ABSTRACT
Cancer-causing herpesviruses infect nearly every human and persist indefinitely in B lymphocytes in a quiescent state known as latency. A hallmark of this quiescence or latency is the presence of extrachromosomal viral genomes with highly restricted expression of viral genes. Silencing of viral genes ensures both immune evasion by the virus and limited pathology to the host, yet how multiple genes on multiple copies of viral genomes are simultaneously silenced is a mystery. In a unifying theme, we report that both cancer-causing human herpesviruses, despite having evolved independently, are silenced through the activities of two members of the Krüppel-associated box (KRAB) domain–zinc finger protein (ZFP) (KRAB-ZFP) epigenetic silencing family, revealing a novel STAT3-KRAB-ZFP axis of virus latency. This dual-edged antiviral strategy restricts the destructive ability of the lytic phase while promoting the cancer-causing latent phase. These findings also unveil roles for KRAB-ZFPs in silencing of multicopy foreign genomes with the promise of evicting herpesviruses to kill viral cancers bearing clonal viral episomes.
IMPORTANCE Despite robust immune responses, cancer-causing viruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) persist for life. This persistence is accomplished partly through a stealth mechanism that keeps extrachromosomal viral genomes quiescent. Quiescence, or latency, ensures that not every cell harboring viral genomes is killed directly through lytic activation or indirectly via the immune response, thereby evicting virus from host. For the host, quiescence limits pathology. Thus, both virus and host benefit from quiescence, yet how quiescence is maintained through silencing of a large set of viral genes on multiple viral genomes is not well understood. Our studies reveal that members of a gene-silencing family, the KRAB-ZFPs, promote quiescence of both cancer-causing human viruses through simultaneous silencing of multiple genes on multicopy extrachromosomal viral genomes.
KEYWORDS: Epstein-Barr virus, KRAB-ZFP, Kaposi's sarcoma-associated herpesvirus, STAT3, SZF1, TRIM28, ZNF557, lytic cycle, viral persistence
INTRODUCTION
Cancer-causing herpesviruses persist in a quiescent or latent state in the host. They belong to a family of ubiquitous DNA viruses that challenge the infected cell with multiple episomal copies of the viral genome. Transcriptional silencing of a large set of viral replicative/lytic genes on these genomes is required for quiescence. This quiescence ensures immune evasion by the virus, thereby favoring persistence of the foreign genomes in the cell. Transcriptional silencing of viral genomes also serves as an antiviral strategy to limit pathology caused by the replicating form of the virus. However, the quiescent state of the virus increases the risk of cancer development by the oncoviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV).
A tightly regulated cascade of replicative gene expression from viral genomes governs the switch from quiescence to the replicative phase that culminates in release of infectious virus particles, thereby terminating quiescence. Quiescence-to-lytic-cycle transition is also regulated by the host. Indeed, not every cell that bears quiescent viral genomes supports transition to the replicative phase (1, 2). As a result, a population of latently infected cells remains at any time to ensure persistence in the host. This property also thwarts efforts to medically eradicate tumor virus-bearing cancer cells, all of which harbor clonal quiescent viral genomes. Therefore, understanding the mechanistic basis for the repressive state of herpesvirus episomes will be essential for eradication of these viruses and diseases caused by them. While data from functional genomic studies suggest combined virus and host regulation of episomes (3), few host proteins that repress episomes of both tumor viruses have been described. Among such proteins that regulate both EBV and KSHV genomes are cohesin subunits and signal transducer and activator of transcription 3 (STAT3) (1–4). Cohesin molecules regulate the formation of higher-order chromatin structures, while STAT3 plays pivotal roles in regulating broad functions, such as immune responses as well as cell growth and differentiation, including in the development of EBV- and KSHV-related and -unrelated cancers (5–7). To understand how STAT3, a transcriptional activator, represses the EBV episome, we intersected the expression profile of B lymphocytes carrying quiescent/latent EBV and the profile of B cells carrying derepressed/lytically active EBV with a chromatin immunoprecipitation sequencing (ChIP-seq) data set of genes bound by STAT3, also in B cells. This strategy identified members of the Krüppel-associated box (KRAB) domain–zinc finger protein (ZFP) (KRAB-ZFP) family of transcriptional repressors as candidates that silence viral genomes (2).
KRAB-ZFPs represent the largest family of transcriptional repressors in tetrapods (8). These repressors harbor a KRAB domain upstream of an array of 2 to 40 C2H2 zinc fingers. The KRAB domain recruits the transcriptional corepressor tripartite motif containing 28 (TRIM28/KAP1/TIF1β) and targets it to DNA via DNA-binding zinc finger domains, ultimately promoting heterochromatinization and repression of the surrounding DNA. Repression is mediated by interactions of TRIM28 with the histone methyltransferase SETDB1, the NuRD histone deacetylase complex, the heterochromatin amplification factor HP1, and DNA methyltransferase 3A (DNMT3A) or DNMT3B (9). Collectively, these factors initiate establishment of heritable gene repression via CpG methylation (9). Previously, KRAB-ZFPs were shown to cause embryonic suppression of endogenous retroviral elements that are integrated into the host genome (10).
In this report, we demonstrate that two members of the KRAB-ZFP repressor family, SZF1 and ZNF557, silence multiple genes on viral episomes to promote virus persistence. This silencing, observed in cultured cells and in virus-infected cells from patients, is achieved via TRIM28 for one of the KRAB-ZFPs. Both ZFPs are regulated by the transcriptional activator STAT3, thereby now assigning STAT3 the unexpected function of epigenome regulation. Furthermore, repression of both EBV and KSHV episomes implicates KRAB-ZFPs in surveillance and silencing of multicopy extrachromosomal foreign genomes. These findings carry implications for evicting herpesviruses and thereby killing viral cancers bearing clonal viral episomes.
RESULTS
STAT3 regulates transcript levels of members of the KRAB-ZFP family.
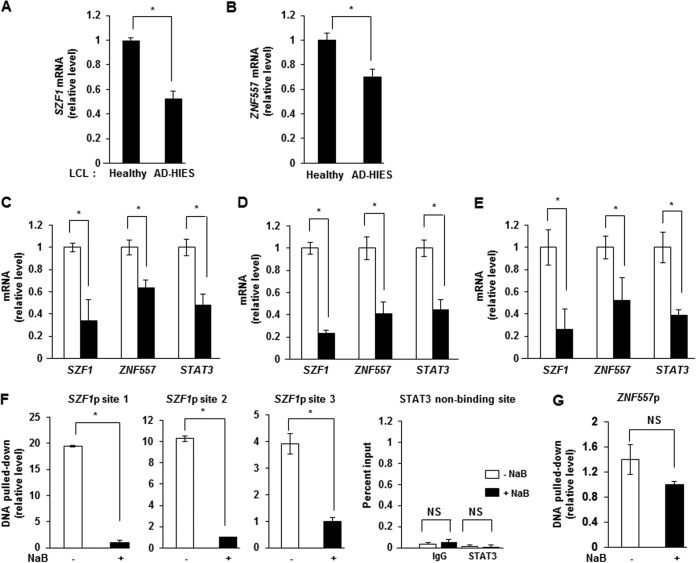
Our investigations into pure populations of B cells with quiescent/latent/episomal EBV genomes versus those with replicative/lytic/linear EBV genomes had indicated that members of the KRAB-ZFP family were expressed at higher levels in cells with episomal genomes (1, 2, 11). Because cells with episomal genomes are known to express high levels of STAT3 (1), we postulated that the KRAB-ZFPs SZF1 and ZNF557 are transcriptional targets of STAT3 and that their expression is subject to STAT3 regulation. Comparison of transcript levels of both KRAB-ZFPs in EBV-positive (EBV+) B lymphoblastoid cell lines (LCLs) derived from healthy humans to those in cells from patients with Job's syndrome with unique mutations in the SH2 or DNA-binding domain of STAT3 revealed that SZF1 and ZNF557 transcript levels were higher in healthy-subject-derived cell lines (Fig. 1A and B). Patients with Job's syndrome (or autosomal dominant hyper-IgE syndrome [AD-HIES]) exhibit a functional knockdown of STAT3 due to a dominant negative mutation in their STAT3 gene (12). Next, introduction of small interfering RNA (siRNA) to STAT3 in 3 types of EBV+ latent B cell lines (a Burkitt lymphoma [BL]-derived cell line, healthy-subject-derived cell lines, and AD-HIES patient-derived cell lines, all carrying episomal EBV) resulted in repression of SZF1 and ZNF557 transcripts (Fig. 1C to E).
FIG 1.
STAT3 localizes to promoters and regulates expression of KRAB-ZFP genes. (A and B) Levels of SZF1 (A) and ZNF557 (B) mRNAs in healthy-donor-derived LCLs and AD-HIES patient-derived LCLs were determined by qRT-PCR. (C to E) Levels of SZF1, ZNF557, and STAT3 mRNAs in EBV+ latent BL cells (C), healthy-donor-derived LCLs (D), and AD-HIES patient-derived LCLs (E) transfected with scrambled siRNA (open bars) or siSTAT3 (black bars) were quantitated by qRT-PCR. Data represent means of three independent experiments (C) or three separate LCLs (A, B, D, and E). (F and G) Relative amounts of SZF1 (F) and ZNF557 (G) promoter DNAs precipitated by anti-STAT3 antibody were determined by qPCR with normalization to input DNA. BL cells were left untreated or treated with NaB for 24 h and examined by qPCR using primers spanning bioinformatically predicted STAT3-binding sites in the SZF1 promoter (site 1, position −1463; site 2, position −1226; site 3, position −74 [relative to transcription start site]) or a non-STAT3-binding site on the SZF1 promoter (position −500 relative to the transcription start site) or the ZNF557 promoter (−255 relative to transcription start site). Data represent the averages of results from three independent experiments; error bars indicate standard errors of the means (*, p ≤ 0.05; NS, not significant; p, promoter).
Examination of SZF1 and ZNF557 genes revealed three and one predicted STAT3 binding sites upstream of the transcriptional start sites of SZF1 and ZNF557, respectively. ChIP using an anti-STAT3 antibody demonstrated preferential enrichment of STAT3 at these sites in tightly latent cells compared to those treated with sodium butyrate (NaB), a chemical inducer of the lytic cycle (Fig. 1F and G). ChIP-PCR of a nearby site on SZFI promoter (SZF1p) that was not predicted to bind STAT3 did not reveal STAT3 enrichment (Fig. 1F). Thus, not only do cells with higher levels of STAT3, i.e., latent cells, express higher levels of SZF1 and ZNF557 transcripts, but STAT3 also regulates SZF1 and ZNF557.
STAT3 regulates transition of EBV from the quiescent/latent state to the replicative/lytic state via SZF1 and ZNF557.
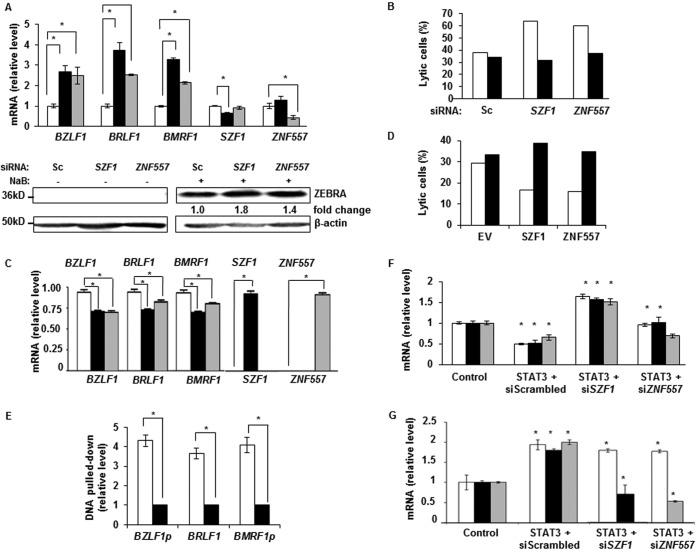
Since latent cells expressed higher levels of STAT3, SZF1, and ZNF557, and STAT3 regulated SZF1 and ZNF557, we investigated the effects of the two KRAB-ZFPs on the transition from latency to lytic state. Knockdown of SZF1 and ZNF557 using two different siRNA sequences resulted in derepression of transcripts from BZLF1, BRLF1, and BMRF1, all of which are EBV lytic genes (Fig. 2A, top, and data not shown), and increased levels of ZEBRA protein (BZLF1 gene product and EBV latency-to-lytic-cycle switch protein) (Fig. 2A, bottom). To assess the effect of gene knockdown on the number of lytic cells, we simultaneously introduced fluorescein isothiocyanate (FITC)-conjugated control siRNA to mark transfected cells. Cells were marked because transfection efficiencies of B cell lines are typically low, ∼20 to 25% (data not shown). There were simultaneous increases (1.6- to 2-fold) in the percentages of lytic cells only in the population with siSZF1/siZNF557 uptake (i.e., FITC+ cells) compared to cells without uptake (i.e., FITC-negative [FITC−] cells) (Fig. 2B). As expected, siRNA to SZF1 did not affect the levels of ZNF557 transcripts and vice versa (Fig. 2A), demonstrating specificity. In contrast to knockdown experiments, overexpression of SZF1 and ZNF557 resulted in smaller amounts of transcripts from BZLF1, BRLF1, and BMRF1 (Fig. 2C). Although moderate, this repression was significant and observed despite the relatively low transfection efficiency. Consistent with repression of lytic transcripts, there were fewer (2.2- to 2.3-fold) lytic cells following uptake of transfected plasmids than cells without uptake of plasmids (Fig. 2D). Although binding sites for most KRAB-ZFPs, particularly in the context of a genome, are not known, consensus binding sites for a few KRAB-ZFPs have been identified by screening random oligonucleotide libraries (13, 14). Based on such a consensus sequence for SZF1, we mapped one putative SZF1-binding site upstream of each of the coding sequences of BZLF1 and BMRF1 and inside the transcribed region of BRLF1. ChIP using an anti-SZF1 antibody showed preferential enrichment of SZF1 at these sites in tightly latent cells compared to those treated with a lytic-cycle-inducing agent (Fig. 2E). A similar experiment for ZNF557 was not possible due to lack of known ZNF557 consensus binding sites. These results implicate the two KRAB-ZFPs in repression of lytic genes.
FIG 2.
STAT3 functions via KRAB-ZFPs to regulate repression of lytic genes on the EBV episome. (A and B) EBV+ latent BL cells transfected with scrambled siRNA (open bars), siSZF1 (black bars), or siZNF557 (gray bars) were treated with NaB, and indicated EBV lytic transcripts were quantitated by qRT-PCR and immunoblot analysis with antibodies against β-actin and EBV lytic protein ZEBRA (A). BL cells transfected with indicated siRNAs and FITC-scrambled (Sc) siRNA (to mark transfected cells) were treated with NaB and percentages of lytic cells in FITC+ (i.e., transfected) cells (open bars) and FITC− (i.e., nontransfected) cells (black bars) were determined by flow cytometry (B). Flow cytometry was performed by using human sera to demarcate lytically active cells, as described previously (11), as well as antibodies against SZF1 or ZNF557. (C) BL cells transfected with empty vector (open bars), HA-SZF1 (black bars), or HA-ZNF557 (gray bars) were treated with NaB, and lytic transcripts were measured by qRT-PCR. (D) BL cells were transfected with indicated plasmids and FITC-scrambled siRNA, as described above for panel B, and treated with NaB, and percentages of lytic cells in FITC+ (i.e., transfected) cells (open bars) and FITC− (i.e., nontransfected) cells (black bars) were determined by flow cytometry, as described above for panel B. EV, empty vector. (E) Relative amounts of DNA precipitated by anti-SZF1 antibody in latent BL cells (open bars) versus NaB-treated BL cells (black bars) were determined by qPCR with normalization to input DNA. PCR primers flanked predicted binding sites for SZF1 on indicated EBV genes. p, promoter. (F) BL cells were transfected with control or STAT3 plasmid and indicated siRNAs, exposed to NaB, and examined by qRT-PCR for relative amounts of EBV lytic transcripts: BZLF1 (open bars), BRLF1 (black bars), and BMRF1 (gray bars). (G) BL cells were transfected with control or STAT3 plasmid and indicated siRNAs and examined by qRT-PCR for relative amounts of STAT3 (open bars), SZF1 (black bars), and ZNF557 (gray bars). Data in panels A, C, E, F, and G represent the averages of results from three independent experiments; error bars indicate standard errors of the means (*, p ≤ 0.05). Data in panels B and D are representative of results from 2 independent experiments.
Since STAT3 regulated the levels of the two KRAB-ZFPs, and both STAT3 and KRAB-ZFPs regulated susceptibility to lytic activation, we addressed whether STAT3 functioned via the KRAB-ZFPs to regulate lytic susceptibility. As expected, overexpression of STAT3 repressed lytic genes despite exposure to a lytic-cycle-inducing agent, but simultaneous knockdown of SZF1 or ZNF557 impaired the ability of STAT3 to suppress lytic susceptibility (Fig. 2F). Figure 2G shows that overexpression of STAT3 led to increased levels of STAT3 mRNA as well as mRNAs from its transcriptional targets SZF1 and ZNF557. Collectively, these experiments demonstrate that STAT3, by transcriptionally modulating the levels of SZF1 and ZNF557, represses lytic genes to maintain the episomal state.
The quiescent/latent state is characterized by high levels of SZF1 and ZNF557 at the single-cell level.
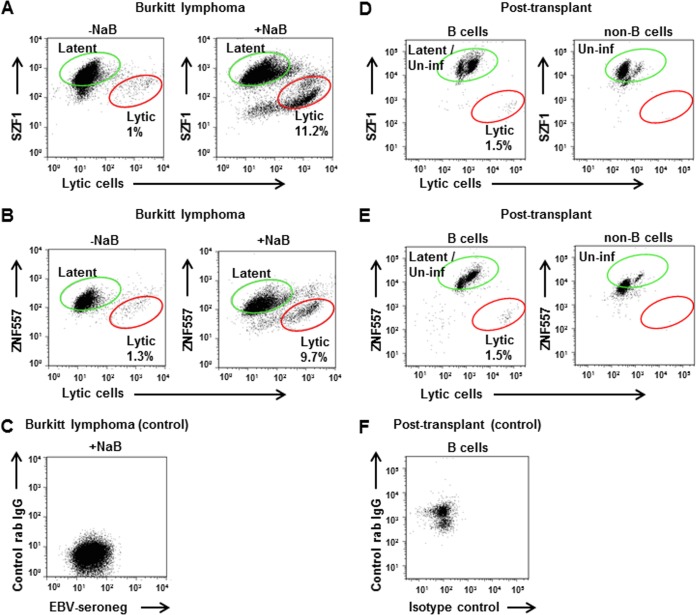
In addressing whether individual cells harboring the episomal form of the viral genome expressed higher levels of KRAB-ZFPs, we separated EBV-infected cells into those that responded to a lytic activation trigger (i.e., lytic cells) from others that did not (i.e., latent/refractory cells bearing episomal DNA) using a sorting strategy that we developed (11). This assay exploits the presence of high titers of EBV lytic-antigen-directed antibodies in EBV-seropositive human sera to identify cells with derepressed lytic genes. We found that compared to lytic cells, refractory cells expressed high levels of both KRAB-ZFPs (Fig. 3A to C). This was true irrespective of whether cells transited to the lytic state spontaneously in culture or after being induced into the lytic phase by a chemical agent (NaB). To determine if this relationship between KRAB-ZFPs and lytic cells held true in vivo, we examined B cells in the blood of an immunosuppressed peripheral blood stem cell transplant recipient whose EBV load was acutely elevated. Of the peripheral B cells, 1.5% expressed an EBV lytic gene while simultaneously demonstrating low levels of KRAB-ZFPs compared to the remainder, i.e., uninfected and latent B cells (Fig. 3D to F). Thus, overall, individual cells with latent/episomal EBV express high levels of KRAB-ZFPs.
FIG 3.
Cells with latent/episomal EBV express higher levels of KRAB-ZFPs than do lytic cells in culture and blood. (A to C) Flow cytometric analysis of latent BL cells that were left untreated (A and B, left) or treated with NaB for 24 h (A and B, right), followed by staining with human serum containing antibodies to EBV lytic antigens (x axis) and anti-SZF1 antibody (y axis) (A) or anti-ZNF557 antibody (y axis) (B), and cells that were stained with human serum lacking antibodies to EBV lytic antigens and isotype control antibody (C). Latent/refractory and lytic cells are indicated; data are representative of results from 3 independent experiments. (D to F) Peripheral blood B cells (D and E, left) and non-B cells (D and E, right) from an immunosuppressed peripheral blood stem cell transplant recipient with EBV viremia were stained with antibodies to EBV lytic antigen ZEBRA (x axis) and KRAB-ZFPs (y axis) (SZF1 [D] and ZNF557 [E]), and B cells were stained with control antibodies (F). Latent (and uninfected) cells as well as lytic cells are indicated.
SZF1 and ZNF557 repress multiple EBV lytic genes in quiescent/latent cells.
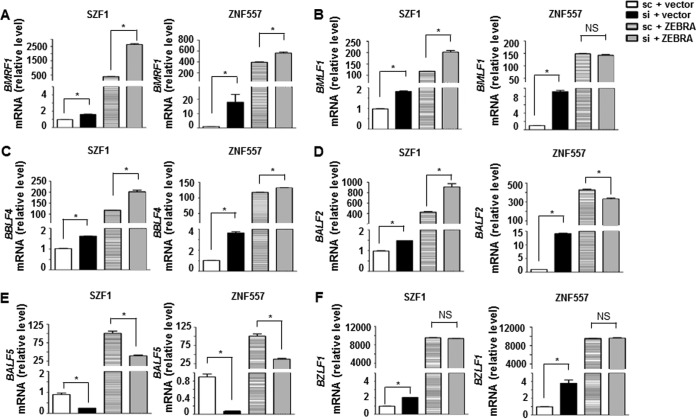
Loss of KRAB-ZFP-mediated repression of BZLF1 resulting from knockdown of KRAB-ZFPs may be sufficient to explain derepression of BRLF1 and BMRF1 (Fig. 2), since both BRLF1 and BMRF1 are transcriptional targets of the BZLF1 gene product ZEBRA. An alternative explanation is that KRAB-ZFPs may simultaneously repress all three (and other) EBV lytic genes. To distinguish between these two possibilities, we induced the EBV lytic cycle by transfecting a plasmid expressing the primary lytic switch gene BZLF1; by not using a lytic-cycle-inducing agent, we avoided the simultaneous induction of lytic genes other than BZLF1. As expected, introduction of BZLF1 resulted in expression of BMRF1, BMLF1, BBLF4, BALF2, and BALF5, all of which are EBV lytic genes (Fig. 4A to E). Simultaneous knockdown of SZF1 resulted in additional increases in levels of transcripts from BMRF1, BMLF1, BBLF4, and BALF2 but not BALF5; simultaneous knockdown of ZNF557 led to a further increase in BMRF1 mRNA (Fig. 4A to E). These lytic genes were chosen because they are early lytic genes that are targets of ZEBRA and because they have predicted SZF1-binding sites upstream of their coding sequences. While ZNF557 appeared to repress only BZLF1, BRLF1, and BMRF1 (Fig. 2A to D and 4A), additional targets cannot be excluded, since the binding sites for ZNF557 are presently unknown. Figure 4F shows expected increases in BZLF1 mRNA following introduction of the BZLF1-expressing plasmid. These experiments support the idea that KRAB-ZFPs simultaneously repress multiple EBV lytic genes.
FIG 4.
KRAB-ZFPs repress multiple EBV lytic genes. BL cells were transfected with empty vector plus scrambled siRNA, empty vector plus siSZF1 (left-hand bar graphs) versus empty vector plus siZNF557 (right-hand bar graphs), BZLF1 plasmid plus scrambled siRNA, or BZLF1 plasmid plus siSZF1 (left-hand bar graphs) versus BZLF1 plasmid plus siZNF557 (right-hand bar graphs). Transfected cells were examined for relative amounts of mRNA from indicated lytic genes that are transcriptional targets of ZEBRA (BZLF1 gene product) (A to E) and from BZLF1 (F). Data represent the averages of results from two independent experiments with 3 technical replicates each; error bars indicate standard errors of the means (*, p ≤ 0.05).
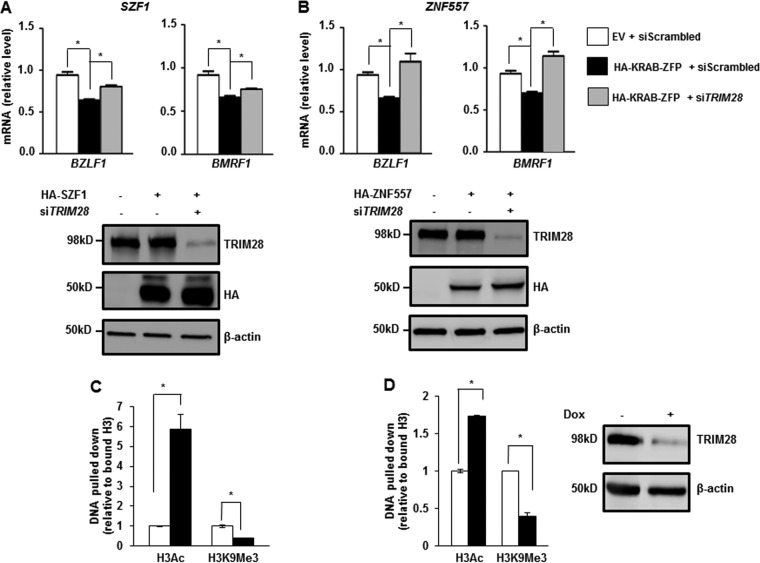
SZF1 and ZNF557 repress lytic genes via TRIM28.
KRAB-ZFPs are thought to mediate transcriptional repression via TRIM28 (9). However, several KRAB-ZFPs have been found to mediate repression independent of TRIM28 (9). To test whether KRAB-ZFPs functioned via TRIM28 to repress the EBV genome, we overexpressed SZF1 or ZNF557 while simultaneously knocking down TRIM28 using a previously validated siRNA to TRIM28 (15). As expected, overexpression of KRAB-ZFPs repressed transcripts from BZLF1 and BMRF1, which were shown in previous experiments to be targets of both KRAB-ZFPs. However, this repressive effect was partially or completely obliterated when TRIM28 was simultaneously knocked down (Fig. 5A and B). Accordingly, short hairpin RNA (shRNA)-mediated knockdown of TRIM28 resulted in accumulation of acetylation on histone 3, with simultaneous reduction of trimethylation at lysine 9 of H3 within the promoters of both EBV lytic genes (Fig. 5C and D). Immunoblot analysis demonstrated efficient knockdown of TRIM28 in experiments shown in Fig. 5C and D (right). Collectively, these experiments indicate that SZF1 and ZNF557 mediate repression of lytic genes via TRIM28.
FIG 5.
KRAB-ZFPs repress EBV lytic genes through the transcriptional corepressor TRIM28. (A and B) BL cells were transfected with empty vector and scrambled siRNA, SZF1 (A) or ZNF557 (B) plasmid and scrambled siRNA, or SZF1 (A) or ZNF557 (B) plasmid and siTRIM28. Cells were exposed to NaB and harvested for determination of relative amounts of transcripts from the EBV latency-to-lytic-cycle switch gene BZLF1 and the early lytic gene BMRF1 by qRT-PCR. Data represent results from two independent experiments with 3 technical replicates each; error bars indicate standard errors of the means (*, p ≤ 0.05). Samples were also harvested for immunoblotting and reacted with indicated antibodies. Representative data from two independent experiments are shown. (C and D) ChIP was performed on BL cells containing doxycycline (dox)-inducible shTRIM28 cassette. Cells were left untreated (open bars) or treated with doxycycline (black bars) for 12 h. DNA precipitated by using anti-H3K9Ac and anti-H3K9Me3 antibodies was analyzed via qPCR using primers spanning predicted SZF1-binding sites in the promoters of lytic genes BZLF1 (C) and BMRF1 (D) and normalized to input. Immunoblot analysis of TRIM28 after doxycycline induction of shTRIM28 was performed to assess knockdown efficiency. Data represent the averages of results from three independent experiments; error bars indicate standard errors of the means (*, p ≤ 0.05).
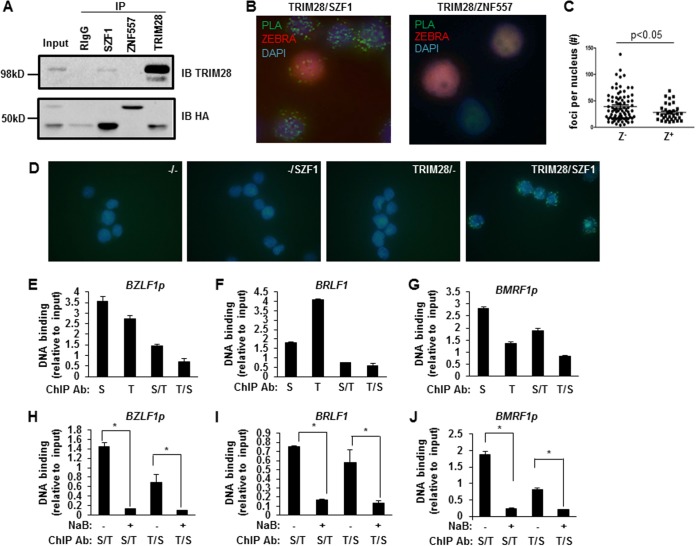
While KRAB domains of KRAB-ZFPs were capable of binding TRIM28 in a mammalian two-hybrid system, full-length KOX1, a prominent KRAB-ZFP originally used to purify TRIM28, demonstrated only weak interaction with endogenous TRIM28 (16). Thus, questions have been raised about the ability of endogenous KRAB-ZFPs to bind to endogenous TRIM28. Moreover, evidence for direct interactions between KRAB-ZFPs and TRIM28 inside cells is lacking. To address whether SZF1 and ZNF557 bound TRIM28, we first performed a coimmunoprecipitation between KRAB-ZFPs and TRIM28 in BL cells. Endogenous TRIM28 coimmunoprecipitated with SZF1 and vice versa following expression of hemagglutinin (HA)-tagged SZF1. However, we were unable to detect a similar coimmunoprecipitation between TRIM28 and HA-tagged ZNF557 (Fig. 6A). To assess if endogenous SZF1 and TRIM28 interacted directly with each other, we used proximity ligation assay (PLA) and detected direct interactions between SZF1 and TRIM28, indicated by the green foci; in contrast, and consistent with lack of coprecipitation of ZNF557 and TRIM28 in Fig. 6A, we did not detect PLA signals when ZNF557 and TRIM28 were examined (Fig. 6B and C). Data from PLA performed with control antibodies are shown in Fig. 6D. Furthermore, there were significantly fewer SZF1-TRIM28 interaction foci in lytic (ZEBRA+) than in latent (ZEBRA−) nuclei (Fig. 6B, left, and C), consistent with loss of SZF1-TRIM28 complex-mediated repression of EBV episomes during the lytic state; of note, latently EBV-infected cells are known to carry multiple episomes per nucleus.
FIG 6.
SZF1 and TRIM28 interact directly and colocalize on EBV lytic promoters preferentially in quiescent/latent cells. (A) Coimmunoprecipitation assays using control rabbit IgG or anti-SZF1, anti-ZNF557, or anti-TRIM28 antibody were performed on BL cells that were cotransfected with HA-SZF1 and HA-ZNF557 plasmids. Precipitates were analyzed by probing with anti-TRIM28 or anti-HA antibody to detect pulled-down endogenous TRIM28 or overexpressed, HA-tagged SZF1 or ZNF557, respectively. IP, immunoprecipitation; IB, immunoblotting. (B) Proximity ligation assays using antibodies targeting SZF1 and TRIM28 (left) or ZNF557 and TRIM28 (right) were performed on NaB-treated BL cells. Lytic cells were identified by immunofluorescence staining with anti-ZEBRA antibody. Representative fields from 3 independent experiments are shown. (C) Thirty lytic (ZEBRA+) and 70 refractory (ZEBRA−) nuclei were examined, and the numbers of PLA foci per ZEBRA+ and ZEBRA− nucleus were determined. (D) PLAs using control goat IgG plus control rabbit IgG (−/−), control goat IgG plus rabbit anti-SZF1 antibody (−/SZF1), goat anti-TRIM28 antibody plus control rabbit IgG (TRIM28/−), or goat anti-TRIM28 plus rabbit anti-SZF1 antibodies (TRIM28/SZF1) were performed on BL cells. Fluorescent green foci indicate in situ interactions between TRIM28 and SZF1. Data are representative of results from 3 independent experiments. (E to G) Chromatin precipitated from BL cells using anti-SZF1, anti-TRIM28, or a control rabbit IgG antibody was subjected to qPCR analysis (S, SZF1; T, TRIM28) using primers spanning bioinformatically predicted SZF1-binding sites within BZLF1p (E), BRLF1 (F), and BMRF1p (G) and normalized to input or subjected to a second round of ChIP using anti-TRIM28 or anti-SZF1 antibody (S/T and T/S), respectively, before qPCR analysis. Negligible amounts of DNA were pulled down by using rabbit IgG control antibody, and values were subtracted from experimental results prior to normalization. (H to J) ChIP–re-ChIP was performed, as described above for panels E to G, on untreated and NaB-treated BL cells. Data represent the averages of results from three independent experiments; error bars indicate the standard errors of the means (*, p ≤ 0.05).
To assess if SZF1 interacts with TRIM28 on the EBV genome, we performed ChIP–re-ChIP experiments in which anti-SZF1 and anti-TRIM28 antibodies were used sequentially. Figures 6E to G show that SZF1 and TRIM28 co-occupied the predicted SZF1-binding sites on all 3 EBV lytic loci. Furthermore, occupancy of the SZF1-TRIM28 complex at these loci was abolished when the latent state was terminated by the addition of the lytic-phase-inducing agent NaB (Fig. 6H to J). Together, these findings indicate that although both KRAB-ZFPs are important for repression of the EBV episome, SZF1 functions by directly recruiting TRIM28 to EBV lytic loci, while ZNF557 may rely on other factors.
Upregulation of SZF1 and ZNF557 is a shared mechanism by which STAT3 prevents transition from the quiescent/latent state of both human oncogenic herpesviruses.
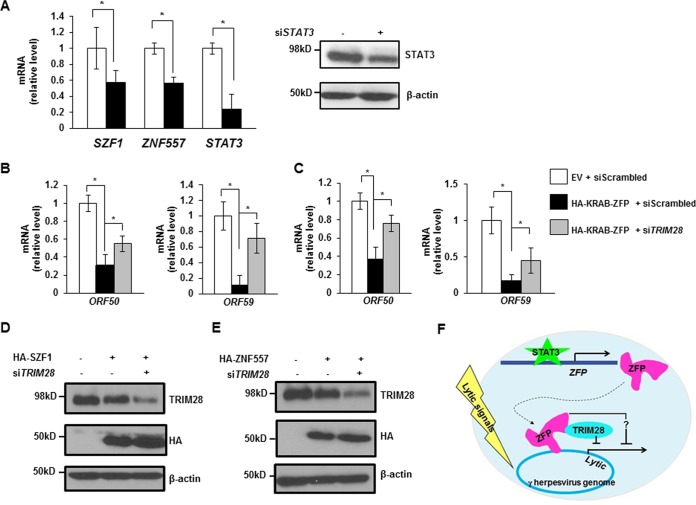
In light of the involvement of SZF1/ZNF557-TRIM28 complexes in STAT3-mediated repression of the EBV genome, we tested if STAT3 similarly regulates these KRAB-ZFPs in latently KSHV-infected primary effusion (B) lymphoma cells. We found that the transcript levels of both KRAB-ZFPs were suppressed upon siRNA-mediated knockdown of STAT3; as expected, siSTAT3 resulted in knockdown of STAT3 (Fig. 7A). Furthermore, overexpression of either KRAB-ZFP resulted in impaired transcript levels of two selected KSHV lytic genes but recovery of both KSHV lytic messages when TRIM28 was simultaneously knocked down by siRNA (Fig. 7B and C); simultaneous changes to SZF1, ZNF557, and TRIM28 proteins are shown in Fig. 7D and E.
FIG 7.
KRAB-ZFPs regulate persistence of KSHV, the other oncogenic human herpesvirus. (A) Levels of SZF1, ZNF557, and STAT3 mRNAs in KSHV+ primary effusion lymphoma (BCBL-1) cells transfected with scrambled siRNA (open bars) or siSTAT3 (black bars) were quantitated by qRT-PCR. BCBL-1 cells were transfected with scrambled siRNA or siSTAT3 and harvested for immunoblotting with indicated antibodies. (B and C) BCBL-1 cells were transfected with empty vector and scrambled siRNA, SZF1 (B) or ZNF557 (C) plasmid and scrambled siRNA, or SZF1 (B) or ZNF557 (C) plasmid and siTRIM28, exposed to VPA, and harvested for determination of the relative amounts of transcripts from the KSHV latency-to-lytic-cycle switch gene ORF50 and the early lytic gene ORF59 by qRT-PCR. All data represent the averages of results from three independent experiments; error bars indicate standard errors of the means (*, p ≤ 0.05). (D and E) BCBL-1 cells were transfected with combinations of empty vector, HA-tagged KRAB-ZFP plasmids, scrambled siRNA, and siTRIM28, treated with VPA, and harvested for immunoblotting with indicated antibodies. Data are representative of results from two independent experiments. (F) Model of oncogenic human herpesvirus persistence depicting that STAT3 transcriptionally activates the KRAB-ZFPs SZF1 and ZNF557, which localize to EBV and KSHV genomes. Bound SZF1 functions directly through TRIM28 to repress lytic genes despite the presence of lytic triggers, while bound ZNF557 functions in a TRIM28-independent manner (?), thereby maintaining latency and promoting virus persistence.
We conclude that STAT3 regulates KRAB-ZFPs to retain KRAB-ZFP with (or without) TRIM28 complexes on lytic genes within the genomes of human oncolytic herpesviruses (oncoherpesviruses), thereby silencing lytic genes and preventing transition of the virus from the quiescent/latent state (Fig. 7F).
DISCUSSION
In a converging theme, we find that both oncogenic human herpesviruses, EBV and KSHV, despite having evolved independently, are silenced by members of the KRAB-ZFP transcriptional repressor family. In so doing, we also reveal a novel STAT3-KRAB-ZFP axis of virus latency. Furthermore, these findings indicate that KRAB-ZFPs survey and silence multicopy extrachromosomal foreign genomes. Mammalian genomes carry hundreds of KRAB-ZFP genes, but the physiological targets and functions of these repressors remain ill-defined. Little is also known about the forces that have driven the number and diversity of KRAB-ZFP genes. Previous studies have shown that KRAB-ZFPs bind to and repress transcription of retroviruses and endogenous retroelements (17, 18). Indeed, recent studies have proposed that the KRAB-ZFP family evolved primarily as an adaptive genomic surveillance system against such retroelements (9). However, unlike in mice, this hypothesis remains unsupported in humans, since evidence for transposition-competent endogenous retroviruses in the human genome is lacking. Yet there continue to be over 300 KRAB-ZFP genes in humans (19), raising the prospect that KRAB-ZFPs in humans have been repurposed during more-recent evolutionary periods to ward off viruses, such as oncogenic and possibly nononcogenic herpesviruses.
Despite the vast number of KRAB-ZFP genes annotated in the mammalian genome, little is known about how KRAB-ZFPs are themselves regulated. Indeed, the promoters of KRAB-ZFP genes are highly divergent. However, when KRAB-ZFP genes are clustered together, they are consistently derepressed following impairment of TRIM28, suggesting that KRAB-ZFPs may themselves be regulated by TRIM28 (20). Here, we show that STAT3 also regulates KRAB-ZFP genes and that a reduction in STAT3 impairs its ability to localize to target KRAB-ZFP genes, resulting in disruption of latency. Reduced enrichment of STAT3 on KRAB-ZFP promoters or KRAB-ZFPs on lytic promoters following NaB treatment may be due to (i) smaller amounts of protein (STAT3 or ZFPs) in the presence of NaB resulting in less occupancy at target promoters and/or (ii) reduced ability of the protein to occupy target promoters due to posttranslational modifications or recruitment of other binding partners in the presence of NaB. In contrast to its well-known function in activating transcription, our findings now reveal an epigenome-modifying repressive role of STAT3 through regulation of two KRAB-ZFPs.
A balance between quiescently infected and lytically infected cells is essential for herpesvirus persistence in the host. Furthermore, for a cell to remain quiescently infected, lytic genes on multiple viral episomes must be repressed. It is therefore not surprising that an orchestrated network of proteins is needed to maintain quiescence and that this equilibrium between STAT3, KRAB-ZFPs, and TRIM28 in the cell may be disturbed by events that influence the levels or functions of any of these constituents. For example, dominant negative mutations in the STAT3 gene in patients with Job's syndrome result in disruption of the quiescent state; similarly, phosphorylation of TRIM28 at S824, which impairs the repressive functions of TRIM28, results in loss of the latent state (2, 15, 21, 22). It is noteworthy that this network thwarts lytic replication of episomal genomes but paradoxically, in so doing, supports the virus life cycle by promoting virus persistence in the cell.
Interestingly, an unrelated KRAB-ZFP, K-RBP, also favored the quiescent/latent state of KSHV without actively repressing lytic genes; in a zinc finger module-independent manner, it prevented access of the key viral transactivator to its targets on the viral genome. In contrast, two other KRAB-ZFPs, ZBRK1 and KZLP, supported the lytic state of EBV and KSHV (23, 24). Furthermore, while silencing of BZLF1 via binding of zinc finger proteins ZEB1 and ZEB2 was described previously, the putative SZF1-binding site does not overlap that of ZEB1 and ZEB2 (positions −55 to −63 for SZF1 versus positions −17 to +10 for ZEB1/2, relative to the transcription start site [TSS] of BZLF1) (25). Our experiments demonstrate not only nonredundant roles for two KRAB-ZFPs in regulating lytic genes but also simultaneous repression of multiple lytic genes by KRAB-ZFPs. Furthermore, only one of these ZFPs functions via TRIM28. This level of complexity that involves KRAB-ZFPs and other cellular proteins may ensure that not all cells at any given time support lytic activation. Indeed, such layers of repression using different proteins may allow fine-tuning of the system and explain the phenomenon of partial susceptibility of herpesviruses to lytic triggers at any point in time. Whether the same mechanisms operate in epithelial cells is presently not known.
TRIM28 influences epigenetic marks to regulate the dynamic organization of chromatin structure. In response to double-strand DNA breaks in heterochromatin, TRIM28 is phosphorylated, resulting in remodeling, relaxation, and repair of damaged DNA (26–28). This very mechanism of repression is exploited to curb the herpesviruses EBV, KSHV, and human cytomegalovirus (CMV) (15, 21, 22, 29–31). However, which KRAB-ZFPs recruit TRIM28 to viral DNA has remained a mystery. While SZF1 interacts physically with TRIM28, ZNF557, the other KRAB-ZFP, appears to function through additional factors. Also notably, SZF1-TRIM28 binding serves as a solitary example of direct in situ interaction between an endogenous KRAB-ZFP and endogenous TRIM28 in a biologically relevant setting, i.e., on DNA inside the cell, while in the natural context of other proteins.
TRIM28 has been found to mediate long-range silencing through the spread of repressive marks beyond regions to which TRIM28 is recruited (32). Whether the KRAB-TRIM28 complex exercises similar long-range effects beyond its immediate binding sites on EBV genes remains to be determined; nonetheless, our data show that the BALF5 gene, which is only 4 kb away from the SZF1-regulated BALF2 gene, is not repressed by SZF1. Repression through histone methylation, histone deacetylation, and HP1-mediated heterochromatinization is generally followed by DNA methylation via the activities of DNMT3A, -3B, and -3L on CpG. DNA methylation has been shown to lock repressive marks on endogenous retroelements so that expression from such foreign DNA is suppressed despite proliferation and differentiation of cells during embryogenesis (33). The extent to which such methylation follows TRIM28-mediated epigenetic changes at lytic promoters needs to be determined, particularly given the agility with which herpesviruses transition from the latent/episomal state to the lytic phase. Indeed, lack of increased demethylase transcripts in pure populations of EBV-infected lytic cells (2) is in line with the lack of DNA methylation of TRIM28-mediated heterochromatin in differentiated adult cells (33). Although the relative contributions of the KRAB-TRIM28 complex and cohesin components to lytic repression on the episome remain to be determined (34), they likely contribute in different ways: KRAB-ZFP/TRIM28 by chromatin compaction and cohesin components through DNA looping.
Taken together, this evidence points to a key role for KRAB-ZFPs in regulating virus infections by silencing multicopy extrachromosomal foreign DNA elements, particularly in the context of oncogenic herpesviruses that infect practically every human. By influencing the balance between quiescence and the lytic state, these repressors effect virus persistence in the host and the population. On the one hand, the STAT3-KRAB-ZFP pathway curbs pathology by restricting the lytic phase. On the other hand, since both EBV and KSHV B lymphomas are dominated by the latent/episomal state, this pathway may contribute to cancer development. Importantly, disrupting this pathway may boost oncolytic efforts to kill viral cancers by evicting herpesviruses.
MATERIALS AND METHODS
Cells and cell lines.
Peripheral blood mononuclear cells were isolated from the blood of a pediatric patient about 1 month after allogeneic stem cell transplantation when EBV load was elevated in the context of fever. Peripheral blood mononuclear cells were isolated as described previously (2, 35). The endemic Burkitt lymphoma cell line HH514-16 (a gift from George Miller, Yale University) and the body-cavity-based KSHV+ B lymphoma cell line BCBL-1 (a gift from Christine King at Upstate Medical University) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). CLIX-shTRIM28 (clone HH514-16 transfected with pLIX_shTRIM28) cells were generated from puromycin (catalog number P8833; Sigma-Aldrich)-selected HH514-16 cells transfected with pLIX_402-ShTRIM28. Lymphoblastoid cell lines (LCLs) generated from healthy subjects or patients with AD-HIES (two with mutations in the SH2 domain and a third with a mutation in the DNA-binding domain of STAT3) were described previously (2, 35).
Ethics statement.
Blood was drawn from the patient after obtaining written informed consent from a parent. The study of human subjects was approved by the institutional review boards at Stony Brook University and the University of Florida.
Chemical treatment of cell lines.
Sodium butyrate (NaB) (3 mM) (catalog number 303410; Sigma-Aldrich) and doxycycline (5 μg/ml) (catalog number D9891; Sigma-Aldrich) were used to treat Burkitt lymphoma cells, except where specific concentrations are indicated. Valproic acid (VPA) (0.6 μM) (catalog number P4543; Sigma-Aldrich) was used to treat BCBL-1 cells.
Plasmids, siRNAs, and transfection.
Plasmid HA-SZF1 was constructed by inserting the SZF1-coding sequence amplified from a cDNA library of HH514-16 cells with forward primer ATACACTCGAGATGTGGGCCCCGCGGGAGCA and reverse primer ATACAGCGGCCGCTTACTATCAATCTCTGCACACATAAG into pCMV-HA (a gift from Christopher A. Walsh [Addgene plasmid 32530]) at the XhoI and NotI sites. Plasmid HA-ZNF557 was generated by inserting the ZNF557 sequence [into the pcDNA3.1(+) plasmid using the NheI and ApaI restriction sites] with an N-terminal HA tag and surrounded by small 5′ and 3′ untranslated regions (UTRs) that were synthetically generated. The 5′ UTR includes a Kozak consensus sequence proximal to the start codon. The 3′ region includes a sequence matching the histone stem loop (HSL) metazoan consensus that was designed for minimal interaction with known human microRNA (miRNA) sequences. 3′ of the HSL is a segment expected to act as a “histone downstream element” (HDE) to facilitate HSL-mediated transcription termination. The resulting plasmid has a CMV promoter at the 5′ end and a bGH polyadenylation signal at the 3′ end. The poly(A) signal ensures that transcripts produced outside S phase are terminated with a poly(A) tail. Plasmid pLIX_402-shTRIM28 was constructed by annealing shTRIM28 sequences using forward primer CTAGCCCATGACCAAGATCCAGAACTTCCTGTCAGATTCTGGATCTTGGTCATGGTTTTTA and reverse primer CCGGTAAAAACCATGACCAAGATCCAGAATCTGACAGGAAGTTCTGGATCTTGGTCATGGG into pLIX_402 (a gift from David Root [Addgene plasmid 41394]) at NheI and AgeI sites. pCMV-STAT3 was a gift from Nancy Reich, Stony Brook University. Plasmid pHD1013-Z was a gift from Ayman El-Guindy at Yale University.
siRNAs targeting human STAT3, SZF1, ZNF557, and TRIM28 transcripts (catalog numbers sc-29493, sc-78092, sc-97365, and sc-38550) were purchased from Santa Cruz Biotechnology. Another set of siRNAs targeting human SZF1 and ZNF557 (catalog numbers J-020953-05-0005 and J-014343-17-0005) was purchased from Dharmacon and reconstituted with nuclease-free water. The second set of siRNAs to SZF1 and ZNF557 was comparable to the first set in the ability to knock down target genes and increase EBV lytic transcript levels. In these experiments, 1 × 106 HH5514-16 cells, LCLs, or BCBL-1 cells were transfected with either 20 μg of plasmid or 200 pmol of siRNA in Ingenio solution (catalog number MIR50117; Mirus) by using an Amaxa Nucleofector II system (program A-024), as described previously (2, 15).
Antibodies.
Rabbit anti-STAT3 (catalog number sc-482) and anti-SZF1 (catalog number sc-100263) antibodies were purchased from Santa Cruz Biotechnology; rabbit anti-ZNF557 antibodies (catalog numbers sc-130005 and 20500-1-AP) were obtained from Santa Cruz Biotechnology and Proteintech, respectively; anti-TRIM28 antibodies (catalog numbers A300-274A, A303-838A, and ab22553) were purchased from Bethyl Laboratories and Abcam; mouse anti-β-actin (catalog number AC-15) and anti-HA (catalog number H3663) antibodies were obtained from Sigma; and anti-H3 (ab1791), anti-acetylated H3 (catalog number ab47915), and anti-trimethylated lysine 9 H3 (catalog number ab8898) antibodies were purchased from Abcam. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG(H+L) (catalog number AP308P) and HRP-conjugated goat anti-rabbit IgG(H+L) (catalog number AP307P) were purchased from EMD Millipore, FITC-conjugated anti-human IgG (whole molecule) (catalog number F53512) was obtained from Sigma-Aldrich, and Alexa Fluor 647-conjugated goat anti-rabbit IgG (catalog number A-21245) was purchased from Thermo Fisher. Mouse anti-ZEBRA antibody was a gift from Paul Farrell at Imperial College, London, United Kingdom.
ChIP and ChIP–re-ChIP.
HH514-16 cells were subcultured to 3 × 105 cells/ml. After 24 h, the cells were counted and treated with 3 mM NaB at a concentration of 5 × 105 cells/ml. Cells were harvested and summarily processed for ChIP, as described previously (36). For each ChIP, 5 μg of antibody was used for lysate derived from 1 × 107 cells. Eluted chromatin was purified by using phenol-chloroform extraction and subjected to quantitative PCR (qPCR) analysis. In ChIP–re-ChIP experiments, 3 × 107 cells were lysed, and 15 μg of antibody was used per reaction. Eluted protein-chromatin complexes from a first round of ChIP were subjected to a second round of ChIP by adding 5 μg of a second antibody. Precipitated chromatin was analyzed by qPCR.
Quantitative reverse transcriptase PCR.
Quantitative reverse transcriptase PCRs (qRT-PCRs) were performed and analyzed by using the ΔΔCT method, as described previously (2). Primer sequences included forward primer GTAACCCGTTGAACCCCATT and reverse primer CCATCCAATCGGTAGTAGCG for 18S rRNA, forward primer TTCCACAGCCTGCACCAGTG and reverse primer GGCAGAAGCCACCTCACGGT for BZLF1, forward primer ACCTGCCGTTGGATCTTAGTG and reverse primer GGCGTTGTTGGAGTCCTGTG for BMRF1, forward primer AACCAGAATAATCTCCCCAATG and reverse primer CGAGGCACCCCAAAAGTC for BFRF3, forward primer CTACCTCGGCATCGTTTG and reverse primer CTTCTCGTCCTCGTCCCT for BMLF1, forward primer AAGGTGCTGATGCTGTGCC and reverse primer GCCCGTTGATGATGTAGTTCTC for BALF2, forward primer CGTCTCATTCCCAAGTGTTTC and reverse primer GCCCTTTCCATCCTCGTC for BALF5, forward primer GCTATGGACCACCAATGTGA and reverse primer AGCCAGGCGTTGAAGAAC for BBLF4, forward primer CCCTGAGCCAGTTTGTCATT and reverse primer ATGGGTTAAAGGGGATGATG for ORF50, forward primer TTAGAAGTGGAAGGTGTGCC and reverse primer TCCTGGAGTCCGGTATAGAATC for ORF59, forward primer TCCAAATCCTCCTAACCCCT and reverse primer GAGCAGCTACTGGGCTGG for SZF1, forward primer GCTCCAGCTGGGAGATCAG and reverse primer TGTACAGGGACGTGATGCTG for ZNF557, and forward primer GCCAGAGAGCCAGGAGCA and reverse primer ACACAGATAAAACTGTCTTCAGGTATG for STAT3.
Primers for ChIP-qPCR analysis.
For ChIP-qPCR analysis, the following primers were used: forward primer TAGCCTCGAGGCCATATTTCAACTGG and reverse primer GCCAAGCTTCAAGAATGTTTAGTGAG for BZLF1p (flanking a putative SZF1-binding site at nucleotide −59 [midpoint] relative to TSS), forward primer ACTGCCCGCTCACCTACAT and reverse primer CCAGAGCAGAGGCAGGCAGG for BMRF1p (provided by Ayman El-Guindy) (putative SZF1-binding site at nucleotide −291 [midpoint] relative to TSS), forward primer TTAGCAATGCCTGTGGCTCA and reverse primer TGGCCATTTGGACGAACTGA for BRLF1 (putative SZF1-binding site at nucleotide +1489 [midpoint] relative to TSS), forward primer GAGAGGGAGGTTGCAGTGAG and reverse primer TGAAGCACAACTTCCCCAAT for SZF1p site 1 (predicted STAT3-binding site at nucleotide −1463 [midpoint] relative to TSS), forward primer CCCTTGGGAAGGGCTATTAT and reverse primer CCCACCTCATCTTCCTTTCA for SZF1p site 2 (predicted STAT3-binding site at nucleotide −1226 [midpoint] relative to TSS), forward primer ATTTTCTCTCGCCCTGCAC and reverse primer AAAGAAGCAGGACTGGGTGA for SZF1p site 3 (predicted STAT3-binding site at nucleotide −74 [midpoint] relative to TSS), and forward primer CTGCTTTTTCATTTATCAGCTTT and reverse primer TGATTATGTTTGGTGAGGAAAA for the ZNF557p site (predicted STAT3-binding site at nucleotide −255 [midpoint] relative to TSS). As a negative control, forward primer GGTGCTCACTCCACGACTCTT and reverse primer GAGAACTGCTTGAACCCGGG were used to amplify a site adjacent to a predicted STAT3-binding site within SZF1p (at position −500 relative to TSS).
Flow cytometry.
Cells were fixed with BD Cytofix/Cytoperm solution (catalog number 554722; BD Bioscience) at room temperature for 15 min, washed with 1× BD Perm/Wash buffer (catalog number 554723; BD Bioscience), and incubated with indicated primary antibodies (or reference EBV-seropositive or EBV-seronegative human sera [Fig. 2B and D and 3A to C]) for 1 h at room temperature. After washing, cells were incubated with the corresponding secondary antibodies (or anti-human IgG [Fig. 2B and D and 3A to C]) conjugated to fluorochrome for another hour at room temperature and then subjected to flow cytometry using a FACSCalibur instrument (BD Bioscience) and analysis of data using FlowJo software (TreeStar). Analysis gates for flow cytometry were determined based on parallel staining with isotype-matched control antibodies (Fig. 3D and E) or reference EBV-seronegative human sera (Fig. 2B and D and 3A to C) (2).
Immunoblotting.
Immunoblotting with indicated antibodies was performed as described previously (2). All immunoblotting was performed by using 10% SDS-PAGE gels, except for Fig. 6A, in which a 12% gel was run.
Proximity ligation assay.
Proximity ligation assay was performed with goat anti-TRIM28 and rabbit KRAB-ZFP antibodies, the Duolink Anti-Goat Minus plus Anti-Rabbit Plus in situ PLA probe (catalog numbers DUO92006 and DUO92002; Sigma-Aldrich), Duolink Green in situ detection reagents (catalog number DUO92014; Sigma-Aldrich), and washing buffers (catalog number DUO82049; Sigma-Aldrich), as described previously (15).
Statistical analysis.
The p values were calculated by comparing the means for two groups of interest using unpaired Student's t test.
ACKNOWLEDGMENTS
We thank Nancy Reich at Stony Brook University for providing plasmid pCMV-STAT3 and Ayman El-Guindy at Yale University for plasmid pHD1013-Z.
S.B.-M. was supported by NIH grants R01 AI113134 and R41 AI115834, the SUNY Research Foundation, and the University of Florida. E.M.B. was supported by NIH grant T32AI007539.
REFERENCES
- 1.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill ER, Koganti S, Zhi J, Megyola C, Freeman AF, Palendira U, Tangye SG, Farrell PJ, Bhaduri-McIntosh S. 2013. Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes. J Virol 87:11438–11446. doi: 10.1128/JVI.01762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, Leslie C, Lieberman PM. 2012. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HS, Wikramasinghe P, Showe L, Lieberman PM. 2012. Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency. J Virol 86:9454–9464. doi: 10.1128/JVI.00787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nepomuceno RR, Snow AL, Beatty PR, Krams SM, Martinez OM. 2002. Constitutive activation of Jak/STAT proteins in Epstein-Barr virus-infected B-cell lines from patients with posttransplant lymphoproliferative disorder. Transplantation 74:396–402. doi: 10.1097/00007890-200208150-00017. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, Jove R. 2004. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Pardoll D, Jove R. 2009. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. 2013. KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genomics 14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ecco G, Imbeault M, Trono D. 2017. KRAB zinc finger proteins. Development 144:2719–2729. doi: 10.1242/dev.132605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. 2016. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev Cell 36:611–623. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhaduri-McIntosh S, Miller G. 2006. Cells lytically infected with Epstein-Barr virus are detected and separable by immunoglobulins from EBV-seropositive individuals. J Virol Methods 137:103–114. doi: 10.1016/j.jviromet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schaffer AA, Puck JM, Grimbacher B. 2007. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 13.Peng H, Zheng L, Lee WH, Rux JJ, Rauscher FJ III. 2002. A common DNA-binding site for SZF1 and the BRCA1-associated zinc finger protein, ZBRK1. Cancer Res 62:3773–3781. [PubMed] [Google Scholar]
- 14.Wagner S, Hess MA, Ormonde-Hanson P, Malandro J, Hu H, Chen M, Kehrer R, Frodsham M, Schumacher C, Beluch M, Honer C, Skolnick M, Ballinger D, Bowen BR. 2000. A broad role for the zinc finger protein ZNF202 in human lipid metabolism. J Biol Chem 275:15685–15690. doi: 10.1074/jbc.M910152199. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Burton EM, Bhaduri-McIntosh S. 2017. Chloroquine triggers Epstein-Barr virus replication through phosphorylation of KAP1/TRIM28 in Burkitt lymphoma cells. PLoS Pathog 13:e1006249. doi: 10.1371/journal.ppat.1006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itokawa Y, Yanagawa T, Yamakawa H, Watanabe N, Koga H, Nagase T. 2009. KAP1-independent transcriptional repression of SCAN-KRAB-containing zinc finger proteins. Biochem Biophys Res Commun 388:689–694. doi: 10.1016/j.bbrc.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 17.Horiba M, Martinez LB, Buescher JL, Sato S, Limoges J, Jiang Y, Jones C, Ikezu T. 2007. OTK18, a zinc-finger protein, regulates human immunodeficiency virus type 1 long terminal repeat through two distinct regulatory regions. J Gen Virol 88:236–241. doi: 10.1099/vir.0.82066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishitsuji H, Sawada L, Sugiyama R, Takaku H. 2015. ZNF10 inhibits HIV-1 LTR activity through interaction with NF-kappaB and Sp1 binding motifs. FEBS Lett 589:2019–2025. doi: 10.1016/j.febslet.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D. 2013. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS One 8:e56721. doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauzlaric A, Ecco G, Cassano M, Duc J, Imbeault M, Trono D. 2017. The mouse genome displays highly dynamic populations of KRAB-zinc finger protein genes and related genetic units. PLoS One 12:e0173746. doi: 10.1371/journal.pone.0173746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang PC, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang DH, Ann DK, Luciw PA, Kung HJ. 2009. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res 69:5681–5689. doi: 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. 2015. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. Elife 4:e06068. doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao G, Huang J, Fixman ED, Hayward SD. 2005. The Epstein-Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J Virol 79:245–256. doi: 10.1128/JVI.79.1.245-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe A, Higuchi M, Fukushi M, Ohsawa T, Takahashi M, Oie M, Fujii M. 2007. A novel KRAB-zinc finger protein interacts with latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus and activates transcription via terminal repeat sequences. Virus Genes 34:127–136. doi: 10.1007/s11262-006-0048-x. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, McCarthy PJ, Wang Z, Gorlen DA, Mertz JE. 2012. Shutoff of BZLF1 gene expression is necessary for immortalization of primary B cells by Epstein-Barr virus. J Virol 86:8086–8096. doi: 10.1128/JVI.00234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 27.White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ III. 2012. The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol Cancer Res 10:401–414. doi: 10.1158/1541-7786.MCR-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 29.Gjyshi O, Roy A, Dutta S, Veettil MV, Dutta D, Chandran B. 2015. Activated Nrf2 interacts with Kaposi's sarcoma-associated herpesvirus latency protein LANA-1 and host protein KAP1 to mediate global lytic gene repression. J Virol 89:7874–7892. doi: 10.1128/JVI.00895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King CA, Li X, Barbachano-Guerrero A, Bhaduri-McIntosh S. 2015. STAT3 regulates lytic activation of Kaposi's sarcoma-associated herpesvirus. J Virol 89:11347–11355. doi: 10.1128/JVI.02008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Zhu C, Guo Y, Wei F, Lu J, Qin J, Banerjee S, Wang J, Shang H, Verma SC, Yuan Z, Robertson ES, Cai Q. 2014. Inhibition of KAP1 enhances hypoxia-induced Kaposi's sarcoma-associated herpesvirus reactivation through RBP-Jkappa. J Virol 88:6873–6884. doi: 10.1128/JVI.00283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. 2012. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep 2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman PM. 2013. Keeping it quiet: chromatin control of gammaherpesvirus latency. Nat Rev Microbiol 11:863–875. doi: 10.1038/nrmicro3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koganti S, Clark C, Zhi J, Li X, Chen EI, Chakrabortty S, Hill ER, Bhaduri-McIntosh S. 2015. Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr virus lytic activation in B lymphocytes. J Virol 89:5002–5011. doi: 10.1128/JVI.00121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, Hearing P. 2014. The use of chromatin immunoprecipitation (ChIP) to study the binding of viral proteins to the adenovirus genome in vivo. Methods Mol Biol 1089:79–87. doi: 10.1007/978-1-62703-679-5_6. [DOI] [PubMed] [Google Scholar]