ABSTRACT
Severe dengue virus (DENV) infection is associated with overactivity of the complement alternative pathway (AP) in patient studies. Here, the molecular changes in components of the AP during DENV infection in vitro were investigated. mRNA for factor H (FH), a major negative regulator of the AP, was significantly increased in DENV-infected endothelial cells (EC) and macrophages, but, in contrast, production of extracellular FH protein was not. This discord was not seen for the AP activator factor B (FB), with DENV induction of both FB mRNA and protein, nor was it seen with Toll-like receptor 3 or 4 stimulation of EC and macrophages, which induces both FH and FB mRNA and protein. Surface-bound and intracellular FH protein was, however, induced by DENV, but only in DENV antigen-positive cells, while in two other DENV-susceptible immortalized cell lines (ARPE-19 and human retinal endothelial cells), FH protein was induced both intracellularly and extracellularly by DENV infection. Regardless of the cell type, there was an imbalance in AP components and an increase in markers of complement AP activity associated with DENV-infected cells, with lower FH relative to FB protein, an increased ability to promote AP-mediated lytic activity, and increased deposition of complement component C3b on the surface of DENV-infected cells. For EC in particular, these changes are predicted to result in higher complement activity in the local cellular microenvironment, with the potential to induce functional changes that may result in increased vascular permeability, a hallmark of dengue disease.
IMPORTANCE Dengue virus (DENV) is a significant human viral pathogen with a global medical and economic impact. DENV may cause serious and life-threatening disease, with increased vascular permeability and plasma leakage. The pathogenic mechanisms underlying these features remain unclear; however, overactivity of the complement alternative pathway has been suggested to play a role. In this study, we investigate the molecular events that may be responsible for this observed alternative pathway overactivity and provide novel findings of changes in the complement system in response to DENV infection in primary cell types that are a major target for DENV infection (macrophages) and pathogenesis (endothelial cells) in vivo. Our results suggest a new dimension of cellular events that may influence endothelial cell barrier function during DENV infection that could expand strategies for developing therapeutics to prevent or control DENV-mediated vascular disease.
KEYWORDS: complement, dengue virus, factor H
INTRODUCTION
Dengue virus (DENV) is currently considered the most important mosquito-borne viral infection of humans worldwide, causing between 50 million and 390 million estimated infections per year in over 100 countries (1, 2). DENV consists of four serotypes (1–4) and belongs to the family Flaviviridae, genus Flavivirus (3). Disease caused by DENV infection ranges from asymptomatic, undifferentiated fever and classical dengue fever to severe forms of the disease that include dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). These clinical descriptions of dengue have been revised as dengue with or without warning signs and severe dengue (4). One life-threatening outcome of DENV infection is increased vascular permeability and plasma leakage, which ultimately can lead to fatal hypovolemic shock (5–8). Although the pathogenic mechanisms underlying the increased vascular permeability remain unclear, a number of studies have demonstrated that DENV infection of macrophages and endothelial cells (EC) plays a critical role in altering cellular responses that control capillary leakage and barrier integrity (9–11). Macrophages are not only the major target for DENV replication in vivo, but they also are important sources of cytokines, chemokines, and vasoactive factors that converge on the endothelium to contribute to vascular permeability (12, 13). The role of the endothelium itself has been debated, and while many studies suggest that EC are not a major site for viral replication, the endothelium is undoubtedly a major site for DENV-mediated pathogenesis. DENV is reported to induce effects that alter the barrier function of the endothelium and that increase EC cytokine and chemokine release and EC inflammatory responses (14–18). Multiple immunomodulatory and vasoactive factors, such as tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1), IL-6, macrophage inhibitory factor, and metalloproteinases, from macrophages or dendritic cells have been implicated in severe dengue or DENV-induced vascular dysfunction (11, 19–21). Recently, the capacity of viral nonstructural protein 1 (NS1) to directly induce vascular leakage and endothelial cell dysfunction has also been shown (22, 23). Thus, the pathogenesis of DENV is clearly multifactorial and overwhelmingly a function of dysregulated immune responses.
The complement system is suggested to be involved in DENV disease and particularly in initiation of vascular leakage (24–27). Complement comprises three pathways, the classical pathway, the lectin pathway, and the alternative pathway (AP), that involve a cascade of proteolytic cleavages forming various vasoactive and immunostimulatory proteins (28, 29). These three pathways converge on C3, which is activated by cleavage to form C3b, with subsequent interactions that lead to the formation of a C3b convertase complex and the terminal membrane attack complex (MAC). The MAC can lyse pathogens and target cells, while other cleavage products of the complement system, such as C3a and C5a, are inflammatory and vasoactive factors (29). While the classical and lectin pathways are stimulated by pathogen recognition, the AP is constitutively active (29, 30) and is tightly controlled to prevent unwanted tissue damage. The AP negative regulatory protein factor H (FH) is considered the master regulator of this pathway that ensures that the AP activity is tightly controlled (30–33). FH can control AP activity by competing with factor B (FB) for C3b binding (30, 31, 34, 35), promoting the decay of the C3 convertase, and stimulating C3b degradation (30, 36). Additionally, FH acts as a cofactor and promotes enzymatic inactivation of C3b by factor I (FI) to form iC3b (37, 38). These actions of FH can occur in the fluid phase but are facilitated at cell surfaces by FH binding to polyanions and glycosaminoglycans (39–41). C3b deposition on surfaces is nondiscriminatory and arises from constitutive AP-driven C3 hydrolysis or “tickover” to form C3b (42). The subsequent binding of FB to C3b forms the C3bB complex, which is cleaved by the serine protease factor D (FD) to produce more AP C3 convertase (C3bBb), which is part of an activation loop that further promotes the complement cascade (29). Alternatively, C3b can be deactivated by the actions of FH, as described above. This balance between the actions of FB and FH can thus determine if a C3b-coated surface is targeted for complement action or not (32, 34, 35).
Overactivity of the AP due to the low activity of FH is associated with human disease, such as atypical hemolytic-uremic syndrome, which shares some common pathogenic features with dengue disease, such as intravascular hemolysis, thrombocytopenia, damage to the endothelium, and increased vascular leakage (43, 44). We postulated that, similarly, AP overactivity due to the low activity of FH might be involved in DENV pathogenesis. Several reports have supported an association of overactivity of the complement AP with DENV disease severity, with complement protein consumption, low serum levels of FH, and high levels of FD being reported in the circulation of severe DENV patients (8, 25–27, 45, 46).
Herein, the molecular events at the cellular level that align with AP activity are defined and provide evidence of dysregulation of FH production locally within macrophages and EC, the major in vivo sites for DENV replication and pathogenesis, respectively. These changes in FH in combination with elevation of other complement components, such as FB and C3b deposition, are associated with increased complement AP activity in vitro, which we propose reflects AP activity in the cellular microenvironment in vivo. Our results raise the possibility of designing strategies, such as those to promote the levels of FH protein, as therapy to prevent complement-mediated vascular dysfunction during DENV infection.
RESULTS
DENV induces FH mRNA but not protein in primary EC and MDM.
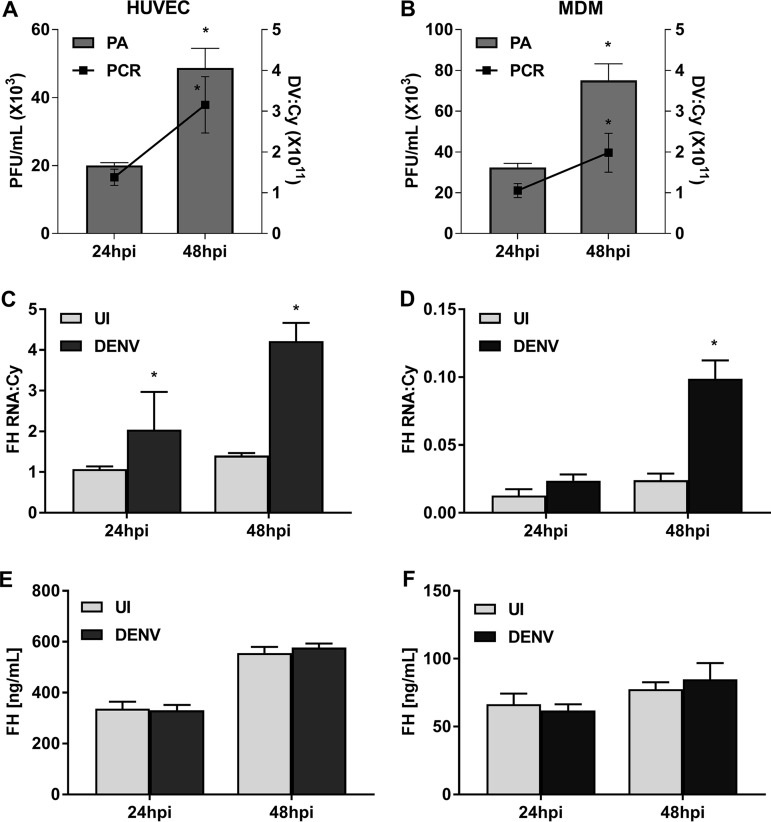
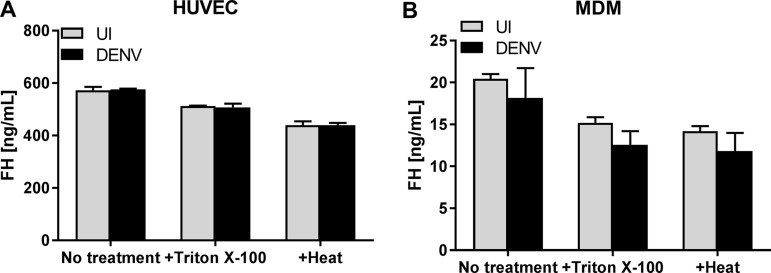
The induction of FH, a negative regulator of the complement AP, was evaluated following DENV infection in two cell types that represent major targets for DENV pathogenesis and replication in vivo: EC and macrophages, respectively. Active virus replication occurred, with increasing DENV RNA and infectious virus release taking place from 24 to 48 h postinfection (hpi) in primary human umbilical vein EC (HUVEC) (Fig. 1A) and monocyte-derived macrophages (MDM) (Fig. 1B). Quantitation of FH mRNA levels demonstrated a significant induction in both HUVEC (Fig. 1C) and MDM (Fig. 1D) following DENV infection. In contrast, the levels of extracellular FH protein showed no significant change in DENV-infected supernatants from either cell type (Fig. 1E and F). Notably, there were much lower levels of FH mRNA and protein in MDM (Fig. 1D and F) than in HUVEC (Fig. 1C and E). The lack of detectable FH protein in cultured supernatant from DENV-infected cells was not due to blocking of FH antibody binding sites required for enzyme-linked immunosorbent assay (ELISA) detection of FH, for example, by a protein-protein interaction, since supernatants collected from DENV-infected cells and treated with either detergent (Triton X-100) or heat (56°C) prior to ELISA showed no difference in FH quantitation (Fig. 2A and B). Analysis of FH protein by Western blotting was attempted, but consistent with the nanogram levels of protein quantitated by ELISA, FH protein in cultured supernatants was undetectable by this method (data not shown).
FIG 1.
DENV infection of HUVEC and MDM induces FH mRNA but not protein. HUVEC and MDM were isolated and were left uninfected or DENV infected at MOI of 1 and 3, respectively. (A, B) At 24 and 48 hpi, supernatants and total cellular RNA were collected from HUVEC (A) and MDM (B) and analyzed by plaque assay (PA; bars) and RT-PCR (lines), with the values being normalized against those for cyclophilin. (C, D) Cell lysates of HUVEC (C) of MDM (D) were analyzed for FH mRNA by RT-PCR, with the values being normalized against those for cyclophilin. (E, F) Supernatants of HUVEC (E) of MDM (F) were analyzed by ELISA for FH protein. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments. *, P < 0.05, Student's unpaired t test. UI, uninfected; DV, dengue virus.
FIG 2.
Treatment with detergent or heat does not increase FH protein detection in DENV-infected samples. HUVEC and MDM were isolated and were left uninfected or DENV infected at MOI of 1 and 3, respectively. At 48 hpi, supernatants of HUVEC (A) and MDM (B) were collected, treated with or without 0.05% Triton X-100 or heat (56°C), and analyzed by ELISA for FH protein. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments.
The discord between DENV induction of FH mRNA and protein is not observed with FB or following TLR3 or TLR4 stimulation of MDM and EC.
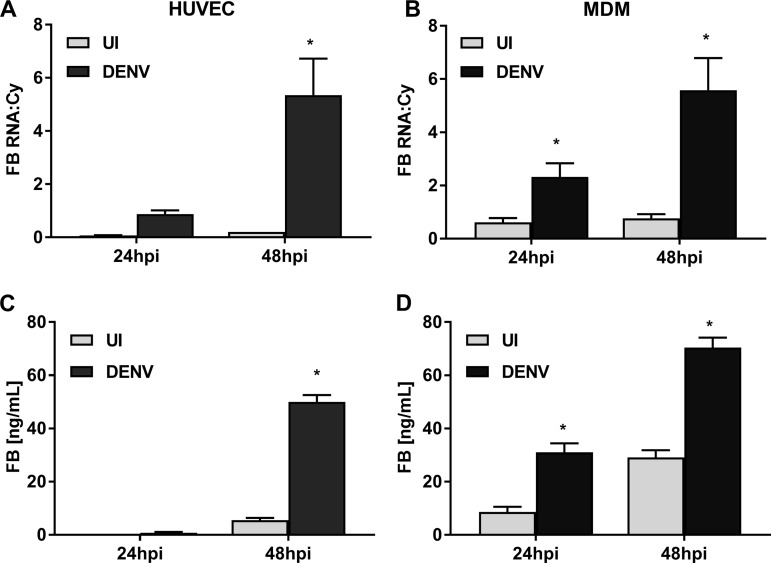
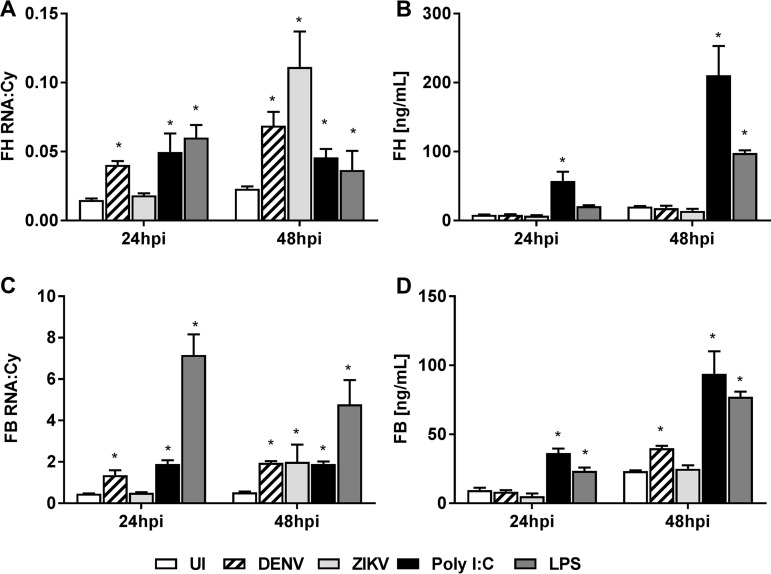
To assess if DENV infection specifically prevents induction of FH protein, changes in FB, another AP complement component, were assessed. Quantitation of FB mRNA levels demonstrated a significant induction in both HUVEC (Fig. 3A) and MDM (Fig. 3B) following DENV infection. Similarly, but in contrast to the results for FH, FB protein levels significantly increased in DENV-infected supernatants from both cell types (Fig. 3C and D). FB mRNA and protein levels were comparable between MDM and HUVEC (Fig. 3). To further assess if the induction of FH mRNA but not protein is specific for infection, cells were stimulated with Toll-like receptor 3 (TLR3) and TLR4 ligands: poly(I·C) and lipopolysaccharide (LPS), respectively. Following TLR stimulation, both FH mRNA (Fig. 4A) and protein (Fig. 4B) were significantly increased at 24 and 48 hpi in MDM. In contrast, DENV infection again induced FH mRNA, but not protein, in the supernatant (Fig. 4A and B). Notably, induction of FH mRNA by DENV was comparable to that by poly(I·C) or LPS, but the increased FH mRNA in DENV-infected cells did not translate into an increase in secreted FH protein (Fig. 4A and B). As expected and consistent with the results presented in Fig. 3, both FB mRNA and protein increased in response to TLR stimulation and DENV infection (Fig. 4C and D). Zika virus (ZIKV) infection induced FH mRNA to a level much higher than that seen with DENV at 48 hpi (Fig. 4A) but still failed to increase FH protein (Fig. 4B). ZIKV infection also induced FB mRNA at 48 hpi (Fig. 4C) but, interestingly, and in contrast to DENV infection, not FB protein (Fig. 4D).
FIG 3.
DENV infection of HUVEC and MDM induces FB mRNA and protein. HUVEC and MDM were isolated and were left uninfected or DENV infected at MOI of 1 and 3, respectively. Infection was verified as described in the legend to Fig. 1A and B. (A, B) At 24 and 48 hpi, total HUVEC (A) and MDM (B) RNA was collected and analyzed for FB mRNA by RT-PCR, and the values were normalized against those for cyclophilin. (C, D) HUVEC (C) and MDM (D) supernatants were collected and analyzed by ELISA for FB protein. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments. *, P < 0.05, Student's unpaired t test.
FIG 4.
Stimulation with TLR ligands but not DENV or ZIKV induces FH and FB mRNA and protein. MDM were isolated and were left uninfected, DENV or ZIKV infected, or stimulated with poly(I·C) or LPS. (A, C) At 24 and 48 hpi, supernatants were collected, the cells were lysed, and total RNA was extracted and analyzed for FH (A) and FB (C) mRNA by RT-PCR. PCR results were normalized against those for cyclophilin. (B, D) Supernatant was collected and analyzed by ELISA for FH (B) and FB (D) proteins. Results represent the mean ± standard deviation for duplicate samples at each time point and are representative of those from three independent infection experiments. Significance was calculated in relation to the results for the uninfected control group. *, P < 0.05, one-way ANOVA/Tukey's test.
Similarly, in HUVEC, poly(I·C) and LPS stimulated both FH mRNA and protein, while DENV induced FH mRNA but not protein (data not shown). ZIKV responses in HUVEC were not reliably defined since ZIKV infection of HUVEC induced a substantial visual cytopathic effect.
Cell surface and intracellular FH is induced in DENV antigen-positive cells.
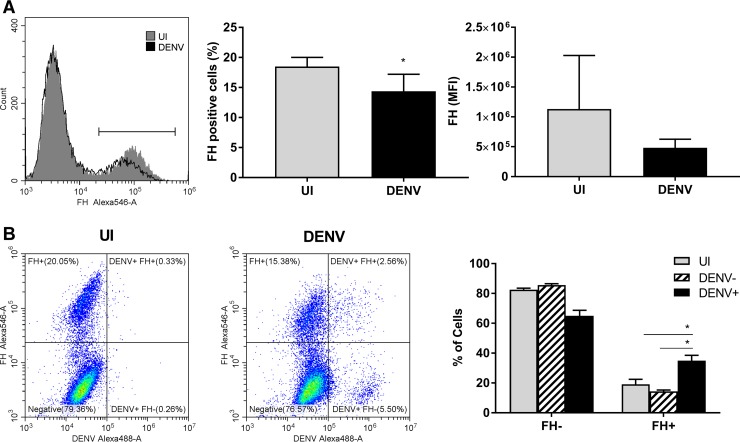
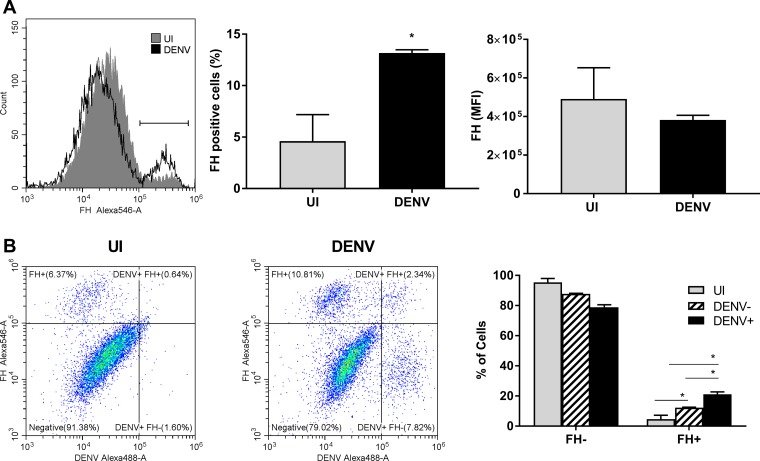
FH also has a cell surface binding capacity (39, 40). To investigate if the lack of an increase in extracellular FH protein is due to FH rebinding to DENV-infected cells, cell surface-bound FH was analyzed by flow cytometry. Surface-bound endogenous FH was lower in DENV-infected HUVEC than in uninfected HUVEC, as determined by the percentage of FH-positive cells when analyzed as combined data from 3 independent experiments (Fig. 5A), although this was significant in only one of three experiments when analyzed individually. Further, as shown in the histogram plot, this finding represents a very minor shift in the FH-positive cell population (Fig. 5A). DENV infection had no significant effect on the mean fluorescent intensity (MFI) of FH binding to cells (Fig. 5A). An increase in FH binding, however, was seen specifically in the DENV antigen-positive but not antigen-negative cells of the DENV-infected HUVEC population (Fig. 5B). In contrast to HUVEC, surface-bound endogenous FH was significantly higher in DENV-infected MDM than in uninfected MDM in terms of the percentage of FH-positive cells but not in terms of the intensity and, thus, the amount of FH bound per cell (Fig. 6A). Again, in contrast to HUVEC, this increase in FH binding was seen in both the DENV antigen-positive and the DENV antigen-negative cells of the DENV-infected MDM population (Fig. 6B). Together, these results confirm that the DENV induction of FH mRNA can result in production of FH protein but that at least some of this protein rebinds back to the infected cell or, in the case of MDM, infected and uninfected cell surfaces.
FIG 5.
Cell surface FH binding is induced in DENV antigen-positive HUVEC. HUVEC were isolated and were left uninfected or DENV infected at an MOI of 1. (A) HUVEC were detached with 5 mM EDTA, stained for FH on cell surfaces and intracellularly for DENV, and analyzed by flow cytometry. Gray and black plots represent FH staining on uninfected (UI) and DENV-infected cells, respectively. The percentage of FH-positive cells was calculated from the gate indicated on the histogram, and the mean florescent intensity (MFI) of the population was determined. *, P < 0.05, Student's unpaired t test. (B) Representative quad plots of FH-Alexa Fluor 546 versus DENV-Alexa Fluor 488 for uninfected and DENV-infected populations. The percentage of FH-positive and -negative cells from uninfected, DENV antigen-negative, and DENV antigen-positive populations was calculated using gates, as shown. Results represent the mean ± standard deviation for triplicate samples, and data from three independent infection experiments were combined. *, P < 0.05, two-way ANOVA/Tukey's test.
FIG 6.
Cell surface FH binding is induced in DENV antigen-positive MDM. MDM were isolated and were left uninfected or DENV infected at an MOI of 3. (A) MDM were detached with 5 mM EDTA, stained for FH on cell surfaces and intracellularly for DENV, and analyzed by flow cytometry. Gray and black plots represent FH staining on uninfected (UI) and DENV-infected cells, respectively. The percentage of FH-positive cells was calculated from the gate indicated on the histogram, and the mean florescent intensity (MFI) of the population was determined, *, P < 0.05, Student's unpaired t test. (B) Representative quad plot graphs of FH-Alexa Fluor 546 versus DENV-Alexa Fluor 488 for uninfected and DENV-infected populations. The percentage of FH-positive and -negative cells from uninfected, DENV antigen-negative, and DENV antigen-positive populations was calculated using gates, as shown. Results represent the mean ± standard deviation for triplicate samples, and data from three independent infection experiments were combined. *, P < 0.05, two-way ANOVA/Tukey's test.
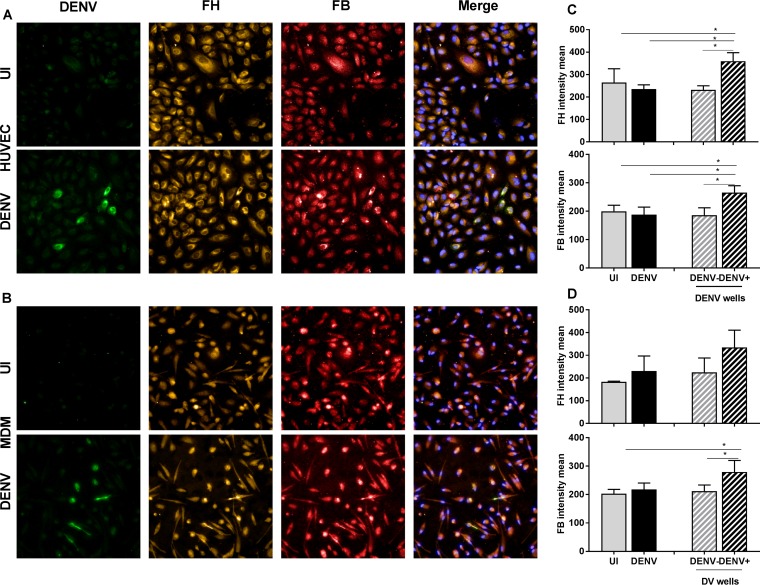
To further confirm the ability of DENV-infected cells to produce FH protein, quantitative immunostaining for intracellular FH and, for comparison, FB was performed. DENV infection of HUVEC (Fig. 7A) or MDM (Fig. 7B) resulted in only a small percentage (3 to 15% of the total cell population) of cells staining DENV antigen positive, as expected from previous studies (11, 18, 47, 48) and evident from the results shown in Fig. 5 and 6. Basal staining for both the FB and FH proteins was detected in uninfected cells. Visually, there was no major change in the overall DENV-infected cell population, but those cells staining for DENV antigen appeared to be more strongly positive for both FH and FB than uninfected cells (Fig. 7A and B). The staining intensity for FB and FH was quantified using an Operetta high-content imaging system. The results showed no increase in the overall FB and FH staining intensity in DENV-infected wells compared to uninfected wells for either cell type (Fig. 7C and D). The FH and FB staining intensities in individual DENV antigen-negative or -positive cells from within the same DENV-infected well were then compared. A significant induction of both the FH and FB proteins in DENV antigen-positive HUVEC compared to antigen-negative cells was apparent (Fig. 7C). In contrast, induction in DENV antigen-negative cells was not significantly different from that in cells in uninfected wells. Similarly, in DENV-infected MDM, FB protein was induced in DENV antigen-positive cells, and although the results were not statistically significant, a similar trend toward increased FH protein was observed (Fig. 7D).
FIG 7.
FH and FB are increased specifically in DENV antigen-positive HUVEC and MDM. (A, B) HUVEC (A) and MDM (B) were isolated and were left uninfected (UI) or DENV infected. At 48 hpi, cells were fixed and immunostained for DENV (green), FH (orange), and FB (red). Nuclei were stained with Hoechst (blue). Cells were imaged with an Operetta high-content imaging system at a ×20 magnification. (C, D) Images of 49 fields of view were analyzed, and the intensity means were calculated using Harmony software. The intensity means for FH and FB were compared in uninfected (gray bars) versus DENV-infected (black bars) wells and in DENV-negative cells (gray bars with a hatching pattern) versus DENV-positive cells (black bars with a hatching pattern) within a well. HUVEC (C) and MDM (D) were analyzed. Results represent the mean ± standard deviation for triplicate samples and are representative of those from three independent infection experiments *, P < 0.05, one-way ANOVA/Tukey's test.
DENV infection can induce FH protein in some immortalized cell lines.
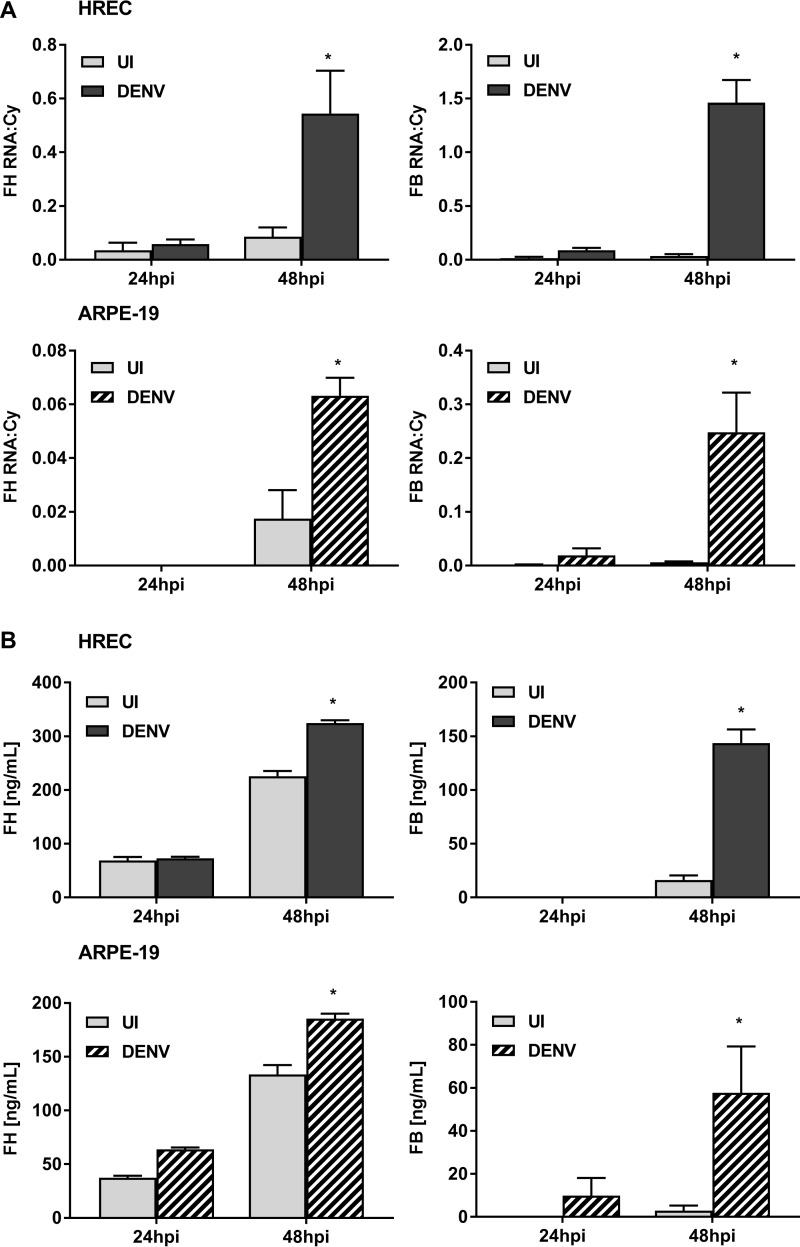
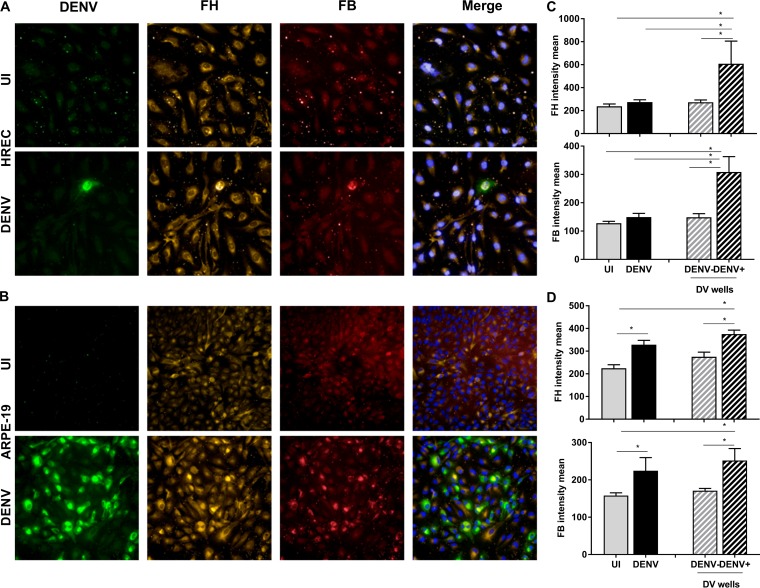
The low susceptibility of HUVEC (18) and MDM (47, 48) to DENV infection in vitro might account for the observed lack of detectable induction of extracellular FH in these cells. To analyze this, responses were evaluated in another EC transformed cell line (HPV E6/E7-transduced human retinal endothelial cells [HREC]), which have low susceptibility to DENV infection, and ARPE-19 retinal pigment epithelial cells, which have high susceptibility to DENV infection (49). Consistent with the results in HUVEC and MDM, the levels of FB and FH mRNA were significantly increased in DENV-infected HREC and ARPE-19 cells at 48 hpi (Fig. 8A). Both FB and, in contrast to HUVEC and MDM, also FH protein were significantly elevated at 48 hpi in HREC and ARPE-19 cells (Fig. 8B). Immunostaining and quantitative analysis for intracellular FH and FB were also performed in HREC and ARPE-19 cells, as shown in Fig. 7 for primary MDM and HUVEC. As expected, DENV infected only a small percentage of HREC but the majority of ARPE-19 cells (Fig. 9A and B). Visually, DENV antigen-positive cells appeared to be more strongly positive for both FH and FB than uninfected cells for both cell types (Fig. 9A and B), and this was confirmed by quantitative imaging, which showed an increase in staining intensity when considered over the entire well or specifically in DENV antigen-positive cells (Fig. 9C and D).
FIG 8.
DENV infection of HREC and ARPE-19 cells induces FH and FB mRNA and protein. HREC and ARPE-19 cells were isolated and were left uninfected (UI) or DENV infected at an MOI of 1. (A) At 24 and 48 hpi, supernatants and total cellular RNA were collected and analyzed for FH and FB mRNA by RT-PCR, and the results were normalized against those for cyclophilin. (B) Supernatants were analyzed by ELISA for FH and FB proteins. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments. *, P < 0.05, Student's unpaired t test.
FIG 9.
DENV infection of HREC and ARPE-19 cells induces intracellular FH and FB proteins. (A, B) HREC (A) and ARPE-19 cells (B) were isolated and were left uninfected (UI) or DENV infected. At 48 hpi, cells were fixed and immunostained for DENV (green), FH (orange), and FB (red). Nuclei were stained with Hoechst (blue). Cells were imaged with an Operetta high-content imaging system at a ×20 magnification. (C and D) Images of 49 fields of view were analyzed, and the intensity means were calculated using Harmony software. The intensity means of FH and FB were compared in uninfected (gray bars) versus DENV-infected (black bars) wells and in DENV-negative cells (gray bars with a hatching pattern) versus DENV-positive cells (black bars with a hatching pattern) within a well. HUVEC (C) and MDM (D) were analyzed. Results represent the mean ± standard deviation for triplicate samples and are representative of those from three independent infection experiments *, P < 0.05, one-way ANOVA/Tukey's test.
DENV infection is associated with indicators of active complement.
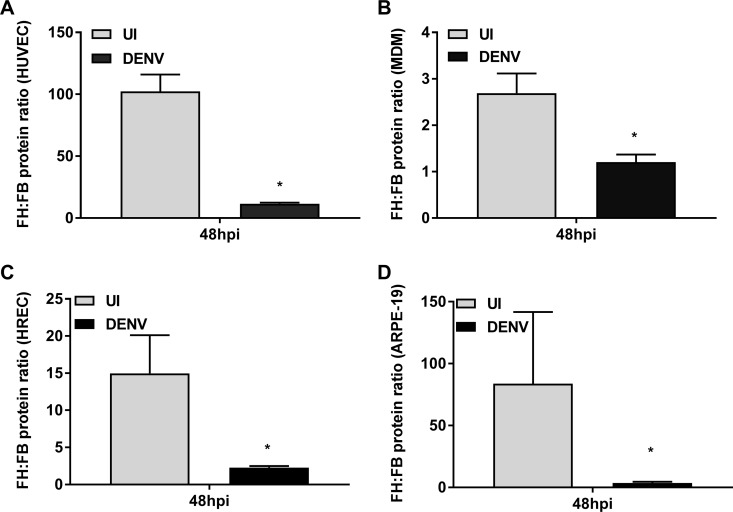
Given that the actions of FH as a negative regulator of the AP oppose complement components, such as FB, that promote AP activity, the ratio of FH/FB protein was calculated. The results demonstrated a significantly lower proportion of FH relative to FB in the supernatant from DENV-infected cells than in that from uninfected cells (Fig. 10). Interestingly, there was a decrease in FH/FB of greater than 90% in HUVEC (Fig. 10A) but only approximately 50% in MDM (Fig. 10B). Additionally, although FH was induced in the supernatant of DENV-infected HREC and ARPE-19 cells, analysis of the FH/FB ratio revealed that, consistent with the results from MDM and HUVEC, FB was induced at higher levels relative to FH protein, resulting in a significant decrease in the FH/FB ratio (Fig. 10C and D).
FIG 10.
DENV induces lower levels of FH than FB. HUVEC (A), MDM (B), HREC (C), and (D) ARPE-19 cells were isolated and were left uninfected (UI) or DENV infected. At 48 hpi, supernatants were analyzed by ELISA for FH and FB proteins, and the ratio between the two was calculated. A lower ratio implies lower levels of FH than FB. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments. *, P < 0.05, Student's unpaired t test.
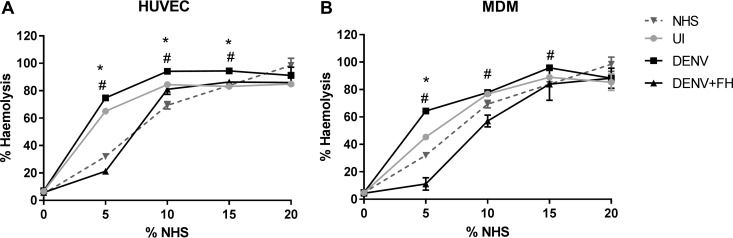
To test the ability of cultured supernatant to promote the complement AP, supernatants were incubated with normal human serum (NHS) under buffer conditions specific for the AP, and lysis of rabbit erythrocytes was quantitated. Supernatant from DENV-infected HUVEC showed a significantly greater ability to promote AP-mediated hemolysis than that from uninfected cells (Fig. 11A). Similar results were obtained with supernatant from DENV-infected MDM (Fig. 11B), suggesting in both cases that DENV-infected cells produce secreted factors that promote AP activity. Importantly, the addition of purified exogenous FH protein to supernatants from DENV-infected HUVEC or MDM significantly impaired the ability of these supernatants to promote the activity of the AP (Fig. 11), supporting a role for FH in controlling complement AP activity in this assay system.
FIG 11.
DENV infection releases factors that promote complement AP activity. HUVEC (A) and MDM (B) were isolated and were left uninfected (UI) or DENV infected. At 48 hpi, supernatants were collected, mixed with different concentrations of normal human serum (NHS) and/or purified FH protein, and analyzed for the ability to promote AP activity by quantitation of hemolysis of rabbit erythrocytes under buffer conditions specific for the AP. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments. *, P < 0.05 for the comparison between uninfected and DENV cell supernatant by one-way ANOVA/Tukey's test; #, P < 0.05 for the comparison between DENV and DENV cell supernatant supplemented with purified FH by one-way ANOVA/Tukey's test.
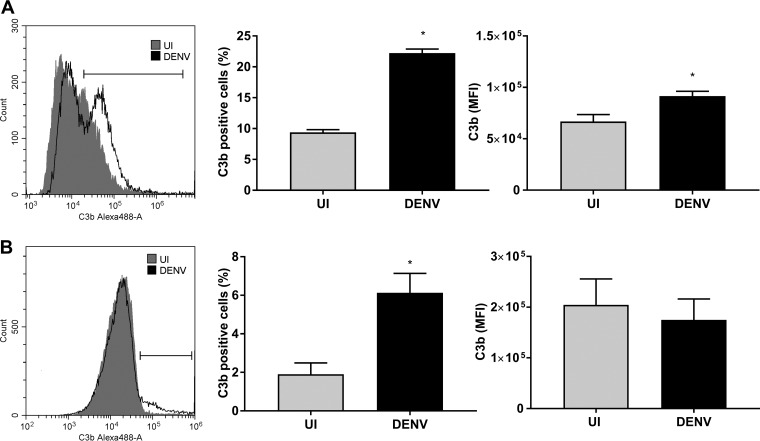
Additionally, during a viral infection and in the presence of the full spectrum of complement proteins, C3b should be deposited onto the surface of infected cells that can be regulated by the opposing roles of FH and FB (30–32). While FB interacts with C3b to promote C3 convertase, FH, acting with FI, cleaved surface-bound C3b to form inactivated C3b (iC3b), avoiding pathogenic C3b deposition (30, 31, 50). Thus, C3b binding to DENV-infected cells was quantitated by flow cytometry, with NHS being used as a source of complement. The results showed a significant increase in C3b binding to DENV-infected EC compared to uninfected cells in terms of the amount of C3b bound and the number of C3b-positive cells seen (Fig. 12A). Consistent with the results in HUVEC, C3b showed a similarly increased binding to DENV-infected MDM (Fig. 12B).
FIG 12.
DENV infection results in increased cellular C3b deposition. HUVEC (A) and MDM (B) were isolated and were left uninfected (UI) or DENV infected at MOI of 1 and 3, respectively. At 48 hpi, cells were incubated with NHS for 30 min, detached with 5 mM EDTA, stained for C3b deposition, and analyzed by flow cytometry. Gray and black plots represent C3b staining on uninfected and DENV-infected cells, respectively. The percentage of C3b-positive cells was calculated from the gate indicated on the histogram, and the mean florescent intensity (MFI) of the population was determined. Results represent the mean ± standard deviation for duplicate samples and are representative of those from three independent infection experiments. *, P < 0.05, Student's unpaired t test.
DISCUSSION
The complement system is a vital part of the body's response to pathogens and is an important player in host defenses against DENV (51–53). In contrast, overactivity of complement, in particular, the AP, is associated with increased DENV disease severity (8, 26, 45); thus, fine control of specific aspects of the complement pathways is needed to trigger a protective but nondetrimental immune response. It is well-known that FH is one of the main negative regulators of AP activity (30, 33, 37). Additionally, macrophages and EC are recognized as major targets for DENV replication and pathogenesis in vivo, and both these cell types may contribute to FH production, antiviral immune responses, and induction of vascular permeability, a hallmark of DENV disease (11–13, 54). Thus, this work focused on defining the DENV-induced changes in FH in macrophages and EC and linking these with changes in other complement components, FB and C3b, to understand the cellular and molecular responses that contribute to the complement AP overactivity reportedly associated with DENV disease.
First, the results showed that DENV infection of HUVEC and MDM effectively induces both FB and FH mRNA. Surprisingly, however, this translates to a significant increase only in FB protein, while the levels of secreted FH protein remain unchanged from those in uninfected cells for both cell types. Induction of FB mRNA has previously been reported in DENV-infected HUVEC, although with a higher 34-fold induction of mRNA (16). Results presented here confirm induction of FB mRNA and extend this to demonstrate an increase in FB protein. This report is the first to describe the DENV induction of FH mRNA and additionally presents several lines of evidence suggesting that the resultant production and/or release of FH protein from the infected cell is compromised. This inability to detect an increase in secreted FH from DENV-infected cells is not due to FH being complexed in the supernatant to other proteins, as validated by detergent (Triton X-100) or heat (56°C) treatment of samples prior to ELISA. Additionally, since FH also exerts its regulatory function on the cell surface via binding to cell surface glycosaminoglycans (39–41), DENV could influence the rebinding of FH to infected cells to reduce apparent levels in the supernatant. Indeed, the results showed increased levels of endogenous FH binding at the surface of DENV antigen-positive cells within a DENV-infected population of either HUEVC or MDM. In the DENV-infected population, endogenous surface-bound FH was also increased in the DENV antigen-negative population of DENV-infected MDM but not HUVEC, suggesting cell type differences in the ability of FH to bind and protect the surfaces of uninfected bystander cells. Additionally, the lack of an increase in FH in supernatants from DENV-infected MDM and EC was not due to an inability per se of our cell system to induce both FH mRNA and secreted protein, with both being induced following stimulation of HUVEC or MDM with TLR3 or -4 agonists. This is consistent with the findings of other studies demonstrating an upregulation of FB mRNA and protein by LPS and poly(I·C) in macrophages (55, 56), increased production of FH protein in U937 cells upon LPS stimulation (57), and emerging evidence indicating extensive cross talk between complement and TLR signaling (58). Since DENV stimulates TLR3 (59) and TLR4 (22, 23) and given that the DENV-induced FH mRNA levels are comparable to or greater than those induced by TLR3/4, our results imply that the induction of FH mRNA but not FH protein in the supernatant of DENV-infected cells is due to a virus-induced defect in the ability to produce more FH protein extracellularly in these primary cell types. This discordance between FH mRNA and extracellular levels of FH was also observed following infection with the related flavivirus ZIKV. Additionally, DENV induced both FH mRNA and secreted protein in the cell lines ARPE-19 and HREC.
To investigate FH protein production further, intracellular FH was quantitated. Although no significant change in intracellular FH protein was detected in MDM, a small but significant increase in intracellular FH protein was detected in DENV-infected HUVEC and a substantial increase in FH was observed following DENV infection of ARPE-19 cells and HREC. For the latter, as with MDM and HUVEC, this was even in the face of a low level of DENV-infected cells in culture. This stronger induction of intracellular levels of FH in HREC than HUVEC correlates with the ability to detect extracellular changes in FH protein levels in HREC. This suggests that DENV infection of only a small number of cells does not of itself preclude detection of increased secreted FH, but the level of intracellular FH protein induction may be restricted in primary MDM and HUVEC compared to that in the cell lines ARPE-19 and HREC. Overall, although significant levels of FH mRNA are induced following DENV infection, there is a poor subsequent increase in FH protein extracellularly. This may, at least in part, be accounted for by increased cell surface binding of FH, as described above, although DENV infection could also influence the production of extracellular FH by blocking intracellular FH translation, inhibiting protein secretion, or inducing FH degradation either inside or outside the cell, and these possible mechanisms remain to be investigated.
One of the primary consequences of poor induction of FH following DENV infection is a reduction in the extracellular negative regulatory capacity for the AP. This is of particular importance in the face of increases in other complement AP components, as described previously for FD (45) and shown here for FB and C3b. The latter finding of increased binding of C3b during DENV infection has also been described previously (60), and the increased C3b binding observed in our study may be related to activation of the AP but could also result from the activity of the classical and lectin pathways. Additionally, the results of our study show for the first time a consistent imbalance in components of the complement AP, with higher relative levels of FB than FH being found in DENV-infected cells from all cell types examined. Notably, this imbalance still occurs even when extracellular FH protein is significantly induced in the HREC and ARPE-19 cell lines. FB activates the AP by binding C3b to form the AP C3 convertase (C3bBb), which stimulates complement consumption and the production of the anaphylatoxins C3a and C5a (28, 30). Likewise, FH may be recruited to infected cell surfaces to regulate C3b deposition and negatively influence the formation of C3 convertase. Additionally, there may be other cell surface complement regulators, such as conserved repeat 1, monocyte chemoattractant protein (CD46), and decay-accelerating factor (CD55) (50), that may be modulated during DENV infection, and these remain to be investigated. The failure to induce local secretion of FH protein, alongside the increased production of FB, elevated C3b binding to DENV-infected cells, and a greater ability of supernatant from DENV-infected cells to promote AP activity, suggests increased complement AP activity in the local microenvironment of DENV-infected cells. This is consistent with previous reports linking high levels of complement consumption and the hyperactivity of the AP to the severity of DENV-associated vascular leakage (8, 16, 25–27, 45). Recently, it has been shown that the AP ability to lyse rabbit erythrocytes is dependent on the levels of FB and FH, among other factors (61). While the imbalance in the ratio of FH/FB in DENV-infected cells may not be directly or solely responsible for this increased AP activity, our observations of FH and FB changes in ZIKV-infected cells suggest that this ratio may be related to pathogenesis. Similar to DENV infection, ZIKV infection induces FH mRNA but not an increase in FH protein, and in contrast to DENV, ZIKV additionally fails to induce FB protein. Thus, for ZIKV-infected cells, the relative levels of FB and FH are unchanged and C3b deposition and complement activity would be predicted to also be unaffected. In terms of clinical disease, DENV is well described to induce effects on the vascular endothelium, but this is not a characteristic of ZIKV disease. Thus, we propose that the dysregulation and imbalance in the complement AP components FB and FH that we have described here are likely of significance to the pathogenesis of DENV-induced vascular leakage.
It is well described that microorganisms can hijack the complement system by binding to regulatory proteins, such as FH, and utilize their negative regulatory function to downregulate complement AP activity and evade complement function (62–65). West Nile virus NS1 binds to FH, although DENV NS1 does not (64). DENV NS1 does, however, bind C1s and C4 to restrict and evade classical/lectin pathway activation (60) and binds to vitronectin to prevent C9 polymerization and MAC formation (66). Our findings here are in contrast to these previously reported pathogen evasion strategies, with induction of FB but not effective extracellular FH protein production and increased C3b binding to cells providing a mechanism supporting overactivity of complement rather than the DENV-FH interaction providing a complement-pathogen evasion strategy.
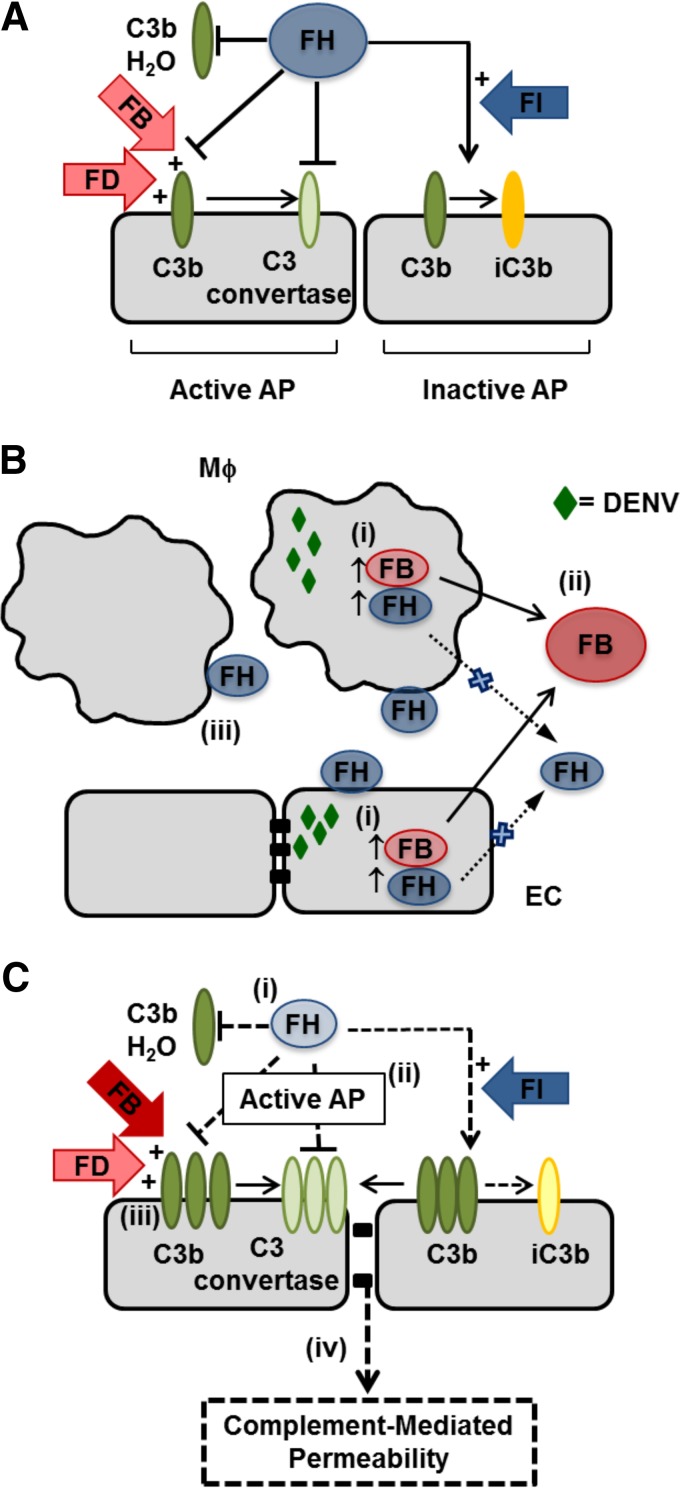
As modeled in Fig. 13, in a normal situation, where FB and FH are produced in a balanced ratio, deposition of C3b on cells is tightly controlled by opposing roles of complement AP regulators, leading to controlled production of inactivated C3b (iC3b) and C3 convertase (Fig. 13A). DENV infection of EC and MDM induces FH and FB mRNA and protein inside DENV-infected cells but fails to produce significant amounts of extracellular secreted FH protein in the fluid phase (Fig. 13B). FH can bind back to the surface of both DENV antigen-positive and antigen-negative cells in MDM but mainly DENV antigen-positive cells for HUVEC (Fig. 13B). In DENV-infected cells, this study has defined a reduced FH/FB in the extracellular space, increased AP activity released from cells, and increased C3b binding to cell surfaces (Fig. 13C). The increased ability of secreted components from DENV-infected EC or MDM to activate the AP is reversed by addition of exogenous FH and thus may relate to our described changes in FB and FH or as yet undescribed factors produced from DENV-infected cells that can promote AP activity (Fig. 13C). These overall changes are predicted to result in increased production of C3 convertase (C3bBb) and reduced C3b decay or conversion to iC3b, leading to increased complement activity. For the endothelium, this excessive fluid phase and cell surface complement activity has the potential to affect EC function and promote vascular permeability (Fig. 13C).
FIG 13.
Summary and proposed complement AP interactions during DENV infection. (A) Normally, FB and FH are produced in a balanced ratio and the cleavage of C3, the deposition of C3b, and the formation of either iC3b or C3 convertase on cells are tightly controlled by opposing roles of FB and FH in conjunction with other regulators, such as FD and FI. (B) DENV infection of macrophages (Mϕ) and EC (i) induces FB and FH mRNA but intracellular protein only in DENV-antigen positive cells, (ii) induces FB protein but little increase in extracellular FH protein production, and (iii) increases FH binding to cell surfaces of both uninfected DENV antigen-negative and DENV antigen-positive MDM but only in DENV antigen-positive HUVEC. (C) In the local environment of DENV-infected cells, this study shows that there is (i) a reduced amount of FH protein relative to FB, (ii) an increased ability of soluble factors to promote the activity of the AP, and (iii) increased binding of C3b at the cell surface. (iv) In the context of the endothelium, this is predicted to promote vascular permeability.
In conclusion, our study has defined changes in regulators of the complement AP using primary cells relevant to the pathology of DENV infection and disease and the cellular mechanisms by which this pathogenic overactivity of the complement AP may be mediated. It has been hypothesized that during DENV infection, the endothelium coordinates a multitude of signals and factors released by immune cells, like macrophages, which ultimately act on the endothelium to contribute to vascular permeability and hemorrhage (6, 11, 67). This study proposes an additional layer of contributing signals that affect the EC barrier function during DENV infection: the AP activity in the cellular microenvironment of both macrophages and EC, with DENV induction of FB and deposition of C3b but limited induction of FH, particularly at the protein level at the surface of an infected cell. These results stimulate interest in complement AP regulators, for example, the development of agents to promote FH protein production from MDM and EC, as therapeutics to alleviate complement hyperactivity and potentially reduce DENV-induced vascular permeability and severe forms of DENV disease.
MATERIALS AND METHODS
Cell lines and cell culture.
HUVEC were isolated from human umbilical cords collected with approval from the Central Northern Adelaide Health Service Ethics Committee. In brief, HUVEC were isolated by collagenase digestion and cultured on 0.2% (wt/vol) gelatin (Sigma)-coated flasks in M199 medium (HyClone) supplemented with 20% (vol/vol) fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% glutamine (all from Gibco) plus 0.3% endothelial cell growth supplement (BD Bioscience). HUVEC were utilized in infection studies at passages 1 to 4. Human MDM were isolated from healthy blood donors at the Australian Red Cross Blood Service and collected under ethics approval from the Southern Adelaide Clinical Human Research Ethics Committee (SACHREC). Monocytes were isolated by adherence and cultured for 4 to 5 days in Dulbecco modified Eagle medium (DMEM) supplemented with 10% (vol/vol) FBS, 10% (vol/vol) human heat-inactivated serum, 1% penicillin-streptomycin, and 1% glutamine to differentiate into MDM, as previously described and validated by CD14 and Wright-Giemsa staining (48). Human serum was collected from healthy donors in accordance with SALHN/HREC ethics approval and was determined to be DENV antibody negative by a diagnostic rapid immunochromatographic IgG and IgM assay. Generation and characterization of the HREC line has been described previously (68). The HREC line was cultured in MCDB-131 medium (Sigma-Aldrich) with 5% FBS and endothelial growth factors (EGM-2 SingleQuots supplement, omitting FBS, hydrocortisone, and gentamicin; Clonetics-Lonza, Walkersville, MD). The ARPE-19 cell line (American Type Culture Collection, Manassas, VA) was cultured in DMEM–F-12 medium supplemented with 5% FBS. All cells were grown in a humidified incubator with 5% CO2 in air and at 37°C.
DENV production and infection.
Mon601, a laboratory clone of the DENV serotype 2 New Guinea C strain, was used for infections (69) and is here referred to as DENV. Virus stocks were produced from in vitro-transcribed RNA that was transfected into baby hamster kidney clone 21 (BHK-21) cells and amplified in Aedes albopictus C6/36 cells. Cell culture supernatants containing virus were harvested, clarified, filtered, and stored at −80°C until use. The titer of infectious virus was determined by plaque assay using African green monkey kidney (Vero) cells and quantitated as the number of PFU per milliliter. HUVEC, ARPE-19 cells, and HREC were either left uninfected or infected with DENV at a multiplicity of infection (MOI) of 1. MDM were infected at an MOI of 3 as previously described (11). After 90 min of infection, the inoculum was removed, the cells were washed with phosphate-buffered saline (PBS; pH 7.4), and fresh medium was added. Supernatants and cells were harvested after 24 and 48 hpi and stored at −80°C until analysis. ZIKV infections utilized the cosmopolitan ZIKV strain PRVABC59, which was amplified in C6/36 cells; stocks were collected, titers were determined, and cells were infected as described above for DENV.
RNA extraction and real-time qRT-PCR.
Total RNA was extracted from cells using the TRIzol reagent (Ambion Life Technologies) according to the manufacturer's instructions. The extracted RNA was DNase treated (Zymo Research) and quantitated by spectrophotometry (NanoDrop Elite; Thermo Scientific). RNA (0.5 μg) was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (NEB) and random hexamers (NEB). The cDNA template was subjected to real-time quantitative reverse transcription-PCR (qRT-PCR) using iTaq SYBER green (Bio-Rad) in a Rotor-Gene 600 real-time PCR cycler (Corbett Research) and primers for DENV and cyclophilin as previously described (18). The cyclophilin gene was used as a normalization gene since its expression did not change in DENV-infected cells in our prior studies (18, 48) and, similarly, in the study described herein, where for the same input amount of RNA, threshold cycle (CT) values for cyclophilin were comparable. For FH PCR, primers were designed to anneal with short consensus repeat 2 (SCR2): forward (f) primer, 5′AGGCCCTGTGGACATC3′; reverse (r) primer, 5′AACTTCACATATAGGAATATC3′. FB PCR primers were as previously reported (70): f primer, 5′ACTGAGCCAAGCAGACAAGC3′; r primer, 5′AGAAGCCAGAAGGACACACG3′. Both FH and FB PCRs were performed under the following conditions: 1 cycle of 95°C for 5 min; 40 cycles of 95°C for 15 s, 59°C for 20 s, and 72°C for 20 s; and 1 cycle of 72°C for 5 min. All PCR mixtures included high- and low-copy-number comparative controls. The results were normalized against those for cyclophilin, and the relative RNA level was determined by the ΔCT method (71).
Flow cytometry.
For FH and DENV staining, HUVEC or MDM were cultured in a 6-well plate and infected as described above. After 48 h, cells were washed twice with PBS and detached by scraping in PBS with 5 mM EDTA. Cells were washed again with PBS and blocked with 1% (wt/vol) bovine serum albumin (BSA) for 30 min at room temperature. Cells were rinsed once with PBS and incubated with a goat anti-human FH (1:25; Calbiochem) for 1 h at room temperature. After three washes with PBS–1% (wt/vol) BSA, cells were incubated for 1 h at room temperature with Alexa Fluor 546-labeled anti-goat immunoglobulin (1:200; Invitrogen). Subsequently, cells were fixed with 2% (wt/vol) paraformaldehyde (Sigma) for 10 min at room temperature. After three washes with PBS–1% (wt/vol) BSA, cells were permeabilized with 0.05% (wt/vol) octylphenoxy poly(ethyleneoxy) ethanol (Igepal CA-630; Sigma) in PBS for 20 min. Cells were washed again with PBS and blocked with 1% (wt/vol) BSA for 30 min at room temperature. Cells were rinsed once with PBS and incubated overnight at 4°C with a mouse anti-DENV (1:10), D1-4G2-4-15 (ATCC HB-112TM). After three washes with PBS–1% (wt/vol) BSA, cells were incubated for 1 h at room temperature with Alexa Fluor 488-labeled donkey anti-mouse immunoglobulin (1:200; Invitrogen). Following a final set of PBS-BSA washes, cells were analyzed by flow cytometry. Flow cytometry was performed with a CytoFlex S flow cytometer (Beckman Coulter Inc.) and analyzed by CytExpert (version 2.0.0.153) software (Beckman Coulter Inc.).
For detection of C3b deposition, HUVEC or MDM were cultured and infected as described above. At 48 hpi, the culture medium was removed and cells were incubated in M199 medium (HyClone) containing 10% NHS (as the complement source) for 30 min at 37°C in a 5% CO2 incubator. Cells were also incubated with the corresponding medium containing 10% heat-inactivated serum as a negative control. Cells were washed and detached with PBS–5 mM EDTA. C3b deposition was detected with a mouse anti-human C3b (1:25; BioLegend) and Alexa Fluor 488-labeled donkey anti-mouse immunoglobulin (1:200; Invitrogen). Cells were fixed and analyzed by flow cytometry as described above.
Treatment of TLR ligands.
HUVEC or MDM cells were treated with 10 μg/ml poly(I·C) (TLR3) or 1 μg/ml LPS (TLR4) for 24 and 48 h. DENV infection was carried out in parallel and as described above. Supernatants and cells were harvested after 24 and 48 hpi and stored at −80°C until analysis. TLR ligands were purchased from Sigma.
Human complement FH purification.
FH was purified from human serum by one-step affinity chromatography using CNBr-activated Sepharose 4B coupled to a sheep anti-human FH polyclonal antibody (pAb) (72). Briefly, total human serum was diluted 1:2 in PBS and loaded onto the column, and the flowthrough was reloaded at least four times. After washing with PBS, FH was eluted with 0.1 M glycine (pH 2.3) and immediately neutralized with 1 M Tris-HCl (pH 8.8). Eluted fractions were analyzed by SDS-PAGE under reducing and nonreducing conditions and Western blotting using the primary goat anti-human FH pAb (1:2,000; Calbiochem) and the secondary anti-goat IgG coupled to horseradish peroxidase (1:30,000; Calbiochem). Bound complexes were detected by chemiluminescence (Clarity Western ECL substrate; Bio-Rad) and visualized with a LAS4000 imaging system (Fuji Imaging Systems). FH-containing fractions were pooled and concentrated using an Amicon Ultra 0.5-ml centrifugal filter (100-molecular-weight cutoff; Millipore), and the purity of FH was reassessed by Western blotting as described above.
Quantitation of FH and FB by ELISA.
FH or FB proteins were quantitated in cell culture supernatant by ELISA. For FH detection, 96-well microtiter plates (Greiner) were coated with goat anti-human FH (Calbiochem) at 10 μg/ml diluted in carbonate-bicarbonate buffer, pH 9.6, and incubated overnight at 4°C. Plates were blocked with 2% (wt/vol) BSA (Sigma) diluted in PBS (pH 7.4) for 1 h at 37°C. Purified FH protein, isolated as described above, was used to generate a standard curve. Standards and supernatant samples were diluted in PBS containing 1% (wt/vol) BSA (PBS-BSA) and incubated on the plates for 2 h at 37°C. The plates were washed five times with PBS containing 0.05% (vol/vol) Tween 20 (PBS-T) and incubated with a mouse anti-human FH monoclonal antibody (Abcam) diluted 1:10,000 in PBS-BSA for 1 h at 37°C. The plates were washed again and incubated with an anti-mouse IgG coupled to horseradish peroxidase (Promega) and diluted 1:10,000 in PBS-BSA for 1 h at 37°C. The plates were washed seven times and developed with tetramethylbenzidine peroxidase substrate (KPL), the reactions were stopped with 1 M sulfuric acid, and the absorbance at 450 nm was quantitated in a microplate reader (Beckman Coulter). A standard curve with purified FH was established using a linear regression curve (R2 > 0.99) with eight standard concentrations. The range of detection was from 20 to 1,000 ng/ml. To disrupt possible interactions between FH and viral protein(s), supernatants were treated with 0.05% Triton X-100 (Sigma) for 30 min at room temperature or incubated for 30 min at 56°C and then evaluated by the FH ELISA as described above.
The levels of FB in the cell culture supernatants were measured using a human FB ELISA kit (Abcam) in accordance with the manufacturer's instructions.
AP in vitro activity assay.
Whole blood was collected under aseptic conditions from a healthy rabbit in accordance with Flinders University Animal Welfare Committee approvals for collection of scavenge material. Blood was collected into a conical flask containing glass beads and gently swirled until a clot formed. The defibrinated blood was decanted and washed three to four times in AP buffer (Veronal-buffered saline containing 0.01 M EGTA and 0.1% gelatin, pH 7.5) until the supernatant was clear. The cells in the cell-containing supernatant were enumerated, and 5 × 107 cells/ml of rabbit erythrocytes were resuspended in AP buffer and used as a master stock for the hemolysis assay.
For the assay, NHS was incubated with AP buffer for 15 min on ice to inactivate the classical and lectin complement pathways. Uninfected and DENV-infected cell supernatants were mixed with treated NHS (to support the AP activity) at from 5 to 20% NHS. To evaluate the possible effect of FH, DENV-infected cell supernatant was supplemented with exogenous purified FH protein at 500 μg/ml and serially diluted with different concentrations of NHS as described above. Fifty microliters of each NHS-supernatant mix was added in duplicate to a flat-bottom 96-well plate (Costar), and 50 μl of the rabbit erythrocyte master stock was overlaid onto the wells. The plate was incubated for 30 min at 37°C with intermittent agitation. Three controls were included in the assay: NHS serially diluted in AP buffer, a 100% hemolysis control consisting of erythrocytes mixed with water 1:1, and an erythrocyte cell blank consisting of erythrocytes mixed with AP buffer 1:1. After incubation, 150 μl ice-cold saline (0.15 M NaCl) was added to all wells except the 100% lysis wells, the plate was centrifuged, and 150 μl of the supernatant was transferred to a new plate. Hemolysis was assessed by measuring the absorbance at 405 nm in a microplate reader (Beckman Coulter), and percent hemolysis was calculated by the following equation: percent lysis = [(OD405 for the sample − OD405 for the blank)/(OD405 for total lysis − OD405 for the blank)] × 100, where OD405 is the optical density at 405 nm.
Immunostaining and high-throughput image analysis.
HUVEC, MDM, ARPE-19 cells, or HREC (1 × 104) were plated in a 96-well plate (Cell Carrier Ultra; PerkinElmer) and allowed to attach for 24 h. Cells were DENV infected as described above, and at 48 hpi cells were fixed with 2% (wt/vol) paraformaldehyde (Sigma) for 10 min at room temperature. After three washes with PBS, cells were permeabilized with 0.05% (vol/vol) Igepal CA-630 (Sigma) in PBS for 20 min. Cells were washed again with PBS and blocked with 1% (wt/vol) BSA and 2% (vol/vol) normal goat serum diluted in Hanks' balanced salt solution (Gibco) for 30 min at room temperature. The cells were rinsed once with PBS and incubated overnight at 4°C with a mouse anti-DENV (1:10), D1-4G2-4-15 (ATCC HB-112); a goat anti-human FH (1:25; Calbiochem); and a rabbit anti-human FB (1:25; Santa Cruz). After three washes with PBS, the cells were incubated for 1 h at room temperature with the corresponding secondary antibodies: Alexa Fluor 488-labeled donkey anti-mouse immunoglobulin (1:75; Invitrogen), donkey anti-sheep immunoglobulin-Cy3 (1:75; Invitrogen), and Alexa Fluor 647-labeled goat anti-rabbit immunoglobulin (1:75; Invitrogen). Nuclei were stained with Hoechst 33342 (5 μg/ml). Following a final set of PBS washes, cells were imaged with an Operetta high-content imaging system with Harmony software (PerkinElmer) at a ×20 magnification. Forty-nine different images, representing approximately 10,000 cells, were taken for each well. Nuclei and the cytoplasm were discriminated using the Hoechst and Cy3 channels, respectively. The mean Alexa Fluor 488, Cy3, and Alexa Fluor 647 intensity in the cell cytoplasm of each individual cell was calculated. Using visual observation and intensity histograms, an Alexa Fluor 488 intensity threshold was set to define DENV-infected cells. Imaging was performed at the Flinders University Cell Screen South Australia (CeSSA) facility.
Statistical analysis.
Results were expressed as the mean ± standard deviation (SD), and statistical analyses were performed using a two-tailed unpaired Student t test or one-way or two-way analysis of variance (ANOVA) in GraphPad Prism (version 7) software (GraphPad Software, La Jolla, CA, USA). Differences were considered statistically significant if P was <0.05.
ACKNOWLEDGMENTS
We thank the following people: Tahlia Hayes for initial establishment of the FH and FB RT-PCR, Yuefang Ma and Andrew J. Stempel for assistance with culture of the retinal cell lines, and Julie Calvert for ongoing technical support.
This research was supported in part by grants from the Flinders Medical Centre Research Foundation and the Australian Research Council (FT130101648 to J. R. Smith). Sheila Cabezas is supported by an Australian international postgraduate research scholarship.
REFERENCES
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. 2010. Dengue: a continuing global threat. Nat Rev Microbiol 8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ. 1998. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11:480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 2009. Dengue: guidelines for diagnosis, treatment, prevention and control: new edition. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 5.Basu A, Chaturvedi UC. 2008. Vascular endothelium: the battlefield of dengue viruses. FEMS Immunol Med Microbiol 53:287–299. doi: 10.1111/j.1574-695X.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trung DT, Wills B. 2010. Systemic vascular leakage associated with dengue infections—the clinical perspective. Curr Top Microbiol Immunol 338:57–66. doi: 10.1007/978-3-642-02215-9_5. [DOI] [PubMed] [Google Scholar]
- 7.Hottz ED, Oliveira MF, Nunes PC, Nogueira RM, Valls-de-Souza R, Da Poian AT, Weyrich AS, Zimmerman GA, Bozza PT, Bozza FA. 2013. Dengue induces platelet activation, mitochondrial dysfunction and cell death through mechanisms that involve DC-SIGN and caspases. J Thromb Haemost 11:951–962. doi: 10.1111/jth.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nascimento EJ, Hottz ED, Garcia-Bates TM, Bozza F, Marques ET Jr, Barratt-Boyes SM. 2014. Emerging concepts in dengue pathogenesis: interplay between plasmablasts, platelets, and complement in triggering vasculopathy. Crit Rev Immunol 34:227–240. doi: 10.1615/CritRevImmunol.2014010212. [DOI] [PubMed] [Google Scholar]
- 9.Halstead SB. 1989. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev Infect Dis 11(Suppl 4):S830–S839. doi: 10.1093/clinids/11.Supplement_4.S830. [DOI] [PubMed] [Google Scholar]
- 10.Green S, Rothman A. 2006. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis 19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- 11.Carr JM, Hocking H, Bunting K, Wright PJ, Davidson A, Gamble J, Burrell CJ, Li P. 2003. Supernatants from dengue virus type-2 infected macrophages induce permeability changes in endothelial cell monolayers. J Med Virol 69:521–528. doi: 10.1002/jmv.10340. [DOI] [PubMed] [Google Scholar]
- 12.Malavige GN, Ogg GS. 2017. Pathogenesis of vascular leak in dengue virus infection. Immunology 151:261–269. doi: 10.1111/imm.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzelnick LC, Coloma J, Harris E. 2017. Dengue: knowledge gaps, unmet needs, and research priorities. Lancet Infect Dis 17:e88–e100. doi: 10.1016/S1473-3099(16)30473-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnamurti C, Peat RA, Cutting MA, Rothwell SW. 2002. Platelet adhesion to dengue-2 virus-infected endothelial cells. Am J Trop Med Hyg 66:435–441. doi: 10.4269/ajtmh.2002.66.435. [DOI] [PubMed] [Google Scholar]
- 15.Azizan A, Sweat J, Espino C, Gemmer J, Stark L, Kazanis D. 2006. Differential proinflammatory and angiogenesis-specific cytokine production in human pulmonary endothelial cells, HPMEC-ST16R infected with dengue-2 and dengue-3 virus. J Virol Methods 138:211–217. doi: 10.1016/j.jviromet.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Dalrymple NA, Mackow ER. 2012. Endothelial cells elicit immune-enhancing responses to dengue virus infection. J Virol 86:6408–6415. doi: 10.1128/JVI.00213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalrymple NA, Mackow ER. 2014. Virus interactions with endothelial cell receptors: implications for viral pathogenesis. Curr Opin Virol 7:134–140. doi: 10.1016/j.coviro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvert JK, Helbig KJ, Dimasi D, Cockshell M, Beard MR, Pitson SM, Bonder CS, Carr JM. 2015. Dengue virus infection of primary endothelial cells induces innate immune responses, changes in endothelial cells function and is restricted by interferon-stimulated responses. J Interferon Cytokine Res 35:654–665. doi: 10.1089/jir.2014.0195. [DOI] [PubMed] [Google Scholar]
- 19.Chen YC, Wang SY. 2002. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J Virol 76:9877–9887. doi: 10.1128/JVI.76.19.9877-9887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assuncao-Miranda I, Amaral FA, Bozza FA, Fagundes CT, Sousa LP, Souza DG, Pacheco P, Barbosa-Lima G, Gomes RN, Bozza PT, Da Poian AT, Teixeira MM, Bozza MT. 2010. Contribution of macrophage migration inhibitory factor to the pathogenesis of dengue virus infection. FASEB J 24:218–228. doi: 10.1096/fj.09-139469. [DOI] [PubMed] [Google Scholar]
- 21.Chuang YC, Chen HR, Yeh TM. 2015. Pathogenic roles of macrophage migration inhibitory factor during dengue virus infection. Mediators Inflamm 2015:547094. doi: 10.1155/2015/547094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. 2015. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med 7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 23.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, Liu L, Hume DA, Stacey KJ, Young PR. 2015. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med 7:304ra142. doi: 10.1126/scitranslmed.aaa3863. [DOI] [PubMed] [Google Scholar]
- 24.Malasit P. 1987. Complement and dengue haemorrhagic fever/shock syndrome. Southeast Asian J Trop Med Public Health 18:316–320. [PubMed] [Google Scholar]
- 25.Bokisch VA, Top FH Jr, Russell PK, Dixon FJ, Muller-Eberhard HJ. 1973. The potential pathogenic role of complement in dengue hemorrhagic shock syndrome. N Engl J Med 289:996–1000. doi: 10.1056/NEJM197311082891902. [DOI] [PubMed] [Google Scholar]
- 26.Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, Auethavornanan K, Jairungsri A, Kanlaya R, Tangthawornchaikul N, Puttikhunt C, Pattanakitsakul SN, Yenchitsomanus PT, Mongkolsapaya J, Kasinrerk W, Sittisombut N, Husmann M, Blettner M, Vasanawathana S, Bhakdi S, Malasit P. 2006. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J Infect Dis 193:1078–1088. doi: 10.1086/500949. [DOI] [PubMed] [Google Scholar]
- 27.Avirutnan P, Malasit P, Seliger B, Bhakdi S, Husmann M. 1998. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J Immunol 161:6338–6346. [PubMed] [Google Scholar]
- 28.Merle NS, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V, Roumenina LT. 2015. Complement system part II: role in immunity. Front Immunol 6:257. doi: 10.3389/fimmu.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walport MJ. 2001. Complement. First of two parts. N Engl J Med 344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 30.Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. 2015. Complement system part I—molecular mechanisms of activation and regulation. Front Immunol 6:262. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meri S. 2016. Self-nonself discrimination by the complement system. FEBS Lett 590:2418–2434. doi: 10.1002/1873-3468.12284. [DOI] [PubMed] [Google Scholar]
- 32.Meri S, Pangburn MK. 1990. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci U S A 87:3982–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pangburn MK. 2000. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology 49:149–157. doi: 10.1016/S0162-3109(00)80300-8. [DOI] [PubMed] [Google Scholar]
- 34.Conrad DH, Carlo JR, Ruddy S. 1978. Interaction of beta1H globulin with cell-bound C3b: quantitative analysis of binding and influence of alternative pathway components on binding. J Exp Med 147:1792–1805. doi: 10.1084/jem.147.6.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pangburn MK, Pangburn KL, Koistinen V, Meri S, Sharma AK. 2000. Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b, and target in the alternative pathway of human complement. J Immunol 164:4742–4751. doi: 10.4049/jimmunol.164.9.4742. [DOI] [PubMed] [Google Scholar]
- 36.Weiler JM, Daha MR, Austen KF, Fearon DT. 1976. Control of the amplification convertase of complement by the plasma protein beta1H. Proc Natl Acad Sci U S A 73:3268–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pangburn MK, Muller-Eberhard HJ. 1984. The alternative pathway of complement. Springer Semin Immunopathol 7:163–192. doi: 10.1007/BF01893019. [DOI] [PubMed] [Google Scholar]
- 38.Lambris JD, Lao Z, Oglesby TJ, Atkinson JP, Hack CE, Becherer JD. 1996. Dissection of CR1, factor H, membrane cofactor protein, and factor B binding and functional sites in the third complement component. J Immunol 156:4821–4832. [PubMed] [Google Scholar]
- 39.Hellwage J, Jokiranta TS, Friese MA, Wolk TU, Kampen E, Zipfel PF, Meri S. 2002. Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J Immunol 169:6935–6944. doi: 10.4049/jimmunol.169.12.6935. [DOI] [PubMed] [Google Scholar]
- 40.Clark SJ, Ridge LA, Herbert AP, Hakobyan S, Mulloy B, Lennon R, Wurzner R, Morgan BP, Uhrin D, Bishop PN, Day AJ. 2013. Tissue-specific host recognition by complement factor H is mediated by differential activities of its glycosaminoglycan-binding regions. J Immunol 190:2049–2057. doi: 10.4049/jimmunol.1201751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jokiranta TS, Cheng ZZ, Seeberger H, Jozsi M, Heinen S, Noris M, Remuzzi G, Ormsby R, Gordon DL, Meri S, Hellwage J, Zipfel PF. 2005. Binding of complement factor H to endothelial cells is mediated by the carboxy-terminal glycosaminoglycan binding site. Am J Pathol 167:1173–1181. doi: 10.1016/S0002-9440(10)61205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison RA. 2018. The properdin pathway: an “alternative activation pathway” or a “critical amplification loop” for C3 and C5 activation? Semin Immunopathol 40:15–35. doi: 10.1007/s00281-017-0661-x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang K, Lu Y, Harley KT, Tran MH. 2017. Atypical hemolytic uremic syndrome: a brief review. Hematol Rep 9:7053. doi: 10.4081/hr.2017.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jokiranta TS. 2017. HUS and atypical HUS. Blood 129:2847–2856. doi: 10.1182/blood-2016-11-709865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nascimento EJ, Silva AM, Cordeiro MT, Brito CA, Gil LH, Braga-Neto U, Marques ET. 2009. Alternative complement pathway deregulation is correlated with dengue severity. PLoS One 4:e6782. doi: 10.1371/journal.pone.0006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shresta S. 2012. Role of complement in dengue virus infection: protection or pathogenesis? mBio 3:e00003-. doi: 10.1128/mBio.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wati S, Soo ML, Zilm P, Li P, Paton AW, Burrell CJ, Beard M, Carr JM. 2009. Dengue virus infection induces upregulation of GRP78, which acts to chaperone viral antigen production. J Virol 83:12871–12880. doi: 10.1128/JVI.01419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wati S, Li P, Burrell CJ, Carr JM. 2007. Dengue virus (DV) replication in monocyte-derived macrophages is not affected by tumor necrosis factor alpha (TNF-alpha), and DV infection induces altered responsiveness to TNF-alpha stimulation. J Virol 81:10161–10171. doi: 10.1128/JVI.00313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carr JM, Ashander LM, Calvert JK, Ma Y, Aloia A, Bracho GG, Chee SP, Appukuttan B, Smith JR. 2017. Molecular responses of human retinal cells to infection with dengue virus. Mediators Inflamm 2017:3164375. doi: 10.1155/2017/3164375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zipfel PF, Skerka C. 2009. Complement regulators and inhibitory proteins. Nat Rev Immunol 9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 51.Avirutnan P, Hauhart RE, Marovich MA, Garred P, Atkinson JP, Diamond MS. 2011. Complement-mediated neutralization of dengue virus requires mannose-binding lectin. mBio 2:e00276-. doi: 10.1128/mBio.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehlhop E, Ansarah-Sobrinho C, Johnson S, Engle M, Fremont DH, Pierson TC, Diamond MS. 2007. Complement protein C1q inhibits antibody-dependent enhancement of flavivirus infection in an IgG subclass-specific manner. Cell Host Microbe 2:417–426. doi: 10.1016/j.chom.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka A, Kosugi S, Konishi E. 2008. Infection-enhancing and -neutralizing activities of mouse monoclonal antibodies against dengue type 2 and 4 viruses are controlled by complement levels. J Virol 82:927–937. doi: 10.1128/JVI.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ripoche J, Mitchell JA, Erdei A, Madin C, Moffatt B, Mokoena T, Gordon S, Sim RB. 1988. Interferon gamma induces synthesis of complement alternative pathway proteins by human endothelial cells in culture. J Exp Med 168:1917–1922. doi: 10.1084/jem.168.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou L, Feng Y, Li Y, Zhang M, Chen C, Cai J, Gong Y, Wang L, Thurman JM, Wu X, Atkinson JP, Chao W. 2013. Complement factor B is the downstream effector of TLRs and plays an important role in a mouse model of severe sepsis. J Immunol 191:5625–5635. doi: 10.4049/jimmunol.1301903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaczorowski DJ, Afrazi A, Scott MJ, Kwak JH, Gill R, Edmonds RD, Liu Y, Fan J, Billiar TR. 2010. Pivotal advance: the pattern recognition receptor ligands lipopolysaccharide and polyinosine-polycytidylic acid stimulate factor B synthesis by the macrophage through distinct but overlapping mechanisms. J Leukoc Biol 88:609–618. doi: 10.1189/jlb.0809588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minta JO. 1988. Regulation of complement factor H synthesis in U-937 cells by phorbol myristate acetate, lipopolysaccharide, and IL-1. J Immunol 141:1630–1635. [PubMed] [Google Scholar]
- 58.Hajishengallis G, Lambris JD. 2010. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol 31:154–163. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang Z, Wu S, Li Y, He L, Wu M, Jiang L, Feng L, Zhang P, Huang X. 2011. Activation of Toll-like receptor 3 impairs the dengue virus serotype 2 replication through induction of IFN-beta in cultured hepatoma cells. PLoS One 6:e23346. doi: 10.1371/journal.pone.0023346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, Youn S, Diamond MS, Atkinson JP. 2010. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med 207:793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blom AM, Volokhina EB, Fransson V, Stromberg P, Berghard L, Viktorelius M, Mollnes TE, Lopez-Trascasa M, van den Heuvel LP, Goodship TH, Marchbank KJ, Okroj M. 2014. A novel method for direct measurement of complement convertases activity in human serum. Clin Exp Immunol 178:142–153. doi: 10.1111/cei.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parente R, Clark SJ, Inforzato A, Day AJ. 2017. Complement factor H in host defense and immune evasion. Cell Mol Life Sci 74:1605–1624. doi: 10.1007/s00018-016-2418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glennie S, Gritzfeld JF, Pennington SH, Garner-Jones M, Coombes N, Hopkins MJ, Vadesilho CF, Miyaji EN, Wang D, Wright AD, Collins AM, Gordon SB, Ferreira DM. 2016. Modulation of nasopharyngeal innate defenses by viral coinfection predisposes individuals to experimental pneumococcal carriage. Mucosal Immunol 9:56–67. doi: 10.1038/mi.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. 2006. Antibodies against West Nile virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J Virol 80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MS, Jones T, Song DY, Jang JH, Jung JU, Gao SJ. 2014. Exploitation of the complement system by oncogenic Kaposi's sarcoma-associated herpesvirus for cell survival and persistent infection. PLoS Pathog 10:e1004412. doi: 10.1371/journal.ppat.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conde JN, da Silva EM, Allonso D, Coelho DR, Andrade ID, de Medeiros LN, Menezes JL, Barbosa AS, Mohana-Borges R. 2016. Inhibition of the membrane attack complex by dengue virus NS1 through interaction with vitronectin and terminal complement proteins. J Virol 90:9570–9581. doi: 10.1128/JVI.00912-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spiropoulou CF, Srikiatkhachorn A. 2013. The role of endothelial activation in dengue hemorrhagic fever and hantavirus pulmonary syndrome. Virulence 4:525–536. doi: 10.4161/viru.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bharadwaj AS, Appukuttan B, Wilmarth PA, Pan Y, Stempel AJ, Chipps TJ, Benedetti EE, Zamora DO, Choi D, David LL, Smith JR. 2013. Role of the retinal vascular endothelial cell in ocular disease. Prog Retin Eye Res 32:102–180. doi: 10.1016/j.preteyeres.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gualano RC, Pryor MJ, Cauchi MR, Wright PJ, Davidson AD. 1998. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J Gen Virol 79(Pt 3):437–446. doi: 10.1099/0022-1317-79-3-437. [DOI] [PubMed] [Google Scholar]
- 70.Timmerman JJ, van der Woude FJ, van Gijlswijk-Janssen DJ, Verweij CL, van Es LA, Daha MR. 1996. Differential expression of complement components in human fetal and adult kidneys. Kidney Int 49:730–740. doi: 10.1038/ki.1996.102. [DOI] [PubMed] [Google Scholar]
- 71.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 72.Ormsby RJ, Jokiranta TS, Duthy TG, Griggs KM, Sadlon TA, Giannakis E, Gordon DL. 2006. Localization of the third heparin-binding site in the human complement regulator factor H1. Mol Immunol 43:1624–1632. doi: 10.1016/j.molimm.2005.09.012. [DOI] [PubMed] [Google Scholar]