Abstract
Immune cell heterogeneity due to the differential expression of RNA splicing variants still remains unexplored. This is mainly because single-cell imaging technologies of splicing variants with precise sequence or base resolution are now not readily available. Herein, we design a splice-junction anchored padlock-probe-mediated rolling circle amplification assay (SpliceRCA) for single-cell imaging of splice isoforms of essential regulatory immune gene (CD45) upon T-cell activation. Two recognition regions in the padlock probe can target the splice-junction sequence, resulting in a close proximity for triggering in situ one-target-one-amplicon amplification. With the read length of ∼30 nucleotides, this method allows discrimination of isoforms with single-base precision and quantification of isoforms with single-molecule resolution. We applied SpliceRCA to single-cell image splice variants of essential regulatory immune gene (CD45) upon T-cell activation. It is found that CD45RO isoform presents a distal nuclear spatial distribution and is coregulated with CD45RB upon activation. Our strategy provides a single-cell analysis platform to investigate the mechanism of complex immune responses and may further guide immunotherapy.
Short abstract
We developed a method for multiplexed profiling of RNA splicing variants in single cells and investigated the role of RNA splicing in eliciting immune responses.
Introduction
Alternative splicing is a fundamental regulatory process of gene expression that allows generation of multiple mRNA isoforms from single genes,1,2 thereby increasing transcriptome complexity.3,4 Splicing variability among individual cells accounts for a large part of gene expression heterogeneity, which plays a significant role in the immune system for the efficient battling rich variability of pathogens.5,6 Isoforms with diversified functions were evolved in the generation of the immune response.7 CD45, the prototypic receptor-like protein tyrosine phosphatase gene, acts as an essential regulator of signal transduction pathways in immune cells.8 The transition from naïve to activated T cells is marked by CD45 pre-mRNA alternative splicing.9 Abnormal CD45 splice variant expressions are associated with autoimmune and infectious diseases.10,11 However, the roles of differential CD45 splicing variants, such as their relative expression level, intracellular localization, expression heterogeneity, and regulation in eliciting immune responses, are underexplored. Thus, visualizing12 intracellular mRNA variants in single cells is essential for understanding differential splicing-mediated immune cell heterogeneity and resolving the complexity and heterogeneity of RNA variant-related immune diseases.
Currently, only a limited number of methods have been explored to detect RNA splicing variants at the single-cell level. Single-molecule fluorescence in situ hybridization (smFISH) is a facile and powerful method to in situ image RNA splice variants in single cells, and brings a significant advance of RNA splicing in single-cell study.13,14 However, the FISH method can only detect RNA sequences with a minimum length of 600 nucleotides (nt) (usually need >30 different hybridization probes with ∼20 nt to ensure that the labeling signals are distinguished from the background).15,16 However, the human exon is with the average length of only 320 nt; thus, large amounts of exons can hardly be detected by smFISH.17,18 Recently, a newly proposed plasmonic in situ hybridization (plasmonic ISH) based on gold nanoparticle (AuNP) labeling is masterly adapted to analyze RNA splicing and is able to differentiate splice variants with ∼300 nt sequence.19 However, limitations on AuNPs remain currently unsolvable, such as their large size for impeding the delivery efficiency, alteration in a real cell state, and easy aggregation, etc.20,21 The visualization of short exon mRNA variants still remains a challenge. Meanwhile, a large amount of short exon mRNA variants are involved in crucial biological processes,22 like CD45 isoforms, of which the average length of the alternative exon is less than 200 nt.23 There is still an urgent need for developing a versatile RNA imaging method with high sequence resolution capable of in situ analyzing splice isoforms with different lengths.
To address these issues, we develop a high base-resolution strategy termed as splice-junction anchored padlockprobe-mediated rolling circle amplification24 (SpliceRCA), enabling single-cell imaging of CD45 splicing variants. Rolling circle amplification (RCA) can achieve localized isothermal amplification, converting the target sequence into a long single-stranded DNA or RNA product with thousands of tandem repeats.25 Attributed to its one-target-one-amplicon amplification process, RCA can achieve target RNA/DNA detection or imaging at the single-molecule level.26 Furthermore, the padlockprobe-based RCA method has the capability to target short RNAs and discriminate highly similar sequences to genotype RNAs with single-nucleotide variations,27 which prompted us to explore the potential of RCA in RNA splicing variant detection. In this study, two recognition regions in the padlock probe are specifically hybridized to a newly formed splice-junction sequence, resulting in a close proximity for triggering in situ one-target-one-amplicon amplification,28 achieving shortening the read length of the imaging method to ∼30 nt. This method allows discrimination of isoforms with single-base precision and quantification of isoforms with single-molecule29 resolution. With this method, we measure the isoform expression and spatial distribution of three critical variants of CD45, and analyze how the isoforms covariation between individual cells changes in T-cell activation. The method provides a single-cell-level RNA splicing analysis platform to explore the differential expression of splicevariant-mediated immune cell heterogeneity in a quantitative manner.
Results and Discussion
Overview of SpliceRCA
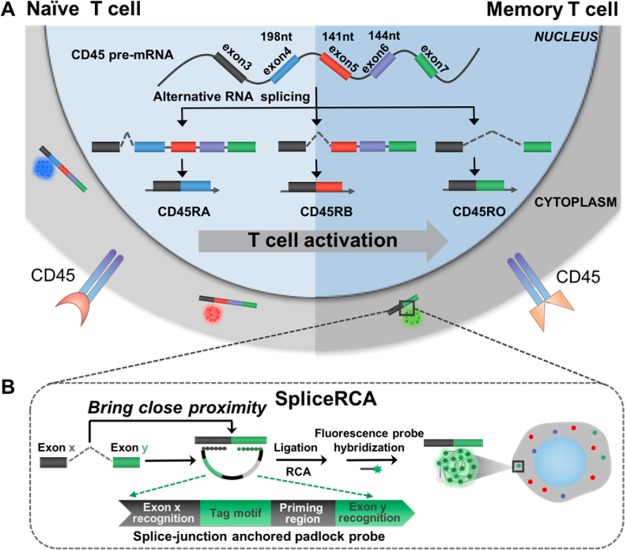
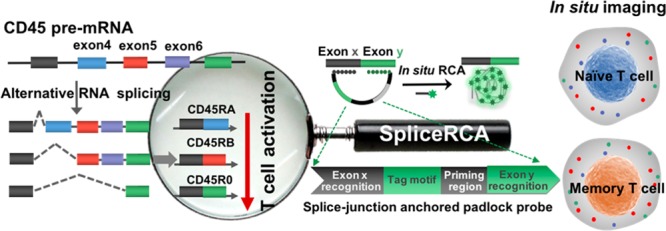
Scheme 1 illustrates the procedures of the direct visualization of RNA splice variants in single cells by SpliceRCA. Alternative splicing of exon 4 (198 nt), 5 (141 nt), and 6 (144 nt) in CD45 pre-mRNA is strictly regulated in T-cell activation: naïve T cells express various larger isoforms whereas memory T cells tend to produce small isoforms.30 We chose three critical alternative splicing variants of the CD45 gene involved in the T-cell activation process which include the largest isoform CD45RA (all exons retained), middle isoform CD45RB (exon skipping, exon 4 deleted), and smallest isoform CD45RO (exon skipping, exon 4, 5, and 6 deleted) (Scheme 1A). SpliceRCA is performed with a splice-junction anchored padlock probe composed of four modules: the recognition of exon junction sites (Rx, Ry), universal priming region (P), and tag motif (T) modules (Scheme 1B). The splice junction in the target splice isoform brings close proximity between Rx and Ry in the padlock probe for circularizing, following triggering in situ RCA upon the hybridization of primer with the P. By tuning the sequence of T corresponding to different fluorophores, three splice isoforms can be simultaneously differentiated, and visualized with single-molecule resolution attributed to the in situ one-target-one-amplicon amplification method. Benefiting from the recognition of the target splice-junction sequence, the read length of the imaging method can be shortened to ∼30 nt. Moreover, the ligation reaction confers high-specificity for discrimination between perfectly matched and mismatched oligonucleotides, which can realize the imaging of RNA isoforms with even single-base resolution.31
Scheme 1. Schematic Diagram of Multiplex Detection of mRNA Variants in Single Cells by SpliceRCA.
(A) Alternative splicing patterns of CD45 during T-cell activation. Isoforms (CD45RA, CD45RB, CD45R0) with decreasing exon inclusion were expressed upon T-cell activation. (B) The procedures of SpliceRCA for detecting splice variants in single cells. The splice-junction anchored padlock probe is composed of four modules: the recognition of exon junction sites (Rx, Ry), universal priming region (P), and tag motif (T) modules. The newly formed splice junction in the target splice isoform brings close proximity between Rx and Ry in the padlock probe for circularizing, following primer hybridized with the P, triggering in situ RCA. Upon tuning of the sequence of T corresponding to different fluorophores, the three RNA splicing variants can thus be simultaneously differentiated, and visualized with single-molecule resolution attributed to the in situ one-target-one-amplicon amplification method.
Multiplex in Situ Imaging of CD45 mRNA Splicing Variants in Single Cells by SpliceRCA
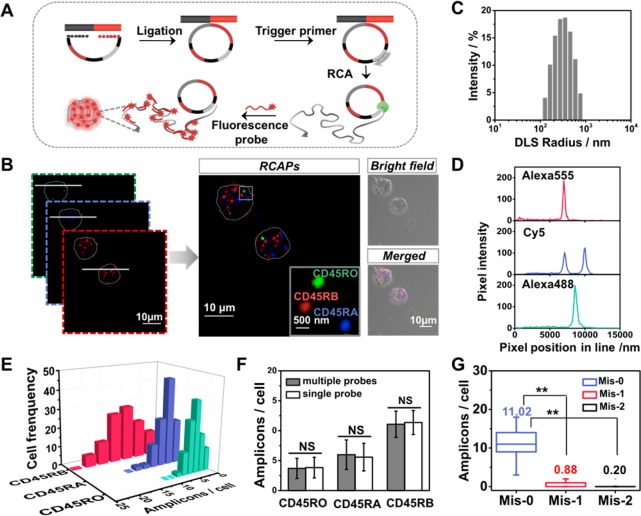
The specific scheme of in situ RCA is shown in Figure 1A. The splice-junction sequence was specifically recognized by the padlock probes and as the template for ligating the padlock probe. Then, RCA is initiated with the help of an additional DNA primer, resulting in a long DNA amplicon with hundreds of copies of the padlock probe.32,33 The RCA amplicon forms as a nanoclew that can be visible as a diffraction-limited fluorescent spot upon hybridization with different detection probes. To demonstrate the feasibility of this principle for detection of RNA splice isoforms, we first performed the assay in vitro to amplify the target synthetic sequence of CD45RO, CD45RA, and CD45RB (Table S1, Supporting Information, SI). This method can effectively discriminate isoforms and exhibit a high-sensitivity performance for in vitro detection (Figures S1 and S2).
Figure 1.
Simultaneous imaging of CD45 splicing variants in single Jurkat T cells. (A) Schematic diagram of SpliceRCA for detection of CD45 RNA isoforms using in situ RCA. (B) Fluorescent image of CD45RO, CD45RA, and CD45RB isoforms visualized by SpliceRCA in Jurkat T cells. Inset: the fluorescent image of the boxed subregion with colored labels indicating potential mRNA isoforms. The outline of the Jurkat T cell is marked with a gray dot line. Scale bars: 10 μm (overview images) and 500 nm (insets). (C) Dynamic light scattering (DLS) analysis for RCA amplicons. (D) Intensity plots of the lines in panel B show that mRNA spot signals are distinguished from background. (E) Frequency histogram of amplicons of three splicing isoforms in single cells (cell number >100). (F) Comparison of SpliceRCA results in simultaneous detection with separate detection (n > 100; NS, not significant). (G) Quantification of the average numbers of CD45RB amplicons detected in Jurkat cells by using Mis-0, Mis-1, and Mis-2 splice-junction anchored padlock probes (n > 100; **P < 0.001).
Next, we explored the potential of the SpliceRCA method for imaging CD45 RNA splicing variants in Jurkat T cells. As illustrated in Figure 1B, the generated superbright dots amplified from the target splice isoform could be clearly distinguished from the background inside cell. These bright dot signals were generated from one-target-one-amplicon in situ amplification, thus presumably corresponding to separated mRNAs. Characterization of RCA amplicons by dynamic light scattering (DLS) and transmitted electron microscopy (TEM) demonstrated that the RCA amplicons are monodisperse particles with a size of ∼300 nm (Figure 1C and Figure S3). The size of the RCA amplicon is larger than the resolution of optical imaging, thus conferring the single-molecular RCA amplicons resolvable by confocal microscopy.16,34 The signal intensities of the RCA amplicon amplified from individual splice isoforms were much larger than the background (Figure 1D). Next, for verification that signals were amplified from the target, several experiments were performed as control. As shown in Figure S4, no bright spot was observed when no padlock probe or trigger primer was used. There was only a rare fluorescence signal when random probes were added. Additionally, to confirm that the signal resulted from the target sequence, we blocked the binding sites with an unlabeled complementary probe before carrying out SpliceRCA. Only less than 0.2 amplicons per cell could be seen, thus suggesting that the bright spots came from the amplification of the target RNA sequence. Furthermore, we also performed a siRNA knockdown experiment to suppress the expression of CD45; the signals of CD45RO, CD45RA, and CD45RB isoforms decreased 24.59%, 31.27%, and 27.01%, respectively, with the knockdown of CD45 (Table S3). The knockdown efficiency was verified by RT-qPCR assay (Figure S5 and Table S4). All of these results suggest that the bright dot signals come from target mRNA splicing isoforms.
As shown in Figure 1E, CD45 isoforms were presented at different levels. The average numbers of amplicons for CD45RO, CD45RA, and CD45RB were 3.70, 5.98, and 11.07 per cell. The in situ detection efficiency of mRNA splice isoform-initiated RCA was estimated to be 10–20% on the basis of a comparison to RT-qPCR data (Table S5 and discussed in the Supporting Information). The obvious variability in the copy numbers of CD45 isoforms indicates that significant cell-to-cell variation in isoform expression would be exhibited even in the same batch cells. In addition, SpliceRCA has been successfully applied to image splicing isoforms of BRCA1, breast cancer susceptibility gene 1.19,35 The alternative splicing of BRCA1 was closely related with the transformation of malignant breast cancer (Figure S6).36,37 As a validation, we performed the reverse transcription quantitative PCR (RT-qPCR) assay for expression comparison. The results of SpliceRCA are in good accordance with the RT-qPCR results in general (Figures S7 and S8). The single-molecule resolution of the SpliceRCA method allows precise quantification of isoform abundances.
Moreover, the assurance of the discrimination among the various splicing variants, especially which have sharing sequences in splice junction, highly depends on the stringent specificity of the RNA profiling method. To assess the accuracy of SpliceRCA for profiling RNA splicing variants inside cells, we compared the isoform expression level measured by SpliceRCA in simultaneous and separate detection. The quantification of the three isoforms by simultaneous and separate detection shows negligible difference (Figure 1F and Figure S9, P < 0.001), indicating minimal crosstalk among the targets simultaneously detected. Thus, SpliceRCA confers low crosstalk for multiplex in situ mRNA isoforms detection. Further, we test the base precision of the SpliceRCA method, in which the splice-junction padlock probe used for imaging CD45RB was altered by one or two bases. The copy number of amplicons per cell decreased sharply after the introduction of a one- or two-base mismatch (Figure S10). The average numbers of amplicons when using one-base mismatched (Mis-1) and two-base mismatched (Mis-2) padlock probes were 0.88 per cell and 0.20 per cell, respectively, much less than the 11.02 per cell measured with the matched probe (Mis-0) (Figure 1G). Thus, the SpliceRCA presents high specificity conferring single-nucleotide resolution, ensuring the precise recognition of mRNA isoforms in situ.
Quantification of Isoform Variability in Jurkat Cells upon T-Cell Activation
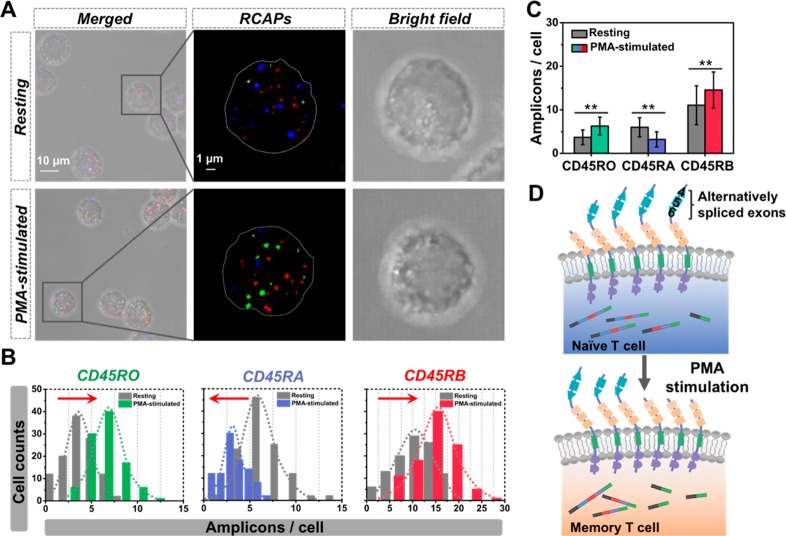
Determining the alternative splicing changes induced by T-cell activation is crucial for understanding the cellular outcomes of the antigen challenge. To characterize the extent of expression variability on CD45 mRNA isoforms and decipher its functional implications, we applied SpliceRCA to profile the isoform expression in the Jurkat T-cell response to phorbol-12-myristate13-acetate (PMA) stimulation. Immunofluorescence imaging of HnRNPLL, one marker of T-cell maturation,38,39 shows a skewed increase in expression level after PMA stimulation (Figure S11), indicating that Jurkat T cells were efficiently stimulated. Fluorescent images reveal a different isoform pattern after stimulation, and the copy numbers of three mRNA isoforms all present a shift upon T-cell activation (Figure 2A,B). Moreover, the expressions of CD45RO and CD45RB isoforms show 70% and 32% increase (Figure 2C). Then, RT-qPCR analysis further confirms the isoform expression patterns and the splicing shift in PMA stimulation (Figure S12). These results indicate that T-cell activation leads to the skipping of three variable exons in CD45 pre-mRNA (Figure 2D). The skipping process would reduce the phosphatase activity of the CD45 molecule and preserve the normal status to prevent autoimmune disease.30,40 Interestingly, cell-to-cell variability in isoform ratios differed between the two cell states before and after PMA stimulation. After stimulus, the three isoforms showed subtle lower cell-to-cell variability [coefficients of variation (CV, SD/mean) of CD45RO, CD45RA, and CD45RB were 0.328, 0.361, and 0.283, respectively] than resting T cells (isoform CV > 0.4) (Table S6). The variability in single-cell isoform expression may reflect functional differences in the stimulated Jurkat T cell population.
Figure 2.
Cell-to-cell variation in isoform expression upon PMA stimulation. (A) Fluorescent images of three CD45 splicing isoforms visualized by SpliceRCA before and after PMA stimulation. (B) Histograms of the copy numbers of three mRNA isoforms in Jurkat T cells measured by SpliceRCA before and after stimulus. The arrow shows the expression shift; arrow to the right (left) means increased (decreased) expression upon PMA stimulation. (C) Quantification of average expression for three CD45 isoforms before and after stimulus. (D) Schematic diagram of the alternative splicing upon T-cell activation.
Mapping the Spatial Distribution of CD45 mRNA Variants in Single Cells
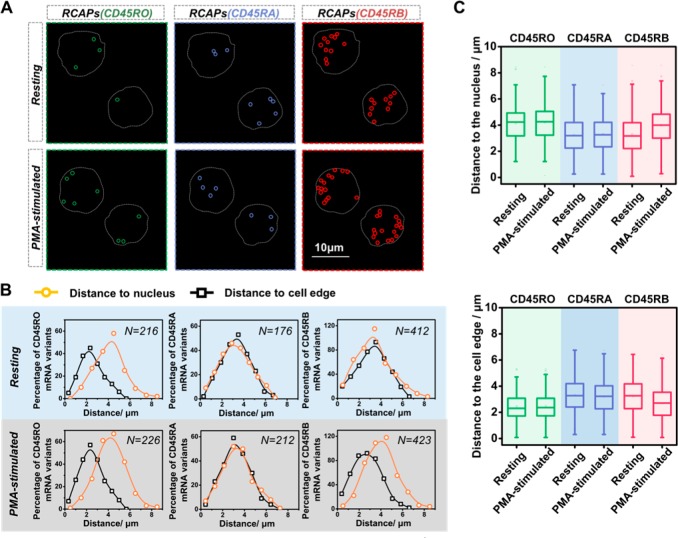
The location distribution of splicing variants from the same gene has not been well-characterized in mammalian cells, let alone suspension cells such as T cells. We then exploit the in situ RNA visualization ability of SpliceRCA to investigate the spatial distributions of multiple splice variants simultaneously. From the visual inspection of each mRNA variant in resting Jurkat T cells, isoform CD45RO appeared enriched near the cell periphery, whereas CD45RA and CD45RB appeared in a random distribution throughout the cell (Figure 3A). Quantitative analysis of the distances between each mRNA splicing variant and the cell nucleus or the cell periphery further confirmed the visual impression (Figure 3B). The average distances to the nucleus for the mRNA variants CD45RO, CD45RA, and CD45RB were 3.25, 3.32, and 4.15 μm, respectively. The average distances to cell edge for mRNA variants CD45RA, CD45RB, and CD45RO were 2.30, 3.24, and 2.47 μm, respectively. Furthermore, the spatial distribution of splice isoforms after T-cell activation was investigated. As shown in Figure 3C, the average distances to the nucleus for CD45RO, CD45RA, and CD45RB increased by 0.07, 0.08, and 0.67 μm, respectively, after T-cell activation. The average distances to the cell edge for CD45RO, CD45RA, and CD45RB decreased by 0.02, 0.08, and 0.54 μm, respectively, after T-cell activation. The results indicate that CD45RO and CD45RA showed a modest change in the distance to the nucleus and cell edge upon T-cell activation, while the CD45RB presented an obvious change to be more likely expressed in the cell periphery after T-cell activation. The different spatial patterns may result from the expression of some RNA binding proteins as splice mRNA localization is closely related with regulation of many interaction proteins.41,42 The ability for spatial mapping of splice isoforms in single cells may offer us new ways to predict and study the rich diversity in splicing functions.
Figure 3.
Mapping the spatial distribution of CD45 isoforms in the Jurkat T cells. (A) Example images of the spatial distributions observed for CD45 isoforms before and after PMA stimulation. The RCA amplicons of CD45RO, CD45RA, and CD45RB are marked with green, blue, and red circles, respectively, and the outlines of cell are marked by a gray dotted line. Scale bars: 10 μm. (B) Single-cell profiles for distance to cell nucleus (yellow dots) and the edge (black dots). N is the number of cell dots per plot. (C) Quantification of average distances for three CD45 isoforms to the cell nucleus or the edge before and after PMA stimulation.
Analysis of Expression Covariation among Different Isoforms
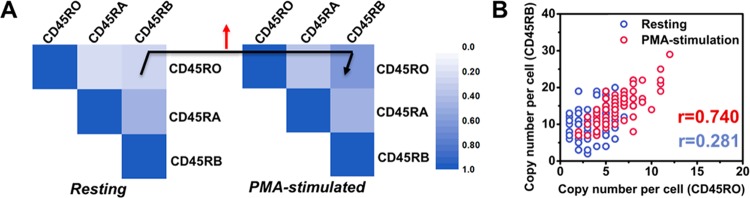
We next examined the expression covariation among different isoforms that might arise from differential activity of immune activation. Pairwise correlation coefficients for the RNA species were calculated as shown in Figure 4A. Upon T-cell activation, the correlation coefficients of CD45RO and CD45RB presented a pronounced increase whereas pair correlations of CD45RO and CD45RA, and CD45RA and CD45RB, presented a modest change. The single-cell correlation coefficients of CD45RO and CD45RB vary from 0.281 to 0.740 upon stimulus (Figure 4B). Covariation across single cells between splicing isoforms of a gene upon T-cell activation would represent a potential regulatory interaction between alternative splicing and T-cell activation. Isoforms CD45RO and CD45RB encode the transcript mRNA which both skip exon 4. T-cell activation would lead to the up-regulation of some regulators, such as HnRNP LL, SR proteins, or other RNA binding protein which can alter alternative splice sites by binding to different exons.43 We suggest that the expression of some RNA binding proteins involved in exon 4 skipping was up-regulated with T-cell activation, which thus may lead to the simultaneous up-regulation of CD45RO and CD45RB, resulting in the increase of their correlation coefficients.44 The normal functioning of the immune system may be associated with the complex interplay of regulators that mediate the appropriate splicing of CD45 exon 4. Analysis of covariations in the expression levels of different isoforms on the single-cell level could reveal coregulated isoforms and help to elucidate splicing regulatory circuits in the T-cell maturation.
Figure 4.
Cell-to-cell pairwise correlations for the RNA species upon T-cell activation. (A) Matrix of the pairwise correlation coefficients of the cell-to-cell variation in expression for CD45 splice isoforms before and after PMA stimulation. (B) CD45RO–CD45RB expression correlation analysis in a single cell before (blue) and after (red) PMA stimulation. (P value <0.001). The Pearson correlation coefficient changes from 0.281 to 0.740 upon PMA stimulation (P value <0.001).
Conclusions
In summary, we develop a facile and high base-resolution strategy for multiplexed profiling of RNA splicing variants in single cells. This imaging method, termed SpliceRCA, has several advantages: (1) Benefitting from recognition of the target splice-junction sequence, the read length of the imaging method can be shortened to ∼30 nt, which resolved the limitations in read length of current imaging technologies. (2) The ligation process and one-target-one-amplicon amplification involved in SpliceRCA confer high specificity with single-base precision and quantification of splice isoforms with single-molecule resolution. (3) Insight into RNA splicing variants on expression level, spatial distribution, and cell heterogeneity was simultaneously acquired by SplicRCA. The method was used to image splice variants of the essential regulatory immune gene (CD45) upon T-cell activation. We have identified that CD45RO shows a distal nuclear spatial distribution, which is different from other isoforms. After T-cell activation, CD45RB presented an obvious change in spatial distribution, which appeared enriched near the cell periphery. Furthermore, it is found that CD45RO and CD45RB coregulate upon PMA stimulation, which has not been observed by previous cell-population-based RNA quantification methods. Our study demonstrates the promise of single-cell alternative splicing analysis in deciphering rich diversity in functional communities and understanding the essential features of immune responses for assisting clinical monitoring.
Materials and Methods
Oligonucleotide Sequences
The DNA sequences (Tables S1 and S2 in the Supporting Information) were purchased from Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). The sequences modified with Alexa488, Alexa555, and Cy5 were purchased from Thermo Fisher Scientific (Beijing, China), and were purified by HPLC. RNA sequences were created by in vitro T7 transcription reactions with ordering single-stranded DNA used as templates.
Cell Culture and Stimulations
The Jurkat cells were maintained in a standard RPMI 1640 medium supplemented with 15% fetal bovine serum, 100 units mL–1 of penicillin and 100 μg mL–1 of streptomycin. The cells were grown at 37 °C, 5% CO2, and 95% air humidity. For stimulations, cells were diluted to 3 × 106 cells mL–1 and incubated in medium with PMA plus ionomycin (100 ng mL–1 and 1 μM, respectively).
Cell Fixation and Permeabilization
Cells were fixed at a proper density on a 22 mm × 22 mm gelatin-coated cover glass (VWR, Radnor, PA) enclosed by a PDMS with a chamber (5 mm in diameter) by being maintained in 4% (w/v) paraformaldehyde in phosphate buffered saline (PBS) for 15 min at room temperature (20–25 °C), washed twice with 1× DEPC-treated PBS (DEPC-PBS). Then, the cells were permeabilized for 5 min with 0.5% v/v Triton-X100 in 1× PBS at room temperature, and washed twice with DEPC-PBS.
In Situ Visualization of Splicing Variants by SpliceRCA
The hybridization of the exon-junction padlock probe with the target mRNA splicing variants was conducted in a volume of 20 μL containing 2 μL 20× saline-sodium citrate buffer (SSC) (Ambion, AM9763), 2 μL of each phosphorylated padlock probe (10 μM), 1 μL of DTT (100 mM), 2 μL of yeast tRNA (10 mg mL–1), and 0.5 μL of RiboLock RNase inhibitor (40 U μL–1) overnight at 37 °C. The sample was then washed twice using PBS-T (DEPC-PBS with 0.05% Tween-20) for 3 min at room temperature. The ligation reaction was carried out in 10 μL of circularization reaction mixture [1 μL 10× ligase reaction buffer, 1 μL of 20× SSC, 1 μL of T4 DNA ligase (5 U μL–1), 0.25 μL of RiboLock RNase inhibitor (40 U μL–1), 6.75 μL of RNase-free water] at 37 °C for 2 h. After ligation, a 20 μL mixture containing 2 μL of 20× SSC, 1 μL of each primer (4 μM), 2 μL of formamide, 1 μL of DTT (100 mM), 13.5 μL of RNase-free water, and 0.5 μL of RiboLock RNase inhibitor (40 U μL–1) were added to the sample and incubated for 60 min at 37 °C, following a wash using PBS-T. RCA was then conducted with a 10 μL mixture containing 1 μL of 10× phi29 DNA polymerase reaction buffer, 0.5 μL of phi29 DNA polymerase, 3 μL of dNTPs (10 mM for each of dATP, dGTP, dCTP, and dTTP), 5.25 μL of RNase-free water, and 0.25 μL of RiboLock RNase inhibitor (40 U μL–1) for 120 min at 37 °C. The incubation was followed by a wash in PBS-T. Then, the hybridization of amplicons with detection probes was conducted in a 20 μL mixture of 100 nM fluorophore-labeled detection probes, 2× SSC, 15% formamide, and 10 ng μL–1 salmon sperm DNA for 30 min at 37 °C, following two washes using PBS-T. After mounting with Fluoromount-G [containing 4′,6-diamidino-2-phenylindole (DAPI) to counterstain the cell nuclei], the slides were ready for imaging.45,46
Image Acquisition and Analysis
Fluorescence imaging was performed using a Zeiss LSM 710 META confocal microscope (Zeiss). The cellular images were acquired with a 63× (oil) objective. Argon/2 (488 nm) was used as excitation source for Alexa488-labeled probe, and a 500–535 nm bandpass filter was used for fluorescence detection. The Alexa555 dye was excited with HeNe1 (561 nm) laser and detected with a 580–620 nm bandpass filter. The Cy5 dye was excited with HeNe2 (633 nm) laser and detected with a 650–750 nm bandpass filter. The DAPI dye was excited with a 405 nm diode laser and detected with a 430–550 nm bandpass filter. Z-stacks were collected at step size of 0.4 μm for 20 slices to image the entire cell. The images were processed by ImageJ version 1.46r software. The outline of cells was determined by the bright field images. The amplicons which had bright fluorescent signals were distinguished from background by setting the threshold value.16,47 For determination of the copy number of amplicons per cell, the number of bright pixels was counted by particle analysis in ImageJ software.48
Real-Time Quantitative PCR (RT-qPCR) Analysis of RNA Splice Variants inside Cells
Total RNA from Jurkat cells was harvested by using TransZol according to the included protocol. The reverse transcription was performed using TransScript one-step gDNA removal and cDNA synthesis kit. In brief, a total volume of 20 μL of mixture containing 10 μL of 2× TS reaction mixture, 2 μL of the total RNA (from 50 ng to 5 μg), 1 μL of RT primer (0.5 μg μL–1), 1 μL of TransScript RT/RI enzyme mix, 1 μL of gDNA remover, and 5 μL of RNase-free water was incubated at 42 °C for 15 min, following heat inactivation of reverse transcriptase at 85 °C for 5 s. The produced cDNA samples were then stored at −80 °C for future use. Upstream primers used for the reverse transcription reaction are listed in Table S1.
For qPCR analysis, following the manufacturer’s instructions on a Bio-Rad C1000TM (Bio-Rad) instrument, the 20 μL reaction solution contained 10 μL of 2× SYBR Select master mix, 2 μL of cDNA sample, 2 μL of forward primer (5 μM), 2 μL of reverse primer (5 μM), and 4 μL of RNase-free water. The qPCR was done by staying at 50 °C for 2 min for the hot start, annealing at 95 °C for 2 min, then followed by 40 cycles of 15 s at 95 °C, 60 s at 60 °C, and 5 min at 60 °C. Ct values were converted into absolute GAPDH copy numbers using a standard curve from a control RNA (human GAPDH mRNA in RevertAid First Strand cDNA synthesis kit). The experiments were performed in triplicate. The copy numbers of target mRNA splicing variants CD45RO, CD45RA, and CD45RB were evaluated by referring to the expression of GAPDH mRNA using the 2−ΔΔCt method. Calculations of mRNA copy numbers were based on the number of counted cells at harvest. No unexpected or unusually high safety hazards were encountered in the experiments.
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (21621003, 21235004, 21327806), and Tsinghua University Initiative Scientific Research Program.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00081.
Additional experimental details including fluorescence analysis of RCA products; TEM image; additional confocal microscopy analysis; siRNA knockdown data; discussion of detection efficiency; immunofluorescence analysis; and RT-qPCR data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Maniatis T.; Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002, 418 (6894), 236–243. 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Hemphill J.; Liu Q.; Uprety R.; Samanta S.; Tsang M.; Juliano R. L.; Deiters A. Conditional control of alternative splicing through light-triggered splice-switching oligonucleotides. J. Am. Chem. Soc. 2015, 137 (10), 3656–3662. 10.1021/jacs.5b00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Manley J. L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10 (11), 741–754. 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M. J.; Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods 2013, 10 (3), 246–248. 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K.; Cooper T. A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012, 18 (8), 472–482. 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay P. K.; Gierahn T. M.; Roederer M.; Love J. C. Single-cell technologies for monitoring immune systems. Nat. Immunol. 2014, 15 (2), 128–135. 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A. K.; Satija R.; Shuga J.; Trombetta J. J.; Gennert D.; Lu D.; Chen P.; Gertner R. S.; Gaublomme J. T.; Yosef N.; Schwartz S.; Fowler B.; Weaver S.; Wang J.; Wang X.; Ding R.; Raychowdhury R.; Friedman N.; Hacohen N.; Park H.; May A. P.; Regev A. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 2014, 510 (7505), 363–369. 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston M. L.; Xu Z.; Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 2003, 21, 107–137. 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- Machura E.; Mazur B.; Pieniazek W.; Karczewska K. Expression of naive/memory (CD45RA/CD45RO) markers by peripheral blood CD4+ and CD8 + T cells in children with asthma. Arch. Immunol. Ther. Exp. 2008, 56 (1), 55–62. 10.1007/s00005-008-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchilian E. Z.; Beverley P. C. Altered CD45 expression and disease. Trends Immunol. 2006, 27 (3), 146–153. 10.1016/j.it.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Yamane H.; Paul W. E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489. 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.; Wu C. S.; Jimenez E.; Zhu Z.; Dajac J. G.; You M.; Han D.; Zhang X.; Tan W. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew. Chem., Int. Ed. 2013, 52 (7), 2012–2016. 10.1002/anie.201209440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas D. Y.; Shah K.; Batish M.; Levandoski M.; Sinha S.; Marras S. A.; Schedl P.; Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 2011, 147 (5), 1054–1065. 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waks Z.; Klein A. M.; Silver P. A. Cell-to-cell variability of alternative RNA splicing. Mol. Syst. Biol. 2011, 7, 506. 10.1038/msb.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E.; Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 2012, 9 (7), 743–748. 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. H.; Boettiger A. N.; Moffitt J. R.; Wang S.; Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348 (6233), aaa6090. 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y.; Liu J.; Irudayaraj J. Beyond quantification: in situ analysis of transcriptome and pre-mRNA alternative splicing at the nanoscale. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9 (4), e1443. 10.1002/wnan.1443. [DOI] [PubMed] [Google Scholar]
- Long X.; Colonell J.; Wong A. M.; Singer R. H.; Lionnet T. Quantitative mRNA imaging throughout the entire Drosophila brain. Nat. Methods 2017, 14 (7), 703–706. 10.1038/nmeth.4309. [DOI] [PubMed] [Google Scholar]
- Lee K.; Cui Y.; Lee L. P.; Irudayaraj J. Quantitative imaging of single mRNA splice variants in living cells. Nat. Nanotechnol. 2014, 9 (6), 474–480. 10.1038/nnano.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. L.; Yin Y.; Kunchakarra S.; Mukherjee B.; Gerion D.; Jett S. D.; Bear D. G.; Gray J. W.; Alivisatos A. P.; Lee L. P.; Chen F. F. A nanoplasmonic molecular ruler for measuring nuclease activity and DNA footprinting. Nat. Nanotechnol. 2006, 1 (1), 47–52. 10.1038/nnano.2006.51. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Wang Y.; Wang H.; Jiang J. H.; Shen G. L.; Yu R. Q.; Li J. Electrochemical DNA biosensor based on the proximity-dependent surface hybridization assay. Anal. Chem. 2009, 81 (5), 1982–1987. 10.1021/ac802512d. [DOI] [PubMed] [Google Scholar]
- Ankenbruck N.; Courtney T.; Naro Y.; Deiters A. Optochemical Control of Biological Processes in Cells and Animals. Angew. Chem., Int. Ed. 2018, 57 (11), 2768–2798. 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K. W.; Weiss A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol. Cell. Biol. 2000, 20 (1), 70–80. 10.1128/MCB.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M. M.; Li F.; Zhang Z.; Zhang K.; Kang D. K.; Ankrum J. A.; Le X. C.; Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43 (10), 3324–3341. 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- Nilsson M.; Malmgren H.; Samiotaki M.; Kwiatkowski M.; Chowdhary B. P.; Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 1994, 265 (5181), 2085–2088. 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- Larsson C.; Grundberg I.; Soderberg O.; Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat. Methods 2010, 7 (5), 395–397. 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- Lizardi P. M.; Huang X.; Zhu Z.; Bray-Ward P.; Thomas D. C.; Ward D. C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998, 19 (3), 225–232. 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Ren X.; Deng R.; Wang L.; Zhang K.; Li J. RNA splicing process analysis for identifying antisense oligonucleotide inhibitors with padlock probe-based isothermal amplification. Chem. Sci. 2017, 8 (8), 5692–5698. 10.1039/C7SC01336A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Gao Y.; Xu Y.; Chao J.; Liu H.; Wang L.; Li D.; Fan C. Real-Time Imaging of Single-Molecule Enzyme Cascade Using a DNA Origami Raft. J. Am. Chem. Soc. 2017, 139 (48), 17525–17532. 10.1021/jacs.7b09323. [DOI] [PubMed] [Google Scholar]
- Zikherman J.; Weiss A. Alternative splicing of CD45: the tip of the iceberg. Immunity 2008, 29 (6), 839–841. 10.1016/j.immuni.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Deng R.; Zhang K.; Li J. Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell.. Acc. Chem. Res. 2017, 50, 1059–1068. 10.1021/acs.accounts.7b00040. [DOI] [PubMed] [Google Scholar]
- Hu R.; Zhang X.; Zhao Z.; Zhu G.; Chen T.; Fu T.; Tan W. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem., Int. Ed. 2014, 53 (23), 5821–5826. 10.1002/anie.201400323. [DOI] [PubMed] [Google Scholar]
- Deng R.; Zhang K.; Wang L.; Ren X.; Sun Y.; Li J.. DNA-Sequence-Encoded Rolling Circle Amplicon for Single-Cell RNA Imaging. Chem., in press, 2018; 10.1016/j.chempr.2018.03.003 [DOI] [Google Scholar]
- Ke R.; Mignardi M.; Pacureanu A.; Svedlund J.; Botling J.; Wahlby C.; Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 2013, 10 (9), 857–860. 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- Budhram-Mahadeo V.; Ndisang D.; Ward T.; Weber B. L.; Latchman D. S. The Brn-3b POU family transcription factor represses expression of the BRCA-1 anti-oncogene in breast cancer cells. Oncogene 1999, 18 (48), 6684–6691. 10.1038/sj.onc.1203072. [DOI] [PubMed] [Google Scholar]
- Orban T. I.; Olah E. Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem. Biophys. Res. Commun. 2001, 280 (1), 32–38. 10.1006/bbrc.2000.4068. [DOI] [PubMed] [Google Scholar]
- Wang H.; Wang H.; Duan X.; Sun Y.; Wang X.; Li Z. Highly Sensitive and Multiplexed Quantification of mRNA Splice Variants by Direct Ligation of DNA Probes at Exon Junction and Universal PCR Amplification. Chem. Sci. 2017, 8, 3635–3640. 10.1039/C7SC00094D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S.; Moita L. F.; Neems D.; Freitas R. P.; Hacohen N.; Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 2008, 321 (5889), 686–691. 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; Jia X.; de la Cruz L.; Su X. C.; Marzolf B.; Troisch P.; Zak D.; Hamilton A.; Whittle B.; Yu D.; Sheahan D.; Bertram E.; Aderem A.; Otting G.; Goodnow C. C.; Hoyne G. F. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity 2008, 29 (6), 863–875. 10.1016/j.immuni.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. M.; Pan Q.; Cole B. S.; Yarosh C. A.; Babcock G. A.; Heyd F.; Zhu W.; Ajith S.; Blencowe B. J.; Lynch K. W. Alternative splicing networks regulated by signaling in human T cells. RNA 2012, 18 (5), 1029–1040. 10.1261/rna.032243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrute-Nayak M.; Bullock S. L. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat. Cell Biol. 2012, 14 (4), 416–423. 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum A. R.; Haimovich G.; Singer R. H. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015, 16 (2), 95–109. 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preussner M.; Schreiner S.; Hung L. H.; Porstner M.; Jack H. M.; Benes V.; Ratsch G.; Bindereif A. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res. 2012, 40 (12), 5666–5678. 10.1093/nar/gks221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp J. D.; Jackson J.; Melton A. A.; Lynch K. W. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 2008, 14 (10), 2038–2049. 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C.; Koch J.; Nygren A.; Janssen G.; Raap A. K.; Landegren U.; Nilsson M. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat. Methods 2004, 1 (3), 227–232. 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- Lubeck E.; Coskun A. F.; Zhiyentayev T.; Ahmad M.; Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 2014, 11 (4), 360–361. 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R.; Zhang K.; Sun Y.; Ren X.; Li J. Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification. Chem. Sci. 2017, 8, 3668–3675. 10.1039/C7SC00292K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battich N.; Stoeger T.; Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods 2013, 10 (11), 1127–1133. 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.