ABSTRACT
Polyprotein processing has an important regulatory role in the life cycle of positive-strand RNA viruses. In the case of alphaviruses, sequential cleavage of the nonstructural polyprotein (ns-polyprotein) at three sites eventually yields four mature nonstructural proteins (nsPs) that continue working in complex to replicate viral genomic RNA and transcribe subgenomic RNA. Recognition of cleavage sites by viral nsP2 protease is guided by short sequences upstream of the scissile bond and, more importantly, by the spatial organization of the replication complex. In this study, we analyzed the consequences of the artificially accelerated processing of the Semliki Forest virus ns-polyprotein. It was found that in mammalian cells, not only the order but also the correct timing of the cleavage events is essential for the success of viral replication. Analysis of the effects of compensatory mutations in rescued viruses as well as in vitro translation and trans-replicase assays corroborated our findings and revealed the importance of the V515 residue in nsP2 for recognizing the P4 position in the nsP1/nsP2 cleavage site. We also extended our conclusions to Sindbis virus by analyzing the properties of the hyperprocessive variant carrying the N614D mutation in nsP2. We conclude that the sequence of the nsP1/nsP2 site in alphaviruses is under selective pressure to avoid the presence of sequences that are recognized too efficiently and would otherwise lead to premature cleavage at this site before completion of essential tasks of RNA synthesis or virus-induced replication complex formation. Even subtle changes in the ns-polyprotein processing pattern appear to lead to virus attenuation.
IMPORTANCE The polyprotein expression strategy is a cornerstone of alphavirus replication. Three sites within the ns-polyprotein are recognized by the viral nsP2 protease and cleaved in a defined order. Specific substrate targeting is achieved by the recognition of the short sequence upstream of the scissile bond and a correct macromolecular assembly of ns-polyprotein. Here, we highlighted the importance of the timeliness of proteolytic events, as an additional layer of regulation of efficient virus replication. We conclude that, somewhat counterintuitively, the cleavage site sequences at the nsP1/nsP2 and nsP2/nsP3 junctions are evolutionarily selected to be recognized by protease inefficiently, to avoid premature cleavages that would be detrimental for the assembly and functionality of the replication complex. Understanding the causes and consequences of viral polyprotein processing events is important for predicting the properties of mutant viruses and should be helpful for the development of better vaccine candidates and understanding potential mechanisms of resistance to protease inhibitors.
KEYWORDS: alphavirus, protease, replicase, polyprotein processing, hyperprocessive mutants
INTRODUCTION
The genus Alphavirus from the Togaviridae family of positive-strand RNA viruses includes several members that are of significant medical importance, such as chikungunya virus (CHIKV) and Ross River virus (RRV), both of which belong to the Semliki Forest virus (SFV) antigenic complex. Relatively nonpathogenic SFV is considered to be one of the best studied alphaviruses, serving as an excellent model in studies of the viral replication strategy. According to current understanding, the foundation for which was established in the early 1990s by experiments performed in the laboratories of Strauss, Rice, and Sawicki, the formation of the alphaviral replication complex depends on the production of its enzymatic and scaffolding components in the form of the nonstructural polyprotein (ns-polyprotein) P1234 precursor (reviewed in references 1 and 2). During the course of infection, the polyprotein-embedded protease nsP2 performs three sequential cleavages, ultimately releasing four mature nonstructural proteins, nsP1 to -4 (3). Cleavage of the processing site between nsP3 and nsP4 (3/4 site) occurs cotranslationally or immediately after polyprotein translation. The recognition of the processing site between nsP1 and nsP2 (1/2 site) is delayed, but its actual cleavage immediately triggers the processing of the last site between nsP2 and nsP3 (2/3 site) (4). It has been shown that the polyprotein stage is essential for the correct assembly and subcellular localization of the viral replicase (5). In early stages of infection, free nsP4 (RNA-dependent RNA polymerase) recognizes the viral RNA template and remains associated with the P123 polyprotein to establish the negative-strand RNA polymerase complex of alphaviruses (6). These complexes localize to membrane invaginations, termed spherules, which are formed on the plasma membranes of vertebrate cells (7), whereas the presence of polyproteins P123 and/or P23 is crucial for spherule formation (8). Subsequent processing of P123 into mature nsP1, nsP2, and nsP3 leads to the formation of stable late replicase complexes synthesizing genomic and subgenomic (SG) RNAs (3). New replication complexes and negative-strand RNAs are made only during early stages of infection, typically up to 3 to 4 h postinfection (hpi) (9), because during later stages of infection the accumulation of free nsP2 in infected cells reroutes processing of ns-polyprotein to the nonproductive P12+P34 pathway (4).
Elegant in its simplicity, the pathway of alphaviral replicase complex transformation demands the presence of efficient mechanisms for regulating the protease activity. The first layer of control may rely on the dissimilar affinities of the protease to three different cleavage sites within the polyprotein. nsP2 is a papain-like cysteine protease, which requires 6 residues upstream of the scissile bond for substrate recognition. The P2 position of the cleavage site (nomenclature is according to Schechter and Berger [10]) is invariably occupied by a Gly residue. For SFV, it was also revealed that the Arg residue in the P4 position of the 3/4 site is the main determinant for its efficient cleavage, whereas His in the P4 position of the 1/2 site is suboptimal for recognition efficiency (11). Accordingly, in vitro cleavage of the SFV 3/4 site is several thousand times more efficient than cleavage of the 1/2 site (4), which coincides with the order of their processing. However, this explanation for processing order is difficult to apply to either Sindbis virus (SINV), where cleavage sites are not recognized in the form of short substrates at all, to Venezuelan equine encephalitis virus (VEEV), where all sites are cleaved with similar efficiency (12), or to CHIKV, where the sequences of all three cleavage sites are similar to each other and yet are processed with different efficiencies (13). Thus, other factors that regulate processing, including site presentation and protease competence, should exist. Thus, it has been found that cleavage of the 2/3 site of SFV depends on long-range interactions involving the N-terminal region of nsP2, the C-terminal protease domain of nsP2, and residues at the C-terminal region of the macrodomain of nsP3 (11); the same phenomenon was also observed for CHIKV (14). Extensive mutagenesis of the cleavage site sequences in the SFV ns-polyprotein revealed a remarkable tolerance to substitutions: even a complete replacement of affinity-determining P6 to P4 residues with alanines in all three cleavage sites could not disrupt correct polyprotein processing (15). These findings clearly indicate that information sufficient to maintain the correct processing pattern is present within the architecture of the ns-polyprotein, and therefore the regulated processing of P1234 is achieved by the interplay of macromolecular assembly and sequence-dependent recognition mechanisms.
Although the mode of cleavage for the 1/2 site is different from that for the 2/3 site, i.e., intramolecular (in cis) versus intermolecular (in trans), respectively (4), they are both mostly dependent on the architecture of the polyprotein itself (15). However, in this case, the seeming unimportance of the amino acid compositions of the 1/2 and 2/3 sites raises the obvious question of why the particular “inefficient” recognition sequences have been evolutionarily selected. Clearly, it is not accidental, as several results indicate that altering the timing of P123 processing by an increased cleavage rate at the 1/2 (and, consequently, also the 2/3) site may have a major impact on the properties of the alphavirus: for both SFV and SINV, the accelerated processing of P123 is associated with a nonneurovirulent phenotype (16, 17). Furthermore, it has been demonstrated that the N614D mutation in nsP2 of SINV results in a hyperactivated enzyme, causing a lethal phenotype of the corresponding mutant virus (3, 18). However, the processing of which of the three cleavage sites is accelerated by that mutation and the reasons why this defect is detrimental for the infectivity of SINV genomes have not been studied in sufficient detail.
In-depth knowledge of the mechanisms responsible for substrate recognition and regulation of protease activity would be an essential prerequisite for elaborating antiviral inhibitors affecting proteolytic function with high potency. Importantly, an additional understanding of the causes, timing, and consequences of proteolytic events may also lead to the identification of critically important points in the regulation of viral replication, potentially laying the groundwork for specific inhibitors with a higher threshold for the development of resistance. Along these lines, in this study, we aimed to understand the consequences of interfering with the correct timing of cleavage events in the SFV ns-polyprotein on viral replication. Two approaches were used in an attempt to accelerate the proteolysis of P123: transfer of efficient cleavage sequence from the 3/4 site into the 1/2 and 2/3 sites and/or modification of the protease domain. It was found that optimization of the 2/3 site had a minor effect, if any, on the infectivity of the corresponding genomes, confirming previous conclusions that this is an assembly-dependent cleavage (19). In contrast, acceleration of the 1/2 site cleavage drastically diminished the infectivity of the corresponding RNA genomes and resulted in the emergence of either compensatory mutations in the cleavage site itself or combinations of mutations in the cleavage site and in the body of the nsP2 protease. The phenotype of the barely viable SINV N614D mutant with hyperactive nsP2 was found to be similar to that of SFV mutants with enhanced processing at the 1/2 site. Consistent with this, the SINV phenotype caused by hyperactivity of nsP2 could be partly compensated for by the mutations in the 1/2 site, which decreased the efficiency of its processing. This finding suggests that the 1/2 cleavage site plays the role of a molecular timer that controls replicase complex formation and/or the succession of viral replication events; for these reasons, the amino acid composition is evolutionally selected to prevent premature cleavage at this boundary. Notably, it was found that responsibility for the recognition of the P4 position in the 1/2 site lies with the amino acid residue M515 of nsP2, in contrast to Q706, which was found to be crucial for recognizing the P4 residue in the 3/4 site (15). Combined with the results of an in vivo pathogenesis study (17), a link between mutation in the P4 residue in the 1/2 site, the residue at position 515 of nsP2, and the attenuation of neurovirulent properties of SFV is proposed. This new knowledge is expected to further improve our understanding of the process of substrate recognition by alphaviral proteases and eventually provide additional means to constrain alphaviral infection.
RESULTS
Sequence optimization at the 1/2 site, but not at the 2/3 site, results in attenuation of mutant SFV genomes.
In our previous study, it was found that the substitution of all residues, except for the invariable Gly in the P2 position, within the minimal, 6-amino-acid-residue-long recognition sequence of the 1/2 site with alanines had little effect on the recovery of viruses from in vitro-synthesized mutant RNA genomes (hereafter “infectivity”) (15). This finding suggested that the polyprotein architecture, rather than primary structure of the cleavage site, is decisive for substrate targeting. However, if the amino acid composition of the cleavage site is not important and only a few small residues surrounding P2 Gly are sufficient for the site to be recognized and cleaved in the polyprotein context, then why does the sequence of the cleavage site remain conserved during the error-prone viral replication process, and for what purpose is it evolutionarily selected? To address these questions, we decided to challenge the seeming unimportance of the actual amino acid composition of the cleavage site sequences by replacing the P6 to P4 residues of the 1/2 and 2/3 sites of SFV4 with the very efficiently recognized Leu-Gly-Arg (LGR) sequence originally found in the 3/4 site (Fig. 1A), and we analyzed the consequences of such mutations.
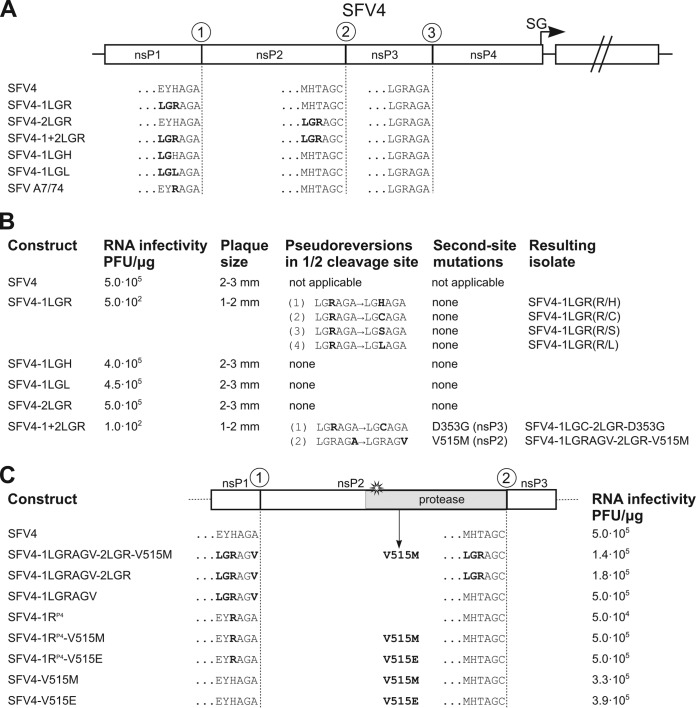
FIG 1.
Effects of the 1/2 and/or 2/3 site optimization mutations in the context of the SFV4 genome. (A) Schematic presentation of the parental SFV4 genome and its mutant versions. Mutated upstream (P-side) residues are shown in boldface. (B) Infectivities of in vitro-synthesized virus RNAs, comparative plaque sizes, and pseudoreversions/second-site mutations identified in the genomes of rescued mutant viruses. (C) Graphical presentation of SFV4 nsP2 with regions flanking 1/2 and 2/3 sites. Mutations and compensatory changes affecting P-side residues and the Val 515 residue of nsP2 in the listed constructs are shown in boldface. The infectivity of in vitro-synthesized virus RNAs is shown for each construct. All experiments were repeated at least twice with similar results.
The infectious-center assay (ICA) revealed that SFV4-2LGR had RNA infectivity equal to that of the parental SFV4. All introduced mutations were preserved, and no second-site mutations were found in the genomes of plaque-purified isolates of SFV4-2LGR (Fig. 1B). These results correlate with our previous findings concerning the SFV(23/RAGC) mutant virus, which, despite having a P4 Arg residue in the 2/3 site, had RNA infectivity and ns-polyprotein processing similar to those of wild-type (wt) SFV4 (11), confirming that optimization of the 2/3 site has no significant effect on the infectivities of the corresponding viruses. In contrast, compared with that of wt SFV4, the infectivities of SFV4-1LGR and SFV4-1+2LGR RNAs were reduced 1,000- and 5,000-fold, respectively (Fig. 1B). This result indicated that although the optimization of the 1/2 site was not completely lethal, the introduced mutations also were not tolerated. Furthermore, such a drop in infectivity often leads to the emergence of compensatory changes in the viral genome, such as reversions, pseudoreversions, and/or second-site compensatory mutations (15, 19). As expected, the sequencing of 30 individual clones, each representing an example of the 1/2 site region of the rescued viruses, revealed that the cleavage-optimized 1/2 site was consistently altered. By far the most common change, which was found in all 30 cases for the SFV4-1LGR and in 29 of 30 cases for SFV4-1+2LGR, was the reversion of P4 Arg to the original His residue or its replacement by a Cys, Ser, or Leu residue (Fig. 1B). The frequencies of all four types of changes, all resulting from a single nucleotide substitution, were nearly equal and were not affected by five passages of SFV4-1LGR and SFV4-1+2LGR at a low multiplicity of infection (MOI). Growth curves of viruses with His, Cys, Ser, or Leu in the P4 position were identical to that of wt SFV4 (data not shown). No additional mutations were detected for plaque-purified SFV4-1LGR isolates, whereas a single SFV4-1+2LGR isolate had an additional D353G change in the hypervariable region of nsP3 (Fig. 1B). Therefore, it was concluded that four (pseudo)reversions in the P4 position of the 1/2 site are functionally equivalent, and each of them is sufficient to restore the infectivity of mutant viruses. This result was directly confirmed by analysis of recombinant SFV4 genomes encoding ns-polyproteins with LGH or LGL sequences in the P6 to P4 positions of the 1/2 site: each of them had an infectivity similar to that of the wt SFV4 (Fig. 1B).
SFV genomes with Arg in the P4 position of the 1/2 site restore infectivity via adaptive mutations.
The only virus which had maintained the Arg residue in the P4 position of the 1/2 site was isolated from the SFV4-1+2LGR progeny. It had two potential adaptive mutations in its genome. First, the P1 residue of the 1/2 site was changed from the original Ala to Val, and second, the Val515-to-Met (V515M) mutation in nsP2 was detected (Fig. 1B). To test the functional effect of these mutations, recombinant virus genomes based on both SFV4-1LGR and SFV4-1+2LGR backgrounds were designed (Fig. 1C). First, the mutation in the P1 residue was analyzed. It was found that in the context of SFV4-1LGR, introduction of the P1 Val residue was sufficient to completely restore RNA infectivity (Fig. 1C). However, in the context of SFV4-1+2LGR, the infectivity remained slightly (3-fold) lower than that of wt SFV4. Thus, the presence of Val in the P1 position of the 1/2 site was sufficient to restore the infectivity of SFV4 bearing the LGR sequence in positions P6 to P4. The combination of the V515M mutation in nsP2 and the P1 Val change in the 1/2 site did not further increase the RNA infectivity; instead, a small decrease was observed (Fig. 1C). Curiously, earlier studies by Russo and colleagues predicted that amino acid residue 513 in nsP2 of VEEV, corresponding to the Val515 residue in nsP2 of SFV4, is implicated in the recognition of the P4 residue of the substrate (20). Moreover, the SFV isolate A7(74), which naturally carries an Arg residue in the P4 position of the 1/2 site, has, unlike SFV4, a Glu515 residue in nsP2. These observations strongly suggest that the V515M substitution should have functional significance. To test this hypothesis, a panel of mutants was constructed and the infectivities of their RNA genomes measured. First, it was found that the P4 His-to-Arg (1RP4) change results in a 10-fold decrease in infectivity (Fig. 1C). This observed decrease was much smaller than the defect caused by triple mutation in the P6 to P4 positions, which suggests that the P5 and/or P6 positions also have a significant role in regulating the processing of the 1/2 site. As expected, the introduction of the V515E mutation into nsP2 fully restored the infectivity of SFV4-1RP4, indicating that the combination of P4 Arg in the 1/2 site and Glu515 in nsP2, which are naturally found in SFV A7(74) and other alphaviruses such as CHIKV and RRV, is also fully functional in the context of the SFV4 genome. Furthermore, the V515M mutation completely restored the infectivity of SFV4-1RP4 (Fig. 1C). However, a V515M or V515E mutations alone had only a minor influence on the recovery of wt SFV4 (Fig. 1C). In all these assays, all introduced mutations were preserved in the rescued virus stocks, and the morphology of the plaques was similar to that of the plaques produced by wt SFV4.
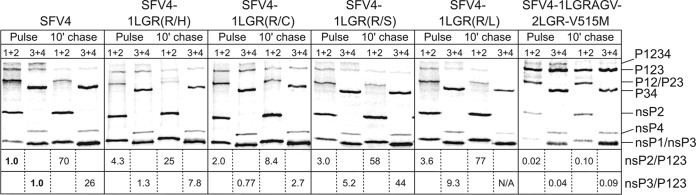
Recombinant viruses harboring compensatory changes have P1234 processing similar to that of wt SFV4.
It is known that the SFV isolate A7(74) with Arg in the P4 position of the 1/2 site has accelerated processing of P123 in infected cells (17). Concomitantly, mutations of the P6 to P4 residues of SFV4, presumably leading to artificial acceleration of 1/2 site processing, immediately induce compensatory changes (Fig. 1B). Therefore, it can be assumed that compensatory mutations act by decreasing the rate of P123 cleavage back to acceptable levels. To confirm this conclusion, P1234 processing of four plaque-purified isolates from SFV4-1LGR progeny [SFV4-1LGR(R/C), SFV4-1LGR(R/H), SFV4-1LGR(R/L), and SFV4-1LGR(R/S)] and one isolate from SFV4-1+2LGR progeny (SFV4-1LGRAGV-2LGR-V515M) was compared to that of wt SFV4 in pulse-chase experiments. In infected BHK-21 cells, the processing of ns-polyproteins of (pseudo)revertants of SFV4-1LGR with a His, Leu, or Ser residue in the P4 position of the 1/2 site was indeed very similar to that for parental SFV4; the only difference was a slight reduction in the amounts of unprocessed polyprotein precursors in the chased samples (Fig. 2). This finding indicated that despite compensatory changes, the 1/2 site cleavage rate remained slightly higher than that in wt virus. All of these differences, however, were minor, consistent with the high infectivity of the SFV4-1LGH and SFV4-1LGL constructs (Fig. 1B). SFV4-1LGR(R/C) displayed a slight delay in P123 processing; simultaneously, no difference in the processing of P1234 and P34 was observed (Fig. 2). The processing of P1234 of SFV4-1LGRAGV-2LGR-V515M was affected to a somewhat greater extent: the release of nsP2 was clearly delayed, and the amounts of the P123 and P12 precursors concomitantly increased (Fig. 2). Nevertheless, it can be concluded that all rescued viruses had a P1234 processing pattern and timing that were rather similar to those of wt SFV4. These results clearly demonstrate that the composition of the 1/2 site is actively selected to enable such timing in the cleavage pattern that is optimal for performing the tasks of viral replication.
FIG 2.
Processing of the ns-polyproteins in BHK-21 cells infected with SFV4 and its mutant variants. Cells were infected with wt SFV4 and five independently plaque-purified isolates of mutants and labeled with [35S]methionine, followed by a 10-min chase with an excess of cold methionine. The proteins were extracted from the infected cells, denatured with 1% SDS, analyzed via immunoprecipitation with antibodies against nsP1 and nsP2 or against nsP3 and nsP4 (shown above the lines) followed by SDS-PAGE, and visualized using a Typhoon imager; quantification was performed using ImageQuant software. The viruses used in the assays are indicated at the top of each panel, and the origin of the samples (pulse or chases) is shown below. The positions of the SFV nsPs and their polyprotein precursors are indicated on the right. The ratios indicate the amount of label that was incorporated into individual nsP2s or nsP3s compared with the amount incorporated into unprocessed P123; ratios for SFV4 in pulsed samples are taken as 1.
The negative effect of hyperprocessing mutation in nsP2 of SINV can be alleviated by deoptimization of the 1/2 site.
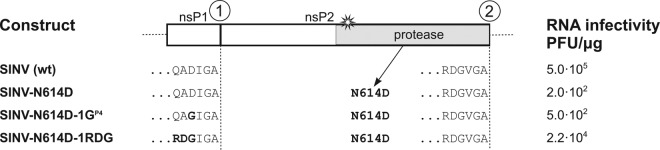
The presumed acceleration of 1/2 site cleavage by cleavage site mutation leads to reduced infectivity and causes the emergence of compensatory mutations (Fig. 1). Interestingly, instead of modifying the cleavage site to optimize it for more efficient cleavage, processing of P1234 can also be accelerated by alteration of protease properties via the introduction of hyperactivating mutations in nsP2. Therefore, we were interested in determining whether hyperactivation of nsP2 would lead to consequences for corresponding virus growth characteristics that were similar to those with cleavage site mutations. The N614D substitution in SINV nsP2 has been reported to accelerate the processing of the SINV ns-polyprotein in cell-free translation experiments and to cause a lethal phenotype for SINV (18). However, earlier studies have not clarified whether the N614D substitution causes a general increase in SINV nsP2 protease activity or causes the enzyme to be abnormally active in performing cleavage at a particular site(s) in P1234. To address this question, we first attempted to copy this mutation into SFV4, which would allow comparative analysis of the properties of hyperactive nsP2 protease with the existing set of cleavage site mutants. Sequence alignment suggested that N614 in SINV nsP2 corresponds to N605 in nsP2 of SFV4; however, introduction of the N605D mutation into the SFV4 genome did not render a hyperprocessing phenotype but caused a severe temperature-sensitive (ts) defect: the infectivity of SFV4-N605D was 1.4 × 102 PFU/μg of RNA at 37°C but was close to wt levels (5.8 × 105 PFU/μg of RNA) at 31°C. The virus was genetically unstable, and the N605D mutation reverted rapidly. Therefore, it was concluded that the specific phenotype of SINV-N614D protease cannot be easily reproduced in SFV4.
For this reason, we returned to the original Toto1101 clone of SINV to analyze the effects caused by the hyperprocessive mutation. Interestingly, in our hands, the N614D mutation in SINV nsP2 was not lethal; instead, an approximately 2,500-fold reduction of RNA infectivity compared with that for wt SINV was observed (Fig. 3). The rescued virus maintained the N614D mutation even after three passages at a low MOI. No second-site adaptive mutation was found in the ns region of the virus, indicating that a compensation of the processing defect was not strictly required for SINV-N614D. Nevertheless, based on the findings that the timing of cleavage of the 1/2 site is important for SFV, we hypothesized that the strongly attenuated SINV-N614D could be rescued by deoptimization of the 1/2 site (QADIGA↓A). We took advantage of the sequence of the 2/3 site of SINV (RDGVGA↓A), which is known to be the least optimal for protease recognition, and replaced the P4 Asp residue of the 1/2 site with a Gly residue (SINV-N614D-1GP4) or replaced the P6 to P4 residues in the 1/2 site with those from the 2/3 site (SINV-N614D-1RDG). Analysis of the recombinant SINV genomes revealed that in comparison to SINV-N614D, SINV-N614D-1GP4 had 6-fold- and SINV-N614D-1RDG more than 100-fold-higher infectivity (Fig. 3). Both rescued viruses replicated to high titers, and all introduced mutations were stably inherited in their progeny (Fig. 3). These data suggest that the enhanced and/or premature cleavage of the 1/2 site is the main molecular defect underlying the low infectivity of SINV-N614D. It also indicates that defects, caused either by the excessive activity of alphavirus protease or by the enhanced cleavage of the 1/2 site, can be compensated for by introducing or selecting amino acid changes that restore wt-like polyprotein processing.
FIG 3.
Effects of hyperactivation of nsP2 protease and compensatory effect of changes in the 1/2 site on the infectivity of SINV. A graphical presentation of the SINV 1/2 site and nsP2 region is shown. The star symbol represents the location of the catalytic Cys residue of the protease active site. The hyperactivating mutation in nsP2 (N614D) and mutated P-side residues of 1/2 site are shown in boldface. The infectivity of the in vitro-synthesized virus RNAs is shown for each mutant SINV. Experiments were repeated at least thrice with similar results.
Effects of the cleavage site and nsP2 protease mutations on the efficiency of SFV P1234 processing.
The data presented thus far represent only indirect proof that mutations introduced into cleavage sites and/or the nsP2 protease affect P1234 processing. Unfortunately, direct confirmation was not possible due to the immediate appearance and accumulation of (pseudo)reversions and/or second-site mutations (Fig. 1). Therefore, the direct effects of introduced mutations were studied using the in vitro transcription/translation/processing assay. This analysis clearly demonstrated that all mutant polyproteins carrying the LGRAGA sequence in the 1/2 site were indeed processed faster than wt P1234 (Fig. 4). As anticipated, no dramatic effect on processing was observed in the case of polyproteins with the LGRAGA sequence in the 2/3 site. Consistent with the data obtained in pulse-chase experiments (Fig. 2), polyproteins containing LGHAGA or LGLAGA in the 1/2 site were processed slower than P1234 with the LGRAGA sequence but still somewhat faster than wt P1234 (Fig. 4). These findings confirmed that the main effect of pseudoreversions present in the progeny of SFV4-1LGR was indeed achieved by slowing down the artificially enhanced processing of the 1/2 site. The processing efficiency of P1234-1LGRAGV-2LGR-V515M was found to be very close to that of wt SFV P1234, whereas elimination of the V515M mutation resulted in faster processing. The processing of P1234-1RP4 was also fast, and it was slightly decelerated by the addition of V515M or V515E mutations. Individually, the V515M and V515E mutations also slowed down the processing of P1234. Thus, taking the data together, we concluded that although the V515M mutation did not cause a drastic change in the infectivity of recombinant virus genomes (Fig. 2), it did, similar to the V515E mutation, slow down the processing of wt P1234 and P1234 with different mutations in the vicinity of the 1/2 site (Fig. 4). As expected, the processing of SINV-P1234-N614D was found to proceed at elevated rates and was slowed down by the introduction of either the 1RDG or 1GP4 mutations (Fig. 4). These data support the idea that the low infectivity of SINV N614D was indeed mostly due to the premature processing of the 1/2 site and that the infectivity of mutant genomes was partially restored by slowing down the abnormally fast processing at the 1/2 site.
FIG 4.
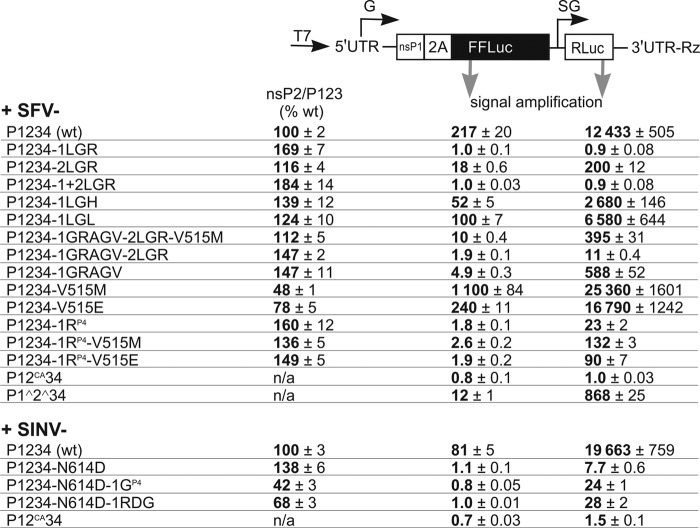
Effects of mutations in P1234 of SFV and SINV on polyprotein processing and their ability to perform minigenome replication and transcription. (Top) Schematic presentation of the construct for production of replication-competent template RNAs (minigenome reporters) for SFV and SINV replicases. T7, minimal promoter for bacteriophage T7 RNA polymerase; G, genomic promoter of virus; SG, subgenomic promoter of virus; nsP1, sequence encoding N-terminal fragment of nsP1; 2A, sequence encoding foot-and-mouth disease virus 2A autoprotease; Rz, HDV negative-strand ribozyme. (Bottom) Results of in vitro transcription/translation/processing and trans-replication assays. In vitro transcription/translation was carried out in triplicate using the TNT-coupled T7 rabbit reticulocyte lysate system; obtained products were separated via SDS-PAGE. Bands corresponding to nsP2 and P123 precursor were quantified using a Typhoon imager. nsP2/P123 ratios were calculated, normalized to those obtained for wt P1234 of SFV or SINV (each was taken as 100), and presented as the mean ± standard deviation (SD). The trans-replication assay was carried out in triplicate using BSR-T7/5 cells transfected with mixtures consisting of plasmid encoding SFV or SINV polyprotein and plasmid for expression of the corresponding minigenome reporter. Cells transfected only with plasmid encoding the minigenome reporter served as controls. Cells were lysed at 24 hpt, and FFLuc and RLuc activities were measured. Signal amplifications were calculated by dividing the corresponding FFLuc and RLuc activities from cotransfected cells by values measured using control cells (corresponding values were taken as 1). The experiments were repeated thrice with similar results. The values are presented as the mean ± SD.
Mutations in the cleavage sites and nsP2 protease domain affect the ability of the alphavirus replicase to perform genome replication and SG RNA synthesis.
Accelerated processing of P1234 most likely hampers crucial viral functions such as the formation and/or functioning of replication complexes. To analyze the effects of introduced mutations on the viral RNA synthesis, a trans-replicase assay that excludes reversion/pseudoreversion of any mutations was used. The assay was carried out using BSR-T7/5 cells cotransfected with expression plasmids for replicase (P1234) and a minigenome reporter, encoding firefly and Renilla luciferases (FFLuc/RLuc). In these cells, the constitutively expressed bacteriophage T7 RNA polymerase utilizes the FFLuc/RLuc reporter plasmid to synthesize noncapped template RNA. If this RNA is recognized in trans and utilized for genomic RNA replication and SG RNA transcription, RLuc would be expressed from the newly made SG RNAs, and the expression of FFLuc from the replicase-made capped genomic RNAs should be enhanced (Fig. 4). The relevance of such an in cellulo testing system to actual events of viral RNA replication has been elucidated previously (21, 22). The additional advantage of the system is the ability to employ the same constructs used for the in vitro transcription/translation assay.
As expected, in cells transfected only with minigenome reporter plasmids, the expression of both markers was low. Cotransfection of minigenome reporter plasmid with the plasmid expressing wt P1234 of SFV4 activated the expression of FFluc and RLuc ∼200- and ∼12,000-fold, respectively; the plasmid expressing wt P1234 of SINV activated the expression of these markers from the corresponding template ∼80 and ∼20,000-fold, respectively (Fig. 4). Much more prominent amplification of the signal of the SG marker (RLuc) was due to a lower background, making this reporter a more reliable indicator of viral replicase activity, especially in cases in which viral RNA synthesis was relatively inefficient; hence, only corresponding values were used to determine the significance of observed differences. As expected, cotransfection with plasmids expressing the P12CA34 polyproteins, which are unable to release functional nsP4 subunit due to the mutation in the active site of nsP2 protease, failed to increase the expression of any marker protein, emphasizing the vital role of 3/4 cleavage and making these constructs suitable negative controls. In contrast, expression of P1^2^34 of SFV4, which is unable to process only the P123 part of polyprotein, activated expression of FFLuc and RLuc ∼12- and ∼870-fold, respectively (Fig. 4).
P1234-1LGR and P1234-1+2LGR, both of which are characterized by significantly faster cleavage at the 1/2 site than wt P1234 (P < 0.01 for both mutants), failed to activate the expression of any markers. The ability of P1234-1RP4, also possessing significantly faster cleavage at the 1/2 site (P < 0.05), to form functional replicase was also significantly (approximately 540-fold, P < 0.001) diminished (Fig. 4). Consistent with the SFV4-1LGL and SFV4-1LGH RNA infectivity data (Fig. 1B) and the results of the in vitro transcription/translation/processing assay, these pseudoreversions that slowed down processing of the 1/2 site restored the ability of the polyprotein to perform replication/transcription of minigenomes (P < 0.01 for both pseudoreversions) (Fig. 4). The V515M and V515E mutations, when introduced into wt P1234 polyprotein, also significantly slowed down its processing (P < 0.01 for both V515M and V515E) and increased its ability to replicate/transcribe the minigenome reporter (P < 0.01 and P < 0.05, respectively) (Fig. 4). In agreement with predictions (20), similar substitutions also clearly and significantly (P < 0.01 for both V515M and V515E) increased the ability of the polyprotein harboring the 1RP4 substitution to activate the expression of the RLuc marker via SG RNA (Fig. 4). We conclude that these effects likely originate from the reduction of general protease activity of nsP2 harboring the V515E substitution, similar to previous findings (17).
Mutant polyprotein containing the LGRAGC sequence in the 2/3 site was able to activate expression of both markers, although with greatly and significantly (P < 0.001) decreased efficiency compared with wt P1234 (Fig. 4). A similar effect was also observed when the ability of P1234-1GRAGV-2LGR-V515M and its split variants (P1234-1GRAGV-2LGR and P1234-1GRAGV) to activate replication/transcription of minigenome reporter was analyzed. P1234-1GRAGV provided relatively efficient activation of template RNA replication/transcription; compared to P1234, only an approximately 20-fold reduction (P < 0.001) was observed. The addition of the 2LGR mutation resulted in approximately 50-fold further reduction (P < 0.01) of replication/transcription activity (Fig. 4). The V515M mutation, after addition to 1GRAGV-2LGR, again increased replication/transcription significantly (approximately 40-fold, P < 0.01) (Fig. 4). Thus, although the 2LGR mutation did not impair the infectivity of the corresponding viruses (Fig. 1B), it did have a prominent negative effect on the template RNA engagement and/or its replication/transcription. The accelerated processing at the 2/3 site was, to an extent, compensated for by decreased general protease activity of nsP2 harboring the V515M substitution (Fig. 4). Taken together, these findings indicate that premature cleavage of the 2/3 site has a negative effect on SFV replication. In addition, these data also clearly show that the trans-replicase assay is more sensitive and informative than the experiments using mutant viral genomes, since the latter failed to reveal that the acceleration of 2/3 cleavage could indeed affect virus replication (11, 19).
As anticipated, SINV-P1234-N614D was very inefficient in the trans-replication assay: the expression of FFLuc was not activated at all, while the expression of RLuc was increased as little as 7.7-fold (an approximately 2,500-fold reduction compared to wt P1234, P < 0.001) (Fig. 4). Thus, the inability to form a functional replicase due to accelerated processing of 1/2 and, possibly, also 2/3 cleavage sites was clearly the cause of the drastically reduced infectivity of the corresponding RNA genomes. No increase in the expression of the marker from genomic RNA was observed when 1GP4 or 1RDG mutations were introduced into the P1234-N614D polyprotein; however, the RLuc reporter signal from SG RNA increased 3- to 4-fold, and these increases were statistically highly significant (P < 0.001 for 1GP4 and P < 0.01 for 1RDG) (Fig. 4). These findings may also indicate that the N614D mutation has an impact on the 2/3 site cleavage, possibly with consequences similar to those for the 2LGR mutation in the context of SFV P1234. Remarkably, such a fairly modest increase in replicative ability in the trans-replicase assay was apparently sufficient to cause more than a 100-fold improvement of RNA infectivity after addition of the 1RDG mutation to the genome of SINV-N614D (Fig. 3). Thus, the correlation between the ability of the polyprotein to activate replication/transcription of the minitemplate and the genome infectivity of the corresponding mutant viruses was not linear. Indeed, the replicase/transcriptase activity of P1234-1RP4 of SFV4 was even lower than that of SINV P1234-N614D-1RDG (Fig. 4), and yet the SFV4 genome harboring such a mutation had an infectivity of 5 × 104 PFU/μg RNA (Fig. 1C), which was even higher than that of SINV-N614D-1RDG (Fig. 2). The conjunction of the results of ICA, polyprotein processing, and minigenome replication analyses allowed to conclude that the cleavages within the wt polyprotein are required to proceed within a rather narrow time window, and therefore even small changes in timing of processing can lead to profound defects in replication capacity and/or virus genome infectivity.
Mutational analysis of the N-terminal region of nsP2 reveals its possible implication in contacts with other viral proteins.
Since the above-described experiments identified the utmost importance of the correct timing of 1/2 site processing, it is now of particular interest to identify why this cleavage is so important. The immediate results of 1/2 site cleavage are the separation of nsP1 from the rest of the polyprotein and liberation of the N terminus of nsP2. The most N-terminal region of nsP2 is conserved among different alphaviruses (Fig. 5A) and has been previously shown to be critically important for 2/3 site recognition (19), as well as for nucleoside triphosphatase (NTPase) and RNA helicase activities of nsP2 (23). Therefore, we decided to analyze whether some residues of the immediate N-terminal region of nsP2 are of particular importance and may also be implicated in regulating the 1/2 site cleavage, since they are part of P′ side of the 1/2 site, or start playing a regulative role in viral replication events as part of the freed N terminus of nsP2.
FIG 5.
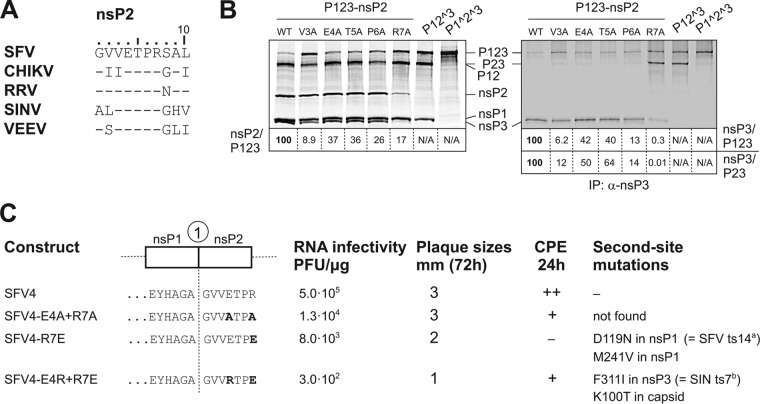
The N-terminal amino acid residues of SFV nsP2 affect polyprotein processing, RNA infectivity, virus growth, and cytotoxicity of the corresponding mutant viruses. (A) Comparison of sequences of first 10 N-terminal amino acid residues of SFV, CHIKV, RRV, SINV, and VEEV nsP2. (B) Left panel, results of in vitro transcription/translation/processing of P123 (wt), P123-V3A, P123-E4A, P123-T5A, P123-P6A, P123-R7A, P12^3, and P1^2^3 of SFV. Right panel, analysis of the same samples after immunoprecipitation with anti-nsP3 antibodies. Proteins were separated by SDS-PAGE and visualized using a Typhoon imager; quantification was performed using ImageQuant software. The ratios under the left panel indicate the amount of label that was incorporated into the individual nsP2 protein compared with the amount incorporated into unprocessed P123. The ratios under the right panel indicate the amount of label that was incorporated into the individual nsP3 protein compared with the amount incorporated into unprocessed P123 or unprocessed P23. In all cases, ratios for wt P123 are taken as 100. (C) Graphical presentation of the 1/2 site region of SFV4 and its mutant versions. Mutated residues of nsP2 are shown in boldface. The results for RNA infectivity, plaque sizes, cytopathic effects (CPE) at 24 hpi, and second-site mutations revealed in the genomes of the rescued mutant viruses are shown. All experiments were repeated at least twice with similar results.
To reveal the possible significance of this region in regulating the 1/2 site cleavage, alanine scanning within amino acid residues 3 to 7 of SFV nsP2 was performed. In vitro transcription/translation/processing revealed that compared with wt P123, mutant P123 polyproteins were less actively processed; the effects were especially prominent for the P123-V3A and P123-R7A mutants (Fig. 5B, left panel). After immunoprecipitation using anti-nsP3 antibody (Fig. 5B, right panel), it became evident that the effects caused by these two mutations were different from each other. The R7A mutation resulted in a prominent reduction in the amounts of released mature nsP2 and nsP3 and in the concomitant accumulation of processing intermediate P23 (Fig. 5B). As the release of mature nsP1 was not affected (Fig. 5B, left panel), it was concluded that the R7 residue of nsP2 was directly involved in the recognition of the 2/3 site and/or sequences in nsP3 required for processing of this site. In contrast, the V3A mutation increased the stability of P123 (Fig. 5B) but caused little or no increase in the amounts of P23 (Fig. 5B). Thus, mutation of the P3′ residue suppressed 1/2 site cleavage. The effect of V3A as well as E4A, T5A, and P6A mutations on 2/3 site processing was mild (barely detectable) and most likely indirect: as 2/3 site cleavage depends on the preceding 1/2 site processing, which releases the N terminus of nsP2 involved in 2/3 site recognition, all the defects in the first site will be passed on to the second cleavage.
In the context of SFV4, all the above-listed alanine mutations in the N terminus of nsP2 had little, if any, effect on virus infectivity (data not shown). Thus, it was unreasonable to expect an accumulation of compensatory changes that could be used to reveal possible genetic interactions of the N-terminal region of nsP2 with other parts of the ns-polyprotein. To enhance the impact of mutations in this region on virus replication, we combined two alanine mutations of charged residues (E4A+R7A) and introduced charge reversal changes, R7E and E4R+R7E, in nsP2 (Fig. 5C). These changes caused a clear reduction of virus infectivity; the effect was most profound for SFV4-E4R+R7E (>1,000-fold) and smallest for SFV4-E4A+R7A (≈40-fold). Viruses containing charge reversion mutations had small-plaque phenotypes, and importantly, sequencing of their genomes revealed different second-site mutations. Surprisingly, none of the changes were found within nsP2 itself. Instead, several mutations were found in the methyltransferase domain of nsP1 (D119N and M241V), one in the zinc-binding domain of nsP3 (F311I), and one in the capsid protein (K100T). Although a detailed analysis of these mutations is beyond the scope of current study, it should be noted that these residues are present in locations that are important for the alphavirus infection process. Thus, the compensatory mutation M241V in nsP1 lies close to the membrane-binding amphipathic peptide (amino acid residues 245 to 264). Most strikingly, however, the compensatory change in D119N in nsP1 is identical to the mutation responsible for the ts phenotype of SFV (ts14) (24, 25), which is known to have reduced P1234 processing and P12 processing intermediate accumulation (25), reinforcing the assumption that the emergence of this change in response to modification of the N terminus of nsP2 is unlikely to be accidental. Interestingly, a recent report by Kumar and colleagues also confirmed direct interactions between the N-terminal region of CHIKV nsP2 and the central part of nsP1 (amino acid residues 170 to 288) that appear to have functional significance for NTPase activity of nsP2 (26). The residue corresponding to F311 in SFV nsP3 is a ts7 mutation site (F312S) in SINV that is known to cause specific defects in negative-strand RNA synthesis (27). The substitution of the charged K100 residue in the capsid protein is also of significant interest because studies with VEEV have shown that the inability of mutated (charge-neutralized) VEEV capsid to package viral RNA can be compensated for by adaptive changes in nsP2, including the modification of conserved N-terminal residues (Val3 and Thr5) (28) in addition to other regions of VEEV nsP2 (29). Therefore, it has been previously suggested that nsP2 may be implicated in coordinating the packaging of alphaviral RNA into nucleocapsids. Taken together, these new findings suggest the intricate involvement of the most N-terminal part of alphaviral nsP2 in regulating various activities of other viral proteins (nsP1, nsP3, and capsid protein) required for viral RNA replication and packaging. It is conceivable that the ability of the N terminus of nsP2 to exert its impact on fellow viral proteins is in turn dependent on its accessibility, which is altered upon 1/2 site processing, thus making this cleavage particularly important and placing the timing of the cleavage event under tight selective pressure.
DISCUSSION
In this study, we aimed to explore the possible impact of speeding up ns-polyprotein processing on alphavirus infection. The first approach, which was used in an attempt to accelerate proteolysis, was based on introducing an efficiently cleaved sequence from the 3/4 site into the 1/2 and 2/3 sites. In the SFV4 ns-polyprotein, the sequences of the 1/2 and 3/4 sites are different (EYHAGA↓G and LGRAGA↓Y, respectively), which under cell-free conditions leads to almost 5,000-fold more efficient cleavage of the latter site (4). The difference in cleavage efficiencies of the 1/2 and 3/4 sites for the related CHIKV, where the sequences of these sites are more similar to each other (EDRAGA↓G and LDRAGG↓Y, respectively), is much smaller, because of both the more efficient cleavage of the 1/2 site and the less efficient cleavage of the 3/4 site (13). Interestingly, the 1/2 site in the natural A7(74) strain of SFV also contains the P4 Arg residue, which has been shown to have consequences for ns-polyprotein processing and virulence of the virus (17). These are just a few examples of findings directly or indirectly indicating the existence of a complex interplay between different factors affecting the processing and functioning of the viral replicase precursor. To obtain more insight into these processes, the second approach, based on the use of SINV harboring an N614D mutation in the protease part of nsP2, rendering the protease hyperprocessive, was implemented.
Although it is known that cleavage of the 1/2 site in the P1234 polyprotein of alphaviruses is postponed during the course of infection, neither the factors delaying this proteolytic event nor the molecular consequences that follow this cleavage occurrence are entirely understood. Our findings revealed that acceleration of 1/2 site cleavage drastically diminished the infectivity of mutant viruses (Fig. 1 and 3). Analysis of the progeny of rescued viruses revealed that they survived with the help of compensatory mutations either in the cleavage site itself or by combinations of mutations in the cleavage site and in the body of nsP2 protease (residue 515) (Fig. 1B); all these changes were associated with decreasing the rate of 1/2 site cleavage (Fig. 4) and restoring the original pattern of ns-polyprotein processing (Fig. 2). These results highlight the importance of the 1/2 site sequence as a subject of evolutionary selection toward not-too-efficient recognition by the ns-protease. The findings also suggest that the 1/2 site performs a function of a molecular time delay relay that controls the succession of crucial events in alphavirus replicase formation and functioning. For a productive viral infection process, the stability of P123 must be sufficient to allow the trafficking of viral protein complexes to the inner surface of the plasma membrane (5), where it is involved in the formation of viral replication compartments (spherules). In addition, the processing of P123 is delayed because only in its unprocessed form does it have specific properties that allow the initiation and completion of the synthesis of negative-strand RNA. Once these tasks are completed, cleavage of P123 is allowed to occur, and it initiates changes in the conformation of the replicase complex which eventually lead to transformation of viral replicase into the genomic and SG RNA-transcribing machine. Since the proteolytic maturation of P123 starts with 1/2 site cleavage, an event of such importance apparently must be regulated by multiple factors. The cis preference of the 1/2 site cleavage (4) represents one such mechanism. A previous study revealed that the 1/2 site targeting is dependent on its presentation to the protease active site that is imposed by the tertiary structure of the viral replication complex (15). Here, we demonstrated that the responsibility for recognizing the P4 position in the 1/2 site lies in amino acid residue V515 in nsP2, in contrast to Q706, which was found to be crucial for recognizing the P4 residue in the 3/4 site (15). Further analysis of the determinants of cis cleavage in the 1/2 site, including the identification of additional viral and cellular factors involved in the regulation of its processing, is the topic of ongoing studies.
In this study, it was found that optimization of the 2/3 site sequence had a minor (if any) effect on the infectivity of the corresponding mutant genomes; this is consistent with a previous conclusion that it is dominantly an assembly-dependent cleavage (15). However, for the first time for this site, we also acquired clear evidence that the timing and/or efficiency of the cleavage is crucial for the alphavirus. First, an interesting double mutant, SFV4-1LGRAGV-2LGR-V515M, was discovered in the progeny of SFV4-1+2LGR. Since Arg in the P4 position of the 1/2 site was found to be strongly counterselected in other isolates, it is reasonable to assume that the adaptation arose by first acquiring the P1 change (LGRAGA to LGRAGV) that aimed to reduce the 1/2 site cleavage efficiency even in the presence of Arg as P4 (Fig. 4); this should also have delayed 2/3 site cleavage due to the decreased release of nsP2 with a free N terminus. This adaptation was, however, followed by the V515M change, which does not offer any advantage for viral RNA infectivity (compare SFV4-1LGRAGV+2LGR and SFV4-1LGRAGV-2LGR-V515M in Fig. 1C), indicating the presence of other flaws that required compensation. As this mutant was rare (found only once among 30 sequences), it could be considered that this failing, almost certainly related to the presence of the LGR sequence in the 2/3 site, was relatively mild yet important for the virus. Indeed, the trans-replicase assay clearly confirmed that the sequence of the 2/3 site is important for virus RNA synthesis (Fig. 4). It has been proposed that when the concentration of free nsP2 in the cell rises, the processing of the ns-polyprotein is switched from the P123+nsP4 to the nonreplicative P12+P34 pathway (4); for polyproteins carrying the 2LGR mutation, this switch likely occurs at lower levels of free nsP2 than it does for wt polyproteins. In addition, recent data on the alphavirus replicase complex assembly process also highlight the crucial role of unprocessed P23, and by extension the slow cleavage of the 2/3 site, in spherule formation (8). Thus, the defect caused by the 2LGR mutation may not provide enough time for such a process to occur. This conclusion is supported by two observations. First, the negative effect of the 2LGR substitution was clearly more prominent in the trans-replicase assay (Fig. 4). During virus infection, the ns-polyprotein and/or its cleavage products (P123 and P23) can directly interact with the viral genome (in cis binding to their own template mRNA) and initiate replication complex assembly. In contrast, in the trans-replicase system this is not possible, and ns-polyproteins should find a suitable template RNA provided in trans. Compared with the binding that occurs in cis, it is obviously less efficient and/or more time-consuming and must therefore also be more vulnerable to premature cleavage of the 2/3 site. Hence, the presence of mutations that reduce the protease activity of nsP2 in general (V515M or V515E) has a more prominent effect in this system. Second, the phenotype of the barely viable SINV-N614D mutant was found to be similar to that of SFV mutants with enhanced processing at the 1/2 and 2/3 sites. Thus, comparison of SINV-N614D and SINV-N614D-1RDG demonstrated that the slowdown of 1/2 site processing (Fig. 4), albeit having a major effect on RNA infectivity (Fig. 3), had a surprisingly small positive effect on the RNA synthesis efficiency in the trans-replicase assay (Fig. 4). Although alternative explanations are certainly possible, this again points toward the importance of 2/3 site processing (P23 stability) for efficient assembly of functional replication complexes.
The conserved N-terminal region of nsP2 has been previously shown to be essential for the ability of the enzyme to recognize the 2/3 junction (19) and for its NTPase and RNA helicase activities (23). The localization of compensatory changes, detected in the progeny of SFV4-E4R+R7E and SFV4-R7E (Fig. 5), also suggests the involvement of the N-terminal part of nsP2 in functional interactions with nsP1, nsP3, and CP, further confirming the central role of nsP2 in alphavirus infection. Dependence of the multiple functions of nsP2 on the liberation of its N terminus points to the conclusion that the conformations of the nsP2 in the precleavage state and in a free form are likely different and that the most N-terminal region of nsP2 may function as a switch. In alphavirus replication, nsP2 also directly interacts with viral RNA, and accordingly, the mutations in the C-terminal part of nsP2 are known to affect the template preference and RNA-synthesizing capabilities of the replication complex (30, 31). nsP4, the polymerase subunit of the replicase complex, has distinct binding sites for recognizing genomic and SG promoters (32), whereas assistance from P123 components is absolutely required for binding to the SG promoter (33). Therefore, it can be hypothesized that after processing, the conformational changes in nsP2 may induce transformations in other viral proteins and alter the properties of nsP4, allowing the presentation of binding sites for the genomic or SG RNA promoter. This assumption is consistent with a previous study in which SINV mutants which are unable to process P123 require only compensatory mutations in nsP4 to obtain replication properties very similar to those of wt SINV (34).
Why is P123 processing needed if the alphavirus can, in principle, survive in its absence (35)? On the one hand, each individual nsP has additional functions that are not related to RNA replication directly. In particular, it has been previously demonstrated that SINV mutants which are unable to produce free nsP2 are also not able to replicate in type I interferon-competent cell lines due to the inability to suppress antiviral responses. This phenomenon occurs because only nsP2 liberated from the membrane-bound nsP complex can become translocated into the nucleus, where it initiates inhibition of cellular transcription by inducing degradation of the catalytic subunit of the RNA polymerase II complex (36). In addition, free nsP2 appears to interact negatively with the translation machinery and interfere with the JAK-STAT signaling pathway (37–39). All these activities of nsP2 (and those of other nsPs released from ns-polyproteins by nsP2) are required during early stages of infection.
On the other hand, deceleration of P123 processing has clear consequences for replicase complex activities. For example, while the slower processing of P123 (caused by the P3 Thr residue in the 1/2 site) in SINV strain S.A.AR86 is associated with increased neurovirulence (40), the opposite applies to RRV, where P1 and P3 Val residues in the 1/2 site slow down P123 processing, increase the synthesis of type I interferon-inducing RNAs, and result in the in vivo attenuation of the virus (45). Taken together, these observations highlight the necessity for evolutionary compromises to balance the delay of polyprotein processing, required for the purposes of proper assembly and functionality of the replication complex, with the requisite for prompt release of viral effectors, needed to antagonize cellular immune defenses as soon as possible, from the viral multiprotein complex.
Therefore, we conclude that the mutational analysis performed here further highlights the distinct roles of nsP2 as a key player during the course of viral replication. To allow ns-polyprotein movement to the site of replication complex formation, template RNA binding, and negative-strand RNA synthesis, the cleavage of the 1/2 site is delayed; this delay is ensured by the cooperative action of multiple factors. The N-terminal region of nsP2 inside the uncleaved P123 polyprotein appears to be involved in regulating negative-strand RNA synthesis through interactions with nsP1 and nsP3, and it is possible that this very engagement deters the protease from cleaving the 1/2 site, as it would simply not be accessible until negative-strand RNA synthesis is completed. Liberation of the N-terminal region leads to conformational changes in nsP2, making protease competent for 2/3 site cleavage (19), allowing for maximal RNA helicase/NTPase activities (23), and presumably contributing to RNA packaging into nucleocapsids (28). In addition to controlling the timing of release of the nsP2 N terminus, cleavage of the 2/3 site is also controlled by its nonoptimal-for-recognition sequence, which ensures sufficient stability of P23 (or the corresponding part of P1234) for the formation of functional replicase complexes. The findings in this study therefore highlight the importance of the timeliness of proteolytic events as an additional layer of regulation of efficient virus replication. An accurate understanding of this property, which appears to be crucial for the virus, may be instructive in the development of better live attenuated vaccine candidates and potential drug discovery processes.
MATERIALS AND METHODS
Cells and media.
BHK-21 cells were grown in Glasgow's minimal essential medium (GMEM) (Gibco) containing 10% fetal calf serum (FCS), 2% tryptose phosphate broth (TPB), 20 mM HEPES (pH 7.2), 100 U/ml penicillin, and 0.1 mg/ml streptomycin in a humidified incubator at 37°C under 5% CO2. BSR-T7/5 cells, a derivative of BHK-21 cells stably expressing bacteriophage T7 RNA polymerase (41), were cultured in the same medium supplemented with 1 mg/ml of G418.
Construction of recombinant viruses.
Mutations altering the P6 to P4 residues of the 1/2 site from EYH to LGR and the P6 to P4 residues the of 2/3 site from MHT to LGR were first introduced into cloned fragments of infectious cDNA (icDNA) of SFV4 using PCR-based mutagenesis. The mutated fragments were used to replace the wild type (wt) counterparts in icDNA clone pSP6-SFV4 (42) utilizing the available restriction sites. The resulting plasmids were designated pSFV4-1LGR and pSFV4-2LGR (Fig. 1A). The icDNA clones containing combinations of these mutations and/or revealed adaptive mutations were constructed using a similar approach; these clones were designated pSFV4-1+2LGR, pSFV4-1LGH, pSFV4-1LGL, pSFV4-1LGRAGV-2LGR-V515M, pSFV4-1LGRAGV-2LGR, pSFV4-1LGRAGV, pSFV4-1RP4, pSFV4-1RP4-V515E, pSFV4-1RP4-V515M, pSFV4-V515E, and pSFV4-V515M (Fig. 1A and C).
The icDNA clones of SFV containing substitutions and their combinations in the N-terminal part of nsP2 or N605D substitution in the protease part of nsP2 were obtained similarly and designated pSFV4-E4A+R7A, pSFV4-R7E, pSFV4-E4R+R7E (Fig. 5C), pSFV4-V3A, pSFV4-E4A, pSFV4-T5A, pSFV4-P6A, pSFV4-R7A, and pSFV4-N605D. For SINV, icDNA clone pToto1101 (43) was used as starting material. Using the approaches described above, mutations affecting residue 614 of nsP2 (N614D), the P4 residue of the 1/2 site D4G (1GP4), or the P6 to P4 residues of the 1/2 site from QAD to RDG (1-RDG) were introduced into the icDNA clone; the resulting clones were designated pSINV-N614D, pSINV-N614D-1GP4, and pSINV-N614D-1RDG (Fig. 3). The sequences of all obtained clones were verified.
Plasmids containing SFV icDNAs were linearized with SpeI and plasmids containing SINV icDNAs with XhoI, followed by in vitro transcription using the mMessage mMachine SP6 transcription kit (Ambion) according to the manufacturer's protocol. Capped RNA transcripts were used for the transfection of BHK-21 cells via electroporation. The ICA was performed as previously described (44). Primary virus stocks were collected following incubation for 24 h at 37°C (or up to 120 h at 31°C for some mutants with a ts phenotype), titrated using plaque assay on BHK-21 cells, and used in subsequent experiments.
Virological methods.
Secondary stocks of mutated viruses and plaque-purified virus isolates were obtained as previously described (15). Briefly, BHK-21 cells were infected with primary viral stocks at an MOI of 0.1. At 24 to 48 hpi, viral stocks were collected and their titers determined through a plaque assay. The procedure was repeated to obtain subsequent passages. For plaque purification, viral stocks were used to infect BHK-21 cells (the amount of virus was selected to obtain 10 to 20 plaques per 35-mm dish), and cells were overlaid with GMEM containing 2% FCS and 0.9% Bacto agar. At 120 hpi, the cells were stained with neutral red, the plaques were picked, and the obtained viruses were amplified in BHK-21 cells.
The presence of the introduced mutations was verified through reverse transcription-PCR (RT-PCR) and sequencing essentially as previously described (15). Briefly, viral RNA extracted from each stock using the RNeasy minikit (Qiagen) was reverse transcribed using the first-strand cDNA synthesis kit (Thermo Fisher Scientific) and PCR amplified using pairs of SFV- or SINV-specific primers amplifying regions containing the mutated residue(s). The PCR products were cloned into pJet1.2 vector (Thermo Fisher Scientific); from the obtained libraries, 30 randomly picked clones were sequenced. To identify second-site mutations, a set of PCR fragments covering either the complete SFV genome or complete ns region of the SINV genome was obtained and sequenced from at least two individual plaque-purified isolates for each mutated genome.
Metabolic labeling and immunoprecipitation.
Pulse-chase labeling was carried out as described previously (11, 15). Briefly, BHK-21 cells were infected at an MOI of 100. At 3 hpi, the cells were starved in methionine- and cysteine-free medium for 30 min and then labeled with 5 μCi of a [35S]methionine-[35S]cysteine mixture (PerkinElmer) for 10 min. In the chased samples, the pulse was followed by a chase for 10 min in the presence of excess unlabeled methionine and cysteine. The cells were then lysed by boiling in 1% SDS, and the material was subjected to immunoprecipitation (IP) analysis using combinations of antisera (nsP1+nsP2 and nsP3+nsP4) against SFV nsPs. The precipitated proteins were separated using SDS-PAGE and visualized using a Typhoon imager.
Construction of plasmids for recombinant polyprotein expression.
The construction of expression plasmids for SFV ns-polyproteins P123, P1234, and P1^2^34 has been previously described (11). All additional ns-polyprotein expression plasmids were obtained using PCR-based site-directed mutagenesis and standard cloning techniques. Constructs designated P123-V3A, P123-E4A, P123-T5A, P123-P6A, and P123-R7A were mutated at residues 3, 4, 5, 6, and 7 of nsP2, respectively; P1^2^3 and P12^3 harbored P2 Gly-to-Val mutations in the 1/2 and/or 2/3 site. Construct P12CA34 expresses nsP2 with the C478A mutation. Constructs designated P1234-1LGR, P1234-2LGR, P1234-1+2LGR, P1234-1LGH, P1234-1LGL, P1234-1LGRAGV-2LGR-V515M, P1234-1LGRAGV-2LGR, P1234-1LGRAGV, P1234-V515M, P1234-V515E, P1234-1RP4, P1234-1RP4-V515E, and P1234-1RP4-V515M were obtained from P1234 by replacement of wt fragments with the corresponding fragments from the above-described icDNA clones. The constructs expressing P1234 of SINV and its mutant forms P1234-N614D, P1234-N614D-1GP4, and P1234-N614D-1RDG were obtained from the corresponding icDNA clones using subcloning procedures and designated SINV-P1234, SINV-P1234-N614D, SINV-P1234-N614D-1GP4, and SINV-P1234-N614D-1RDG. The clone expressing SINV P1234 with mutation C481A in nsP2 was obtained using PCR-based mutagenesis and designated SINV-P12CA34.
Construction of vectors for expression of template RNAs for SFV and SINV replicases and the trans-replication assay.
The cassettes for expression of SFV and SINV replication-competent RNA templates were constructed using the following elements (Fig. 4, top panel): (i) the minimal bacteriophage T7 RNA promoter, (ii) the 5′ untranslated region (UTR) of the virus followed by 74 (SFV) or 66 (SINV) codons from the region encoding the N-terminal part of nsP1, (iii) sequence encoding the FFLuc reporter fused in frame with sequence encoding the N-terminal fragment of nsP1, (iv) sequence encoding foot-and-mouth disease virus 2A autoprotease separating elements ii and iii, (v) the SG promoter of virus spanning from position −150 to +51 (SFV) or −97 to +43 (SINV) with respect to the start site of SG RNA transcription, (vi) sequence encoding the RLuc reporter, (vii) the full-length 3′ UTR of the virus (264 bp for SFV and 322 bp for SINV) followed by 30 A residues, and (viii) the negative-strand ribozyme of hepatitis delta virus (HDV). The constructs were assembled from synthetic DNA fragments (GenScript) and PCR products and verified by sequencing.
The trans-replication assay was performed in triplicate on BSR-T7/5 cells grown on 24-well plates (105 cells per well) via cotransfection with mixtures consisting of 320 ng of plasmid encoding SFV or SINV replicase (wt or mutant) and 160 ng of the corresponding FFLuc/RLuc reporter plasmid using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocols. Control cells were transfected only with plasmid for expression of the RNA template. At 24 h posttransfection (hpt), the cells were lysed in passive lysis buffer (Promega), and FFLuc and RLuc activities were analyzed using the dual-luciferase reporter assay kit and Glomax SIS luminometer (Promega). Signals measured in cells cotransfected with template and replicase expression plasmids were normalized to those from corresponding control cells.
In vitro translation followed by immunoprecipitation.
In vitro translation reactions were carried out using the TNT-coupled T7 rabbit reticulocyte lysate system (Promega) according to the manufacturer's protocol. Reaction mixtures (10 μl) containing 4 μCi of [35S]methionine (PerkinElmer), 5 μM MG132 (proteasome inhibitor; Sigma), and 400 ng of plasmid encoding P1234, P1234-1LGR, P1234-2LGR, P1234-1+2LGR, P1234-1LGH, P1234-1LGL, P1234-1LGRAGV-2LGR-V515M, P1234-1LGRAGV-2LGR, P1234-1LGRAGV, P1234-V515M, P1234-V515E, P1234-1RP4, P1234-1RP4-V515E, P1234-1RP4-V515M, P12CA34, P1^2^34, SINV-P1234, SINV-P1234-N614D, SINV-P1234-N614D-1GP4, SINV-P1234-N614D-1RDG, or SINV-P12CA34 were incubated for 50 min at 30°C. Thereafter, translation mixtures were treated with 10 ng of RNase A for another 5 min and denatured in Laemmli buffer at 70°C for 10 min. Translation/processing products were separated via SDS-PAGE and visualized using a Typhoon imager. The densities of bands corresponding to nsP2 and P123 precursor were quantified from 3 independent experiments using ImageQuant software (GE Healthcare); the processing efficiency was calculated as the ratio of label that was incorporated into individual nsP2 compared to the amount incorporated into the unprocessed P123 precursor.
In another setup, in vitro transcription/translation mixtures for plasmids encoding P123, P1^2^3, P12^3, P123-V3A, P123-E4A, P123-T5A, P123-P6A, or P123-R7A were obtained as described above. Samples corresponding to 9/10 of the reaction volume were first denatured by boiling in lysis buffer (50 mM Tris-HCl [pH 8.0], 2% lithium dodecyl sulfate, 150 mM NaCl, 5 mM EDTA, and 1% NP-40) and then diluted 100-fold with IP buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, and 1% NP-40) and incubated for 1 h at 4°C with rabbit polyclonal antiserum against SFV nsP3 (in house). Immunocomplexes were precipitated with protein A–Sepharose CL-4B (Sigma-Aldrich) overnight at 4°C and then washed four times with IP buffer containing 400 mM NaCl. The precipitated proteins were denatured by heating in Laemmli buffer. Proteins from untreated reaction mixtures (1/10 of the reaction volume per lane) as well as those obtained from IP were separated by SDS-PAGE, visualized using a Typhoon imager, and quantified using ImageQuant software (GE Healthcare).
Statistical analysis.
Statistical analysis was done with the Microsoft Excel software. Student's paired two-tailed t test was used for comparison of two sets of data.
ACKNOWLEDGMENTS
We acknowledge Karl-Klaus Conzelmann for the BSR-T7/5 cell line.
This work was supported by the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering, Estonia (2014-2020.4.01.15-013 to A.M.), institutional research funding (IUT20-27) from the Estonian Research Council (to A.M.), and Estonian Science Foundation grant 9421 (to V.L.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JVI.00152-18.
REFERENCES
- 1.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawicki DL, Sawicki SG. 1994. Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes. Arch Virol Suppl 9:393–405. [DOI] [PubMed] [Google Scholar]
- 3.Lemm JA, Rümenapf T, Strauss EG, Strauss JH, Rice CM. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13:2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kääriäinen L, Ahola T. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem 278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 5.Salonen A, Vasiljeva L, Merits A, Magden J, Jokitalo E, Kääriäinen L. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J Virol 77:1691–1702. doi: 10.1128/JVI.77.3.1691-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemm JA, Rice CM. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol 67:1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spuul P, Balistreri G, Kaariainen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J Virol 84:7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellström K, Kallio K, Utt A, Quirin T, Jokitalo E, Merits A, Ahola T. 2017. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J Virol 91:e00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawicki DL, Sawicki SG. 1980. Short-lived minus-strand polymerase for Semliki Forest virus. J Virol 34:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schechter I, Berger A. 1967. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 27:157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- 11.Lulla A, Lulla V, Tints K, Ahola T, Merits A. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J Virol 80:5413–5422. doi: 10.1128/JVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D, Tözsér J, Waugh DS. 2009. Molecular cloning, overproduction, purification and biochemical characterization of the p39 nsp2 protease domains encoded by three alphaviruses. Protein Expr Purif 64:89–97. doi: 10.1016/j.pep.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausalu K, Utt A, Quirin T, Varghese FS, Žusinaite E, Das PK, Ahola T, Merits A. 2016. Chikungunya virus infectivity, RNA replication and non-structural polyprotein processing depend on the nsP2 protease's active site cysteine residue. Sci Rep 6:37124. doi: 10.1038/srep37124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utt A, Das PK, Varjak M, Lulla V, Lulla A, Merits A. 2015. Mutations conferring a noncytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of nonstructural protein 2. J Virol 89:3145–3162. doi: 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lulla V, Karo-Astover L, Rausalu K, Merits A, Lulla A. 2013. Presentation overrides specificity: probing the plasticity of alphaviral proteolytic activity through mutational analysis. J Virol 87:10207–10220. doi: 10.1128/JVI.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heise MT, Simpson DA, Johnston RE. 2000. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J Virol 74:4207–4213. doi: 10.1128/JVI.74.9.4207-4213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saul S, Ferguson M, Cordonin C, Fragkoudis R, Ool M, Tamberg N, Sherwood K, Fazakerley JK, Merits A. 2015. Differences in processing determinants of nonstructural polyprotein and in the sequence of nonstructural protein 3 affect neurovirulence of Semliki Forest virus. J Virol 89:11030–11045. doi: 10.1128/JVI.01186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauss EG, De Groot RJ, Levinson R, Strauss JH. 1992. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology 191:932–940. doi: 10.1016/0042-6822(92)90268-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lulla A, Lulla V, Merits A. 2012. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J Virol 86:553–565. doi: 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo AT, Malmstrom RD, White MA, Watowich SJ. 2010. Structural basis for substrate specificity of alphavirus nsP2 proteases. J Mol Graph Model 29:46–53. doi: 10.1016/j.jmgm.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spuul P, Balistreri G, Hellstrom K, Golubtsov AV, Jokitalo E, Ahola T. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J Virol 85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utt A, Quirin T, Saul S, Hellström K, Ahola T, Merits A. 2016. versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One 11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das PK, Merits A, Lulla A. 2014. Functional cross-talk between distant domains of chikungunya virus non-structural Protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. doi: 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lulla V, Merits A, Sarin P, Kaariainen L, Keranen S, Ahola T. 2006. Identification of mutations causing temperature-sensitive defects in Semliki Forest virus RNA synthesis. J Virol 80:3108–3111. doi: 10.1128/JVI.80.6.3108-3111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lulla V, Sawicki DL, Sawicki SG, Lulla A, Merits A, Ahola T. 2008. Molecular defects caused by temperature-sensitive mutations in Semliki Forest virus nsP1. J Virol 82:9236–9244. doi: 10.1128/JVI.00711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Kumar A, Mamidi P, Tiwari A, Kumar S, Mayavannan A, Mudulli S, Singh AK, Subudhi BB, Chattopadhyay S. 2018. Chikungunya virus nsP1 interacts directly with nsP2 and modulates its ATPase activity. Sci Rep 8:1045. doi: 10.1038/s41598-018-19295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YF, Sawicki SG, Sawicki DL. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J Virol 68:6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DY, Atasheva S, Frolova EI, Frolov I. 2013. Venezuelan equine encephalitis virus nsP2 protein regulates packaging of the viral genome into infectious virions. J Virol 87:4202–4213. doi: 10.1128/JVI.03142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lulla V, Kim DY, Frolova EI, Frolov I. 2013. The amino-terminal domain of alphavirus capsid protein is dispensable for viral particle assembly but regulates RNA encapsidation through cooperative functions of its subdomains. J Virol 87:12003–12019. doi: 10.1128/JVI.01960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayuri, Geders TW, Smith JL, Kuhn RJ. 2008. Role for conserved residues of Sindbis virus nonstructural protein 2 methyltransferase-like domain in regulation of minus-strand synthesis and development of cytopathic infection. J Virol 82:7284–7297. doi: 10.1128/JVI.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicki DL, Sawicki SG. 1993. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J Virol 67:3605–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M-L, Stollar V. 2007. Distinct sites on the Sindbis virus RNA-dependent RNA polymerase for binding to the promoters for the synthesis of genomic and subgenomic RNA. J Virol 81:4371–4373. doi: 10.1128/JVI.02672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubach JK, Wasik BR, Rupp JC, Kuhn RJ, Hardy RW, Smith JL. 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384:201–208. doi: 10.1016/j.virol.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. 2008. A new role for ns polyprotein cleavage in Sindbis virus replication. J Virol 82:6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhrymuk I, Kulemzin SV, Frolova EI. 2012. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol 86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorchakov R, Frolova E, Frolov I. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol 79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamm K, Merits A, Sarand I. 2008. Mutations in the nuclear localization signal of nsP2 influencing RNA synthesis, protein expression and cytotoxicity of Semliki Forest virus. J Gen Virol 89:676–686. doi: 10.1099/vir.0.83320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fros JJ, Liu WJ, Prow NA, Geertsema C, Ligtenberg M, Vanlandingham DL, Schnettler E, Vlak JM, Suhrbier A, Khromykh AA, Pijlman GP. 2010. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-Stimulated JAK-STAT signaling. J Virol 84:10877–10887. doi: 10.1128/JVI.00949-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heise MT, White LJ, Simpson DA, Leonard C, Bernard KA, Meeker RB, Johnston RE. 2003. An attenuating mutation in nsP1 of the Sindbis-group virus S.A.AR86 accelerates nonstructural protein processing and up-regulates viral 26S RNA synthesis. J Virol 77:1149–1156. doi: 10.1128/JVI.77.2.1149-1156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol 65:4107–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polo JM, Davis NL, Rice CM, Huang HV, Johnston RE. 1988. Molecular analysis of Sindbis virus pathogenesis in neonatal mice by using virus recombinants constructed in vitro. J Virol 62:2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorchakov R, Frolova E, Williams BRG, Rice CM, Frolov I. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol 78:8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Mutso M, Utt A, Lepland A, Herrero L, Taylor A, Bettadapura J, Rudd P, Merits A, Mahalingam S. Decreased virulence of Ross River virus harboring mutation in the first cleavage site of non-structural polyprotein is caused by a novel mechanism leading to increased production of interferon-inducing RNAs. mBio, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]