiquitination of MAVS and the recruitment of TBK1 to MAVS, thereby enhancing the innate immune response to viral infection.
DISCUSSION
To discover novel regulatory factors that enhance innate immune responses against viral infection, this study reveals for the first time that TRIM21 promotes the K27-linked polyubiquitination of MAVS to positively regulate the IFN signaling pathway. Previous studies suggested that the E3 ligases TRIM31 and TRIM44 positively regulate MAVS activity (36, 37). Our study provides five concrete pieces of evidence that TRIM21 also takes part in positively regulating MAVS. First, the expression of TRIM21 in the context of the JAK/STAT pathway was induced by NDV, SeV, or HCV infection, suggesting that TRIM21 may play a role in the IFN signaling pathway. Second, during NDV or SeV infection, the induction of IRF3- and NF-κB-dependent genes by viral infection was significantly increased or attenuated by ectopic expression or silencing of TRIM21, respectively. Third, the replication capacities of NDV, SeV, and HCV were enhanced in virus-infected cells when TRIM21 was silenced. Fourth, TRIM21 targeted and interacted with MAVS, and notably, this interaction was enhanced by SeV, NDV, or HCV infection. Moreover, the PRY-SPRY domain of TRIM21 was critical for its interaction with MAVS. Fifth, TRIM21 induced the K27-linked ubiquitination of MAVS and promoted the recruitment of TBK1 to MAVS for antiviral activity. The RING domain of TRIM21 was the functional region that performed the E3 ligase activity. Moreover, deletion of RING and PRY-SPRY mutants caused the loss of TRIM21 functions in suppressing viral infection.
TRIM21 was originally identified as an autoantigen in autoimmune diseases, such as rheumatoid arthritis, SLE, and Sjogren's syndrome (39). TRIM21 acts as an intracellular antibody receptor that recognizes the antibody-coated virus and inhibits viral infection (57–59). Moreover, TRIM21 may facilitate the replication of DNA viruses by inducing the K48-linked ubiquitination degradation of DDX41, known as a DNA virus sensor (60). In this study, we found that TRIM21 can promote the K27-linked ubiquitination of MAVS, but not K48-linked ubiquitination. Two other studies reported that TRIM21 may interact with IRF3 to regulate IFN signaling upon RNA virus infection, although their results are controversial (43, 44). Higgs et al. claimed that TRIM21 interacts with and catalyzes the ubiquitination of IRF3 to promote its degradation (43). However, Yang et al. found that TRIM21 interacts with and prevents the degradation of IRF3 by removing its ubiquitination (44). Our study demonstrated no interaction between TRIM21 and IRF3.
This controversy can be explained by several factors. Previous studies were mainly performed using HEK293 cell lines, which were derived from human embryos, while highly differentiated human hepatocytes were used in the present study. As described by Yang et al., TRIM21 may remove the polyubiquitination degradation of IRF3 by interfering with the interaction between IRF3 and Pin1 (61). However, the expression of Pin1 is dramatically lower in cell lines derived from liver tissues than in kidney tissues (62). The lower expression of Pin1 in liver cells indicated that the interaction between Pin1 and IRF3 might be weaker under viral infection. Of note, Pin1 is reported to interact with the phosphorylated IRF3 and promotes the ubiquitin degradation of IRF3 (61). Given the fact that TRIM21 is distributed in the cytoplasm and the nucleus (63), it is tempting to speculate that TRIM21 in the cytoplasm may catalyze the K27-linked polyubiquitination of MAVS in the early phase of viral infection. In the later phase, TRIM21 in the nucleus may participate in the antiviral activity by impairing the Pin1-mediated polyubiquitination of phosphorylated IRF3. Nevertheless, this hypothesis has yet to be verified by future studies.
Our study demonstrated that TRIM21 restricts viral infection by positively regulating innate immunity. Specifically, HCV infection triggers the hepatic innate immune response through recognition of the cytosolic sensor RIG-I or MDA5, subsequently inducing the expression of hundreds of ISGs to restrict viral replication. For instance, ISG12a is one of the ISGs that suppress HCV replication by promoting the K48-linked ubiquitination of viral NS5A protein (51). Other ISGs, such as ISG15 and IFI6, can also restrict HCV infection (64, 65). It is possible that some ISGs may be modified or regulated by TRIM21 to eliminate viral infection. In summary, we found another host factor, TRIM21, restricting HCV infection.
Upon viral infection, the adaptor MAVS is responsible for transmitting the recognition signal from the upstream to the downstream pathway. Therefore, regulation of MAVS activity is important for innate immune response to viral infection. Among various regulatory mechanisms, ubiquitination among the posttranslational modifications is one of the major mechanisms that regulate various cellular events, such as the cell cycle, cell division, DNA repair, and immune responses (66). Some E3 ligases, like AIP4, RNF5, RNF125, Smurf1, Smurf2, TRIM25, and MARCH5, negatively regulate the activity of MAVS by catalyzing K48-linked polyubiquitin for its degradation (20, 21, 67–70). However, only two E3 ligases, TRIM31 and TRIM44 (36, 37), have been found to positively regulate MAVS activity. Our study strongly supported the idea that TRIM21 promotes the K27-linked ubiquitination of MAVS to restrict RNA virus infection. K27-linked ubiquitination plays an important role in innate immune response to viral infection. For example, upon DNA virus infection, K27-linked polyubiquitination of cGAS and STING can restrict the replication of viruses (54, 55). In contrast, K27-linked polyubiquitination of dengue virus NS3 protein enhances the cleavage of STING by the NS2B3 protease complex, promoting viral replication (71).
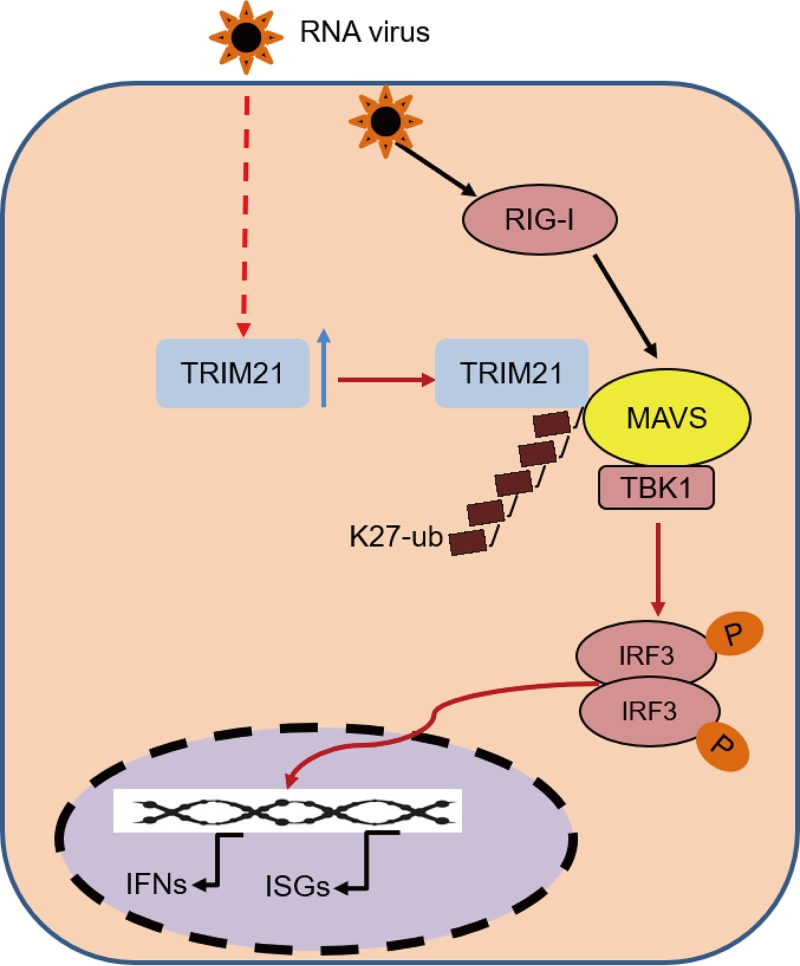
Overall, we identified TRIM21 as a positive regulator of the RNA virus-triggered MAVS-mediated signaling pathway that restricts viral infection. Of note, TRIM21 interacts with MAVS to promote the K27-linked polyubiquitination of MAVS, thus leading to the recruitment of TBK1 to MAVS to enhance the innate immune response to viral infection (Fig. 11). Moreover, the PRY-SPRY domain of TRIM21 is required for its interaction with MAVS, while the RING domain of TRIM21 is essential for catalyzing the polyubiquitination of MAVS. Taken together, our findings shed light on a novel mechanism by which TRIM21 promotes the K27-linked polyubiquitination of MAVS to positively regulate innate immune response, thereby inhibiting viral infection.
FIG 11.
Schematic model of TRIM21-mediated K27 polyubiquitination of MAVS during viral infection. TRIM21 targets and promotes the K27-linked polyubiquitination of MAVS, leading to the recruitment of TBK1 to MAVS to enhance the innate immune response to viral infection.
MATERIALS AND METHODS
Cell culture and reagents.
The HLCZ01 cell line, a novel hepatoma cell line supporting the entire life cycle of HCV and HBV, was previously established in our laboratory (49). HEK293T cells were purchased from Boster. Huh7 cells were purchased from the American Type Culture Collection. Huh7.5.1(MAVS-C508R) cells were kindly shared by Jin Zhong of the Institute Pasteur of Shanghai (45). HLCZ01 cells were cultured in collagen-coated tissue culture plates containing Dulbecco's modified Eagle's medium (DMEM)/F12 medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco), 40 ng/ml of dexamethasone (Sigma), insulin-transferrin-selenium (ITS; Lonza), penicillin, and streptomycin. Other cells were propagated in DMEM supplemented with 10% FBS, l-glutamine, nonessential amino acids, penicillin, and streptomycin.
Plasmid construction.
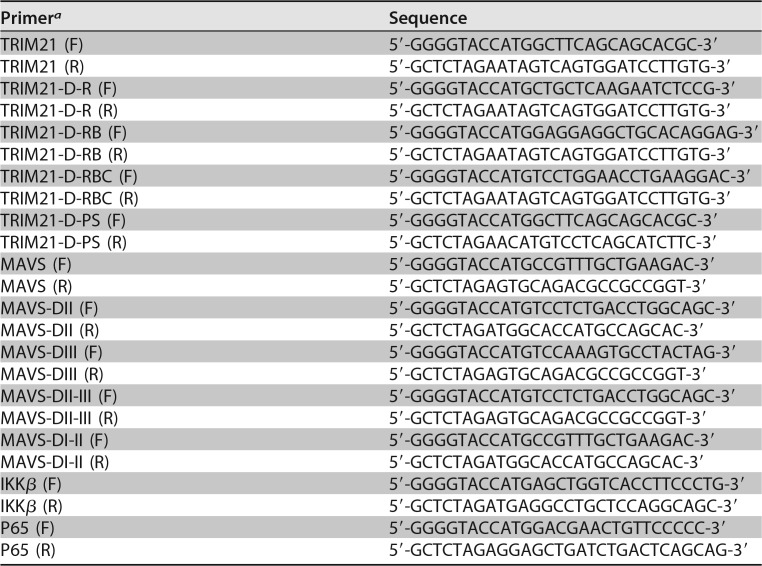
The short hairpin RNA (shRNA) targeting TRIM21 was inserted into the pSilencer-neo plasmid (Ambion). The target sequence of TRIM21 shRNA was 5′-AAGAATCTCCGGCCCAATCGA-3′. TRIM21, MAVS, IKK-β, and p65 cDNAs were synthesized from the total cellular RNA isolated from HLCZ01 cells by standard reverse transcription-PCR (RT-PCR). Subsequently, they were cloned into the pcDNA3.1a vector and the p3×FLAG-CMV vector. Multiple domains of TRIM21 and MAVS were amplified from the templates of full-length TRIM21 and MAVS, which were then cloned into the p3×FLAG-CMV or pcDNA3.1a vector. The primers for amplifying these genes are listed in Table 1. The IFN-β–luciferase and ISRE-Luciferase plasmids were purchased from InvivoGen. Flag-MDA5 and pGL3-NF-κB-Luciferase were kindly shared by Jianguo Wu (Wuhan University, Wuhan, China). The Flag-IRF3-5D plasmid was a gift from Deyin Guo (Wuhan University, Wuhan, China). The pHA-Ub (K7, K11, K27, K29, and K33) plasmids were kindly shared by Hongbing Shu (Wuhan University, Wuhan, China). The Flag-TBK1 and pHA-Ub (K48 and K63) plasmids were kindly provided by Zhengfan Jiang (Peking University, Beijing, China). The Myc-MAVS (WT, K7R, K10R, K136R, K270R, K297R, K311R, K325R, K331R, K348R, K362R, K371R, K420R, K461R, and K500R) plasmids were kindly shared by Chengjiang Gao (Shandong University, Jinan, China).
TABLE 1.
Primers for amplification of domains of TRIM21 and MAVS
aF, forward; R, reverse.
Virus.
The pJFH1 and pJFH1/GND plasmids were gifts from Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan). The linearized DNAs from the pJFH1 and pJFH1/GND plasmids were purified and used as the templates for in vitro transcription with a MEGAscript kit (Ambion, Austin, TX). In vitro-transcribed genomic JFH1 or JFH1/GND RNA was delivered into Huh7.5 cells by electroporation. The transfected cells were cultured for the indicated periods. The cells were passaged every 3 to 5 days, while the corresponding supernatants were collected and filtered with a 0.45-μm filter device. The viral titers were presented as focus-forming units (FFU) per milliliter, determined as the average number of NS5A-positive foci detected in Huh7.5 cells. SeV, VSV, and HSV were kindly shared by Xuetao Cao (Second Military Medical University, Shanghai, China).
Antibodies.
Monoclonal antibodies against β-actin, Flag tag, and HA tag were obtained from Sigma. The V5 tag monoclonal antibody was purchased from Invitrogen. The monoclonal antibody against MAVS was purchased from Santa Cruz Biotechnology. The antibodies for TRIM21, TBK1, p-IRF3, IRF3, p-p65, p65, and Myc tag and the secondary antibody goat anti-rabbit IgG-horseradish peroxidase (HRP) were purchased from Cell Signal Technology. Mouse monoclonal anti-HCV core antibody was a gift from Chen Liu. Goat anti-mouse IgG-HRP secondary antibody was purchased from Merck.
Real-time PCR assay.
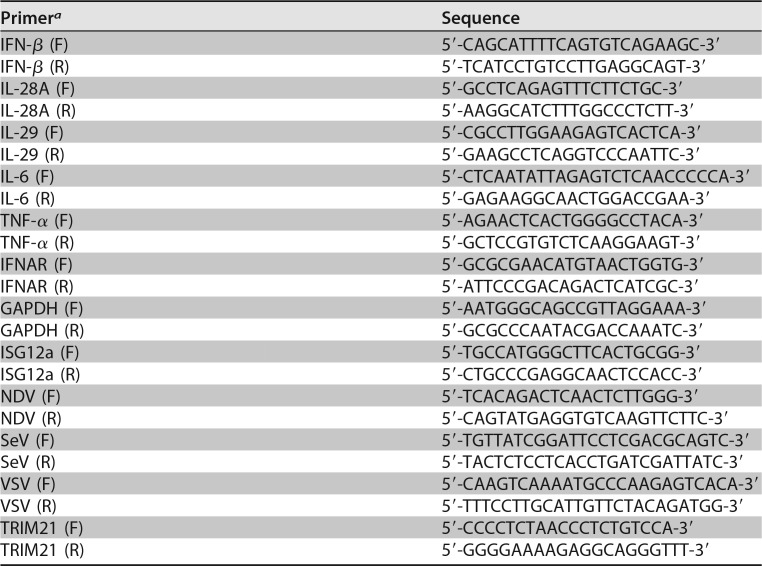
Total cellular RNA was extracted by using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The superscript III first-strand synthesis kit for reverse transcription of RNA was purchased from Invitrogen. After RQ1 DNase (Promega) treatment, the extracted RNA was used as the template for reverse transcription-PCR. The real-time PCR was performed as described previously (51). For absolute quantification analysis of HCV RNA, the standard curve was established by in vitro-transcribed JFH1 RNA. The primers for TRIM21, SeV, VSV, NDV, IFN-β, IL-28A, IL-29, TNF-α, IL-6, and ISG12a are listed in Table 2. The primers for detection of genomic HCV RNA were described previously (72).
TABLE 2.
Primers for real-time PCR
aF, forward; R, reverse.
Western blotting.
Cells were washed with PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer containing 150 mM sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris-HCl (pH 8.0) and supplemented with 2 μg/ml of aprotinin, 2 μg/ml of leupeptin, 20 μg/ml of phenylmethylsulfonyl fluoride, and 2 mM dithiothreitol (DTT). Proteins were resolved on SDS-PAGE gels, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with appropriate primary and secondary antibodies. The bound antibodies were detected by using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Luciferase assay.
The luciferase activity assay results were measured with the Dual-Luciferase Reports assay system (Promega) according to the manufacturer's instructions. The luciferase activity in cells was normalized to the protein concentration determined by Bradford assays.
IP and immunoblotting.
Cells were washed thrice with ice-cold PBS and lysed in lysis buffer supplemented with protease inhibitor cocktail. The cell lysates were incubated at 4°C for 30 min and centrifuged at 12,000 × g at 4°C for 15 min. The lysates were diluted to a concentration of 2 μg/μl with PBS before IP. The lysates (200 μg) were immunoprecipitated with the indicated antibodies. The immunocomplex was captured by adding protein G agarose bead slurry. The protein binding to the beads was boiled in 2× Laemmli sample buffer and was then subjected to SDS-10% PAGE.
Ubiquitination assays and native PAGE.
For analysis of the ubiquitination of MAVS in HEK293T cells, the cells were cotransfected with V5-TRIM21, HA-ub WT or HA-ub mutants, or Flag-MAVS/Myc-MAVS or Myc-MAVS mutants. Whole-cell lysis extracts were immunoprecipitated with anti-Flag or anti-Myc and then analyzed by immunoblotting with the anti-HA antibody. For analysis of the ubiquitination of endogenous MAVS, HLCZ01 cells were infected by NDV and SeV. Huh7.5.1(MAVS-C508R) cells were infected by HCV after transfection with HA-ub WT or HA-ub mutants. Cell lysates were immunoprecipitated with anti-MAVS and analyzed by immunoblotting with the anti-HA antibody.
Native PAGE for the detection of IRF3 dimerization was performed on a 7.5% acrylamide gel without SDS. Cells were harvested with 60 μl ice-cold lysis buffer, including 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.5% NP-40-containing protease inhibitor cocktail. After centrifugation at 13,000 × g for 15 min, proteins in the supernatant were quantified and diluted with 5× native PAGE sample buffer (312.5 mM Tris-HCl, pH 6.8, 75% glycerol, 0.25% bromophenol blue). The gel was prerun for 30 min at 40 mA on ice with 25 mM Tris-HCl (pH 8.4), and 192 mM glycine with and without 1% deoxycholate in the cathode chamber and anode chamber, respectively. The unboiled total protein was added to the gel for 80 min at 25 mA on ice.
Bioinformatics analysis of protein-protein interactions.
Bioinformatics analysis was described in our previous study (19).
Statistical analysis.
All results are presented as means ± standard deviations. Comparisons between two groups were performed using Student's t test, as indicated by asterisks in some of the figures.
ACKNOWLEDGMENTS
We thank Charles M. Rice for the Huh7.5 cells; Jin Zhong for Huh7.5.1 (MAVS WT) and Huh7.5.1(MAVS-C508R) cells; Takaji Wakita for pJFH1 and pJFH1/GND; and Xuetao Cao for SeV, VSV, and HSV. We also thank Chen Liu, Hongbing Shu, Jianguo Wu, Zhengfan Jiang, Deyin Guo, and Chengjiang Gao for kindly sharing research materials.
This work was supported by the National Natural Science Foundation of China (81730064, 81571985, and 31571368), the National Science and Technology Major Project (2017ZX10202201), and the Project of Innovation-Driven Plan of Central South University (2016CX031).
REFERENCES
- 1.Liu J, Cao X. 2016. Cellular and molecular regulation of innate inflammatory responses. Cell Mol Immunol 13:711–721. doi: 10.1038/cmi.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao X. 2016. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol 16:35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Shi H, Wu J, Chen ZJ, Chen C. 2015. Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proc Natl Acad Sci U S A 112:8947–8952. doi: 10.1073/pnas.1507317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 11.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 12.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 15.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellecave P, Sarasin-Filipowicz M, Donze O, Kennel A, Gouttenoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T, Moradpour D, Heim MH. 2010. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology 51:1127–1136. doi: 10.1002/hep.23426. [DOI] [PubMed] [Google Scholar]
- 17.Ding Q, Huang B, Lu J, Liu YJ, Zhong J. 2012. Hepatitis C virus NS3/4A protease blocks IL-28 production. Eur J Immunol 42:2374–2382. doi: 10.1002/eji.201242388. [DOI] [PubMed] [Google Scholar]
- 18.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Y, Xue B, Liu C, Wang X, Tian R, Xie Q, Guo M, Li G, Yang D, Zhu H. 2017. NLRX1 mediates MAVS degradation to attenuate hepatitis C virus-induced innate immune response through PCBP2. J Virol 91:e01264-17. doi: 10.1128/JVI.01264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. 2009. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Tong X, Ye X. 2012. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol 189:5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 22.Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, Kim TH, Sun Kim Y, Lee Y, Kim CJ, Jung JU, Lee JS, Cho H. 2015. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun 6:7910. doi: 10.1038/ncomms8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura T, Jain A, Choi SW, Mandell MA, Johansen T, Deretic V. 2017. TRIM-directed selective autophagy regulates immune activation. Autophagy 13:989–990. doi: 10.1080/15548627.2016.1154254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. 2010. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci U S A 107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bharaj P, Wang YE, Dawes BE, Yun TE, Park A, Yen B, Basler CF, Freiberg AN, Lee B, Rajsbaum R. 2016. The matrix protein of Nipah virus targets the E3-ubiquitin ligase TRIM6 to inhibit the IKKepsilon kinase-mediated type-I IFN antiviral response. PLoS Pathog 12:e1005880. doi: 10.1371/journal.ppat.1005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 27.Lascano J, Uchil PD, Mothes W, Luban J. 2016. TRIM5 retroviral restriction activity correlates with the ability to induce innate immune signaling. J Virol 90:308–316. doi: 10.1128/JVI.02496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, Mothes W. 2013. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol 87:257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J, Li Q, Mao AP, Hu MM, Shu HB. 2014. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J Mol Cell Biol 6:154–163. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 31.Ye W, Hu MM, Lei CQ, Zhou Q, Lin H, Sun MS, Shu HB. 2017. TRIM8 negatively regulates TLR3/4-mediated innate immune response by blocking TRIF-TBK1 interaction. J Immunol 199:1856–1864. doi: 10.4049/jimmunol.1601647. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Jia X, Xue Q, Dou Z, Ma Y, Zhao Z, Jiang Z, He B, Jin Q, Wang J. 2014. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc Natl Acad Sci U S A 111:E245–E254. doi: 10.1073/pnas.1316941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu MM, Yang Q, Zhang J, Liu SM, Zhang Y, Lin H, Huang ZF, Wang YY, Zhang XD, Zhong B, Shu HB. 2014. TRIM38 inhibits TNFalpha- and IL-1beta-triggered NF-kappaB activation by mediating lysosome-dependent degradation of TAB2/3. Proc Natl Acad Sci U S A 111:1509–1514. doi: 10.1073/pnas.1318227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, Sun S, Xiang C, Sun B. 2008. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol 9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Q, Hou J, Zhou Y, Yang Y, Xie B, Cao X. 2015. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res 25:1121–1136. doi: 10.1038/cr.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, Zhang L, Gao C. 2017. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol 18:214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 37.Yang B, Wang J, Wang Y, Zhou H, Wu X, Tian Z, Sun B. 2013. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol 190:3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 38.Ozato K, Shin DM, Chang TH, Morse HC III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oke V, Wahren-Herlenius M. 2012. The immunobiology of Ro52 (TRIM21) in autoimmunity: a critical review. J Autoimmun 39:77–82. doi: 10.1016/j.jaut.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Rajsbaum R, Stoye JP, O'Garra A. 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol 38:619–630. doi: 10.1002/eji.200737916. [DOI] [PubMed] [Google Scholar]
- 41.Gao X, Xu F, Zhang HT, Chen M, Huang W, Zhang Q, Zeng Q, Liu L. 2016. PKCalpha-GSK3beta-NF-kappaB signaling pathway and the possible involvement of TRIM21 in TRAIL-induced apoptosis. Biochem Cell Biol 94:256–264. doi: 10.1139/bcb-2016-0009. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen JQ, Irby RB. 2017. TRIM21 is a novel regulator of Par-4 in colon and pancreatic cancer cells. Cancer Biol Ther 18:16–25. doi: 10.1080/15384047.2016.1252880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol 181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang K, Shi HX, Liu XY, Shan YF, Wei B, Chen S, Wang C. 2009. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol 182:3782–3792. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 45.Cao X, Ding Q, Lu J, Tao W, Huang B, Zhao Y, Niu J, Liu YJ, Zhong J. 2015. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J Hepatol 62:771–778. doi: 10.1016/j.jhep.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uzri D, Gehrke L. 2009. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol 83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, Yu R, Qin Y, Gao Y, Wang Q, Hu J, Wang L, Zhou Z, Liu B, Tan D, Guan Y, Zhu H. 2014. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci U S A 111:E1264–E1273. doi: 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu N, Long Y, Liu B, Yang D, Li C, Chen T, Wang X, Liu C, Zhu H. 2014. ISG12a mediates cell response to Newcastle disease viral infection. Virology 462–463:283–294. doi: 10.1016/j.virol.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Xue B, Yang D, Wang J, Xu Y, Wang X, Qin Y, Tian R, Chen S, Xie Q, Liu N, Zhu H. 2016. ISG12a restricts hepatitis C virus infection through the ubiquitination-dependent degradation pathway. J Virol 90:6832–6845. doi: 10.1128/JVI.00352-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li K, Lemon SM. 2013. Innate immune responses in hepatitis C virus infection. Semin Immunopathol 35:53–72. doi: 10.1007/s00281-012-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell S, Vargas J, Hoffmann A. 2016. Signaling via the NFkappaB system. Wiley Interdiscip Rev Syst Biol Med 8:227–241. doi: 10.1002/wsbm.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q, Huang L, Hong Z, Lv Z, Mao Z, Tang Y, Kong X, Li S, Cui Y, Liu H, Zhang L, Zhang X, Jiang L, Wang C, Zhou Q. 2017. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog 13:e1006264. doi: 10.1371/journal.ppat.1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, Yu H, Pan Y, Wang C. 2014. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41:919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 56.Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, Liuyu T, Zhang ZD, Xiong TC, Wu Y, Zhu QY, Yao J, Shu HB, Lin D, Zhong B. 2017. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat Commun 8:15534. doi: 10.1038/ncomms15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McEwan WA, Hauler F, Williams CR, Bidgood SR, Mallery DL, Crowther RA, James LC. 2012. Regulation of virus neutralization and the persistent fraction by TRIM21. J Virol 86:8482–8491. doi: 10.1128/JVI.00728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. 2013. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol 14:327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaysburd M, Watkinson RE, Cooper H, Reed M, O'Connell K, Smith J, Cruickshanks J, James LC. 2013. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proc Natl Acad Sci U S A 110:12397–12401. doi: 10.1073/pnas.1301918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. 2013. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol 14:172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol 7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 62.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, Von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pourmand N, Blange I, Ringertz N, Pettersson I. 1998. Intracellular localisation of the Ro 52kD auto-antigen in HeLa cells visualised with green fluorescent protein chimeras. Autoimmunity 28:225–233. doi: 10.3109/08916939808995370. [DOI] [PubMed] [Google Scholar]
- 64.Kim MJ, Yoo JY. 2010. Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J Immunol 185:4311–4318. doi: 10.4049/jimmunol.1000098. [DOI] [PubMed] [Google Scholar]
- 65.Zhu H, Zhao H, Collins CD, Eckenrode SE, Run Q, McIndoe RA, Crawford JM, Nelson DR, She JX, Liu C. 2003. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology 37:1180–1188. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]
- 66.Shaheen M, Shanmugam I, Hromas R. 2010. The role of PCNA posttranslational modifications in translesion synthesis. J Nucleic Acids 2010:761217. doi: 10.4061/2010/761217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D. 2012. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 10:44. doi: 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia X, Zhou H, Wu C, Wu Q, Ma S, Wei C, Cao Y, Song J, Zhong H, Zhou Z, Wang J. 2017. The ubiquitin ligase RNF125 targets innate immune adaptor protein TRIM14 for ubiquitination and degradation. J Immunol 198:4652–4658. doi: 10.4049/jimmunol.1601322. [DOI] [PubMed] [Google Scholar]
- 69.Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. 2014. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol 192:4758–4764. doi: 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 70.Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. 2010. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol 184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 71.Liu H, Zhang LL, Sun J, Chen W, Li SL, Wang Q, Yu HS, Xia ZX, Jin X, Wang C. 2017. Endoplasmic reticulum protein SCAP inhibits dengue virus NS2B3 protease by suppressing its K27-linked polyubiquitylation. J Virol 91:e02234-. doi: 10.1128/JVI.02234-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang D, Meng X, Xue B, Liu N, Wang X, Zhu H. 2014. MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One 9:e94501. doi: 10.1371/journal.pone.0094501. [DOI] [PMC free article] [PubMed] [Google Scholar]