ABSTRACT
Rotaviruses (RVs), which cause severe gastroenteritis in infants and children, recognize glycan ligands in a genotype-dependent manner via the distal VP8* head of the spike protein VP4. However, the glycan binding mechanisms remain elusive for the P[II] genogroup RVs, including the widely prevalent human RVs (P[8], P[4], and P[6]) and a rare P[19] RV. In this study, we characterized the glycan binding specificities of human and porcine P[6]/P[19] RV VP8*s and found that the P[II] genogroup RV VP8*s could commonly interact with mucin core 2, which may play an important role in RV evolution and cross-species transmission. We determined the first P[6] VP8* structure, as well as the complex structures of human P[19] VP8*, with core 2 and lacto-N-tetraose (LNT). A glycan binding site was identified in human P[19] VP8*. Structural superimposition and sequence alignment revealed the conservation of the glycan binding site in the P[II] genogroup RV VP8*s. Our data provide significant insight into the glycan binding specificity and glycan binding mechanism of the P[II] genogroup RV VP8*s, which could help in understanding RV evolution, transmission, and epidemiology and in vaccine development.
IMPORTANCE Rotaviruses (RVs), belonging to the family Reoviridae, are double-stranded RNA viruses that cause acute gastroenteritis in children and animals worldwide. Depending on the phylogeny of the VP8* sequences, P[6] and P[19] RVs are grouped into genogroup II, together with P[4] and P[8], which are widely prevalent in humans. In this study, we characterized the glycan binding specificities of human and porcine P[6]/P[19] RV VP8*s, determined the crystal structure of P[6] VP8*, and uncovered the glycan binding pattern in P[19] VP8*, revealing a conserved glycan binding site in the VP8*s of P[II] genogroup RVs by structural superimposition and sequence alignment. Our data suggested that mucin core 2 may play an important role in P[II] RV evolution and cross-species transmission. These data provide insight into the cell attachment, infection, epidemiology, and evolution of P[II] genogroup RVs, which could help in developing control and prevention strategies against RVs.
KEYWORDS: P[6] rotavirus, P[19] rotavirus, glycan binding specificity, VP8* structure, mucin core 2, lacto-N-tetraose (LNT)
INTRODUCTION
Species A rotaviruses (RAVs) are a major cause of acute gastroenteritis in humans and animals, resulting in ∼215 000 deaths annually in children under 5 years of age worldwide (1). The rotavirus (RV) genome contains 11 double-stranded RNA segments, which encode 12 proteins, including 6 structural and 6 nonstructural proteins (2). RAVs have been divided into G and P genotypes according to the VP7 and VP4 genes, respectively (3). VP4 is posttranslationally cleaved into VP5* and VP8*. RAVs consist of 50 P genotypes so far (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/7th-RCWG-meeting), some of which have been grouped into five genogroups (P[I] to P[V]) by the phylogeny of VP8* sequences (4). VP8* is important for cell attachment, playing a critical role in virus infection (5). Previous studies showed that while some animal RVs are sialidase sensitive, the majority of human and animal RVs are sialidase insensitive (6). It was found by nuclear magnetic resonance (NMR) spectroscopy that the sialidase-insensitive human RV Wa could interact with ganglioside GM1 (7), indicating that sialic acids (Sia) may also be involved in receptor binding of some sialidase-insensitive RVs. Several potential RV receptors, including heat shock protein (8, 9), gangliosides (10–12), and integrins (13, 14), have also been reported; however, their roles in RV attachment and penetration remain elusive. Thus, our understanding of the interactions between RVs and their receptors remains limited.
Recently, histo-blood group antigens (HBGAs) have been reported to interact with RVs as cell attachment factors (4, 15, 16), which significantly advanced our knowledge of RV evolution and epidemiology and provided a P-type-based vaccine approach. HBGAs are highly polymorphic, with different ABO, Lewis, and secretor types (17). The major human RV genotypes in genogroups P[II], P[III], and P[IV] recognize HBGAs in a genotype-dependent manner (15). P[11] VP8* in the P[IV] genogroup binds to type 1 (lacto-N-tetraose [LNT]) and type 2 (lacto-N-neotetraose [LNnT]) precursors (18, 19). The VP8* proteins of P[14], P[9], and P[25] RVs in the P[III] genogroup interact with A-type HBGA (4, 16). Human P[8] and P[4] VP8*s in the P[II] genogroup were previously reported to recognize Lewis b and H type 1 HBGAs, while the P[6] VP8* binds only H type 1 (15, 20). However, as shown in a saturation transfer difference (STD) NMR-based study, the VP8* proteins of human P[4] (strain DS-1) and P[6] (strain RV-3) RVs interact with the A-type HBGAs via α1-2 fucose, whereas the P[8] VP8* (strain Wa) does not bind the A or Lewis b/H type 1 antigens (21). These results indicate that more studies are needed to elucidate the exact interaction between P[II] genogroup RVs and HBGAs.
P[6] and P[19] RVs are grouped into the P[II] genogroup, together with P[4] and P[8] (4). P[4] and P[8] RVs are the most common P genotypes in humans, accounting for over 90% of RV infections (22–24). P[6] RVs are prevalent in both humans and pigs, and some human P[6] RVs were found to be porcine like (25–28), while P[19] RVs are common in pigs and infect humans sporadically (29). In view of the prevalence of P[6]/P[19] RVs in humans and pigs, they may be good models to study RV cross-species transmission. A recent study (30) showed that, unlike human P[6], human P[19]/P[4]/P[8] RV VP8*s bind to mucin cores of mucin O-glycans with the disaccharide structure GlcNAcβ1-6GalNAc, particularly mucin core 2 (30), which further complicated the receptor binding mechanisms of P[II] genogroup RVs.
Determining the VP8*-glycan complex structures is crucial to understanding the mechanism of glycan binding specificity of RVs. Previous structural studies have revealed different binding patterns among human RV P genotypes. Structural analysis showed that P[9] (31) and P[14] (16) RVs bind to A-type HBGA using a binding site at the same location as that of sialic acid-binding P[3] (32). Meanwhile, a relatively wide binding pocket for LNT/LNnT close to the former site of A-type HBGA and sialic acid was observed in P[11] (33). During the preparation of our manuscript, a paper reported a distinct binding cavity in human P[19] RV (34). In this study, we focused on characterization of the glycan binding specificities of human and porcine P[6]/P[19] RV VP8*s and explored the structural basis for the glycan binding specificity of P[II] genogroup RVs.
RESULTS
Glycan binding specificities of P[6] and P[19] RV VP8*s.
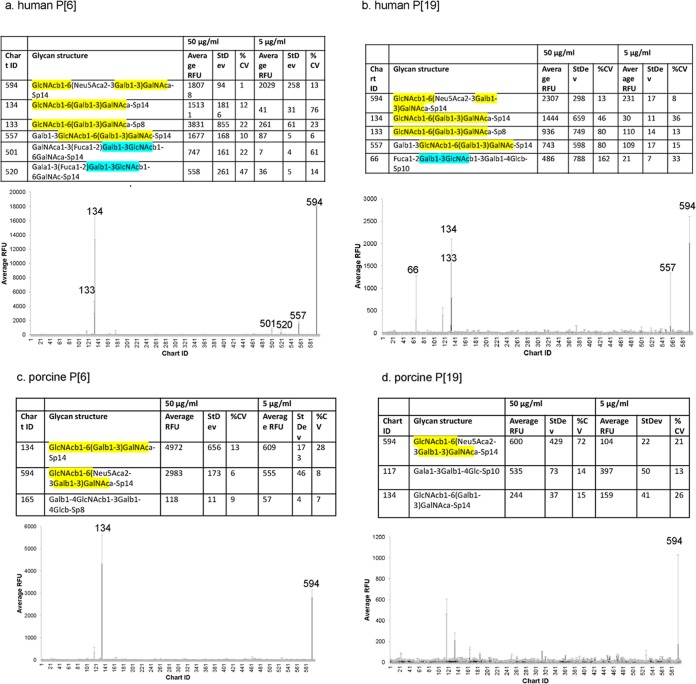
To explore glycan binding specificity, VP8* proteins of human and porcine P[6] and P[19] RVs were subjected to glycan microarray analysis against a library containing 600 glycans. Porcine-like human P[6] RV (strain LL3354) (Fig. 1a) bound to mucin core 2 with a trisaccharide sequence, GlcNAcβ1-6(Galβ1-3)GalNAc (GlcNAc, Gal, and GalNAc stand for N-acetylglucosamine, galactose, and N-acetylgalactosamine, respectively), which is α-linked to Thr (Sp14, probe 134) or to propylamine (Sp8, probe 133). Core 2 extended by a sialic acid (Neu5Ac) from the 3-linked Gal (probe 594) or by a galactose from the 6-linked GlcNAc (probe 557) also showed good binding signals. Human P[19] RV (Mc345) VP8* showed a binding mode similar to that of mucin core 2 (probes 133 and 134) or core 2 extensions (probes 557 and 594) (Fig. 1b). In addition, LL3354 P[6] RV VP8* recognized blood group H pentasaccharide (type I HBGA) with a Galβ1-3GlcNAc motif (probes 501 and 502) (Fig. 1b, cyan). Binding to type I HBGA with a Galβ1-3GlcNAc motif (probe 66) was also detected for human P[19] VP8*. VP8* proteins of the porcine P[6] RV (strain z84) (Fig. 1c) bound to core 2 (probe 134) (Fig. 1, yellow) and core 2 extended by a Neu5Ac from the 3-linked Gal (probe 594), while porcine P[19] showed binding to probe 594 (Fig. 1d).
FIG 1.
P[6] and P[19] VP8*s interact with mucin core 2 in the glycan microarray assay. The VP8*-GST fusion proteins of porcine-like human P[6] (LL3354) (a), human P[19] (b), porcine P[6] (z84) (c), and porcine P[19] (d) were subjected to glycan microarray assay at 50 μg/ml and 5 μg/ml against 600 glycans. The glycan binding profile at 50 μg/ml and a list of glycans recognized by the proteins with the highest RFUs are shown. The numbers on the x axis are probe numbers from the array used for the screen. StdDev represents the standard deviations from four replicate wells. %CV = 100 × standard deviation/mean. Mucin core 2 with a trisaccharide sequence of GlcNAcβ1-6(Galβ1-3)GalNAc is shaded in yellow. Glycans with a Galβ1-3GlcNAc (type 1 HBGA core) motif are shaded in cyan.
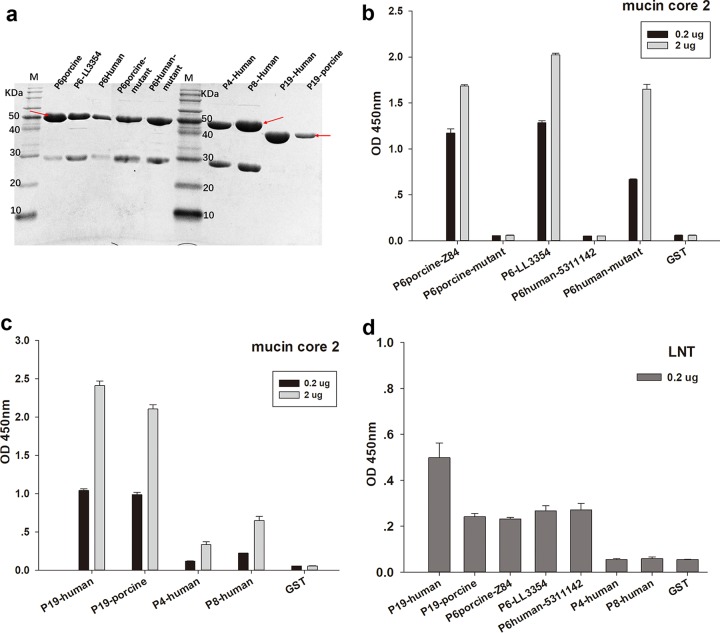
We obtained the purified glutathione S-transferase (GST)-VP8* fusion proteins with a molecular weight of ∼52 kDa (∼46 kDa for GST-P[19] VP8* core proteins) (Fig. 2a). Subsequent enzyme-linked immunosorbent assay (ELISA)-based glycan binding assays showed that P[6] RV VP8*s of porcine strain z84 and porcine-like human strain LL3354 recognized mucin core 2 with good binding signals, while human P[6] RV (5311142) VP8* did not. Meanwhile, VP8* of a porcine P[6] mutant showed no binding to mucin core 2, and a human P[6] mutant bound to mucin core 2 with high intensity (Fig. 2b). Two concentrations of mucin core 2 were subjected to the assay to further confirm the binding. VP8* proteins of human P[19], porcine P[19], human P[4] (11151099), and human P[8] (11221075) also showed good binding to mucin core 2 (Fig. 2c). In addition, VP8* proteins of human P[6], LL3354 P[6], porcine P[6], human P[19], and porcine P[19] displayed binding to a type I HBGA precursor with a Galβ1-3GlcNAc motif (LNT), whereas human P[4] and P[8] did not (Fig. 2d).
FIG 2.
Glycan binding specificities of P[6]/P[19]/P[4]/P[8] VP8*s. VP8* proteins of human P[19], porcine P[19], porcine P[6] (z84), a porcine P[6] mutant, porcine-like human P[6] (LL3354), the prevalent human P[6] (5311142), a human P[6] (5311142) mutant, human P[4] (11151099), and human P[8] (11221075) were subjected to the microtiter plate binding assay. GST protein was used as the negative control. (a) SDS-PAGE of the GST-VP8* fusion proteins. The red arrows indicate the proteins of interest. The 26-kDa protein was free GST. (b) ELISA-based assay of P[6] VP8* protein binding to two concentrations of mucin core 2 (0.2 μg and 2 μg). (c) ELISA-based assay of P[19]/P[4]/P[8] VP8* protein binding to mucin core 2 (0.2 μg and 2 μg). (d) ELISA-based assay of VP8* protein binding to LNT (Galβ1-3GlcNAcβ1-3Galβ1-4Glc) at 0.2 μg. The error bars indicate standard errors from triplicate repeats.
Structural characterization of porcine P[6] VP8*.
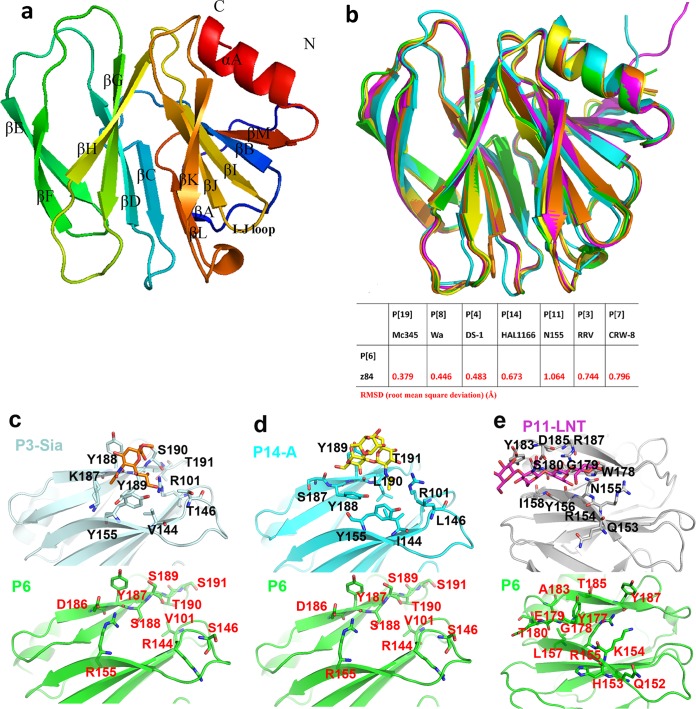
VP8* crystal structures of P[4] (35), P[8] (36, 37), and P[19] (34, 38) in the P[II] genogroup have been solved previously. Here, we determined the crystal structure of a porcine P[6] (z84) VP8* for the first time at a high resolution of 1.8 Å (Protein Data Bank [PDB] identifier [ID] 5YMU) (Table 1). Similar to other known VP8* structures of the P[II] genogroup, porcine P[6] VP8* showed a typical galectin-like fold (32) consisting of two twisted antiparallel β-sheets comprising strands A, L, C, D, G, and H and M, B, I, J, and K, respectively (Fig. 3a). The two β-sheets were separated by a shallow cleft. According to the structural superimposition, P[6] VP8* most closely resembled the P[19] VP8* (Fig. 3b), with a root mean square deviation (RMSD) value of 0.379; it also resembled P[8] and P[4] VP8*s, with RMSD values of 0.446 and 0.483, respectively (Fig. 3b), which is consistent with the sequence phylogenetic analysis (4). Structural analysis showed that the corresponding amino acids involved in Sia binding (Arg101, Val144, Thr146, Tyr155, Lys187, Tyr188, Tyr189, Ser190, and Thr191) in P[3] VP8* (Fig. 3c) and A-type HBGA binding (Arg101, Ile144, Leu146, Tyr155, Ser187, Tyr188, Tyr189, Leu190, and Thr191) in P[14] VP8* (Fig. 3d) were altered to different amino acids in P[6] VP8* (Val101, Arg144, Ser146, Arg155, Asp186, Tyr187, Ser188, Ser189, Thr190, and Ser191) and displayed different conformations in P[6] VP8* (Fig. 3c and d). Amino acids involved in LNT binding (153- to 156QRNY, 158I, 178- to 180WGS, 183Y, 185D, and 187R) in P[11] VP8* were altered in P[6] VP8* (152- to 155QHKR, 157L, 177- to180 GET, 183A, 185T, and 187Y) and displayed various orientations (Fig. 3e). These results showed that the previously identified receptor binding sites may not be used in P[6] VP8*.
TABLE 1.
Crystallographic X-ray diffraction and refinement statistics
| Parameter | Valuea |
||
|---|---|---|---|
| P[19] VP8*-core 2 | P[19] VP8*-LNT | P[6] VP8* | |
| Data collection | |||
| Space group | C121 | P32 | H3 |
| Cell dimensions (Å) | |||
| a | 179.776 | 131.667 | 131.396 |
| b | 129.346 | 131.667 | 131.396 |
| c | 86.365 | 150.530 | 123.592 |
| α, β, γ (°) | 90, 116, 90 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50.00–2.10 (2.18–2.10) | 50.00–2.20 (2.28–2.20) | 50.00–1.80 (1.86–1.80) |
| Rmerge (%)b | 0.164 (1.059) | 0.115 (1.029) | 0.115 (1.156) |
| I /σI | 10.575 (1.717) | 15.231 (2.060) | 21.895 (2.750) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.7) | 100.0 (100.0) |
| Redundancy | 6.3 (6.4) | 6.0 (5.7) | 10.5 (10.6) |
| Refinement | |||
| Resolution (Å) | 49.68–2.10 | 39.91–2.20 | 41.86–1.80 |
| No. of reflections | 98,627 | 132,870 | 73,566 |
| Rwork/Rfree | 0.1812/0.2130 | 0.2261/0.2669 | 0.1663/0.1892 |
| No. of atoms | |||
| Protein | 10,270 | 17,914 | 5,260 |
| Ligand/ion | 164 | 144 | 0 |
| Water | 982 | 664 | 692 |
| B-factors | |||
| Protein | 31.06 | 35.34 | 22.44 |
| Water | 36.76 | 33.92 | 35.81 |
| Ligand | 58.60 | 54.03 | 0 |
| RMSDs | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.005 |
| Bond angles (°) | 1.056 | 0.905 | 0.921 |
| Ramachandran plot | |||
| Favored (%) | 97.0 | 95.84 | 96.2 |
| Allowed (%) | 3.00 | 4.16 | 3.64 |
| Disallowed (%) | 0.00 | 0.00 | 0.16 |
Values in parentheses are for the highest-resolution shell.
Rmerge = Σhkl |I − <I>|/ΣhklI, where I is the intensity of unique relfection hkl and <I> is the average over symmetry-related observations of unique reflection hkl; hkl are the reflection indices.
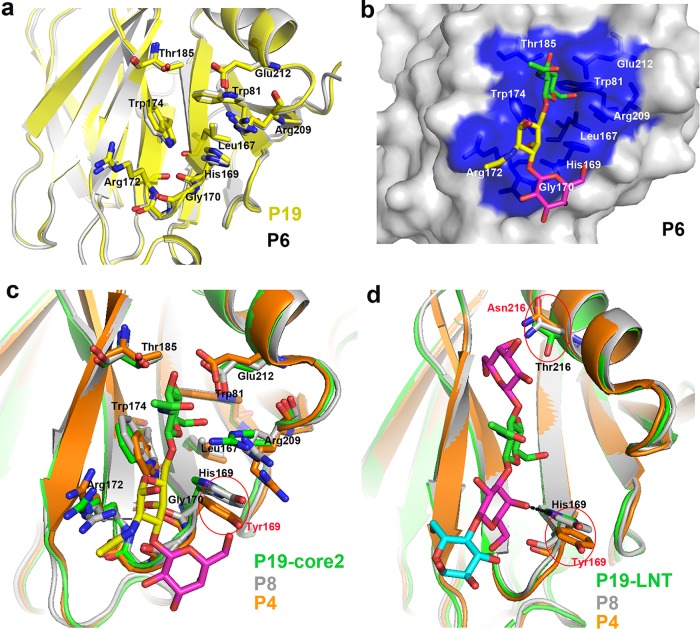
FIG 3.
Structural characteristics of P[6] VP8*. (a) Overall structure of porcine P[6] VP8* with two twisted antiparallel sheets consisting of strands A, L, D, G, and H and M, B, I, J, and K. (b) (Top) Superimposition of porcine P[6] VP8* structure (PDB ID 5YMU; green) on those of P[19] (PDB ID 5GJ6; pale cyan), Wa P[8] (PDB ID 2DWR; yellow), DS-1 P[4] (PDB ID 2AEN; orange), and HAL1166 P[14] (PDB ID 4DRV; cyan). (Bottom) Structure comparison of porcine P[6] (5YMU), RRV P[3] (1KQR), CRW-7 P[7] (2I2S), HAL1166 P[14] (4DRV), DS-1 P[4] (2AEN), Wa P[8] (2DWR), HRV P[11] (4YG0), andMc345 P[19] (5GJ6). The values in the table represent RMSDs (in Å) of the Cα atoms of one VP8* monomer. VP8* structures are shown as a cartoon. (c) Comparison of the residues involved in sialic acid binding in P[3] VP8* (PDB ID 1KQR) (top; pale cyan) and that of porcine P[6] VP8* structure (PDB ID 5YMU) (bottom; green). The amino acids involved in the interaction are shown in stick representation. (d) Comparison of the residues involved in A-type HBGA binding in P[14] VP8* (PDB ID 4DRV) (top; cyan) and that of porcine P[6] VP8* structure (PDB ID 5YMU) (bottom; green). (e) Comparison of the residues involved in LNT binding in P[11] VP8* (PDB ID 4YFZ) (top; gray) and that of porcine P[6] VP8* structure (PDB ID 5YMU) (bottom, green).
Structural basis for binding of human P[19] VP8* to mucin core 2 and LNT.
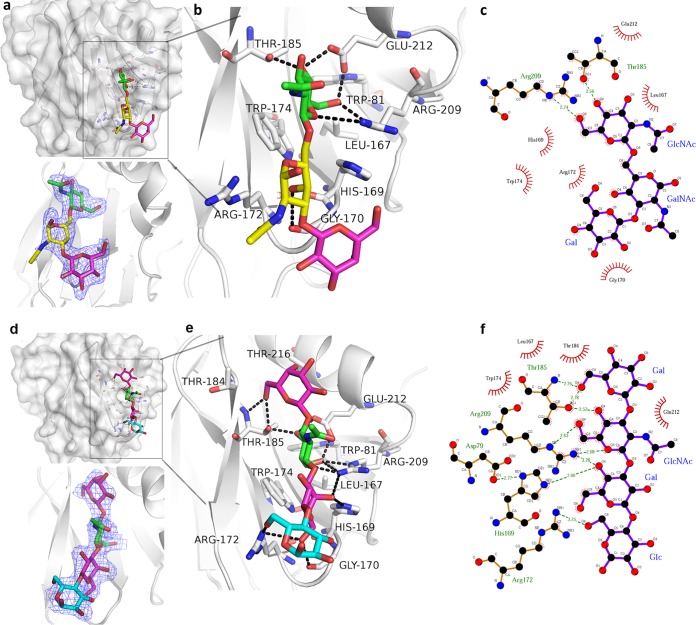
To understand the structural basis of the observed glycan binding specificity of RV VP8*, we crystalized the P[19] VP8* complex with core 2 and LNT, followed by X-ray crystallography. The P[19] VP8*-core 2 structure was determined at 2.1 Å (PDB ID 5YMS) (Table 1), and the density of the bound core 2 was clearly observed in the simulated annealing omit map (Fig. 4a). The glycan binding site was clearly seen. Its edge was composed of β-strand K, the I-J loop, and the 210 loop (residues 209 to 212, which links β-strand M and α-helix A), while the base was formed by 3 residues, Trp81, Leu167, and Trp174 (Fig. 4b). Core 2 was stabilized by a network of hydrogen bonds and hydrophobic interactions (Fig. 4b and c). Core 2 interacted with residues Gly170, Thr185, Arg209, and Glu212 via hydrogen bonds. The main interaction focused on GlcNAc, which formed three hydrogen bonds. GlcNAc also interacted with Trp81, Leu167, His169, Trp174, and Glu212 by hydrophobic interactions. GalNAc formed a hydrogen bond with Gly170 and hydrophobic interactions with Gly170/Arg172. The terminal Gal residue of core 2 formed hydrophobic interactions with Gly170.
FIG 4.
Structures of P[19] VP8*-core 2 and P[19] VP8*-LNT complex. (a and d) Surface representation of the P[19] VP8*-core 2 (a) and P[19] VP8*-LNT (d) complex structures. The monosaccharide residues of core 2 and the amino acids involved in the interaction are shown in stick representation. The binding cavity is colored red in the surface representation. Electron density maps of core 2 (a) and LNT (d) in P[19] VP8* structure, contoured at 1σ level are shown at the bottom. VP8*, gray; GlcNAc, green; GalNAc, yellow; Gal, magenta; Glc, cyan. (b and e) Detailed interactions among the amino acids of VP8*-core 2 (b) and VP8*-LNT (e) with the hydrogen bonds represented by dotted lines. The interactions were first searched by CCP4 (http://www.ccp4.ac.uk/) with a distance of less than 5 Å, verified by LIGPLOT (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/), and then calculated with PyMOL (https://pymol.org/2/). (c and f) Schematic diagrams of the interactions. The details of the interaction between core 2 (c)/LNT (f) and P[19] VP8* were calculated with LIGPLOT (https://www.ebi.ac.uk/thornton-srv/software/LIGPLOT/). The interactions shown are those mediated by hydrogen bonds and by hydrophobic contacts. Hydrogen bonds are indicated by dashed lines between the atoms involved, while hydrophobic contacts are represented by arcs with spokes radiating toward the ligand atoms they contact. The contacted atoms are shown with spokes radiating back. Atoms colored black, red, and blue represent carbon, oxygen, and nitrogen, respectively.
The structure of the P[19] VP8*-LNT complex was solved at 2.2 Å (PDB ID 5YMT) (Table 1), and the density for LNT was clearly visible (Fig. 4d). P[19] VP8* bound to LNT using the same binding cavity. Residues Trp81, Leu167, His169, Gly170, Arg172, Trp174, Thr184, Thr185, Arg209, Glu212, and Asn216 were involved in the interactions (Fig. 4e and f). For the nonreducing terminal Galβ1-3GlcNAc sequence, the Gal1 residue interacted with Thr185 via a hydrogen bond, and GlcNAc formed intensive hydrogen bonds with Thr185, Glu212, and Arg209. Hydrophobic interactions were observed in Gal1 with Thr184, Thr185, and Thr216 and in GlcNAc with Trp81, Leu167, Trp174, and Thr185 (Fig. 4e). The Gal3 of Galβ1-4Glc formed hydrophobic interactions with Gly170 and Trp174, further stabilized by hydrogen bond interactions with His169, Gly170, Arg172, and Arg209 (Fig. 4e). The Glc4 moiety formed one hydrogen bond with Arg172.
Structural characteristics of the glycan site in P[II] genogroup RV VP8*s.
Since P[6] VP8* was closest to P[19] VP8*, structural comparison was performed to explore the characteristics of the glycan binding site in P[6] VP8*. Structural analysis showed that the amino acids involved in ligand binding (Trp81, Leu167, His169, Gly170, Arg172, Trp174, Thr184, Thr185, Arg209, and Glu212) in P[6] VP8* displayed the same conformation as those in P[19] VP8* (Fig. 5a). Furthermore, core 2 was superimposed on the putative binding site of porcine P[6] VP8* in the surface representation (Fig. 5b), showing characteristics similar to those of P[19] VP8*-core 2. P[4]/P[8] VP8*s were also clustered in the same genogroup with P[19] VP8*. Structural superimposition of P[19]VP8*-core 2 and P[4]/P[8] VP8*s suggested that 9 residues (Trp81, Leu167, Gly170, Arg172, Trp174, Thr184, Thr185, Arg209, and Glu212) involved in core 2 binding displayed the same conformation, while 1 residue, His169, was altered to Tyr169 in P[4]/P[8] VP8*s (Fig. 5c). Structural analysis of P[19]-LNT/P[4]/P[8] showed that 2 residues (His169 and Thr216) were altered in P[4]/P[8] (Tyr169 and Asn216) (Fig. 5d).
FIG 5.
Structural characteristics of the glycan binding site in P[6]/P[4]/P[8] VP8*s. (a) Comparison of the conformations of the residues involved in glycan binding in P[19] (PDB ID 5GJ6; yellow) and that of porcine P[6] VP8* structure (PDB ID 5YMU; gray). The amino acids involved in the interaction are shown in stick representation. (b) Superimposition of core 2 on the putative binding site of porcine P[6] VP8* structure (PDB ID 5YMU; gray). P[6] VP8* is shown in surface representation, and the putative binding site is colored blue. (c) Structure superimposition of P[8] and P[4] VP8*s on P[19] VP8*-core 2. Core 2 is shown as sticks (GlcNAc, green; GalNAc, yellow; Gal, magenta;). Residues involved in the core 2 binding are shown as sticks. Tyr169 in P[4]/P[8] is circled and colored red. VP8*s of P[19], P[8], and P[4] are colored green, gray, and orange, respectively. (d) Structure superimposition of P[8] and P[4] VP8*s on P[19] VP8*-LNT. LNT is shown as sticks (Gal, magenta; GlcNAc, green; Glc, cyan). Residues 169 and 216 are shown as sticks. The hydrogen bond between His169 and LNT is shown as a black dotted line. Tyr169/Asn216 in P[4]/P[8] are circled and colored red. P[19], P[8], and P[4] are colored as in panel c.
Structural comparison of the glycan binding sites among RVs.
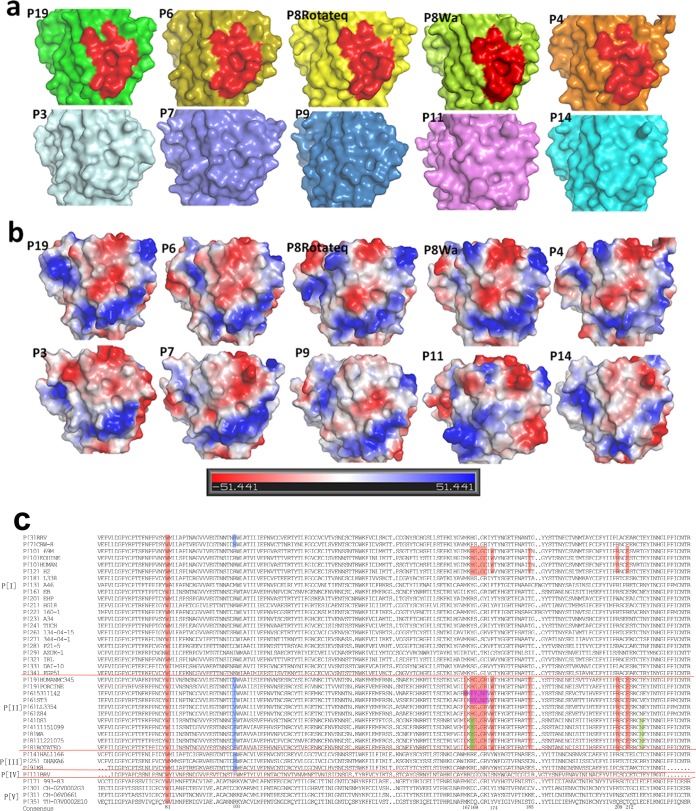
The VP8* crystal structures of several RV P genotypes, including P[3], P[7], P[9], P[11], P[14], P[4], P[8], and P[19], are known. Structural superimposition revealed that the glycan binding site identified in the P[19] RV VP8* was also observed in the corresponding sites of P[4] (DS-1), P[8] (Rotateq and Wa), and P[6] (z84) VP8*s (Fig. 6a, red). P[19] VP8* exhibited negative charge in the base of the glycan binding cavity. A portion of its wall also presented negative charge, while the remaining bottom part presented positive charge (Fig. 6b). P[6], P[8], and P[4] VP8*s showed similar charge configurations. The simian P[3] VP8* showed a distinct glycan binding site with a neutral upper portion, while porcine P[7] VP8* presented a partial cavity with strong negative charge on the upper portion. The VP8*s of the P[9], P[11], and P[14] genotypes displayed no obvious cavity in the corresponding sites, and their electrostatic potentials differed significantly (Fig. 6b).
FIG 6.
Structural comparison and sequence alignment of VP8*s of various P genotypes. (a and b) Surface (a) and electrostatic potential (b) representations of the putative binding cavities in the VP8* structures of different P genotypes. VP8*s of the following genotypes are included: P[19] (5GJ6), P[6] (5YMU), Rotateq P[8] (5JDB), Wa P[8] (2DWR), DS-1 P[4] (2AEN), P[3] (1KQR), CRW-7 P[7] (2I2S), P[9] (5CAZ), P[11] (4YG0), and P[14] (4DRV). The electrostatic potential was generated by vacuum electrostatics protein contact potential in PyMOL, and the calibration bar represents the intensity of the electrostatic potential. Red indicates negative charge, whereas blue indicates positive charge. (c) Sequence alignment of VP8* proteins (residues 61 to 230) among various P genotypes. The alignment was done with DNAMAN. The amino acids involved in the binding cavity with high conservation are shaded in red. The residues altered in prevalent human P[6] strains are highlighted in magenta. The Tyr169 and Asn216 in human P[4] and P[8] are colored green.
Sequence alignment of RV VP8* proteins.
Sequence alignments of VP8* proteins of different RV P genotypes were carried out with DNAMAN software (https://www.lynnon.com/). It showed that the residues involved in mucin core 2 binding in P[19] VP8* were totally different from those of P[11] (P[IV] genogroup) (Fig. 6c). For the VP8*s of other P genotypes, residue Trp81 was highly conserved and Arg209 was moderately conserved, while other residues varied between P[9] and P[14] (P[III] genogroup) and in P[3] and P[7] (P[I] genogroup) (Fig. 6c), indicating that the new glycan binding site observed in the P[19] RV is not present in these RV genotypes, consistent with the structural presentation in Fig. 6a. All of the key residues were rather conserved in VP8*s of human P[8], P[4], and P[19]; porcine-like human P[6]; porcine P[19]; and porcine P[6] (Fig. 6c, red). Moreover, P[10], which was reported to possess the same glycan binding specificity as human P[19] (30) and P[12] in the P[I] genogroup of animal origin, also shared conserved amino acids in the corresponding sites (Fig. 6c). Human P[6] VP8* displayed significant amino acid alterations in residues 169 to 172 (Fig. 6c, magenta). Try169 and Asn216 in human P[8] and P[4] VP8*s that were different from His169/Thr216 in P[19] VP8* are colored green in the figure.
DISCUSSION
Recent reports regarding RV recognition of HBGAs as potential receptors (4, 15, 16, 30) have been further supported by elucidations of different VP8* crystal structures in complex corresponding glycans. This broadened our understanding of RV-host interactions and RV diversity, evolution, and epidemiology. The P[14], P[9], and P[25] RVs of the P[III] genogroup, which have been detected frequently in humans and several wild and domestic animals, were found to interact with type A HBGAs, suggesting that the A antigens shared between humans and animals may play a role in the cross-species transmission of these RVs (4, 16). P[11] RVs of the P[IV] genogroup that commonly infect neonates and young infants recognize evolutionarily conserved type 1 (LNT) and type 2 (LNnT) precursors, providing evidence of an age-specific host range and cross-species transmission via shared HBGAs (33). In this study, we characterized glycan binding specificities of VP8* proteins of several RVs in the P[II] genogroup (P[8], P[4], P[6], and P[19]) and solved the crystal structures of P[6] VP8* and P[19] VP8* complexes to explore the interaction mechanisms.
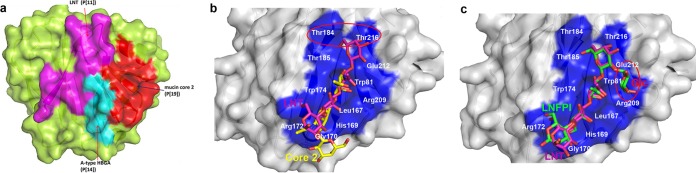
The glycan binding sites of RV VP8*s clustered mainly in the cleft region between the two β-sheets (βH and βK) (16, 32, 33). In P[11] VP8* (33), the ligand LNT/LNnT spanned almost the entire length of the cleft (Fig. 7a). In contrast, the glycan was bound at one corner of the cleft region in P[3] (32) and P[14] (16) VP8*s (Fig. 7a). These residues differed markedly from those in the P[II] genogroup RVs. In particular, the key residue Arg101 was replaced by Val, Ile, or Phe (Fig. 6c, blue), suggesting a new mechanism of the binding mode. Actually, P[19] VP8*-core 2 structure revealed a binding cavity located adjacent to the cleft (Fig. 7a) (34). The porcine P[6] VP8* presented the same characteristics of the putative glycan binding site as P[19] VP8*, implying a similar glycan binding mechanism in P[6] VP8*. Sequence alignment and structural comparison indicated that the VP8*s of RVs in the P[II] genogroup may have a common binding site, as shown in Fig. 6.
FIG 7.
Analysis of the glycan binding site in P[19] VP8*. (a) Surface representation of the corresponding binding sites of type A HBGA in P[14] (cyan), LNT in P[11] (magenta), and mucin core 2 in P[19] (red). VP8*s of P genotype P[19] are shown in the surface representation. (b) Binding sites of core 2 and LNT in P[19]. The amino acids involved in the interaction are shown in stick representation. Thr184 and Thr216 are circled. (c) Structure superimposition of P[19] VP8* with LNT and LNFP1. LNT and LNFP1 are shown as sticks (Gal, magenta; GlcNAc, green; Glc, cyan; fucose, blue). Glc of LNFP1 is circled in red.
Human and porcine P[6] and P[19] VP8*s showed similarities and differences of the glycan binding characteristics in the glycan array assay. Porcine-like human P[6] RV (LL3354) exhibited relatively strong binding signals to mucin core 2 and other glycans with the Galβ1-3GlcNAc motif compared to porcine P[6] (z84) (Fig. 1). Sequence alignment (data not shown) showed 6 amino acids, though these residues were not in the glycan binding site and were altered, which may contribute to the variation in glycan binding specificity. Likewise, the human and porcine P[19] VP8* proteins both recognized the mucin core 2 in the glycan binding assay. Sequence alignment of the human and porcine P[19] (data not shown) revealed identical amino acids in the glycan binding sites; however, nine residue alterations were found, which probably led to the relatively high binding signals of the human P[19] VP8* to mucin core 2 and glycans with Galβ1-3GlcNAc (Fig. 1). These results indicated that the binding patterns may vary even in the same genotype.
A previous study showed that the human P[6] RV strain (ST-3) did not bind to mucin core 2 but bound to the type 1 precursor (LNT) (30); we reported a similar profile in the prevalent human P[6] RV strain (5311142). We found, moreover, that the VP8* proteins of porcine-like human P[6] RV (LL3354) and the porcine P[6] (z84) strains bound to mucin core 2. Sequence alignment revealed that the residues of the I-J loop, Leu167, His169, Gly170, Gly171, and Arg172, of the VP8*s in porcine (z84) and porcine-like human (LL3354) P[6] RV strains were the same as those in human P[19] VP8* (Fig. 6c); however, these residues were significantly altered in the human P[6] RVs, strain ST-3 (Tyr170, Asn171, and Ser172), and strain 5311142 (Met167, Phe169, Tyr170, Asn171, and Ser172) (Fig. 6c, magenta). The porcine and human P[6] VP8* mutants with residues 169 to 172 of the I-J loop exchanged revealed that the porcine P[6] VP8* mutant with the human I-J loop lost the ability to bind to mucin core 2, while the human P[6] VP8* mutant with the porcine I-J loop showed strong binding signals (Fig. 2b), further confirming the vital role of these residues in glycan binding.
Considering the various binding specificities of different P[6] RV strains, we proposed that porcine P[6] bound to mucin core 2 and the porcine-like human P[6] maintained this binding capacity, while the prevalent human P[6] lost it with significant amino acid alterations in the I-J loop (residues 167 to 172). Therefore, the various glycan binding specificities of P[6] RV VP8*s of different origins provided a valuable model to explore potential HBGA-associated host ranges and the evolution of RVs between human and animal hosts. The human P[19] strain in our study was reported to be of porcine origin and did not gain the ability to be prevalent among humans (29). The human and porcine P[19] VP8*s also possessed the same residues in the glycan binding site. Thus, the human P[19] maintained the ability to bind to mucin core 2.
While P[19] VP8* bound to core 2 and LNT via the same cavity (Fig. 7a), we noted that two further residues, Thr184 and Thr216, were involved in the LNT interaction (Fig. 7b). A recent paper reported the complex structures of P[19] VP8* with core 2 and type 1 HBGA (pentasaccharide lacto-N-fucopentaose I [LNFP I]) (34). We noted that VP8* bound to the glycans via the same site. Compared to the structure of VP8*-LNT, fucose of the glycan LNFP I was not involved in the interaction (Fig. 7c), explaining why P[19] can interact with both LNT and LNFP I. This indicated that fucose did not play a key role in glycan binding.
Unlike human P[19] and human P[6], which bound LNT, human P[4] and P[8] did not exhibit binding to LNT in the oligosaccharide binding assay. This was consistent with the previous results showing that human P[4] and P[8] did not bind LNT but recognized a hexasaccharide containing the Lewis epitope (fucose linked to GlcNAc by α1-4) (30). This may explain the high prevalence of P[4] and P[8] RVs in humans, since 90% of the general population are Lewis epitope positive. Sequence alignment of P[8], P[4], and P[19] VP8*s revealed that residues 169 and 216 in the ligand binding cavity were different (Fig. 5c, green). Structural superimposition suggested that Tyr169 in P[4]/P[8] could not form hydrogen bonds with LNT and that Asn216 lost the hydrophobic interaction (Fig. 7c), which probably caused the lack of binding to LNT and contributed to the new pattern of binding to the Lewis epitope. A previous study reported that the neonate-specific bovine-human reassortant P[11] also interacted with LNT (33). The structure comparison of P[11] VP8* with LNT revealed that P[19] RV VP8* bound to LNT in a distinct site, which implied that various RVs could bind to the same ligand via completely different mechanisms and complicated the interactions between RV VP8*s and ligands.
In this study, we found that porcine and porcine-like human P[6] and porcine P[19] recognized mucin core 2; human P[19], P[8], and P[4] of the P[II] genogroup also interacted with mucin core 2. As mucin core 2 is widely distributed in a variety of cells and tissues in both humans and animals (39), it may be crucial for the infection and evolution of P[II] genogroup RVs and other RVs, such as P[10] (30) in the P[I] genogroup. It is proposed that animal RVs of the P[II] genogroup also recognize mucin core 2. While their original interaction with mucin core 2 might have been maintained or altered over the course of evolution, additional interaction with other glycans might have been introduced when these RVs adapted to human hosts.
Our study delineated the glycan binding specificities of human and porcine P[6]/P[19] and determined the glycan binding site in P[19], further confirming a third RV-glycan binding pattern revealed by structural analysis (34). We solved the porcine P[6] VP8* structure. The same amino acid residues and conformation in the putative receptor binding site of P[6] VP8* (Fig. 4c and d) indicated the conservation of the glycan binding cavity in P[6], which was also revealed in P[8] and P[4] VP8*s by structural superimposition. P[19], together with P[6], P[8], and P[4] in the P[II] genogroup, could interact with mucin core 2. Although further studies to solve complex structures of P[6], P[8], and P[4] VP8*s with core 2 are needed, these results provide new insights into the glycan binding mechanism of the P[II] genogroup RVs and lay the foundation for understanding RV evolution and epidemiology and the performances of current RV vaccines in different populations, which may also help in new RV vaccine approaches, such as VP8*-based subunit vaccines.
MATERIALS AND METHODS
VP8* protein expression and purification.
The VP8* fragment (amino acid residues 64 to 223) of human P[19] strain Mc345 (RVA/Human-tc/THA/Mc345/1989/G9P[19]; GenBank accession no. D38054) was cloned into pGEX4T-1 and pET30a vectors with a GST and a His tag, respectively. The recombinant proteins were expressed and purified as described previously (38). The GST fusion and VP8*-His protein were used for functional analysis and crystallization, respectively.
VP8* proteins of porcine P[19] (GenBank accession no.KX455847) (38) and the prevalent human P[6] (5311142) (GenBank accession no. KT162986), human P[4] (11151099) (GenBank accession no. KT162987), and human P[8] (11221075) (GenBank accession no. KT162984) were described previously (20). The VP8* proteins (amino acid residues 1 to 231) of a porcine P[6] strain (z84) (GenBank accession no. MG570048) and a porcine-like human P[6] RV strain (LL3354) (GenBank accession no. EF159567) (40) were expressed and purified with a GST tag as described above. A porcine P6 (z84) mutant (169- to 172HGGR mutated to FYNS) and a human P[6] (5311142) mutant (169- to 172FYNS mutated to HGGR) were also expressed and purified with a GST tag. The VP8* core region (amino acid residues 64 to 223) of porcine P6 (strain z84) was cloned into pET30a with a His tag, expressed, purified by gel filtration chromatography, and analyzed by SDS-PAGE.
Glycan microarray screening analysis.
Glycan ligand screenings for porcine-like human P[6] RV (LL3354), porcine P[6] (z84), human P[19], and porcine P[19] VP8*-GST fusion proteins were performed by the Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics (CFG) (http://www.functionalglycomics.org/) against a library containing 600 glycans. The recombinant GST-VP8* protein was used at protein concentrations of 50 μg/ml and 5 μg/ml. The bound GST-VP8* proteins were detected using a fluorescence-labeled anti-GST monoclonal antibody (Sigma). Fluorescence was measured and quantified with a microarray scanner, and relative fluorescent units (RFU) for binding to each glycan were calculated (41).
ELISA-based glycan binding assay.
Glycan binding specificity was characterized as described previously (15). A plate was coated with VP8*-GST fusion proteins at 20 μg per well. After blocking with 5% nonfat milk, the synthetic glycan-polyacrylamide (PAA)-biotin conjugates mucin core 2 (GlycoTech, Inc., Gaithersburg, MD) and LNT (kindly provided by Jason Xi Jiang) were added (mucin core 2, 0.2 μg or 2 μg per well; LNT, 0.2 μg per well). Horseradish peroxidase-conjugated streptavidin (Abcam, Cambridge, UK) was then added at 0.1 μg per well. The plates were incubated at 37°C for 1 h and washed five times with phosphate-buffered saline (PBS)-0.05% Tween 20 buffer at each step. The reaction was developed using a 3,3′,5,5′-tetramethylbenzidine kit (BD Biosciences, San Diego, CA), and the absorbance was measured at 450 nm.
Crystallization.
The human P[19] VP8* protein in buffer (20 mM Tris-HCl, 50 mM NaCl, pH 8.0) was concentrated to approximately 10 mg/ml, and crystallization screening was carried out using the sitting-drop vapor diffusion method at 18°C with 1 μl of protein mixed with 1 μl of reservoir solution. Human P[19] VP8* was crystallized under conditions of 0.2 M ammonium sulfate, 0.1 M MES (morpholineethanesulfonic acid) monohydrate, pH 6.5, and 30% (wt/vol) polyethylene glycol MME 5000. For preparation of the protein-glycan complex, the tetrasaccharide LNT was purchased from Dextra Laboratories (Reading, UK), and the threonine-linked core 2 trisaccharide was prepared from Fmoc-protected peracetylated core 2-Thr (Sussex Research Laboratories, Ottawa, Canada) after deprotection (42). Core 2-Thr was purified by high-performance liquid chromatography (HPLC) and analyzed by electrospray ionization mass spectrometry (ESI-MS) before use. To obtain crystals of the P[19] VP8*-LNT complex, VP8* was cocrystallized with LNT, using a protein/ligand ratio of 1:50 under the same conditions as for the native crystal described above (38). The P[19]VP8*-core 2 complex was obtained by coincubation of the native crystal with 25 μM core 2-Thr for 5 h at room temperature before X-ray diffraction studies. VP8* of the porcine P[6] strain (z84) at 15 mg/ml was crystallized under conditions of 0.1 M MES monohydrate, pH 6.0, 25% (wt/vol) polyethylene glycol 4000. We have tried to cocrystalize P[6] VP8* with core 2 at protein/ligand ratios of 1:10, 1:20, 1:50, and 1:100 and to coincubate the native crystal with core 2 at concentrations of 25, 50, and 100 μM. Unfortunately, we have been unable to obtain crystal structures of P[6]VP8*-glycan complexes so far.
Data collection and processing.
Crystals were flash-frozen in liquid nitrogen after being dipped briefly in cryoprotectant solution containing 20% (vol/vol) glycerol. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility BL17U. Original data were processed using HKL2000 software (43). The VP8* and ligand structures were solved by molecular replacement using Phaser software implemented in the CCP4 program suite (44) with the structure of P[19] VP8* (PDB ID 5GJ6) as the search model. The initial model was refined using REFMAC5 (45), COOT (46), and PHENIX (47) software. The final statistical analyses for P[19] VP8*-LNT, VP8*-core 2, and P[6] VP8* are presented in Table 1. The structural analysis was performed using the PyMOL software package (https://pymol.org/2/). The RMSD values of the matching Cα atoms between the P[19]/P[6] VP8* and other representative VP8* structures were calculated with the align function in PyMOL.
Accession number(s).
The structures of P[19] VP8*-core 2, VP8*-LNT, and z84 P[6] VP8* have been deposited in the PDB under PDB ID 5YMS, 5YMT, and 5YMU, respectively.
ACKNOWLEDGMENTS
We acknowledge the participation of the Protein-Glycan Interaction Resource of the CFG (supporting grant R24 GM098791) and the National Center for Functional Glycomics (NCFG) at Beth Israel Deaconess Medical Center, Harvard Medical School (supporting grant P41 GM103694), for the analysis of samples by glycan microarray. We are grateful to Jason Xi Jiang for kindly providing the LNT-PAA-biotin oligosaccharide. We thank Ming Tan for revision of the manuscript. We are grateful to Yi Shi and Jinghua Yan in the Institute of Microbiology, Chinese Academy of Sciences, for their advice on the manuscript. We thank Lei Dang, Mengxuan Wang, Xin Cong, Jinjin Wang, Yuanyun Ao, and Huiying Li in our laboratory for their help in performance of the experiments. We thank Yuan Yuan, Min Zhao, Yan Li, Xu Yang, and Yingzi Cui at the Institute of Microbiology for their help in X-ray data collection. We thank Jingyu Yan at the Dalian Institute of Chemical Physics for her help in preparing the figures. The assistance of the staff at the Shanghai Synchrotron Radiation Facility (SSRF beamline 17U) is acknowledged.
We declare no potential conflict of interest.
Zhaojun Duan, Xiaoman Sun, and Dandi Li designed the experiments; Xiaoman Sun, Luyao Wang, Lihong Wang, Dandi Li, Qing Zhang, and Hong Wang performed the experiments; Han Wang and Ruchao Peng collected the X-ray data; and Jianxun Qi solved the structures. Xiaoman Sun, Ruchao Peng, Han Wang, and George F. Gao analyzed the structural data, Dandi Li, Wengang Chai, Miao Jin, Lili Pang, and Xiangyu Kong interpreted the glycan binding data; Xiaoman Sun wrote the draft manuscript. Zhaojun Duan, Xiaoman Sun, Wengang Chai, and George F. Gao revised the manuscript.
This research was supported by grants from the Special National Project on Research and Development of Key Biosafety Technologies (2016YFC1201900) and the National Natural Science Foundation of China (NSFC) (no. 81472003, no. 31500139, and no. 81601813).
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. 2016. Global, Regional, and national estimates of rotavirus mortality in children <5 years of age, 2000-2013. Clin Infect Dis 62(Suppl 2):S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- 2.Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Huang P, Tan M, Liu Y, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. 2012. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J Virol 86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiore L, Greenberg HB, Mackow ER. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553–563. doi: 10.1016/0042-6822(91)90888-I. [DOI] [PubMed] [Google Scholar]
- 6.Kuhlenschmidt TB, Hanafin WP, Gelberg HB, Kuhlenschmidt MS. 1999. Sialic acid dependence and independence of group A rotaviruses. Adv Exp Med Biol 473:309–317. doi: 10.1007/978-1-4615-4143-1_33. [DOI] [PubMed] [Google Scholar]
- 7.Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, von Itzstein M. 2009. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol 5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 8.Zarate S, Cuadras MA, Espinosa R, Romero P, Juarez KO, Camacho-Nuez M, Arias CF, Lopez S. 2003. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J Virol 77:7254–7260. doi: 10.1128/JVI.77.13.7254-7260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Vargas J, Romero P, Lopez S, Arias CF. 2006. The peptide-binding and ATPase domains of recombinant hsc70 are required to interact with rotavirus and reduce its infectivity. J Virol 80:3322–3331. doi: 10.1128/JVI.80.7.3322-3331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo CT, Nakagomi O, Mochizuki M, Ishida H, Kiso M, Ohta Y, Suzuki T, Miyamoto D, Hidari KI, Suzuki Y. 1999. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J Biochem 126:683–688. doi: 10.1093/oxfordjournals.jbchem.a022503. [DOI] [PubMed] [Google Scholar]
- 11.Fleming FE, Bohm R, Dang VT, Holloway G, Haselhorst T, Madge PD, Deveryshetty J, Yu X, Blanchard H, von Itzstein M, Coulson BS. 2014. Relative roles of GM1 ganglioside, N-acylneuraminic acids, and alpha2beta1 integrin in mediating rotavirus infection. J Virol 88:4558–4571. doi: 10.1128/JVI.03431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez MA, Lopez S, Arias CF, Isa P. 2013. Gangliosides have a functional role during rotavirus cell entry. J Virol 87:1115–1122. doi: 10.1128/JVI.01964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero CA, Mendez E, Zarate S, Isa P, Lopez S, Arias CF. 2000. Integrin alpha(v) beta(3) mediates rotavirus cell entry. Proc Natl Acad Sci U S A 97:14644–14649. doi: 10.1073/pnas.250299897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham KL, Halasz P, Tan Y, Hewish MJ, Takada Y, Mackow ER, Robinson MK, Coulson BS. 2003. Integrin-using rotaviruses bind alpha2beta1 integrin alpha2 I domain via VP4 DGE sequence and recognize alphaXbeta2 and alphaVbeta3 by using VP7 during cell entry. J Virol 77:9969–9978. doi: 10.1128/JVI.77.18.9969-9978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, Morrow A, Jiang X. 2012. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol 86:4833–4843. doi: 10.1128/JVI.05507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storry JR, Olsson ML. 2009. The ABO blood group system revisited: a review and update. Immunohematology 25:48–59. [PubMed] [Google Scholar]
- 18.Ramani S, Cortes-Penfield NW, Hu L, Crawford SE, Czako R, Smith DF, Kang G, Ramig RF, Le Pendu J, Prasad BV, Estes MK. 2013. The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. J Virol 87:7255–7264. doi: 10.1128/JVI.03518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Huang P, Jiang B, Tan M, Morrow AL, Jiang X. 2013. Poly-LacNAc as an age-specific ligand for rotavirus P[11] in neonates and infants. PLoS One 8:e78113. doi: 10.1371/journal.pone.0078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, Li DD, Sun XM, Guo YQ, Xiang JY, Wang WH, Zhang LX, Gu QJ, Duan ZJ. 2015. Binding patterns of rotavirus genotypes P[4], P[6], and P[8] in China with histo-blood group antigens. PLoS One 10:e0134584. doi: 10.1371/journal.pone.0134584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohm R, Fleming FE, Maggioni A, Dang VT, Holloway G, Coulson BS, von Itzstein M, Haselhorst T. 2015. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat Commun 6:5907. doi: 10.1038/ncomms6907. [DOI] [PubMed] [Google Scholar]
- 22.Liu N, Xu Z, Li D, Zhang Q, Wang H, Duan ZJ. 2014. Update on the disease burden and circulating strains of rotavirus in China: a systematic review and meta-analysis. Vaccine 32:4369–4375. doi: 10.1016/j.vaccine.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Gentsch JR, Hull JJ, Teel EN, Kerin TK, Freeman MM, Esona MD, Griffin DD, Bielfelt-Krall BP, Banyai K, Jiang B, Cortese MM, Glass RI, Parashar UD, collaborating laboratories of the National Rotavirus Strain Surveillance System. 2009. G and P types of circulating rotavirus strains in the United States during 1996-2005: nine years of prevaccine data. J Infect Dis 200(Suppl 1):S99–S105. doi: 10.1086/605038. [DOI] [PubMed] [Google Scholar]
- 24.Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 25.Seheri LM, Magagula NB, Peenze I, Rakau K, Ndadza A, Mwenda JM, Weldegebriel G, Steele AD, Mphahlele MJ. 2017. Rotavirus strain diversity in eastern and southern African countries before and after vaccine introduction. Vaccine 2017:S0264-410X(17)31679-1. doi: 10.1016/j.vaccine.2017.11.068. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko M, Do LP, Doan YH, Nakagomi T, Gauchan P, Agbemabiese CA, Dang AD, Nakagomi O. 5 April 2018. Porcine-like G3P[6] and G4P[6] rotavirus A strains detected from children with diarrhoea in Vietnam. Arch Virol. doi: 10.1007/s00705-018-3836-4. [DOI] [PubMed] [Google Scholar]
- 27.Ouermi D, Soubeiga D, Nadembega WMC, Sawadogo PM, Zohoncon TM, Obiri-Yeboah D, Djigma FW, Nordgren J, Simpore J. 2017. Molecular epidemiology of rotavirus in children under five in Africa (2006–2016): a systematic review. Pak J Biol Sci 20:59–69. doi: 10.3923/pjbs.2017.59.69. [DOI] [PubMed] [Google Scholar]
- 28.Komoto S, Maeno Y, Tomita M, Matsuoka T, Ohfu M, Yodoshi T, Akeda H, Taniguchi K. 2013. Whole genomic analysis of a porcine-like human G5P[6] rotavirus strain isolated from a child with diarrhoea and encephalopathy in Japan. J Gen Virol 94:1568–1575. doi: 10.1099/vir.0.051011-0. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S, Urushibara N, Taniguchi K, Kobayashi N. 2012. Whole genomic analysis reveals the porcine origin of human G9P[19] rotavirus strains Mc323 and Mc345. Infect Genet Evol 12:471–477. doi: 10.1016/j.meegid.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Ramelot TA, Huang P, Liu Y, Li Z, Feizi T, Zhong W, Wu FT, Tan M, Kennedy MA, Jiang X. 2016. Glycan specificity of P[19] rotavirus and comparison with those of related P genotypes. J Virol 90:9983–9996. doi: 10.1128/JVI.01494-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Mishra R, Holloway G, von Itzstein M, Coulson BS, Blanchard H. 2015. Substantial receptor-induced structural rearrangement of rotavirus VP8*: potential implications for cross-species infection. Chembiochem 16:2176–2181. doi: 10.1002/cbic.201500360. [DOI] [PubMed] [Google Scholar]
- 32.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu L, Ramani S, Czako R, Sankaran B, Yu Y, Smith DF, Cummings RD, Estes MK, Venkataram Prasad BV. 2015. Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus. Nat Commun 6:8346. doi: 10.1038/ncomms9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Xu S, Woodruff AL, Xia M, Tan M, Kennedy MA, Jiang X. 2017. Structural basis of glycan specificity of P[19] VP8*: implications for rotavirus zoonosis and evolution. PLoS Pathog 13:e1006707. doi: 10.1371/journal.ppat.1006707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monnier N, Higo-Moriguchi K, Sun ZY, Prasad BV, Taniguchi K, Dormitzer PR. 2006. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J Virol 80:1513–1523. doi: 10.1128/JVI.80.3.1513-1523.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchard H, Yu X, Coulson BS, von Itzstein M. 2007. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*). J Mol Biol 367:1215–1226. doi: 10.1016/j.jmb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 37.Sun X, Guo N, Li D, Jin M, Zhou Y, Xie G, Pang L, Zhang Q, Cao Y, Duan ZJ. 2016. Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens. Virology 495:129–135. doi: 10.1016/j.virol.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Sun X, Li D, Peng R, Guo N, Jin M, Zhou Y, Xie G, Pang L, Zhang Q, Qi J, Duan ZJ. 2016. Functional and structural characterization of P[19] rotavirus VP8* interaction with histo-blood group antigens. J Virol 90:9758–9765. doi: 10.1128/JVI.01566-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik YPS, Chandrashekar KM, Sharma K, Prasad M, Prasad G. 2011. Evidence for occurrence of human group B rotavirus in central India based on characterization of NSP2 gene. Indian J Virol 22:98–103. doi: 10.1007/s13337-011-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rha B, Tate JE, Payne DC, Cortese MM, Lopman BA, Curns AT, Parashar UD. 2014. Effectiveness and impact of rotavirus vaccines in the United States—2006–2012. Expert Rev Vaccines 13:365–376. doi: 10.1586/14760584.2014.877846. [DOI] [PubMed] [Google Scholar]
- 41.Cholleti SR, Agravat S, Morris T, Saltz JH, Song X, Cummings RD, Smith DF. 2012. Automated motif discovery from glycan array data. Omics 16:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johannes M, Reindl M, Gerlitzki B, Schmitt E, Hoffmann-Roder A. 2015. Synthesis and biological evaluation of a novel MUC1 glycopeptide conjugate vaccine candidate comprising a 4′-deoxy-4′-fluoro-Thomsen-Friedenreich epitope. Beilstein J Org Chem 11:155–161. doi: 10.3762/bjoc.11.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Read RJ. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57:1373–1382. doi: 10.1107/S0907444901012471. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]