ABSTRACT
Adeno-associated viruses (AAVs) encode a unique assembly-activating protein (AAP) within their genomes that is essential for capsid assembly. Studies to date have focused on establishing the role of AAP as a chaperone that mediates the stability, nucleolar transport, and assembly of AAV capsid proteins. Here, we map structure-function correlates of AAP using secondary structure analysis, followed by deletion and substitutional mutagenesis of specific domains, namely, the N-terminal hydrophobic region (HR), conserved core (CC), proline-rich region (PRR), threonine/serine-rich region (T/S), and basic region (BR). First, we establish that the centrally located PRR and T/S are flexible linker domains that can either be deleted completely or replaced by heterologous functional domains that enable ancillary functions such as fluorescent imaging or increased AAP stability. We also demonstrate that the C-terminal BR domains can be substituted with heterologous nuclear or nucleolar localization sequences that display various abilities to support AAV capsid assembly. Further, by replacing the BR domain with immunoglobulin (IgG) Fc domains, we assessed AAP complexation with AAV capsid subunits and demonstrate that the hydrophobic region (HR) and the conserved core (CC) in the AAP N terminus are the sole determinants for viral protein (VP) recognition. However, VP recognition alone is not sufficient for capsid assembly. Our study sheds light on the modular structure-function correlates of AAP and provides multiple approaches to engineer AAP that might prove useful toward understanding and controlling AAV capsid assembly.
IMPORTANCE Adeno-associated viruses (AAVs) encode a unique assembly-activating protein (AAP) within their genomes that is essential for capsid assembly. Understanding how AAP acts as a chaperone for viral assembly could help improve efficiency and potentially control this process. Our studies reveal that AAP has a modular architecture, with each module playing a distinct role and can be engineered for carrying out new functions.
KEYWORDS: adeno-associated virus, assembly-activating protein, capsid assembly, chaperones, gene therapy, nucleolus, scaffold
INTRODUCTION
Adeno-associated virus (AAV) is a nonenveloped, single-stranded DNA virus belonging to the Dependoparvovirus genus within the Parvoviridae family (1). The AAV capsid consists of 60 capsid monomers of VP1, VP2, and VP3 at a ratio of 1:1:10 that packages a 4.7-kb single-stranded genome (2, 3). The AAV genome encodes replication (Rep), capsid (Cap), and assembly-activating protein (AAP) open reading frames (ORFs) flanked by inverted terminal repeats (ITRs), which are the sole requirements for genome packaging (4, 5). As such, the majority of the genome can be replaced by exogenous DNA sequences and packaged inside the AAV capsid to create a recombinant vector for DNA delivery both in vitro and in vivo (6). Unlike other viruses that are replication competent, AAVs are partially defective since they require a helper virus (such as adenovirus or herpes simplex virus) for replication (7). The lack of pathogenicity and ease of genome manipulation have enabled extensive evaluation of recombinant AAV vectors as candidates for clinical gene therapy (8).
AAV encodes a unique protein, AAP, which is not found in other autonomous parvoviruses and is required for AAV capsid assembly. AAP is predicted to be a 20- to 24-kDa protein, with an actual size ranging from 27 to 34 kDa, which may be due to posttranslational modifications (5, 9). AAP is encoded from a +1 frame within the Cap ORF overlapping the junction between VP2 and VP3 (5). Introduction of a stop codon within AAP without affecting the coding frame of VP2/3 prevents capsid assembly and virus/vector production (10). While capsid assembly can be restored by providing AAP in trans, overexpression of wild-type (WT) AAP does not increase the vector yield (10). This suggests that AAP is necessary and sufficient for capsid assembly but is not a limiting factor for vector production. Cellular localization of AAPs overlaps with the location of capsid assembly, supporting a direct role for AAP (11). Most AAPs show strong nucleolar localization, while AAP5 and AAP9 are predominantly nuclear and excluded from the nucleolus (9, 12). Although the exact mechanism by which AAP supports AAV assembly is still elusive, several studies have convincingly shown that AAP is important for intracellular capsid expression and localization.
For instance, the steady-state level of capsid is dramatically reduced in the absence of AAP (13). Such regulation must occur at the translational or posttranslational level as Cap mRNA expression remains the same regardless of the presence or absence of AAP (10). In addition, the AAV2 VP have been shown to change their cellular localization from cytoplasmic/nuclear to nucleolar in the presence of AAP2 (12). A previous study has shown that the N-terminal region of AAP might interact with the C terminus of the VP, albeit weakly (13). Interaction with AAP2 also appears to alter VP conformation as supported by the lack of binding to several conformational specific antibodies (13). All of the above observations support the notion that AAP2 acts as a chaperone to stabilize and translocate VP to the site of assembly. However, it should be noted that significant differences in cellular localization and cross-complementation have been reported (9).
AAPs encoded by different serotypes are closely related, with sequence identity ranging from 48% (AAV4) to 82% (AAV7) relative to AAP1 (11). Phylogenetic analysis of AAP shows the same relationships as AAV VP, where the serotypes 4, 5, 11, and 12 are distinct from the others (11). The phylogenetic distance is also reflected in the biology of AAP4, -5, -11, and -12, which are unable to complement capsid assembly of other AAV serotypes (9). Furthermore, in the absence of AAP, AAV4, AAV5, and AAV11 are capable of producing 20 to 40% of AAV particles compared to the WT level; these have been termed “AAP independent” (9). At the secondary structure level, using previously defined nomenclature, AAP can be separated into multiple functional regions, N to C terminus: the hydrophobic region (HR), the conserved core (CC), the proline-rich region (PRR), the threonine/serine-rich region (T/S), and the basic region (BR) (13). In the present study, we undertake a systematic dissection of these different domains and generate novel chimeric and artificial AAPs that shed light on the structure-function correlates of this unique viral assembly protein.
RESULTS
The T/S of AAP is a flexible linker that can be replaced by different structures.
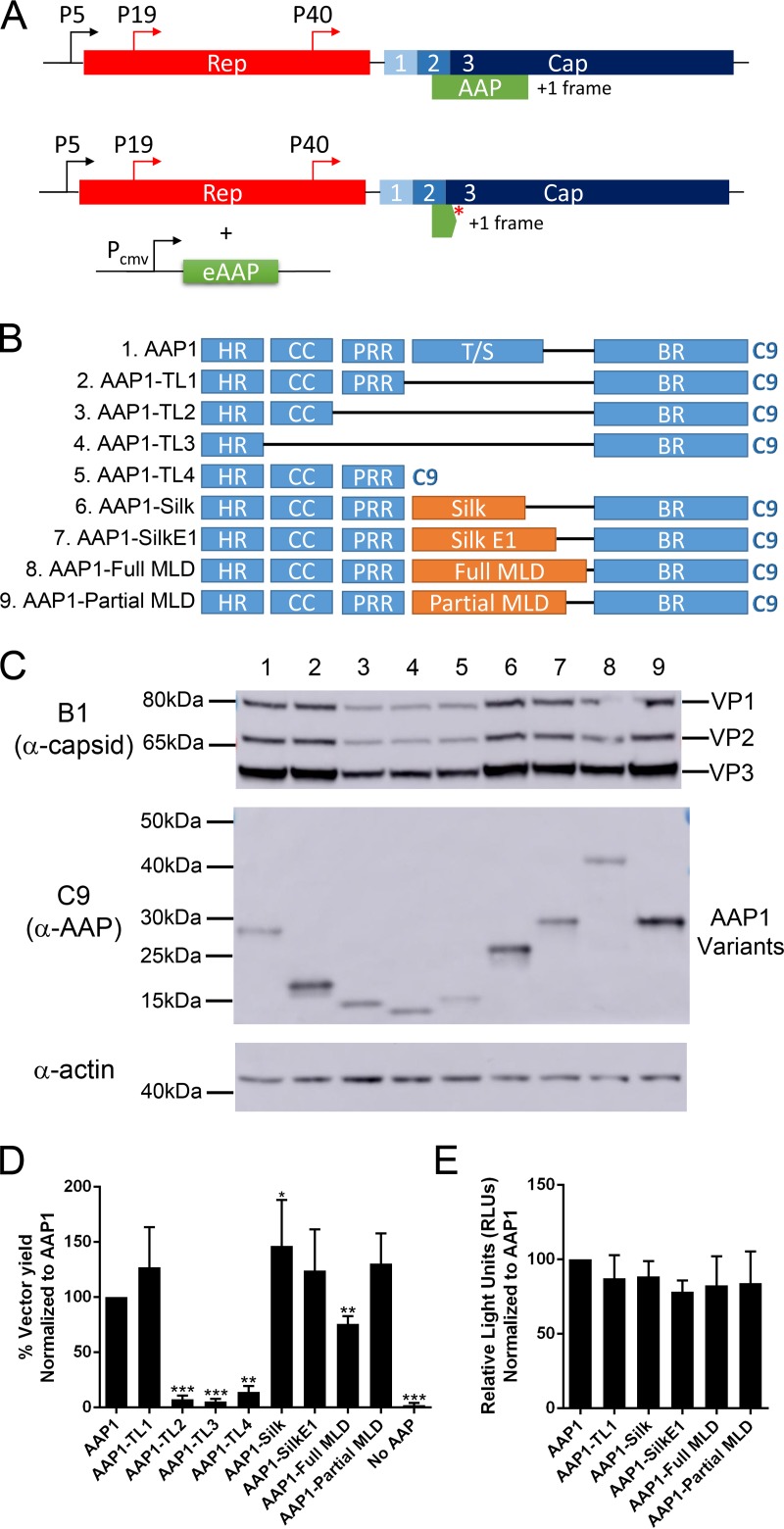
In order to study AAP independent of the VP ORF, we generated an AAP-null construct by introducing a stop codon early in the AAP ORF without affecting the VP coding sequence and provided different AAP constructs in-trans driven by the CMV promoter (Fig. 1A and Table 1). A series of deletion mutants were generated based on AAP domains and tested for the function of AAP using four parameters: steady-state AAP levels, VP levels, vector yield, and transduction efficiency. All AAP constructs have a C9 tag derived from bovine rhodopsin at the C terminus for immunodetection (Fig. 1B). In brief, experiments were performed by transfecting pXX680, pTR-CBA-Luc, pXR1ΔAAP, and a pCDNA3.1-AAP construct into HEK293 cells. All deletion and T/S replacement constructs produced AAP at different levels. Although truncated linker 2 (TL2), truncated linker 3 (TL3), and the T/S replaced with Bombyx mori silk heavy chain repeats (Silk and SilkE1) (14) or the entire mucin-like domain from murine alpha dystroglycan mucin-like domain (Full MLD) (15) showed similar steady levels of AAP, truncated linker 1 (TL1) and T/S replaced by partial mucin-like domain (Partial MLD) produce significantly more AAP. Truncated linker 4 (TL4) shows a reduced level of AAP expression (Fig. 1C and Table 2). Multiple reports have shown that one of the observable functions of AAP is to help stabilize the expression of VP; therefore, we next determined the steady-state VP level in the transfected cells. The level of VP3 correlates proportionally with AAP protein level, except for mutants TL2 and TL3, which have a significant reduction in VP3 level compared to WT AAP, although they produced similar amounts of AAP (Fig. 1C). Despite the production of both AAP and VP proteins in all condition tested, deletion of the HR, CC, or PRR rendered AAP incapable of supporting AAV capsid assembly (Fig. 1D and Table 2). However, deletion of the entire T/S or replacing the region with other flexible protein domains does not affect any tested function of AAP (including capsid assembly), strongly suggesting the T/S is nonessential for the assembly function of AAP (Fig. 1D and Table 2). The transduction efficiency of the recombinant AAV vectors produced using different AAP constructs were similar to control (Fig. 1E). Based on these findings, we concluded that the T/S can potentially be engineered with other heterologous sequences to generate engineered AAPs (eAAPs) with new functions.
FIG 1.
The threonine/serine-rich region (T/S) can be deleted or replaced by heterologous sequences without compromising AAP functions. (A) Original AAV genome showing Rep and Cap genes with AAP encoded in an alternative reading frame (top) and modified AAV genome with a stop codon that truncates endogenous AAP, with eAAP plasmid provided in trans (bottom). (B) Different AAP mutants, truncated linkers 1 to 4 (TL1 to TL4) have different internal regions deleted. For others, the T/S was replaced with Bombyx mori silk heavy chain repeats (Silk and SilkE1) or the entire (Full MLD) or partial (Partial MLD) mucin-like domains from murine alpha dystroglycan mucin-like domain. All AAP1 constructs have a C9 epitope tag at the C terminus for immunodetection. (C) Representative Western blot image of VPs, AAPs, and actin expression level in HEK293 cells transfected by quadruple transfection for rAAV production at 3 days posttransfection. (D) Relative vector yield of different AAP mutants normalized to wild-type AAP1 (AAP1). (E) Transduction of rAAV1 packaging luciferase produced from different AAP1 mutants on HEK293 cells at 10,000 vg/cell. Relative light units (RLUs) were normalized to AAP1. Error bars represent one standard deviation from at least three independent experiments. The data were analyzed by a two-tailed Student t test compared to AAP1 (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
TABLE 1.
Amino acid sequences of different T/S and BR module replacement constructs
| Constructa | Amino acid sequence (5′–3′) |
|---|---|
| T/S replacements | |
| Silk (14) | GAGAGAGQGAGAGAGQGAAAGAGAGAGQT |
| Silk E1 (14) | GAGAGQGAGAGAGQGAAAGAGAGAGQGAGAGAGQGAGAGAGQGAAGAGAGAGQT |
| Full MLD (15) | PTPVTAIGPPTTAIQEPPSRIVPTPTSPAIAPPTETMAPPVRDPVPGKPTVTIRTRGAIIQTPTLGPIQPTRVSEAGTTVPGQIRPTLTIPGYVEPTAVITPPTTTTKKPRVSTPKPATPSTDSSTTTTRRPTKKPRTPRPVPRVTTK |
| Partial MLD (15) | TLTIPGYVEPTAVITPPTTTTKKPRVSTPKPATPSTDSSTTTTRRPTKKPRTPRPVPRVTT |
| Collagen (21) | GSSGVRLWATRQAMLGQVHEVPEGWLIFVAEQEELYVRVQNGFRKVQLEARTPLPR |
| Tetramer (22) | AEIEQAKKEIAYLIKKAKEEILEEIKKAKQEIA |
| WSN | KRMENLNKKVDDGFLDIWTYNAELLVLLENERTLDFHDLNVKNLYEKVKSQLK |
| BR replacements | |
| Rpp29 (16) | RHKRKEKKKKAKGLSARQRRELR |
| AP3D1 (17) | RRHRQKLEKDKRRKKRKEKEERTKGKKKSKK |
| SV40 (18, 19) | KRTADGSEFESPKKKRKVE |
| HIV Rev (20) | RQARRNRRRRWRERQR |
| 4BR | STSRSRRSRRRTARQRWLITLPARFRSLRTRRTNCRT |
| 5BR | STFKSKRSRCRTPPPPSPTTSPPPSKCLRTTTTSCPTSSATGPRDACRPSLRRSLRCRSTVTRR |
| 9BR | STFRSKRLRTTMESRPSPITLPARSRSSRTQTISSRTCSGRLTRAASRRSQRTFS |
The numbers in parentheses indicate references for the sequences.
TABLE 2.
Summary of biological properties of T/S and N-terminal constructsa
| Constructb | AAP level | VP level | Vector yield | Interaction with VP |
|---|---|---|---|---|
| T/S region | ||||
| AAP1 WT | WT | WT | WT | Yes |
| TL1 | ++ | WT | + | NA |
| TL2 | WT | − | −−− | NA |
| TL3 | WT | − | −−− | NA |
| TL4 | − | − | −−− | NA |
| Silk | ++ | WT | + | NA |
| SilkE1 | + | WT | + | NA |
| Full MLD | WT | − | WT | NA |
| Partial MLD | +++ | WT | + | NA |
| AAP1E | ++++ | + | WT | Yes |
| AAP1-Collagen (21) | ++++++ | ++ | +++ | NA |
| AAP1-Tetra (22) | +++++ | + | + | NA |
| AAP1-WSN | ++ | WT | − | NA |
| N terminus | ||||
| AAP1 WT | WT | WT | WT | Yes |
| AAP1E | ++++ | + | WT | Yes |
| AAP5E | ++++ | − − | −−− | Yes |
| ΔHR | ++++++ | − − | −−− | No |
| 5HR | ++++ | − − | −−− | Yes |
| ΔCC | ++++ | − − | −−− | No |
| 5CC | + | − | − − | Yes |
| ΔPRR | ++++ | WT | − − | Yes |
| 5PRR | WT | WT | WT | Yes |
WT represents similar properties, “−” represents reduced levels, and “+” represents increased levels compared to AAP1 WT. The numbers of “+” and “−” are proportional to the increase or decrease in expression levels or titer. NA, not applicable.
The numbers in parentheses indicate references for the sequences.
The BR of AAP is not serotype specific.
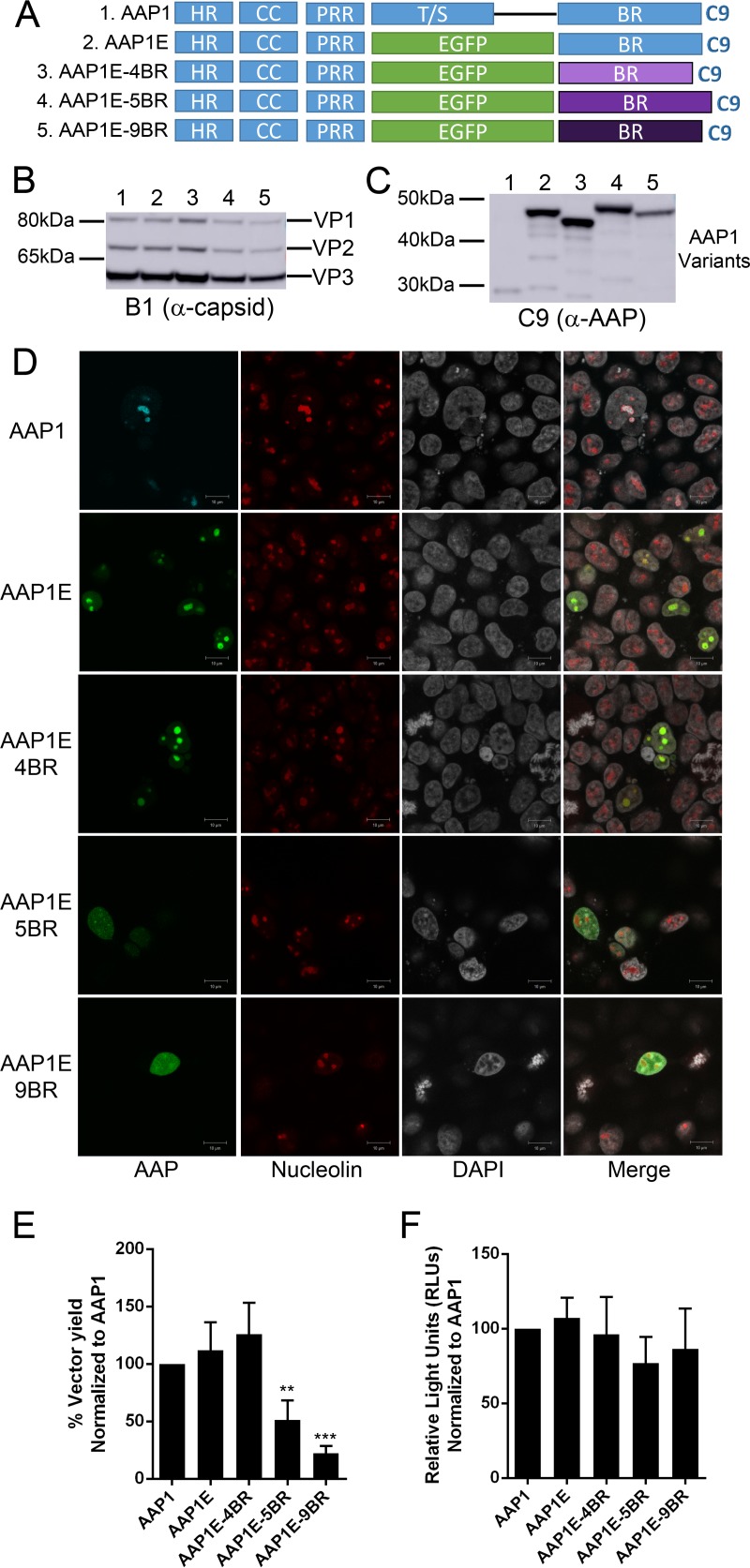
We first created a traceable AAP1 protein by replacing the T/S with enhanced green fluorescent protein (EGFP) to create AAP1-EGFP (AAP1E) for fluorescent tracking (Fig. 2A). This newly engineered AAP1E shows the same level of VP expression and capsid assembly, but a higher level of AAP expression compared to AAP1 (Fig. 2B, C, and E; Table 2). Furthermore, the AAP1E mutant localizes within the nucleolus similar to the WT AAP1, suggesting that AAP1E can be useful for further mechanistic studies (Fig. 2D). Using the AAP1E construct, we swapped the BR of AAP1E with the BR from AAP4, -5, and -9 (Fig. 2A and Table 1). The VP and AAP expression profiles of the BR substitution mutants are generally similar to AAP1E. However, 5BR and 9BR show a slight decrease in VP expression and 9BR shows a slight decrease in AAP expression (Fig. 2B and C; Table 3). Notably, BR1 and BR4 strongly localized to the nucleolus, while BR5 and BR9 only shows partial nucleolus localization as reported earlier (Fig. 2D and Table 3) (9). Intracellular localization appears to correlate with vector titers; for instance, BR5 and BR9 only restore about 50% of assembly (Fig. 2F and Table 3). In contrast, BR4 is able to restore AAP1 function to 120% compared to WT AAP1. The transduction efficiency of the rAAVs generated using different AAPs was not affected (Fig. 2F). Since it is known that AAP4 cannot rescue AAV1 capsid assembly, our data suggest that serotype-specific transcomplementation is not due to the BR but rather to other regions of AAP.
FIG 2.
The BR is not the determinant for serotype specificity of AAV capsid assembly. (A) Schematic of different AAP mutants. The T/S of AAP1 was replaced by EGFP (AAP1E). The BR was replaced with those of other serotypes, including AAP4 (4BR), AAP5 (5BR), and AAP9 (9BR). (B and C) Representative Western blot images of VP (B) and AAP (C) expression as described in Fig. 1B. (D) Confocal microscopy of different AAP1E derivatives. AAP1E derivatives (pCDNA3.1-AAPs) were transfected as described in Materials and Methods and visualized by their native EGFP fluorescence. AAP1 was immunostained with α-C9 antibody, and nucleoli were immunostained with C23 antibody. Nuclei were stained with DAPI. (E) Relative vector yields using different AAP constructs normalized to wild-type AAP1 (AAP1). (F) Transduction of rAAV1 packaging luciferase produced from different AAP1 constructs as described above. RLUs were normalized to AAP1. Error bars were determined and statistical analyses were performed as described above.
TABLE 3.
Summary of biological properties of BR constructsa
| BR construct | AAP level | VP level | Vector yield | AAP localization |
|---|---|---|---|---|
| AAP1 WT | WT | WT | WT | Nucleolus |
| DBR | ++++ | WT | − − | Cytoplasmic |
| 4BR | +++++ | + | + | Nucleolus |
| 5BR | +++++ | WT | − | Nucleus |
| 9BR | +++ | − | − − | Nucleus |
| Rpp29 | ++++ | − | − | Nucleus/nucleolus |
| AP3D1 | ++++ | WT | WT | Nucleolus |
| SV40 | ++++ | + | − − | Nucleus |
| HIV Rev | ++++ | − | − | Nucleolus |
WT represents similar properties, “−” represents reduced levels, and “+” represents increased levels compared to AAP1 WT. The numbers of “+” and “−” are proportional to the increase or decrease in expression level or titer.
The BR of AAP1 can be replaced by other nucleolar localization signals (NoLS) without disrupting function.
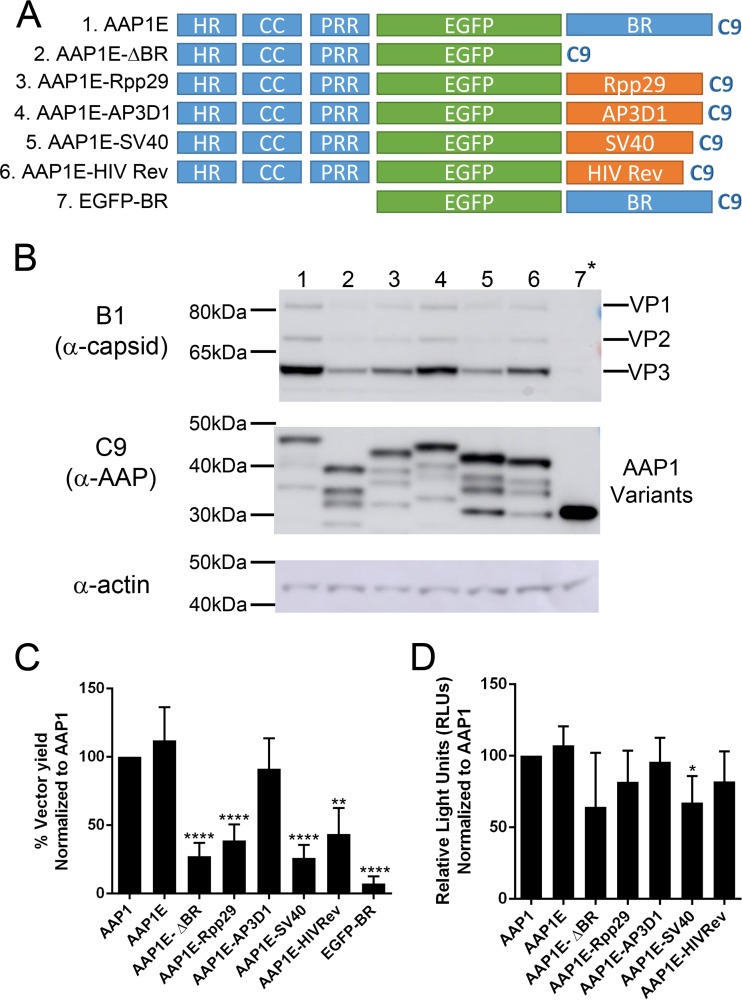
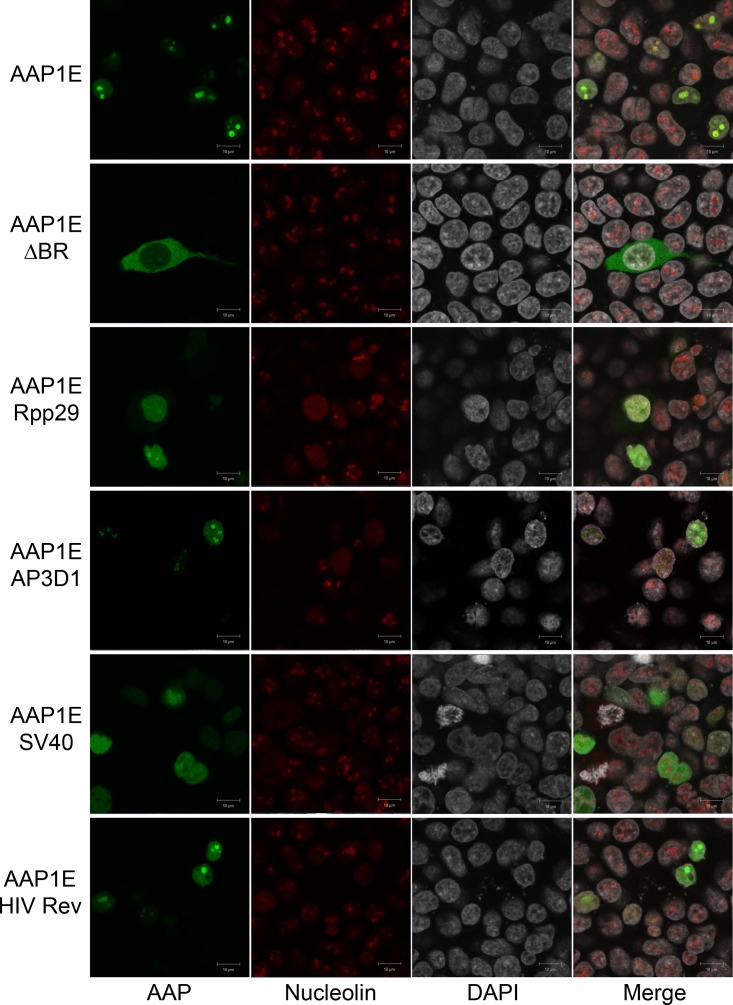
Based on the results presented above, we hypothesized that the BR can be replaced completely by other NoLS without affecting AAP function. Therefore, we engineered AAP1E by replacing the BR with other known nuclear or nucleolar localization signals (NLS/NoLS). The amino acid sequences and annotation of each NLS/NoLS are described in Table 1. In brief, we picked four different NLS/NoLS peptides from the RNase P subunit NoLS (Rpp29) (16), adaptor protein in AP-3 complex (AP3D1) (17), virus-derived simian virus 40 NLS (SV40) (18, 19), and HIV Rev NoLS domain (HIV Rev) (20) (Fig. 3A). Although AAP expression levels are comparable to AAP1E from all the BR replacement mutants, VP expression was moderately reduced for some (Fig. 3B and Table 3). This decrease correlates with assembly based on vector yield, where the various BR mutants show a spectrum of activity. SV40 is unable to restore AAP assembly function, while Rpp29, HIV-Rev, and AP3D1 restore AAP assembly function at 40, 50, and 80%, respectively (Fig. 3C and Table 3). All rAAVs generated with these constructs show similar transduction efficiencies (Fig. 3D). Since the NoLS/NLS mutants showed a spectrum of AAP function, we performed confocal microscopy to investigate trafficking efficiency compared to AAP. AAP1E shows clear colocalization with the nucleolar marker CD23 (Fig. 4). AAP1E-ΔBR shows cytoplasmic localization, as expected, and is not able to restore function (Fig. 3C and 4). AAP1E-SV40 shows nuclear localization, which only partially rescues AAV assembly. AAP1E-Rpp29 shows moderate (40%) rescue of AAV assembly and has partial nucleolar localization (Fig. 3C and 4). HIV-Rev and AP3D1 show nucleolar localization, and AP3D1 restores assembly function completely (Fig. 3C and 4). Therefore, the ability of AAP to drive AAV1 assembly correlates with its nuclear/nucleolar localization, and the BR region can be replaced by other NoLS without affecting AAP function.
FIG 3.
The C-terminal BR is important for AAP1 function but can be replaced by heterologous NoLS. (A) Schematic of different AAP mutants. The T/S of AAP1 was replacing by EGFP (AAP1E). The BR was replaced by RNase P subunit 29 NoLS (Rpp29), adaptor protein in AP-3 complex NoLS (AP3D1), virus-derived SV40 NLS (SV40), and HIV Rev NoLS domain (HIV Rev). (B) Representative Western blot images of VP and AAP expression as described in Fig. 1B. (C) Relative vector yields using different AAP constructs normalized to wild-type AAP1 (AAP1). (D) Transduction of rAAV1 packaging luciferase produced from different AAP1 constructs as described above. RLUs were normalized to AAP1. Error bars were determined and statistical analyses were performed as described above.
FIG 4.
Confocal microscopy of different AAP1E derivatives. AAP1E derivatives (pCDNA3.1-AAPs) are transfected as described in Materials and Methods. The AAP1E derivatives were visualized by their native EGFP fluorescence, the nucleoli were immunostained with C23 antibody, and the nuclei were stained with DAPI.
Functionally mapping the structural domains at the N terminus of AAP1E.
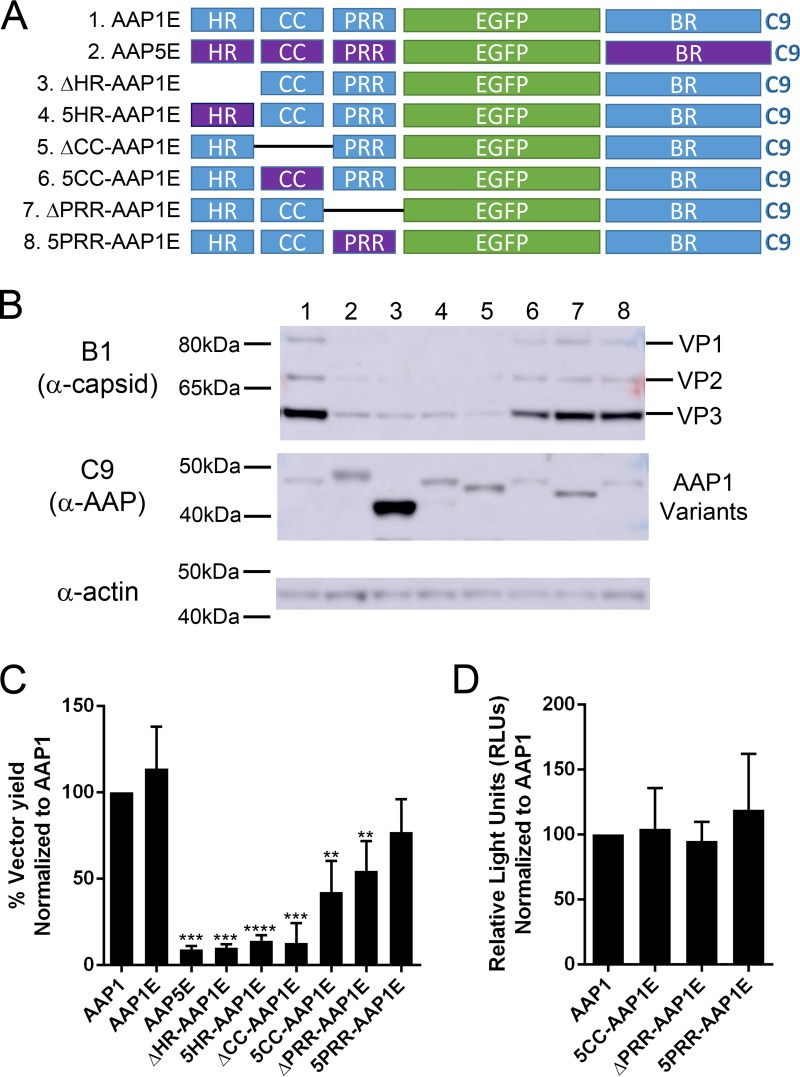
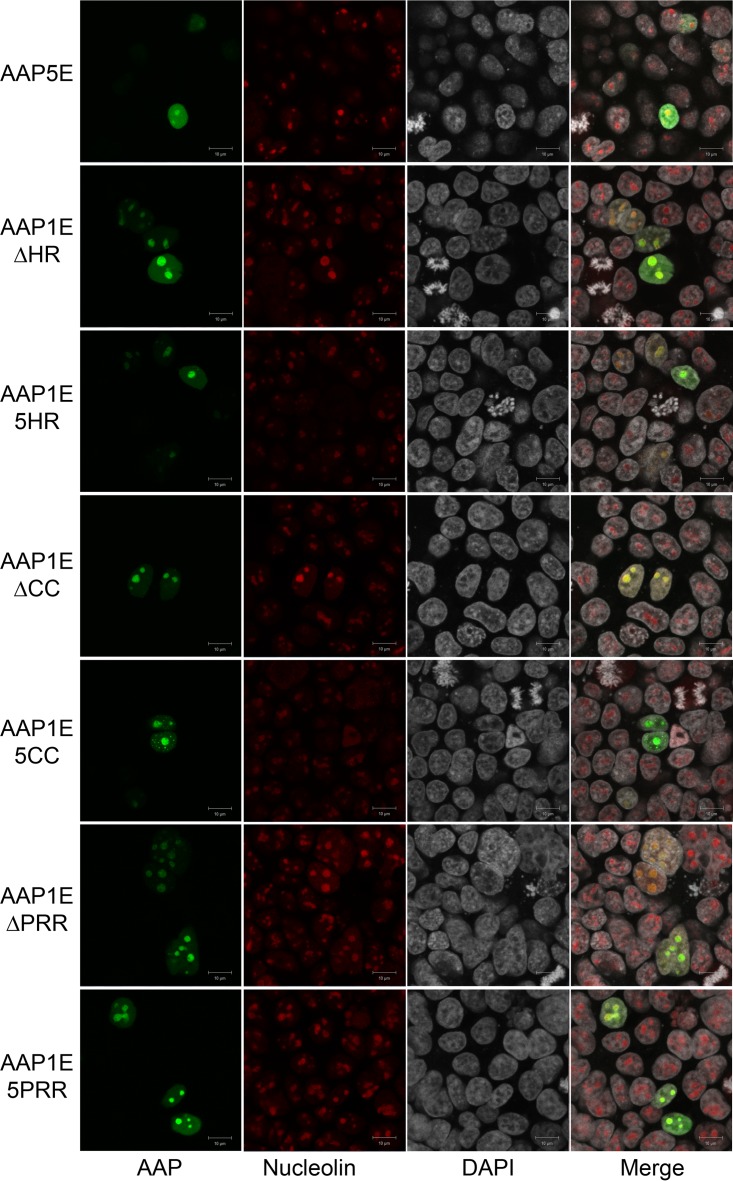
Currently, there is no structural information on AAP. Based on a modular homology modeling, the N-terminal HR is the only region that can be modeled among all AAP tested and CC from AAP1, -6, -7, and -9 can be modeled. The HR is modeled as an alpha-helix, and the CC region is modeled as either a loop or a beta-strand. Deletion or point mutations in these regions completely abolish the ability of AAP to activate capsid assembly, suggesting that these secondary structures play an important role in AAP function. Furthermore, previous reports have shown that AAP5 is unable to support capsid assembly of AAV1, despite the sequence similarity between the PRR, the T/S, and the BR (9). We investigated the function of each subdomain by deletion analysis and by replacement with the corresponding domain of AAP5 (Fig. 5A). While all the constructs were able to produce AAP, the functional efficiencies were varied. The expression level of the AAP1EΔHR construct is higher than that of AAP1E; however, the level of VP is significantly reduced (Fig. 5B and Table 2). In addition, capsid formation is completely abrogated to background levels (Fig. 5C and Table 2). Replacing the AAP1 HR domain with the AAP5 HR domain shows a similar effect as the deletion, although the AAP expression level is only slightly increased compared to AAP1E (Fig. 5B and C; Table 2). Deletion of the CC region resulted in a similar phenotype, where the expression level of AAP was normal, but VP expression and capsid formation were reduced to the background level (Fig. 5B and C; Table 2). Replacing 1CC with 5CC domains restored the level of VP expression and 50% viral titer, suggesting a link between the CC region and capsid protein stability that is not serotype specific (Fig. 5B and C; Table 2). Deletion of the PRR shows a moderate effect on capsid expression and assembly. Replacing 1PRR with 5PRR only slightly affects AAP function and allows 75% viral yield (Fig. 5B and C; Table 2). All rAAVs generated with AAP show similar transduction efficiencies (Fig. 5D). Confocal microcopy studies further confirmed that the N-terminal modules localized at the nucleolus, similar to the AAP1E control (Fig. 6). These results corroborate the notion that the HR and CC domains are essential for VP interactions and maintaining stability.
FIG 5.
Domain deletion analysis of the N terminus of AAP1 reveals that the HR and CC are important for AAP functions. (A) Schematic of different AAP mutants. The N-terminal domains are either deleted or replaced with their AAP5 counterparts (purple). All AAP mutants have a C9 epitope tag at the C terminus. (B) Representative Western blot image of viral proteins, AAPs, and actin expression of HEK293 cells transfected by quadruple transfection for rAAV production after 3 days posttransfection. (C) Relative vector yields using different AAP constructs normalized to wild-type AAP1 (AAP1). (D) Transduction of rAAV1 packaging luciferase produced from different AAP1 constructs as described above. RLUs were normalized to AAP1. Error bars were determined and statistical analyses were performed as described above.
FIG 6.
Confocal microscopy of different AAP5E and AAP1E derivatives. AAP5E and AAP1E derivatives were transfected as described in Materials and Methods. AAP5E and AAP1E derivatives were visualized by their native EGFP fluorescence, nucleoli were immunostained with C23 antibody, and nuclei were stained with DAPI.
The HR and CC domains are the major structural determinants driving AAP-capsid interactions.
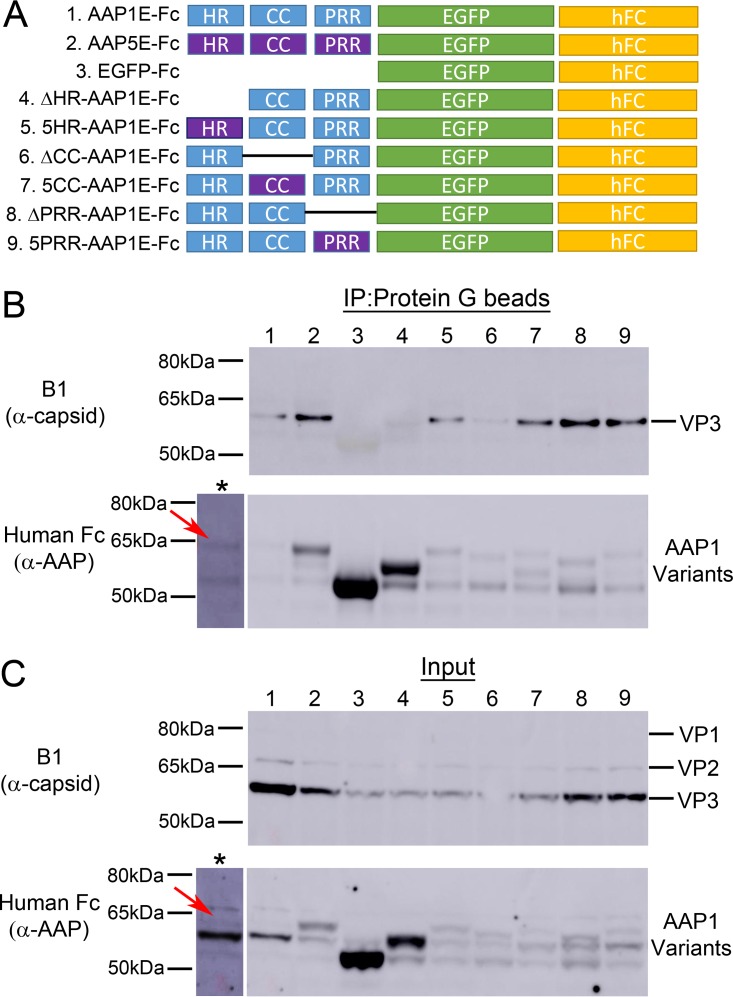
Based on deletion and AAP1/5 chimera analysis, we established that capsid assembly and VP expression levels are tightly correlated. We utilized the same deletion and replacement constructs as above and replaced the BR with a human Fc domain to facilitate VP complexation and immunoprecipitation studies (Fig. 7A). The expression level of both VP and AAP varies between constructs. For instance, EGFP, ΔHR, 5HR, ΔCC, and 5CC show a reduced level of AAP expression, while AAP5E and ΔHR show an increase in AAP expression (Fig. 7C and Table 2). To verify our system, we first showed that AAP1E can pull down the AAV1 VP. Surprisingly, although AAP5E was unable to support AAV1 capsid assembly (Fig. 6C), this protein was able to pull down AAV1 VP similar to AAP1E (Fig. 7B and Table 2). Deletion of the HR or CC regions abolishes the ability of AAP to pull down VP. Replacing the HR and CC regions with AAP5 restores the ability of AAP to pull down AAV1 VP (Fig. 7B and Table 2). Deletion of PRR or replacing 1PRR with 5PRR has no significant effect on AAV1 capsid interaction (Fig. 7B and Table 2). It is worth noting that the lack of immunoprecipitation in some of these constructs cannot be explained by expression level differences. For instance, 5HR and 5CC both have low levels of AAP and VP expression but were able to pull down VP in our assay (Fig. 7B and C; Table 2). On the other hand, ΔHR and the EGFP control are unable to pull down any VP protein despite having the highest levels of AAP expression (Fig. 7B and C; Table 2). However, this interaction is either transient or weak, which is further supported by our observation that majority of the capsid protein is still found in the flowthrough fraction of the pulldown (L. V. Tse and A. Asokan, unpublished data). These results demonstrate that the interactions between AAP and VP are driven by the N-terminal modules.
FIG 7.
Immunoprecipitation of VPs by different N-terminal mutants of AAP1. (A) Schematic of different AAP mutants. The N-terminal domains are either deleted or replaced with AAP5 counterparts (purple). All AAP constructs have their BRs replaced by the human Fc domain at the C terminus. (B and C) Representative Western blot images of immunoprecipitation (B) and input (C) of VPs by AAP1E mutants. The asterisk (*) lane represents an overexposure of lane 1, and red arrows denote the position of AAP1E-hFc. Samples were collected at 3 days posttransfection of HEK293 cells with pXX680, pXR1ΔAAP1, pCDNA3.1-AAP1E-Fc, and pTR-CBA-Luc. Detailed protocols are described in Materials and Methods. AAP1E-Fc was pulled down using magnetic protein G beads and is immunostained with goat α-human-HRP antibody; VP is stained by mouse α-capsid antibody (B1).
Engineering AAP for increased stability and vector production.
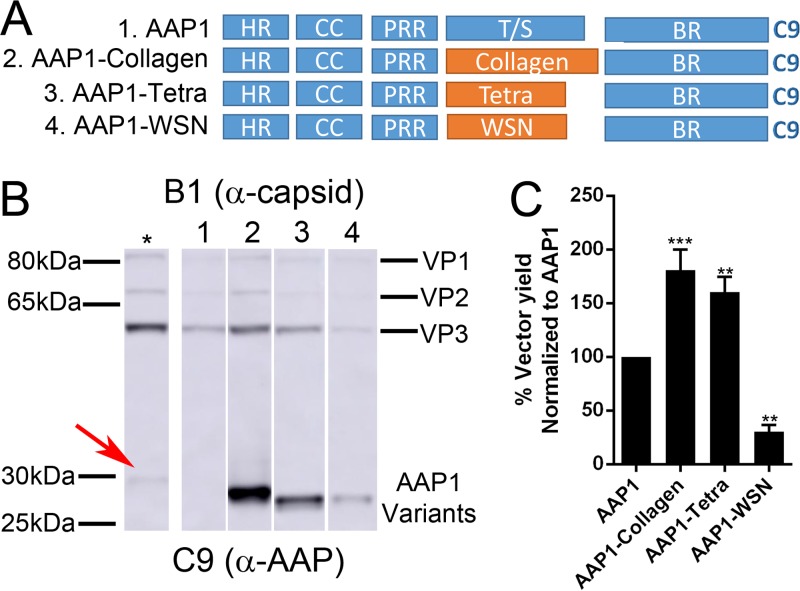
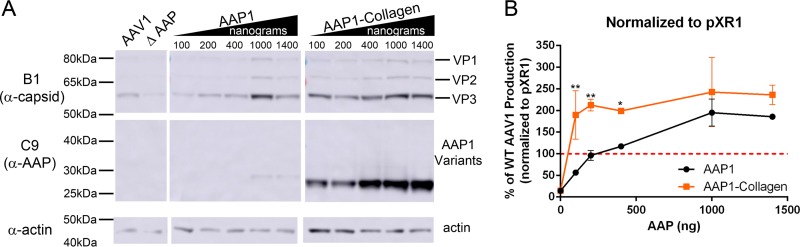
We hypothesized that the T/S of AAP is the most amenable to manipulation for imparting novel/improved functionality. By introducing EGFP in place of T/S, steady-state AAP levels were likely increased probably due to increased protein stability (Fig. 2B). Thus, we attempted to further increase the stability of AAP by replacing the T/S with oligomerization domains. We picked three different oligomerization domains, including (i) Collagen (21), which forms trimers, (ii) influenza A/WSN/1933 hemagglutinin (HA), which is a coiled-coil trimerization domain (WSN), and (iii) a synthetic tetramerization domain (Tetra) (22) to replace the T/S of AAP1 (Fig. 8A). Steady-state AAP protein levels were increased among all the AAP variants. The increase was most pronounced in AAP1-Collagen and AAP1-Tetra, while AAP1-WSN shows a moderate increase in AAP level (Fig. 8B and Table 2). VP levels also showed an increase in case of AAP1-Collagen, remained the same in AAP1-Tetra, and were slightly decreased in AAP1-WSN (Fig. 8B and Table 2). Both AAP1-Collagen and AAP1-Tetra increased the rAAV vector yield by 1.5- and 2-fold, respectively, compared to WT AAP1 (Fig. 8C and Table 2). The AAP1-WSN-mediated vector yield is 60% less than that for WT AAP1, suggesting a potential functional incompatibility with AAP1-WSN, despite increased stability (Fig. 8C and Table 2). To further evaluate the impact of engineered AAPs on vector yield, we attempted to establish a dose-response relationship for natural and engineered AAP constructs in supporting rAAV vector production by varying the amount of AAP DNA during transfection. As expected, the AAP levels increased proportionally to the amount of transfected cDNA. AAP1-Collagen showed a significant increase in AAP levels compared to wild-type AAP1 (Fig. 9A). VP levels were also increased proportional to the amount of AAP DNA, with peak titers observed at 1,000 ng of AAP plasmid transfected. However, it is important to note that titers declined at higher levels potentially due to toxicity of AAP overexpression (Fig. 9A [see also Tse and Asokan, unpublished]).
FIG 8.
Replacement of T/S with oligomerization domains. (A) Schematic of different AAP mutants. The T/S was replaced by different oligomerization domains: the collagen trimerization domain (Collagen), a synthetic tetramerization domain (Tetra), or the influenza A/WSN/33 HA coiled-coil domain (WSN) and AAP1 as a control. All AAP mutants have a C9 epitope tag at the C terminus. (B) Representative Western blot image of VP and AAP expression as described in Fig. 1B. The asterisk (*) lane is transfected with 2× AAP1 plasmid amount compared to lane 1, and the red arrow denotes the position of AAP1. (C) Relative vector yield using different AAP constructs normalized to wild-type AAP1 (AAP1). Error bars were determined and statistical analyses were performed as described above.
FIG 9.
Dose-dependent increase in vector production by AAP1-Collagen. (A) Representative Western blot image of VP, AAP, and actin expression of HEK293 cells at different doses of AAP1 and AAP1-Collagen provided in trans at 3 days posttransfection. (B) Relative vector yield of different doses of AAP1 and AAP1-Collagen normalized to standard AAV1 production (pXR1). Error bars represent one standard deviation from at least three independent experiments. The data were analyzed by two-way ANOVA compared to pXR1 standard production (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
DISCUSSION
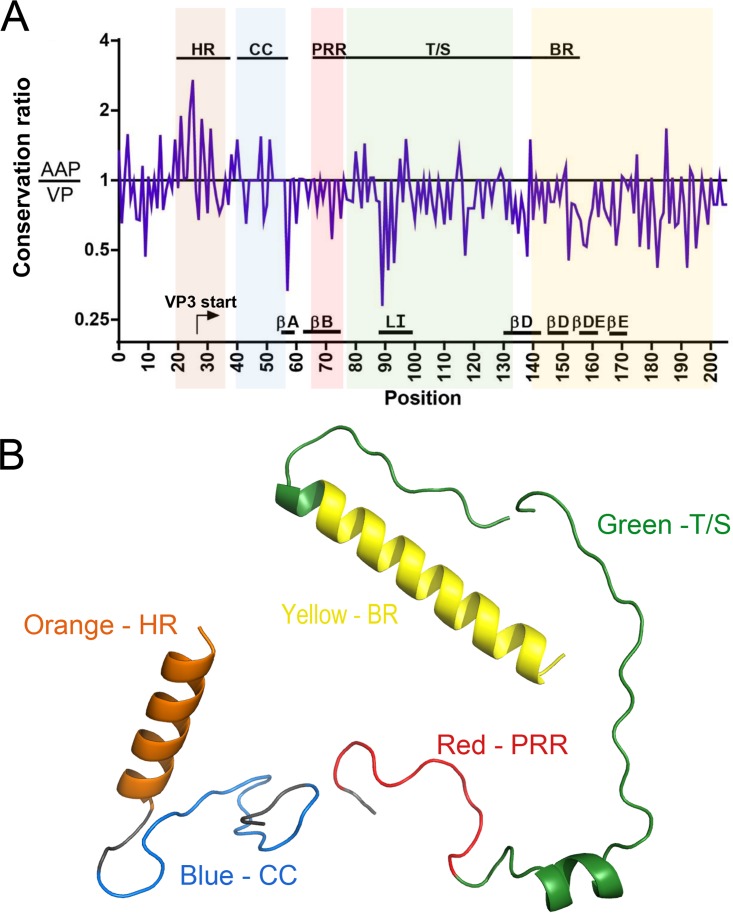
Adeno-associated virus (AAV) encodes different proteins within its 4.7-kb genome using alternative splicing, alternative start codons, and overlapping reading frames (23). AAP is expressed from an alternative reading frame overlapping the VP2/3 sequences (5). We attempted to further understand our observations by calculating the relative conservation of AAP and VP overlapping regions in the Cap gene (24). Briefly, we plotted the ratio (AAP/VP) of the conservation scores such that a ratio of >1 denotes higher AAP conservation and a ratio of <1 implies higher VP conservation. Most regions show a preference for VP over AAP, with values of <1 (Fig. 10A). For instance, the sequences forming the beta-strand regions that form the jelly roll structure of VP and the highly conserved loop I have AAP/VP conservation ratios ranging from 0.3 to 0.5 due to a higher level of VP sequence conservation. These regions correspond to the nonessential T/S and PRR linker domains of AAP, respectively. Thus, in these regions VP function is favored over that of AAP (Fig. 10A). In contrast, the HR and CC regions show a conservation ratio of >1, indicating a preference for AAP function (Fig. 10A). In corollary, this region corresponds to the VP2 N-terminal domain, which has been shown to be nonessential for AAV capsid infectivity and essentially serves as a linker between the unique VP1 N-terminal phospholipase A2 (PLA2) domain and VP3 major subunit. Further, homology modeling of different AAP modules shows that the HR is the only region that has a strong secondary structure requirement in the form of an alpha-helix. Although AAP4 BR is also modeled as an alpha-helix, no other AAP BR can be modeled. All other domains either form undefined loop structures or are unable to be modeled (Fig. 10B).
FIG 10.
Bioinformatic analysis of AAP and VP conservation and homology modeling of AAP. A multiple-sequence alignment of AAP and the corresponding VP amino acids was performed and used for further analysis. (A) The ratio of AAP conservation to VP conservation was calculated for each amino acid position and plotted along a positional axis (see Materials and Methods for a detailed bioinformatic analysis). Ratios of >1 indicate greater conservation in AAP and vice versa. Overlaid are various regions and structural elements of each protein. The functional groups in VP are the VP3 start site (arrow), beta-strands A, B, D, and E (βA, βB, βD, and βE), and loop I (LI). Functional regions of AAP are highlighted in different colors: orange (HR), blue (CC), red (PRR), green (T/S), and yellow (BR). (B) Homology modeling of AAP1 (HR and CC), AAP2 (T/S), and AAP4 (BR). Detailed modeling parameters are presented in Materials and Methods.
The latter theoretical observations corroborate our functional characterization of AAP. Indeed, functional analysis showed that the T/S is dispensable and can be replaced by exogenous sequences. All of our T/S deletion or replacement constructs had higher steady-state levels than the WT AAP1. However, we cannot rule out that the T/S could have other potential, regulatory functions. Serine and threonine are the common amino acid substrates for phosphorylation; there are examples showing that multiple phosphorylation events could mark the protein for degradation via phosphodegron modules by the proteasome (25, 26). As previously shown, degradation of AAP can be inhibited by addition of proteasome inhibitor, MG132 (10). Therefore, the increase in the steady level of the T/S deletion constructs could be the result of decreased phosphorylation and, consequently, proteasomal degradation. Among other possible roles for the T/S that we cannot rule out at this writing is transcriptional regulation. These attributes might have been evolved to exploit the host cell machinery to regulate AAP levels and thus influence AAV infection. These latter aspects are the subject of ongoing investigations.
Similar to the T/S, the PRR does not appear to have any assigned function. Deletion of the PRR and T/S together impairs capsid assembly. However, deletion of the PRR in AAP1E retains 60% of capsid assembly compared to the wild type. Replacement with the PRR from AAP5 further rescues assembly to 80%. These data suggest that the PRR plays a relatively minor role in capsid assembly and serotype specificity but may act as a key structural linker that physically separates the critical HR and CC modules from the T/S. Unlike the PRR and T/S, multiple studies have shown that the BR contains an important NLS/NoLS signal that is responsible for AAP localization and subsequent translocation of the capsid to the assembly site (5, 11, 12). We tested whether other AAP BRs can functionally replace the AAP1 BR for capsid assembly. Surprisingly, only the AAP4 BR is able to support AAV1 capsid assembly function. Since the whole AAP4 protein is unable to rescue AAP1 capsid assembly function (9), our data clearly demonstrate that serotype specificity is independent of the BR. We further corroborate that BR is solely acting as a NoLS in the context of AAP1 by replacing the AAP1 BR with other heterologous NLS/NoLS. Among all of the NLS/NoLS tested, AP3D1, which is supposed to be an endosomal marker as its native form, shows the best nucleolar localization and completely supports capsid assembly. The localization pattern and percent rescue of capsid assembly are highly correlated, where increased nucleolar localization indicates greater restoration of capsid assembly for AAV1. Different BR sequences have been reported for different AAPs, supporting the notion that AAP exerted a functional, rather structural evolutionary constraint in this region compared to VP.
The HR and the CC are the functional domains for AAV capsid assembly and the determinants of serotype specificity. Deleting the HR or CC led to an inability to pull down VP or support capsid assembly. Replacement of the HR and CC from AAP5 rescued interaction with VP; however, only the 5CC replacement was able to restore capsid assembly (to 50%). Since there are only two residues different between 1CC and 5CC (T44M and Q50R), it is not surprising that the 5CC replacement had relatively little effect on AAV1 capsid assembly compared to other N-terminal mutations. However, the ability of 5HR to pull down AAV1 VP was unexpected, since there are 10 amino acid residue differences between the two serotypes. Furthermore, binding of 5HR is not sufficient for function, since capsid assembly was defective. By replacing the T/S with three different oligomerization domains, our experiments suggest that artificially promoting oligomerization in the linker region of AAP does not correlate with the capsid assembly function. However, we note that the HR has been predicted by homology modeling to form an alpha-helix (Fig. 10B). Since amphipathic helices are often found in oligomerization domains, our data does not rule out the possibility that AAP potentially forms an oligomer. Based on our findings, we speculate that AAP interaction with the capsid is bipartite, where CC binds to a VP structural domain that is conserved among various serotypes (e.g., beta-strand), and HR either binds another site or oligomerizes to facilitate formation of a VP oligomer. Accordingly, capsid assembly only occurs when the two domains act together in a manner similar to a “lock and key” mechanism, where CC is the backbone and HR is the gear of the key which confers serotype specificity. Alternatively, since AAP interaction with the VP subunit is required to be transient, it is also possible that these domains play a role in releasing AAP from VP subunits during capsid assembly.
Using our new knowledge of AAP structure and function, we engineered additional properties onto AAP by replacing the nonessential T/S linker region. The fluorescently traceable AAP1E, which retains the same function as the wild-type counterpart, can potentially be utilized for real-time, live-cell imaging to study intracellular trafficking of AAP and capsid assembly events. Further, we engineered an AAP1-Collagen construct that shows improved stability and, by providing this eAAP in trans, we observed a 2-fold increase in vector yield compared to the wild-type counterpart. Such engineered, hyperstable AAPs could also be utilized to solve the structure of this intriguing protein as is or in complex with AAV capsid proteins. These latter observations suggest that with careful dissection of the mechanisms involving AAP biology, we can potentially develop strategies to improve the efficiency of rAAV packaging and rAAV vector yield. Several other questions remain to be addressed. For instance, how are the building blocks for AAV capsid assembly generated: as dimers, trimers, or pentamers? How is AAP involved in facilitating the oligomerization process? Another intriguing question is why do autonomous parvoviruses not appear to require an AAP-like chaperone for capsid assembly? Although these questions provide topics for ongoing and future investigations, the present study constitutes an important step in further understanding and controlling AAV capsid assembly.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HEK293 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher, Waltham, MA), as well as 100 U/ml of penicillin and 10 μg/ml of streptomycin (P/S; Thermo Fisher), in 5% CO2 at 37°C. Hybridoma supernatant of anti-AAV monoclonal antibodies B1 and A20 were produced in-house and have been described earlier (27). Mouse anti-rhodopsin (1D4; ab5417) and mouse anti-actin (ab3280) antibodies were purchased from Abcam (Cambridge, United Kingdom). Mouse anti-CD23 antibody (D-6; sc-17826) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Homology modeling of AAP.
The amino acid sequences of AAP1 to AAP9 were used in structural prediction using SWISS-MODEL (https://swissmodel.expasy.org/). The templates used for modeling AAP is Arabidopsis G protein-coupled receptor 2, GCR2 (PDB 3T33), for the HR and CC regions. The sequence identity of the modeling region is 12.20%, and the global model quality estimation (GMQE) and QMEAN Z-score are 0.08 and −1.42, respectively. Different domains of AAP were also predicted individually; the AAP2 PRR and T/S are modeled with cellobiohydrolase (PDB 1Q9H) (28), and the QMEAN Z-score is −2.08. The AAP4 BR is modeled with HIV-Rev (PDB 4PMI) (29), and the QMEAN Z-score is −3.25. The HR domain can also be modeled with bacterial RNase ligase (PDB 4XRU) and Escherichia coli topoisomerase (PDB 1YUA), with QMEAN Z-scores of −1.81 and −2.08, respectively (30).
Bioinformatic analysis.
Sequences from AAP and VP (only the residues corresponding to those in AAP) were aligned using MUSCLE, followed by manual adjustment. Alignments were done such that residues and gaps directly correspond in AAP and VP. The percent identities and similarities of AAPs from other serotypes compared to AAP1 were calculated with the Sequence Manipulation Suite (31) using the following groups for similarity: GAVLI, FYW, CM, ST, KRH, DENQ, and P. Amino acid conservation scores at each position of AAP and VP were calculated using the prediction tool developed by Capra and Singh (32). Analysis was run using the property entropy scoring method, sequence weighting, the BLOSUM62 background and scoring matrix, and a window size of 0.
Generation of different AAP constructs.
All AAP constructs were cloned into pCDNA3.1 using the EcoRI and NotI sites. Chimera constructs were cloned using either overlapping PCR or Gibson Assembly (NEBuilder HiFi; New England BioLabs, Ipswich, MA). Detailed designs for each construct are illustrated in each figure, and the amino acid sequences are given in Table 1.
AAP and capsid expression of different AAP constructs evaluated by Western blotting.
HEK293 cells at 60 to 70% confluence on a six-well plate were transfected with 600 ng of pXX680 (adenoviral helper plasmid containing E2A, E4orf6, and VA RNA from human adenovirus 5), 400 ng of pTR-CBA-Luc (AAV packaging plasmid containing CBA-Luciferase flanked by ITRs from AAV2), 600 ng of pXR-AAV1-no AAP (AAV1 Rep and Cap plasmid without ITRs; the AAP ORF is terminated by introducing a stop codon at position 64), and 400 ng of pCDNA3.1 AAP constructs (different AAP substitution, deletion, and fusion constructs driven by a cytomegalovirus promoter) using polyethylenimine (PEI) as the transfection reagent. At 3 days posttransfection, cell pellets were washed two times with 1× Dulbecco-modified phosphate-buffered saline (DPBS) and lysed with 200 μl of 1× passive lysis buffer (Promega, Madison, WI) for 30 min on ice with Halt protease inhibitor (Thermo Fisher). Supernatants were collected after centrifugation at 13,000 × g for 5 min at 4°C. The sample were prepared for Western blotting in 1× LDS loading dye (Thermo Fisher) and 100 mM dithiothreitol (DTT), then boiled at 95°C for 5 min, and loaded on a NuPAGE 4 to 12% Bis-Tris SDS-page gel (Thermo Fisher). The protein bands were transferred to nitrocellulose membrane (Thermo Fisher) using a semidry Xcell Surelock module (Thermo Fisher). VP, AAP, and actin proteins were detected by using B1 hybridoma supernatant at a 1:50 dilution, mouse α-rhodopsin antibody (ID4; ab5417) at a 1:2,000 dilution, and α-actin antibodies (ab3280) at a 1:1,000 dilution, respectively; goat α-mouse antibody labeled with horseradish peroxidase (HRP) at a 1:10,000 dilution was used as the secondary antibody. A chemiluminescence reaction was initiated with enhanced chemiluminescent substrate (SuperSignal West Femto maximum sensitivity; Thermo Fisher), and the membrane was developed on an AI600RGB system (Amersham Biosciences, Little Chalfont, United Kingdom).
AAP-dependent capsid assembly/vector production evaluated by qPCR.
HEK293 cells at 60 to 70% confluence on a six-well plate were transfected with pXX680 (600 ng), pTR-CBA-Luc (400 ng), pXR-AAV1-no AAP (600 ng), and pCDNA3.1 AAP constructs (400 ng) using PEI. The transfection medium was replaced with fresh media after 24 h, and the supernatant was harvested at 5 days posttransfection. Supernatants were collected after centrifugation at 13,000 × g for 2 min. The supernatants were used directly for standard quantitative PCR (qPCR) analysis to determine the vector yield and for transduction assays. Subsequent steps involved the harvesting of media, polyethylene glycol precipitation, iodixanol ultracentrifugation, and buffer exchange. Supernatants were treated with DNase (90 μg/ml) for 1 h at 37°C. DNase was inactivated by the addition of EDTA (13.2 mM), followed by proteinase K (0.53 mg/ml) digestion for 2 h at 55°C. Recombinant AAV vector titers were determined by qPCR with primers that amplify AAV2 inverted terminal repeat (ITR) regions (5′-AACATGCTACGCAGAGAGGGAGTGG-3′ and 5′-CATGAGACAAGGAACCCCTAGTGATGGAG-3′). The relative vector yields from different AAP constructs were normalized to wild-type AAP1 unless specifically indicated otherwise in figure legend.
In vitro AAV transduction assays.
AAV vectors produced with different AAP constructs packaging ssCBA-Luc transgenes were prediluted in DMEM plus 5% FBS plus P/S. Portions (50 μl) of recombinant AAV vectors (1,000 to 10,000 vector genomes [vg]/cell) were mixed with 50 μl of 5 × 104 HEK293 cells and added to tissue culture-treated, black, transparent bottom 96-well plates (Corning, Corning, NY). The plates were incubated in 5% CO2 at 37°C for 48 h. The cells were then lysed with 25 μl of 1× passive lysis buffer (Promega) for 30 min at room temperature. The luciferase activity was measured on a Victor 3 multilabel plate reader (Perkin-Elmer) immediately after the addition of 25 μl of luciferin (Promega). All readouts were normalized to wild-type AAP1 or AAV1 controls.
Immunofluorescence and confocal microscopy.
HEK293 cells were seeded on a 12-mm-diameter poly-lysine-treated glass coverslip (GG-12-1.5-PDL; NeoVitro, Vancouver, WA) in a 24-well plate. Cells were transfected at 60 to 70% confluence with pCDNA3.1 AAP alone (250 ng) using PEI as the transfection reagent. At 2 days posttransfection, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min. The nucleolus was stained using α-C23 (D-6) antibody, followed by goat α-mouse IgG H+L Alexa Fluor 594 (Thermo Fisher). The nucleus was stained with DAPI (4′,6′-diamidino-2-phenylindole; Thermo Fisher). Coverslips were mounted onto microscope slides using ProLong Diamond mountant (Invitrogen, Carlsbad, CA). Fluorescence images were taken by using a Zeiss LSM 710 spectral confocal laser scanning microscope at the UNC Microscopy Service Laboratory.
Immunoprecipitation assays.
HEK293 cells at 60 to 70% confluence on a 15-cm plate were transfected with pXX680 (3,000 ng), pTR-CBA-Luc (2,500 ng), pXR-AAV1-no AAP (3,000 ng), and pCDNA3.1 AAP constructs (7,500 ng) using PEI as the transfection reagent. At 3 days posttransfection, the cells were harvested from the plate in cold 1× DPBS, followed by more two washes in 1× DPBS. The pellets were resuspended in 400 μl of buffer D (20 mM HEPES/KOH [pH 7.9], 25% glycerol, 0.1 M KCl, 0.2 mM EDTA) and lysed on ice for 30 min with Halt protease inhibitor (Thermo Fisher). Lysates were spun at 13,000 × g for 2 min. Then, 5% of the supernatant was retained as input and prepared in 1× LDS buffer and 100 mM DTT. Next, 10 μl of Protein G Mag Sepharose Xtra beads washed three times in buffer D (GE Healthcare, Chicago, IL) was added to the remaining lysate; samples were then placed on a rotator at 4°C for 2 h. Next, the beads were washed twice for 30 min each in buffer D, followed by resuspension in 1× LDS buffer and 100 mM DTT. Samples were denatured at 95°C for 5 min and then loaded onto a precast 10% Bis-Tris gel (Thermo Fisher) and run in MOPS-SDS buffer. The protein was transferred to a 0.45-μm-pore size nitrocellulose membrane (Thermo Fisher) in a wet transfer apparatus. AAP was detected using α-human-HRP at a 1:10,000 dilution and visualized after reaction with Femto Western blot substrate (Thermo Fisher) on an Amersham AI600RGB system (Amersham Biosciences). Membranes were incubated in 30% peroxide for 30 min at room temperature and then reblocked. VP, AAP, and actin proteins were detected by using B1 hybridoma supernatant at a 1:50 dilution and α-actin antibody (ab3280) at a 1:2,000 dilution, respectively; goat anti-mouse-HRP at a 1:20,000 dilution was used as the secondary antibody. The membranes were again visualized as described above.
Statistical analysis.
All error bars shown represent one standard deviation. Statistical analysis was carried out in GraphPad Prism software using an unpaired, two-tailed Student t test or two-way analysis of variance (ANOVA) in Fig. 9 (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
ACKNOWLEDGMENTS
We acknowledge grant support from the National Institutes of Health (P01HL112761 and R01HL089221; awarded to A.A.).
L.V.T, S.M.-T., and A.A. designed and executed the overall study and carried out data analysis. L.V.T. and A.A. wrote the manuscript. R.M.M. carried out bioinformatic analysis and edited the manuscript.
A.A. is a founder of StrideBio, Inc.
REFERENCES
- 1.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atchison RW, Casto BC, Hammon WM. 1965. Adenovirus-associated defective virus particles. Science 149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 3.Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol Biol 807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 4.Xiao X, Xiao W, Li J, Samulski RJ. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol 71:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonntag F, Schmidt K, Kleinschmidt JA. 2010. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasileva A, Jessberger R. 2005. Precise hit: adeno-associated virus in gene targeting. Nat Rev 3:837–847. [DOI] [PubMed] [Google Scholar]
- 7.Berns KI. 1990. Parvovirus replication. Microbiol Rev 54:316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daya S, Berns KI. 2008. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earley LF, Powers JM, Adachi K, Baumgart JT, Meyer NL, Qing X, Chapman MS, Nakai H. 2017. Adeno-associated virus (AAV) assembly-activating protein is not an essential requirement for capsid assembly of AAV serotypes 4, 5, and 11. J Virol 91:1–21. doi: 10.1128/JVI.01980-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grosse S, Penaud-Budloo M, Herrmann A-K, Börner K, Fakhiri J, Laketa V, Krämer C, Wiedtke E, Gunkel M, Ménard L, Ayuso E, Grimm D. 2017. Relevance of assembly-activating protein for adeno-associated virus vector production and capsid protein stability in mammalian and insect cells. J Virol doi: 10.1128/JVI.01198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonntag F, Kother K, Schmidt K, Weghofer M, Raupp C, Nieto K, Kuck A, Gerlach B, Bottcher B, Muller OJ, Lux K, Horer M, Kleinschmidt JA. 2011. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol 85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earley LF, Kawano Y, Adachi K, Sun X-X, Dai M-S, Nakai H. 2015. Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein. J Virol 89:3038–3048. doi: 10.1128/JVI.03125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naumer M, Sonntag F, Schmidt K, Nieto K, Panke C, Davey NE, Popa-Wagner R, Kleinschmidt JA. 2012. Properties of the adeno-associated virus assembly-activating protein. J Virol 86:13038–13048. doi: 10.1128/JVI.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grove TZ, Regan L. 2012. New materials from proteins and peptides. Curr Opin Struct Biol 22:451–456. doi: 10.1016/j.sbi.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Moller-Tank S, Albritton LM, Rennert PD. 2014. Characterizing functional domains for TIM-mediated enveloped virus entry. J Virol 88:6702–6713. doi: 10.1128/JVI.00300-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S. 1999. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol 146:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. 2010. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res 38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalderon D, Roberts BL, Richardson WD, Smith AE, Hill M. 1984. A short amino acid sequence able to specify nuclear location. Cell 39(3 Pt 2):499–509. [DOI] [PubMed] [Google Scholar]
- 19.Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. 2010. Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochrane AW, Perkins A, Rosen CA. 1990. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol 64:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boudko SP, Sasaki T, Engel J, Lerch TF, Nix J, Chapman MS, Bächinger HP, Sam SW, Park J. 2009. Crystal structure of human collagen XVIII trimerization domain: a novel collagen trimerization fold. J Mol Biol 392:787–802. doi: 10.1016/j.jmb.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sales M, Plecs JJ, Holton JM, Alber T. 2007. Structure of a designed, right-handed coiled-coil tetramer containing all biological amino acids. Protein Sci 16:2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stutika C, Botschen L, Mietzsch M, Weger S, Feldkamp M, Chen W, Heilbronn R. 2016. A comprehensive RNA sequencing analysis of the adeno-associated virus (AAV) type 2 transcriptome reveals novel AAV transcripts, splice variants, and derived proteins. J Virol 90:1278–1289. doi: 10.1128/JVI.02750-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawano Y, Neeley S, Adachi K, Nakai H. 2013. An experimental and computational evolution-based method to study a mode of co-evolution of overlapping open reading frames in the AAV2 viral genome. PLoS One 8:e66211. doi: 10.1371/journal.pone.0066211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221–230. [DOI] [PubMed] [Google Scholar]
- 26.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209–219. [DOI] [PubMed] [Google Scholar]
- 27.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt JA. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol 71:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grassick A, Murray PG, Thompson R, Collins CM, Byrnes L, Birrane G, Higgins TM, Tuohy MG. 2004. Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii. Eur J Biochem 271:4495–4506. doi: 10.1111/j.1432-1033.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 29.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. 2014. RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. Elife 3:e04120. doi: 10.7554/eLife.04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benkert P, Tosatto SC, Schomburg D. 2008. QMEAN: a comprehensive scoring function for model quality assessment. Proteins 71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 31.Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. [DOI] [PubMed] [Google Scholar]
- 32.Capra JA, Singh M. 2018. Sequence analysis predicting functionally important residues from sequence conservation. Bioinformatics 23:1875–1882. [DOI] [PubMed] [Google Scholar]