Abstract
The muscle-specific ubiquitin ligase atrogin-1 (MAFbx) has been identified as a critical regulator of pathologic and physiological cardiac hypertrophy; it regulates these processes by ubiquitinating transcription factors [nuclear factor of activated T-cells and forkhead box O (FoxO) 1/3]. However, the role of atrogin-1 in regulating transcription factors in aging has not previously been described. Atrogin-1 cardiomyocyte-specific transgenic (Tg+) adult mice (α-major histocompatibility complex promoter driven) have normal cardiac function and size. Herein, we demonstrate that 18-month-old atrogin-1 Tg+ hearts exhibit significantly increased anterior wall thickness without functional impairment versus wild-type mice. Histologic analysis at 18 months revealed atrogin-1 Tg+ mice had significantly less fibrosis and significantly greater nuclei and cardiomyocyte cross-sectional analysis. Furthermore, by real-time quantitative PCR, atrogin-1 Tg+ had increased Col 6a4, 6a5, 6a6, matrix metalloproteinase 8 (Mmp8), and Mmp9 mRNA, suggesting a role for atrogin-1 in regulating collagen deposits and MMP-8 and MMP-9. Because atrogin-1 Tg+ mice exhibited significantly less collagen deposition and protein levels, enhanced Mmp8 and Mmp9 mRNA may offer one mechanism by which collagen levels are kept in check in the aged atrogin-1 Tg+ heart. In addition, atrogin-1 Tg+ hearts showed enhanced FoxO1/3 activity. The present study shows a novel link between atrogin-1–mediated regulation of FoxO1/3 activity and reduced collagen deposition and fibrosis in the aged heart. Therefore, targeting FoxO1/3 activity via the muscle-specific atrogin-1 ubiquitin ligase may offer a muscle-specific method to modulate aging-related cardiac fibrosis.
Aging is described by the National Institute of Aging as the changes that occur within a lifetime. The capability to maintain those changes to a minimum is conferred in the symbiosis between our intrinsic and extrinsic milieu. There are many physiological processes in humans that evidence potential for pitfalls in a time-dependent manner and that could benefit from a much-needed leverage in the process of aging. There is normal deterioration in many systems, as evidenced in decrease of cardiac function and tissue regeneration as life progresses.1 Potential instigators, like diabetes, dyslipidemia, and hypertension, may accelerate the cardiac aging process and disrupt homeostasis and functionality.2 Such processes of homeostasis are maintained by targeted degradation via the ubiquitin proteasome system.
The muscle-specific ubiquitin ligase atrogin-1 (encoded by the gene FBXO32) decreases with age in the heart (GSE112913), quadriceps,4 and vastus lateralis muscle5 in mice and humans. Atrogin-1 (Fbxo32 or MAFbx) plays protective role in the pathophysiology of common heart diseases.6, 7 Atrogin-1 interacts with several components to form the S-phase kinase associated protein (Skp) 1/Cullin 1/F-box protein complex, which conjugates ubiquitin onto specific proteins for targeted degradation. Atrogin1 is upregulated in skeletal muscle atrophy and cachectic syndromes.8, 9 Denervation and dexamethasone have been proved to influence distinctive ubiquitin proteasome system gene activity and post-translational modifications in atrophy models, specifically of the forkhead box O (FoxO) transcription factors.10, 11 Because the FoxO transcription factors and atrogin-1 both play a major role in muscle cell atrophy and hypertrophy, they hold important therapeutic potential to modulate a variety of gene programs.12, 13, 14, 15, 16
Recently, stress—induced either physiologically or pathologically—has been shown to result in adaptations that link the autophagy-lysosome system and the ubiquitin proteasome system, for which atrogin-1 mediation is associated.17, 18 Different substrates for atrogin-1 have been identified to have a role in muscle differentiation, contractility, and hypertrophy. Charged multivesicular body protein 2B (CHMP2B), part of the endosomal sorting complex, has an important role in myocardial interstitial remodeling and development of diastolic dysfunction.17 As well, components of the extracellular matrix (ECM), like matrix metalloproteinase (MMP)-9, may serve as an age-dependent regulator of cardiac aging via alteration of ECM collagen deposit, which has been shown to occur in humans.19 The increased activity of MMP-9 is associated with elevated risk for cardiac-associated mortality because of cardiac fibrosis development.20 Although the ubiquitin proteasome system may regulate such remodeling processes, there is also a parallel increase in inflammatory cells that regulate ECM activity and are a function of age.21 One of the most important cellular components in cardiac tissues are fibroblasts, which are identified as targets for treating age-dependent cardiomyopathies.19, 22, 23 We hypothesized that chronic cardiac-specific overexpression of atrogin-1 would regulate age-dependent changes in fibrosis, cell size/number, and cardiac function.
Materials and Methods
Animals
Adult cardiomyocyte-specific (constitutive α-major histocompatibility complex promoter) atrogin-1 transgenic (Tg+) mice, aged 4 to 18 months, were used throughout this study, as previously described.24 Mice were subject to a light/dark cycle of 12 hours and had access to water and standard chow ad libitum (Harlan Laboratories, Inc., Indianapolis, IN). All animal studies were approved by the Institutional Care and Use Committee for animal research at the University of North Carolina at Chapel Hill.
Echocardiographic Analysis
High-resolution transthoracic echocardiography was performed on loosely restrained conscious atrogin-1 Tg+ and litter-matched wild-type mice to phenotypically characterize them at 4, 6, 12, and 18 months using a Vevo 2100 Biomicroscopy system (VisualSonics, Inc., Toronto, ON, Canada), as previously described.25, 26 Briefly, two-dimensional guided M-mode echocardiography analysis of the left ventricle was performed in the parasternal long axis at the level of the papillary muscle. Distances from the edges of the epicardium and endocardium were used to measure anterior wall thickness (interventricular septal wall thickness, end diastolic, and interventricular septal wall thickness, end systolic), posterior wall thickness (posterior wall thickness in diastole and posterior wall thickness in systole), and left ventricular internal diameters [left ventricular end-diastolic dimension (LVEDD) and left ventricular end-systolic dimension (LVESD)]. LV volume in diastole (LV VolD) was calculated as follows: [LV VolD: (7/2.4 + LVEDD) × LVEDD3 × 1000]. LV volume in systole (LV VolS) was calculated as follows: [LV VolS: (7/2.4 + LVESD) × LVESD3 × 1000]. Left ventricular systolic function was assessed by ejection fraction, [Ejection Fraction %: (LV VolD-LV VolS)/LV VolD × 100], and fractional shortening, [Fractional Shortening %: (LVEDD/LVESD)/LVEDD × 100].
M-mode measurements represent the average of three consecutive cardiac cycles from each mouse, as previously described.25, 26 Data were analyzed and normalized to tibia length while blinded to mouse genotype (T.L.P., M.S.W.).
Western Immunoblot Analysis of Protein
Frozen heart ventricles from atrogin-1 Tg+ and wild-type mice were homogenized in SDS lysis buffer (10% SDS, 10 mmol/L tris with pH 7.5, 10 mmol/L NaF, 5 mmol/L dithiothreitol, and 2 mmol/L EGTA) supplemented with protease and phosphatase inhibitors (5870S; Cell Signaling Technology, Danvers, MA) using an electric homogenizer pulsed five times for 20 seconds at a high speed. Lysates were boiled for 3 minutes and centrifuged at 13,400 × g for 20 minutes at 4°C. Lysates were precipitated using chloroform/method (using a methanol/chloroform/water mixture consistent of a 4:1:3 ratio, respectively), as previously described,27 and the resulting aggregates were solubilized in NuPAGE LDS Sample Buffer (NP0007; Thermo Fisher Scientific, Waltham, MA). Protein concentrations were determined using Pierce 660-nm protein assay (22660; Thermo Fisher Scientific). For immunoblot, 20 μg of sample was resolved with NuPAGE Reducing Agent (NP0004; Thermo Fisher Scientific) and incubated at 65°C for 10 minutes. Then, sample was separated by 4% to 12% SDS-PAGE gel (NP0322; Thermo Fisher Scientific) at 200V in 3-(N-morpholino)propanesulfonic acid (MOPS) SDS running buffer (NP0001; Thermo Fisher Scientific) for cardiac myosin binding protein C (cMyBP-C), c-myc, glucocorticoid receptor (GR), phosphorylated FoxO1/3a, total FoxO1, and total FoxO3a or 2-(N-morpholino)ethanesulfonic acid (MES) SDS running buffer (NP0002; Thermo Fisher Scientific) for vimentin, CHMP2B, calcineurin A, myogenic differentiation 1 (MyoD), muscle ringer finger-1 (MuRF1), and atrogin-1. Protein was then transferred onto polyvinylidene difluoride membrane using NuPAGE transfer buffer with 20% methanol (NP0006; Thermo Fisher Scientific). The polyvinylidene difluoride membrane was blocked with 5% dry milk in tris-buffered saline with Tween 20 for 1 hour at room temperature and incubated at 4°C overnight with the primary antibody.
Primary Antibodies
Primary antibodies included the following: anti–atrogin-1 (sc-166806; 1:500 in 5% milk; mouse; Santa Cruz Biotechnologies, Dallas, TX); anti–calcineurin Aα (07-1492; rabbit; 1:1000; EMD Millipore, Burlington, MA), anti-CHMP2B (ABC292; rabbit; 1:800; EMD Millipore), peroxidase-conjugated anti–c-myc (A5598; rabbit; 1:5000; Sigma-Aldrich, St. Louis, MO), anti-FoxO1 (2880; rabbit; 1:250; Cell Signaling Technology), anti-FoxO3a (2497; rabbit; 1:250; Cell Signaling Technology), anti–phosphorylated FoxO1/3a (9464; rabbit; 1:500; Cell Signaling Technology), anti–glyceraldehyde-3-phosphate dehydrogenase antibody (G8795; mouse; 1:4000; Sigma-Aldrich), anti–glucocorticoid receptor (sc-8992; rabbit; 1:500; Santa Cruz Biotechnology, Inc.), anti-MuRF1 (sc-27642; goat; 1:200; Santa Cruz Biotechnologies), anti-cMyBP-C (AXL-215-057-R050; rabbit; 1:5000; Enzo Life Sciences, Farmingdale, NY), anti-MyoD (ab64159; rabbit; 1:500; Abcam, Cambridge, MA), anti-vimentin (ab92547; rabbit; 1:1000; Abcam), anti–MMP-8 (Ab53017; rabbit; 1:1000; Abcam), anti–tissue inhibitor of metalloproteases-1 (TIMP-1; Ab38978; rabbit; 1:1000; Abcam), anti–TIMP-2 (Ab1828; mouse; 1:1000; Abcam), anti–TIMP-3 (710404; rabbit; 6 μg/uL; Thermo Fisher Scientific), and anti-puromycin (MABE343; mouse; 1:10,000; EMD Millipore). The membranes were washed 3× for 10 minutes in tris-buffered saline with Tween 20, and secondary antibody was incubated in 5% dry milk in tris-buffered saline with Tween 20 for 1 hour at room temperature.
Secondary Antibodies
Secondary antibodies included the following: horseradish peroxidase–anti-goat (sc-2768; 1:10,000; Santa Cruz Biotechnology, Inc.), horseradish peroxidase–anti-mouse (A9917; 1:10,000; Sigma-Aldrich), and horseradish peroxidase–anti-rabbit (A9169; 1:15,000; Sigma-Aldrich). Mouse monoclonal anti–glyceraldehyde-3-phosphate dehydrogenase antibody (G8795; 1:4000; Sigma-Aldrich) or mouse anti–β-actin (A2228; 1:1000; Sigma-Aldrich) was used for our loading control and used for normalization for the densitometry analysis. Membranes were then washed with tris-buffered saline with Tween 20 for 10 minutes (×3), followed by the application of ECL+ Plus (RPN2132; GE Healthcare Life Sciences, Pittsburg, PA). The electrochemiluminescence signal was then visualized for densitometry analysis performed on a UVP Bioimaging System with VisionWorksLS software version 6.1 (Ultra-Violet Products Ltd, Upland, CA). Regions of interest were selected using the rectangular image tool, and the background was defined for analysis (R.M.). Both the target proteins of interests and glyceraldehyde-3-phosphate dehydrogenase were detected within an optimized linear dynamic range; the limits of the UVP Bioimaging System have standardized lower/upper limits of quantitation. Images were previewed with exposure compensation while optimizing the exposure and determining the appropriate final exposure settings. Analyzed images were obtained by exposures from 30 seconds up to 3.5 minutes at 2 × 2 binning.
qPCR Analysis of Gene Expression
Total RNA was isolated from atrogin-1 Tg+ and wild-type ventricles using Trizol, according to manufacturer's instructions (15596026; Ambion by Life Technologies, Carlsbad, CA) and resuspended in RNAse-free water. Real-time quantitative PCR (qPCR) analysis was then performed using a two-step reaction. cDNA was first made using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA), then quantified in a second reaction, described below.
Quantification Using TaqMan Probes
One microliter of cDNA product was then amplified using 0.6 μL of TaqMan probes and TaqMan Universal PCR Master Mix (final volume, 12 μL). The exon-spanning TaqMan probes included the following: Anf (Mm01255747_g1), Bnp (Mm00435304_g1), Acta1 (Mm00808218_g1), Myh7 (myosin, heavy chain 7, cardiac muscle, β; Mm00600555_m1), Nr3c1 (Mm00433832_m1), Ryr (Mm00465877_m1), Ptgds (Mm01330613_m1), Dmd (Mm01216951_m1), Timp3 (Mm00441826_m1), and Klf15 (Mm00517792_m1). The 18S TaqMan probe (Hs99999901_s1) was used as a reference gene.
SYBR Green Quantification
Each reaction consisted of cDNA, 2× Power SYBR Green Master mix, or 2× QuantiNova SYBR Green Master mix, primers (for Power SYBR green reactions, 200 nm; for QuantiNova SYBR green, 0.7 μmol/L), and RNase-free water. Relative mRNA expression was determined using 18S as a loading control. The following primers were designed to detect exon spanning collagen and MMP mRNAs, as follows: Col1a1, 5′-GAGCGGAGAGTACTGGATCG-3′ (forward) and 5′-GCTTCTTTTCCTTGGGGTTC-3′ (reverse); Col6a1, 5′-TGCCCTGTGGATCTATTCTTCG-3′ (forward) and 5′-CTGTCTCTCAGGTTGTCAATG-3′ (reverse); Col6a2, 5′-TGCCCTGTGGATCTATTCTTCG-3′ (forward) and 5′-CTGTCTCTCAGGTTGTCAATG-3′ (reverse); Col6a3, 5′-AACCCTCCACATACTGCTAATTC-3′ (forward) and 5′-TCGTTGTCACTGGCTTCATT-3′ (reverse); Col6a4, 5′-ATGACAAGTGCCGACCAGCC-3′ (forward) and 5′-ACTAGCCGCAAAGCCCCAAG-3′ (reverse); Col6a5, 5′-TGCTCTGTTGGTGGTGTCCC-3′ (forward) and 5′-TGCCCAGGTCTAGCATCCCA-3′ (reverse); Col6a6, 5′-TTCAGTGCACAGAGGGGCAG-3′ (forward) and 5′-ACAGCTGCCTTGGTCACGT-3′ (reverse); Mmp8, 5′-TCAACCAGGCCAAGGTATTG-3′ (forward) and 5′-ATGAGCAGCCACGAGAAATAG-3′ (reverse); Mmp9, 5′-TTGGTTTCTGCCCTAGTGAGAGA-3′ (forward) and 5′-AAAGATGAACGGGAACACACAGG-3′ (reverse); Mmp12, 5′-TGACATACGTAACATTCAGTCCC-3′ (forward) and 5′-TCTCTCCCACTGTTGTGACAG-3′ (reverse); Timp1, 5′-CCCTTCGCATGGACATTTATTC-3′ (forward) and 5′-TCTGCTCTGGTGTGTCTCTA-3′ (reverse); Timp2, 5′-ACATCGAGGACCCGTAAGA-3′ (forward) and 5′-TTCCAGGAAGGGATGTCAAAG-3′ (reverse); and 18S, 5′-AGAAACGGCTACCACATCCA-3′ (forward) and 5′-CTCGAAAGAGTCCTGTATTGT-3′ (reverse). All reactions were run in a LightCycler 480 II System (04707494001; Roche, Basel, Switzerland) and quantified by the LightCycler 480 Software version 1.5.0 SP4 (Roche). Power SYBR Green PCR gene melting curves were performed for each reaction to ensure a single product was detected in each reaction. Raw threshold cycle (CT) values were analyzed using the ΔΔCT method, where CT values were first normalized to 18S. Fold change values (calculated by the formula 2−ΔΔCT) were used as final expression data.
Cardiac Perfusion and Histologic Preparation
Eighteen-month–old mice were euthanized using isoflurane (open-drop method)28 and secondary cervical dislocation, followed by gravity perfusion with 4% paraformaldehyde, fixed for 24 hours, followed by storage in 75% ethanol. Fixed cardiac tissue was processed for paraffin embedding, sectioned, and stained with standard hematoxylin and eosin (H&E) and Masson's trichrome (MT) staining. Additional slides were cut and used for immunofluorescence studies. Stained slides were scanned on a Leica Aperio VERSA ScanScope XT epifluorescent microscope digital scanner (DM6000 B; Leica Biosystems Inc., Buffalo Grove, IL) at 20× with an Andor Zyla sCMOS camera and were visualized/analyzed using Aperio ImageScope version 12.1.0.5029 (Aperio Technologies Inc., Vista, CA), including export of TIFF images used in publication (R.M.).
Algorithmic Analysis of Collagen and Cross-Sectional Area Analysis of Masson's Trichrome–Stained Slides
Scanned MT-stained slides were analyzed in Aperio ImageScope using the Aperio positive pixel v9 algorithm using standard parameter settings (hue value = 0.66; hue width = 0.45 to 0.55 set and confirmed visually using the markup feature for each slide). The amount of collagen was determined as a percentage of tissue. Three biological replicates (each with three sections) were analyzed per genotype. MT-stained slides were analyzed for cardiomyocyte cross-sectional area quantification, as previously described.25 Briefly, ventricular sections were visualized using the Aperio ImageScope version 12.1.0.5029 (Aperio Technologies) and exported as TIFF files with scale bars (at ×20 magnification). Cardiomyocyte length was quantified in a blinded manner (R.M.) using ImageJ software version 1.51 (NIH, Bethesda, MD; http://imagej.nih.gov/ij) across four to six histologic sections per mouse from a total of three hearts per genotype, analyzing 100 cells per heart. Data represent mean cardiomyocyte length (μm).
Algorithmic Analysis of Total Nuclei in H&E-Stained Cardiac Sections
Nuclear quantification analysis was performed on H&E-stained slides with Definiens Architect 2.7 Build 59337 (Biocompare, San Francisco, CA) using the Tissue Studio version 4.4.1 portal. The images were first imported into the application along with their respective ventricular tissue annotations. Each tissue section was independently analyzed, and the output was parsed accordingly. The regions of interest were further segmented into tissue and glass regions, with only the tissue class being analyzed for nuclear detection. The results included Analyzed Tissue Area, Total Nuclei, and Nuclear Density on the basis of the nuclear size values. The H-score was calculated as follows:
H-Score: (3 × % Nucleus Large) + (2 × % Nucleus Medium) + (1 × % Nucleus Small).
The analysis output included all quantitative results as well as color-coded overlays that represented the stained nuclei. Data are represented as sum of nuclei density (n/mm2) and total nuclei/area (μm2) normalized to analyzed tissue area. Analysis was performed in a blinded manner (B.M.; University of North Carolina Tissue Pathology Laboratory core facility). A secondary nuclear quantification analysis was performed using the Aperio ImageScope system in the user-defined regions of interests in the ventricular tissue and analyzed with the Aperio Nuclear v9 algorithm configured as a custom macro for this assay. To generate a customized analysis macro with the Aperio Nuclear v9 algorithm, the principal colors of the H&E stain were sampled to obtain representative OD values for the red–green–blue color space. These values were used as inputs to mark the appropriate stains. Other input parameters (eg, minimum and maximum nuclear size and intensity threshold) were adjusted to achieve nuclear segmentation. Data represent nuclei/μm2. Histologic analysis was performed in a blinded manner (R.M. and B.M.; University of North Carolina Tissue Pathology Laboratory core facility).
Immunofluorescence Staining of Cardiac Histologic Sections for Vimentin and Collagen I
Immunostaining was performed as described previously.25 Briefly, cardiac sections were stained with antibodies against the following: anti-vimentin (ab137321; 1:500; Abcam), anti–collagen I (ab34710; 1:100; Abcam), and rabbit IgG (as a negative control) at 4°C overnight. Sections were then treated with Alexa Fluor 488–conjugated or Alexa Fluor 594–conjugated secondary antibodies and counterstained with DAPI. Total positive vimentin stained cells were identified and counted in each of the four cross-sectional defined areas using Meta-Morph software version 7.8 (Molecular Devices, San Jose, CA). Images were taken using Axio Imager 2 (Carl Zeiss Microscopy, LLC, Thornwood, NY).
Protein Synthesis
The surface sensing of translation (SUnSET) technique was used to determine protein synthesis in 6-month–old atrogin1 Tg+ (n = 5) and litter-matched wild-type (n = 4) mice and in 16-month–old atrogin1 Tg+ (n = 2) and litter-matched wild-type (n = 2) mice, as previously described.29 Briefly, mice were injected intraperitoneally with 0.04 μmol/L puromycin dihydrochloride (61-385-RA; Corning Inc., Corning, NY) diluted in phosphate-buffered saline 30 minutes before harvest. Hearts were harvested and flash frozen in liquid nitrogen. Heart lysates were prepared and blotted for puromycin (MABE343; mouse; 1:10,000; EMD Millipore), as described earlier in Western Immunoblot Analysis of Protein. Total protein levels were assessed by staining the membrane with Ponceau S stain (7170; Sigma-Aldrich) for 5 minutes.
Serum Chemistry
Blood was collected from 13- to 18-month–old atrogin1 Tg+ (n = 8) and litter-matched wild-type (n = 3) mice via submandibular bleed, and serum was collected via serum separator tubes (365978; BD Microtainer; Becton Dickinson, Franklin Lakes, NJ). Serum was next analyzed for alanine aminotransferase (SA1046; Alfa Wassermann Diagnostic Technologies, LLC, Caldwell, NJ), alkaline phosphatase (RX2002; Alfa Wassermann Diagnostic Technologies, LLC), blood urea nitrogen (SA2024; Alfa Wassermann Diagnostic Technologies, LLC), and creatinine (SA1012; Alfa Wassermann Diagnostic Technologies, LLC) via Ace Axcel clinical chemistry system (Alfa Wassermann Diagnostic Technologies, LLC) by the University of North Carolina Clinical Chemistry laboratory.
Cell Culture, Adenoviral Transduction
HL-1 cells were maintained, as published previously.30 Tissue culture dish was coated with a mixture of 0.02% gelatin (w/v)–0.5% fibronectin (v/v) for 60 minutes. Cells were trypsinized (0.05% trypsin + EDTA) and seeded in 6-well dishes to adhere overnight in Claycomb medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 1% norepinephrine, and 1% l-glutamine. After culturing for at least 24 hours, medium was changed and cells were transduced with increasing multiplicities of infection of a bicistronic adenovirus construct expressing myc–atrogin-1 [Ad-Atrogin-1-green fluorescence protein (GFP)] or GFP control (Ad-GFP), as previously described.24, 30 Twenty-four hours after plating, cells were transfected at different multiplicities of infection (0, 10, 20, 50, 100, and 200) in triplicate with both Ad-Atrogin-1-GFP and Ad-GFP for 24 to 48 hours. Fluorescence microscopy imaging was performed on an Eclipse TS100 microscope (Nikon, Inc., Melville, NY) with a QIClick camera (model 74-0083-A0; QImaging, Surrey, British Columbia, Canada) to verify presence of increased GFP and GFP-Atrogin-1 transduction in HL-1 cells as multiplicity of infection increased. Post-transduction cells were harvested for protein using radioimmunoprecipitation assay buffer [10 mmol/L tris-Cl (pH 8.0), 1 mmol/L EDTA, 0.5 mmol/L EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 140 mmol/L NaCl] in the presence of protease and phosphatase inhibitors (5870S; Cell Signaling Technology).
Statistical Analysis
Statistics were computed by either a two-tailed t-test when comparing means from two groups or a one-way analysis of variance to compare means from multiple groups. If the one-way analysis of variance was significant, a subsequent all-pairwise multiple comparison procedure (Holm-Sidak method) was then performed to compare all of the groups with each other in a pairwise manner. P ≤ 0.05 was considered significant. GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA) was used for all statistical analyses, except echocardiography, which used Microsoft Excel 2016 (Microsoft, Redmond, WA) and SigmaPlot version 11.0 build 11.0.0.77 (Systat Software, Inc., San Jose, CA). Data represent means ± SEM, unless otherwise noted.
Results
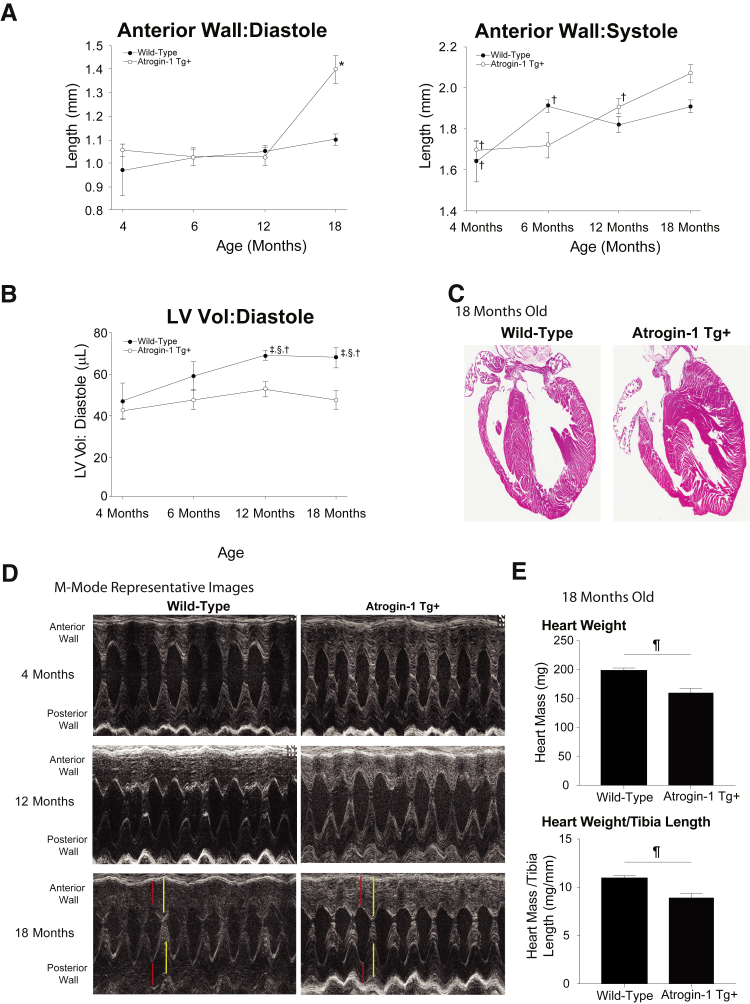
Cardiomyocyte-specific atrogin-1 Tg+ mice were analyzed for cardiac function longitudinally to 18 months of age by conscious echocardiography. The atrogin-1 Tg+ hearts do not significantly differ from litter-matched wild-type mice at 4 or 6 months of age by conscious echocardiography (Table 1), as previously described in 12-week–old atrogin-1 Tg+ mice.31 Significantly increased anterior wall thickness was seen in atrogin-1 Tg+ hearts at 18 months of age (Figure 1A), as was significantly decreased LV volume in diastole (Figure 1B). Significant increases in the left ventricular posterior wall in diastole were also seen in the atrogin-1 Tg+ hearts (1.38 ± 0.03 mm) compared with litter-matched wild-type mice (1.15 ± 0.04 mm) (Table 1). Representative M-mode images illustrate the atrogin-1 Tg+ increased anterior and posterior wall thickness at 18 months, increased anterior wall thickness in diastole, and increased posterior wall in systole in the atrgoin-1 Tg+ anterior wall (Figure 1D). On low-power analysis, the atrogin-1 Tg+ hearts did not grossly appear different (Figure 1C), whereas the measured atrogin-1 Tg+ heart weights at harvest were significantly decreased with and without normalization to tibia length at 18 months of age (Figure 1E). No changes in systolic function were identified in the atrogin-1 Tg+ hearts (ie, ejection fraction and fractional shortening) throughout the course of the study (Table 1), consistent with previous studies of no functional changes for up to 8 months of age.24, 31
Table 1.
High-Resolution Transthoracic Echocardiography Performed on Conscious 4- and 6-Month–Old Atrogin-1 Tg+ and Sibling-Matched Mice
| Echocardiographic measure/calculation (one-way analysis of variance P value) | Wild type at 4 months (n = 3) | Atrogin-1 Tg+ at 4 months (n = 8) | Wild type at 6 months (n = 3) | Atrogin-1 Tg+ at 6 months (n = 8) | Wild type at 12 months (n = 11) | Atrogin-1 Tg+ at 12 months (n = 11) | Wild type at 18 months (n = 11) | Atrogin-1 Tg+ at 18 months (n = 11) |
|---|---|---|---|---|---|---|---|---|
| AWTD, mm (P < 0.001) | 0.97 ± 0.11 | 1.05 ± 0.02 | 1.02 ± 0.03 | 1.03 ± 0.04 | 1.05 ± 0.03 | 1.20 ± 0.03 | 1.10 ± 0.02 | 1.40 ± 0.06∗ |
| AWTS, mm (P < 0.001) | 1.64 ± 0.10 | 1.70 ± 0.05† | 1.91 ± 0.03 | 1.72 ± 0.06† | 1.82 ± 0.04† | 1.91 ± 0.03 | 1.91 ± 0.03 | 2.07 ± 0.04 |
| PWTD, mm (P < 0.001) | 0.94 ± 0.10†‡ | 1.10 ± 0.03† | 1.26 ± 0.13 | 1.09 ± 0.03†‡ | 1.05 ± 0.03†‡ | 1.28 ± 0.05 | 1.15 ± 0.04† | 1.38 ± 0.03 |
| PWTS, mm (P = 0.011) | 1.60 ± 0.15 | 1.78 ± 0.07 | 1.94 ± 0.10 | 1.78 ± 0.03 | 1.90 ± 0.06 | 1.98 ± 0.05 | 1.96 ± 0.09 | 2.08 ± 0.07§ |
| LVEDD, mm (P < 0.001) | 3.37 ± 0.25 | 3.23 ± 0.12 | 3.71 ± 0.19 | 3.38 ± 0.15 | 3.96 ± 0.06†§¶ | 3.53 ± 0.11 | 3.93 ± 0.11†§ | 3.37 ± 0.14 |
| LVESD, mm (P = 0.021) | 1.55 ± 0.19 | 1.49 ± 0.13 | 1.67 ± 0.11 | 1.55 ± 0.11 | 1.88 ± 0.04 | 1.64 ± 0.05 | 1.81 ± 0.04 | 1.58 ± 0.10 |
| LV volume: diastole, μL (P < 0.001) | 47.2 ± 8.4 | 42.6 ± 4.1 | 59.0 ± 7.1 | 47.8 ± 4.9 | 68.8 ± 2.4†¶‖ | 52.8 ± 3.7 | 67.8 ± 4.8†¶‖ | 47.7 ± 4.6 |
| LV volume: systole, μL | 7.0 ± 2.0 | 6.7 ± 1.8 | 8.2 ± 1.3 | 7.0 ± 1.2 | 10.9 ± 0.7 | 7.8 ± 0.7 | 10.0 ± 0.6 | 7.5 ± 1.2 |
| EF, % | 85.7 ± 2.3 | 85.7 ± 2.1 | 86.2 ± 1.3 | 85.9 ± 1.4 | 84.2 ± 0.5 | 85.2 ± 0.7 | 84.8 ± 1.0 | 84.7 ± 1.70 |
| FS, % | 54.2 ± 2.8 | 54.3 ± 2.3 | 54.9 ± 1.5 | 54.6 ± 1.8 | 52.7 ± 0.6 | 53.5 ± 1.0 | 53.6 ± 1.3 | 53.2 ± 1.9 |
| LV mass index, mg (P < 0.001) | 114.0 ± 5.7 | 130.0 ± 7.0 | 175.4 ± 25.5 | 134.9 ± 8.2 | 170.1 ± 7.9§¶ | 183.1 ± 7.2§¶‖ | 185.4 ± 8.6§¶‖ | 204.7 ± 10.3§¶‖ |
| LV mass index/body weight, mg/g | 3.6 ± 0.2 | 4.3 ± 0.3 | 4.7 ± 0.9 | 3.7 ± 0.2 | 3.5 ± 0.2 | 3.8 ± 0.2 | 3.7 ± 0.2 | 4.2 ± 0.3 |
| LV mass index/tibia length, mg/mm∗∗ | 7.0 ± NA (n = 1) | 7.1 ± 0.3 (n = 6) | 12.3 ± NA (n = 1) | 7.8 ± 0.9 (n = 6) | 9.8 ± 0.8 | 9.9 ± 0.5 | 9.9 ± 0.6 | 11.6 ± 0.6¶‖ |
| Body weight, g | 38.4 ± 2.6 | 31.1 ± 2.2 | 38.4 ± 2.7 | 37.0 ± 2.6 | 49.7 ± 2.4§¶‖ | 48.7 ± 2.5§‖ | 51.4 ± 2.3§¶‖ | 50.9 ± 3.4§¶‖ |
| Tibia lengths, mm | ND | ND | ND | ND | ND | ND | 18 ± 0.3 | 17.8 ± 0.3 |
| Heart rate, bpm | 669 ± 48 | 622 ± 22 | 667 ± 10 | 668 ± 10 | 567 ± 20 | 584 ± 26 | 599 ± 24 | 609 ± 21 |
Data are expressed as means ± SEM. A one-way analysis of variance was performed (P < 0.05), followed by an all-pairwise multiple comparison procedure (Holm-Sidak method) to compare means of groups with all other groups.
AWTD, anterior wall thickness in diastole; AWTS, anterior wall thickness in systole; bpm, beats/minute; EF, ejection fraction [calculated as (end Simpson's diastolic volume – end Simpson's systolic volume)/end Simpson's diastolic volume * 100]; ExLVD, external left ventricle dimension; FS, fractional shortening [calculated as (LVEDD-LVESD)/LVEDD × 100]; LV, left ventricular; LVEDD, LV end-diastolic dimension; LVESD, LV end-systolic dimension; NA, not applicable; ND, not determined; PWTD, posterior wall thickness in diastole; PWTS, posterior wall thickness in systole.
P < 0.05 versus all other groups.
P < 0.05 versus atrogin-1 Tg+ at 18 months.
P < 0.05 versus atrogin-1 Tg+ at 12 months.
P < 0.05 versus wild type at 4 months.
P < 0.05 versus atrogin-1 Tg+ at 4 months.
P < 0.05 versus atrogin-1 Tg+ at 6 months.
LV mass (index) = [1.055 ∗ ((ExLVD;d)3 − (LVEDD;d)3)]. Eighteen-month tibia lengths used throughout for normalization.
Figure 1.
Conscious echocardiographic analysis of aging atrogin-1 Tg+ hearts. A and B: High-resolution transthoracic echocardiography measurements of atrogin-1 Tg+ and litter-matched wild-type mouse hearts identified significant alterations in the anterior wall thickness at 4, 6, 12, and 18 months of age (A) and left ventricular volume (LV Vol) in diastole (B). C: Four-chamber view of representative wild-type and atrogin-1 Tg+ hearts at 18 months of age. D: Representative M mode of atrogin-1 Tg+ hearts at 4, 12, and 18 months of age. E: Heart weight at 18 months of age at harvest and heart weight at 18 months of age at harvest normalized to tibia length. A one-way analysis of variance was performed, followed by an all-pairwise multiple comparison procedure (Holm-Sidak method) to compare means of groups with all other groups. A t-test was performed. Red: left ventricular wall thickness in diastole. Yellow: left ventricular wall thickness in systole. Data are expressed as means ± SEM (A, B, and E). ∗P < 0.05 versus all other groups (analysis of variance); †P < 0.05 versus atrogin-1 Tg+ at 18 months (analysis of variance); ‡P < 0.05 versus atrogin-1 Tg+ at 4 months (analysis of variance); §P < 0.05 versus atrogin-1 Tg+ at 12 months (analysis of variance); ¶P < 0.05 versus wild-type (t-test).
Age-related changes in cardiac homeostasis are observed in the heart. The hallmarks of these changes are progressive cardiomyocyte hypertrophy, inflammation, and the gradual development of fibrosis.21 Therefore, measures of these hallmarks were studied to determine whether the increased atrogin-1 in cardiomyocytes affected these aging changes. First, the amount of collagen deposition was analyzed by MT-stained histologic sections using a nonbiased logarithmic analysis of digitally scanned glass slides. The atrogin-1 Tg+ hearts exhibited significantly less (68.8% less) collagen deposition (1.45 ± 0.26%) compared with the collagen staining consistently seen in the wild-type hearts (4.68 ± 0.72%) (Figure 2A). Cross-sectional analysis of atrogin-1 Tg+ hearts was found to be significantly increased 40.1% in cross-sectional area (189.1 ± 19.0 μm2) compared with age-matched wild-type mice (134 ± 1.7 μm2) (Figure 2B). To identify if this apparent increase in cardiomyocyte size (hypertrophy) was a result of pathologic hypertrophy signaling pathways, skeletal muscle actin, Myh7, Anf, and Bnp mRNAs were assayed in atrogin-1 Tg+ hearts compared with wild-type hearts, and no differences were observed by qPCR analysis (Figure 2C). Analysis of the H&E-stained slides indicated that there might be an increase in nuclei (Figure 2D), so the slides were analyzed using two different algorithms (Definiens and Aperio) that unbiasedly recognized nuclei (Figure 2, E and F). Analysis of the atrogin-1 Tg+ hearts revealed a significant increase in nuclei in LV cross sections normalized to area using the Definiens (Figure 2E) and Aperio (Figure 2F) algorithms. Together, these studies illustrate that the atrogin-1 Tg+ hearts have significantly less fibrosis and an increase in cardiomyocyte area not related to pathologic hypertrophy (evidenced by no increase in fetal gene expression), with an increase in nuclei using a systemic analysis of the histology.
Figure 2.
Histologic analysis of perfused atrogin-1 Tg+ and litter-matched wild-type hearts at 18 months of age. A: Determination of the percentage collagen in atrogin-1 Tg+ hearts at 18 months of age using an algorithm-based determination of collagen with representative low-power images. Histologic analysis was performed in a total of three histologic sections per mouse heart (three biological replicates per genotype). B: Cardiomyocyte cross-sectional area determined on three biological replicate assays in three left ventricular cross sections with representative low-power images. C: Real-time quantitative PCR analysis of 18-month–old atrogin-1 Tg+ ventricles for skeletal muscle actin (Acta1), β-major histocompatibility complex (MHC; Myh7), Anf, and Bnp mRNA associated with pathologic cardiac hypertrophy. D: Representative hematoxylin and eosin–stained histologic sections of atrogin-1 Tg+ hearts. E: Quantification of nuclei density (number per area) and total nuclei per section in atrogin-1 Tg+ hearts using Definiens software. Histologic analysis was performed in a total of three histologic sections per mouse heart (three biological replicates per genotype). F: Histologic analysis of nuclei number using Aperio software. Quantification of nuclei density (number per area) and total nuclei per section in atrogin-1 Tg+ hearts using Aperio software. Histologic analysis was performed in a total of three histologic sections per mouse heart (three biological replicates per genotype). A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM (A–C, E, and F). n = 3 biological replicates per group (C). ∗P < 0.05, ∗∗P < 0.01 versus wild type. Scale bars: 300 μm (A); 100 μm (B and D).
Cardiac fibroblasts are the major producer of the ECM, including collagen. The aging effects on cardiac fibroblasts include an increase in collagen production and an increase in fibroblast senescence.32 We, therefore, hypothesized that the significantly greater atrogin-1 Tg+ nuclei number (Figure 2, D and F) represented an increase in cardiac fibroblasts. Immunohistochemistry was performed on atrogin-1 Tg+ hearts for fibroblast-associated vimentin, and the area was quantified to identify the fibroblast areas in different regions of the heart (Figure 3, A and B, and Supplemental Figure S1). There appear to be regional differences in the atrogin-1 heart fibroblast areas, with significantly less in quadrant 1 compared with wild-type hearts and significantly more in quadrant 3 (Figure 3B). Overall, atrogin-1 Tg+ hearts did not have significantly different fibroblast areas compared with wild-type hearts (Figure 3C). Immunoblot analysis of heart lysates similarly found no significant differences in atrogin-1 Tg+ hearts compared with wild-type controls at 18 months of age (Figure 3D). Together, these findings demonstrate that atrogin-1 Tg+ hearts have cardiac fibroblast numbers equivalent to wild-type hearts at 18 months of age.
Figure 3.
Analysis of atrogin-1 Tg+ cardiac fibroblasts by vimentin immunofluorescence analysis. A: Atrogin-1 fibroblast areas in a perivenular distribution were accessed for (vimentin-positive) cells colocalized by immunohistochemistry in four quadrants (Q1–Q4) as oriented to the right and left ventricles (RV and LV). B: Nonbias quantitative logarithmic analysis of vimentin (+) cell staining in quadrants 1 to 4 by immunofluorescence microscopy of 18-month–old atrogin-1 Tg+ hearts. Representative immunofluorescence images can be found in Supplemental Figure S2. C: Whole section vimentin-positive fibroblast area (Q1–Q4). D: Vimentin immunoblots (IBs) of atrogin-1 Tg+ heart lysate. A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM (B–D). n = 3 biological replicates per group (B); n = 4 per group (D). ∗P < 0.05, ∗∗P < 0.01 versus age-matched wild-type controls. AU, arbitrary unit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The significant reduction in atrogin-1 Tg+ heart fibrosis identified in Figure 2A may be the result of either decreased collagen 1/3 or increased degradation. Because the collagen subtypes can contribute significantly to the heart phenotype,33 collagen and Mmp mRNA expression was investigated by qPCR. Unexpectedly, atrogin-1 Tg+ hearts did not have decreased Col1 mRNA (Figure 4A), which is a major contributor to the MT-stained blue signal in Figure 2A. Type 6 collagen has been reported to induce cardiac myofibroblast differentiation,34 with the three novel subtypes (collagen 6α-4, 6α-5, and 6α-6) expressed in fetal tissues.35 Atrogin-1 Tg+ hearts exhibited significant increases in Col6a4 and Col6a5, whereas no significant differences were seen in the other subtypes compared with wild-type hearts (Figure 4, B–G). Furthermore, atrogin-1 Tg+ hearts had significantly increased Mmp8 and Mmp9 mRNA (Figure 4, H and I), but not Mmp12 mRNA (Figure 4J). No significant differences were found in Timp1, Timp2, and Timp3 mRNA levels (Figure 4, K–M). Collagen 1 protein levels were determined by immunohistochemistry (Figure 5) and illustrated the significant decrease in collagen 1 in atrogin-1 Tg+ hearts compared with wild-type hearts, where it was found distributed at much higher levels throughout the tissue (Figure 5). Together, these findings suggest that the atrogin-1 Tg+ hearts have alterations in collagen 1 degradation, given their significantly decreased collagen 1 protein with nonsignificant changes in Col1 mRNA. Furthermore, the identification of significantly increased Col6a4, Col6a5, Mmp8, and Mmp9 mRNA illustrates altered remodeling of distinct collagen subtypes, with the collagen 6α subtypes relatively unknown in the context of the aging heart.
Figure 4.
Real-time quantitative PCR analysis of extracellular matrix genes in 18-month–old atrogin-1 Tg+ hearts. Quantitative analysis of ventricular collagen 1 (Col1; A), collagen 6α-1 (Col6a1; B), collagen 6α-2 (Col6a2; C), collagen 6α-3 (Col6a3; D), collagen 6α-4 (Col6a4; E), collagen 6α-5 (Col6a5; F), collagen 6α-6 (Col6a6; G), matrix metalloproteinase 8 (Mmp8; H), matrix metalloproteinase 9 (Mmp9; I), matrix metalloproteinase 12 Mmp12; J), tissue inhibitor of metalloproteinase 1 (Timp1; K), tissue inhibitor of metalloproteinase 2 (Timp2; L), and tissue inhibitor of metalloproteinase 3 (Timp3; M) mRNA. A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM (A–M). ∗P < 0.05, ∗∗P < 0.01 versus age-matched wild-type controls.
Figure 5.
Immunofluorescence (IF) staining of collagen 1 in atrogin-1 Tg+ hearts at 18 months of age. Representative collagen 1 staining (red) with DAPI (blue) counterstaining of atrogin-1 Tg+ hearts versus litter-matched wild-type hearts. Original magnification, ×20.
Aging has been associated with a decrease in GR expression in the heart. Decreases up to 70% in nuclear receptor subfamily 3 group C member 1 (Nr3c1) mRNA encoding GR have been reported in old hearts compared with young hearts (from published data sets: GSE112913 and GSE814636). Inducing GR deficiency in adult mice results in the development of increased cardiomyocyte size,37, 38 as seen in the atrogin-1 Tg+ hearts. Because GR degradation occurs in a proteasome-dependent manner39 and the ubiquitin ligase mediating its degradation is unknown, we hypothesized that atrogin-1 might mediate this degradation. HL-1 cardiomyocyte-derived cells were transduced with increasing atrogin-1 using adenovirus (Ad-Atrogin-1) to identify its effect on steady-state GR. Increasing atrogin-1 resulted in a decreased GR protein level at the highest atrogin-1 level (Supplemental Figure S2). Eighteen-month–old atrogin-1 Tg+ hearts were next assayed by qPCR for the expression of reported GR-regulated genes,38 including dystrophin (Dmd), Kruppel-like factor 15 (Klf15), ryanodine receptor 2 (Ryr2), GR (Nr3c1), and prostaglandin D2 synthase (Ptgds) (Supplemental Figure S3). We identified that atrogin-1 Tg+ did not have any changes in GR activity, as identified by the expression of these GR-regulated genes (Supplemental Figure S3). Further assay of atrogin-1 Tg+ hearts for atrogin-1 levels demonstrated a 43.0% increase in atrogin-1 protein (Supplemental Figure S4), which may explain the lack of effect of apparent GR activity because much higher atrogin-1 levels (65.3-fold endogenous levels) were needed to identify an effect on GR protein levels (Supplemental Figure S2). Together, these findings ruled out atrogin-1's effect on GR as a mechanism of the observed cardiac phenotype in vivo.
The ubiquitin ligase activity of the muscle-specific atrogin-1 protein has multiple substrates that have been reported as substrates, including calcineurin A,24 FoxO1/3a,31 CHMP2B,17 MyoD,40, 41 and cMyBP-C.42 Because these activities have been described in the context of different disease states, including pathologic24 and physiological31 cardiac hypertrophy, cardiomyopathies,17, 42 and skeletal muscle atrophy,40, 41 their roles in cardiac aging were not anticipated. Consistent with previous studies demonstrating that atrogin-1 monoubiquitination preserved calcineurin A protein levels in pathologic cardiac hypertrophy,24 immunoblot analysis revealed that atrogin-1 Tg+ hearts had significantly increased calcineurin A protein levels at 18 months of age (Figure 6A). Prior studies identified that increasing atrogin-1 resulted in the interaction and monoubiquitination of FoxO1/FoxO3a substrates, resulting in increased FoxO1/3a activity in physiological cardiac hypertrophy.31 Consistent with this, decreases were identified in phosphorylated FoxO1 and phosphorylated FoxO3a (Figure 6B), which are the protein species excluded from the nucleus. Recent studies identified CHMP2B as a critical protein regulating endosomal sorting complex in autophagy, with atrogin-1−/− hearts failing to degrade CHMP2B, resulting in increased CHMP2B proteins levels.17 On the basis of these findings, it was anticipated that increased atrogin-1 in atrogin-1 Tg+ hearts would have significantly less CHMP2B, resulting from enhanced protein degradation. However, significantly greater CHMP2B protein levels were identified by immunoblot (Figure 6C). Another atrogin-1 substrate found in skeletal muscle atrophy is MyoD, which has been reported to be degraded after atrogin-1 ubiquitination.40, 41 However, like CHMP2B, MyoD levels were not decreased, as expected (Figure 6D). In fact, atrogin-1 Tg+ heart MyoD protein levels trended (P = 0.0752) toward being increased compared with aged wild-type control hearts (Figure 6D). Limited previous studies in cardiomyopathies have identified cMyBP-C as an atrogin-1 substrate targeted for degradation, making it expected to be decreased in atrogin-1 Tg+ hearts.42 However, atrogin-1 Tg+ heart cMyBP-C was not significantly different from wild-type hearts (Figure 6E). Last, it was investigated if the other muscle-specific ubiquitin ligase MuRF1 protein expression was affected by increased atrogin-1, because MuRF1 expression is regulated by FoxO1/3a in skeletal muscle atrophy,9, 43, 44 and changes in MuRF1 expression are altered in specific disease contexts.30, 45, 46 Atrogin-1 Tg+ hearts were not significantly different from wild-type control hearts at 18 months of age (Figure 6F). There were no significant differences in protein synthesis between groups, as evidenced by incorporation of the amino acid analog puromycin, detected by immunoblot in 6-month–old atrogin-1 Tg+ hearts (Supplemental Figure S5A) or in 16-month–old (aged) atrogin-1 Tg+ hearts (Supplemental Figure S5B). No significant differences were identified in atrogin-1 Tg+ serum alanine aminotransferase, alkaline phosphatase, blood urine nitrogen, and creatinine (Supplemental Figure S6) compared with wild-type controls. Overall, it was identified that atrogin-1 Tg+ hearts had significantly increased calcineurin A and CHMP2B expression, along with significantly decreased phospo-FoxO1/FoxO1 and phospo-FoxO3a/FoxO3a.
Figure 6.
Immunoblot analysis of previously reported atrogin-1 substrates (see Results) in atrogin-1 Tg+ hearts at 18 months of age. Densitometric analysis of atrogin-1 Tg+ heart protein levels of calcium and calmodulin-dependent serine/threonine protein phosphatase calcineurin Aα (A), p-forkhead box protein O1 (FoxO1)/total FoxO1 (B, left graph), p-forkhead box protein O3 (FoxO3)/FoxO3 (B, right graph), charged multivesicular body protein 2B (CHMP2B; C), myogenic differentiation 1 (MyoD; D), cardiac myosin binding protein C (cMyBP-C; E), and muscle ring finger-1 (MuRF1; F). All immunoblots (IBs) were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM. n = 4 per group. ∗P < 0.05 versus age-matched wild-type controls. AU, arbitrary unit.
Discussion
Aging is associated with a decrease in cardiac (GSE112913), quadriceps,4 and vastus lateralis muscle5 atrogin-1 (encoded by the gene FBXO32) in mice and humans. Atrogin-1 loss-of-function mutations have recently been identified as a cause of dilated cardiomyopathy in human patients,47 and atrogin-1−/− animals exhibited increased fibrosis with aging, recently identified to be related to the accumulation of CHMP2B, a purported atrogin-1 substrate, linked to an observed impaired autophagy.17 In the present study, we determined how prevention of this age-associated loss of atrogin-1 in cardiomyocytes may affect the age-related changes that occur in the heart. Aged atrogin-1 Tg+ hearts had a significant decrease in cardiac fibrosis by trichrome staining (Figure 2A) and collagen 1 immunofluorescence (Figure 5), in the absence of increased collagen 1 mRNA (Figure 4A). These findings are consistent with an increase in collagen degradation. Significant increases in Mmp8 and Mmp9 mRNA (Figure 4, H and I) with no corresponding increase in Timp1-3 mRNA (Figure 4, K–M) offer one potential mechanism by which this may occur. MMPs, including MMP-8 and MMP-9, are elevated with cardiac aging and oversee signaling during the aging process by modulating cytokines, chemokines, growth factors, hormones, and angiogenic factor expression and activity.21 Pro–MMP-9 forms a tight complex with TIMP-1 and TIMP-3.48 Because all TIMPs are known to interact with MMP-9 and inhibit its activity,49 the ratio of MMP-9/TIMPs is also critical to the accumulation of collagen and age-associated fibrosis. Although cardiomyocyte transgenic MMP-9 mice have not been generated, to our knowledge, macrophage transgenic MMP-9 mice exhibit decreases in fibrosis in age-related cardiac changes.50 MMP-8 is a collagenase that cleaves interstitial collagens I, II, and III into characteristic fragments, while digesting other ECM molecules and proteins.48 These activities may explain why elevated MMP-8 has antifibrotic roles in the liver51 and may be responsible for the antifibrotic phenotype identified in the atrogin-1 aged hearts in the present study.
The FoxO1/FoxO3 transcription factors play a role in aging and longevity.52, 53 The FoxO1/FoxO3 homolog FoxO (encoded by Daf-16) in Caenorhabditis elegans is pivotal in aging, as demonstrated by microarray, proteomics, and DNA adenine methyltransferase identification studies.54, 55, 56, 57 More than 100 ortholog of the FoxO family of transcription factors in the nematode Caenorhabditis elegans (DAF-16) direct targets were identified,58 with more than a dozen targets involved in longevity.52 These targets include ACOnitase, glyceraldehyde 3-phosphate dehydrogenase, leucyl amino-acyl tRNA synthetase, PeRoxireDoXin, and sarcoplasmic-endoplasmic reticulum calcium ATPase, among others.52 The ubiquitin proteasome system determines the stability of FoxO proteins, the ubiquitin ligases reported to mouse double minute 2, SKP2, constitutive photomorphogenesis protein 1, and C terminus of Hsp70-interacting protein responsible for the ubiquitination and degradation of FoxOs.59, 60, 61, 62 Moreover, phosphorylation of FoxOs by extracellular signal–regulated kinase or IκB kinase contributes to FoxO degradation.61, 63 The arginine methyltransferase protein arginine methyltransferase 1 methylates DAF-16/FoxO, which can block AKT phosphorylation of DAF-16, resulting in longevity and stress tolerance.64 This parallels findings in mammalian cell.65 Previous studies by our group have found that cardiac atrogin-1 does not mediate ubiquitin-dependent degradation of FoxO transcription factors.31 In contrast, atrogin-1 was found to polyubiquitinate (via K63 linkages) FoxO and enhance its activity.31 In these studies, atrogin-1 was found to inhibit Akt-dependent (physiological) cardiac hypertrophy by ubiquitin-dependent coactivation of FoxO1/FoxO3 in vivo, including the atrogin-1 Tg+ mouse model described in the present study.31 Consistent with these findings, the present study identified that increased cardiomyocyte atrogin-1 enhanced FoxO1/FoxO3a activity in aged mice (Figure 6, B and C). Because modest increases in FoxO levels have been reported to be cardioprotective in age-associated functional declines in Drosophila,66 increasing atrogin-1 may be one way to support FoxO1/FoxO3a activity to protect against age-associated declines in cardiac function.
Age-related sarcopenia is the decline of muscle mass and strength with age and involves decrease in insulin-like growth factor-1 (IGF-1) signaling, which is required for maintenance of muscle mass in the aging process.67 Age-related decreases in IGF-1 promote phosphatidylinositol 3-kinase, mitogen-activated protein kinase, and calcineurin pathways, and exercise and injury induction of IGF-1 protect against age-related sarcopenia.67 The IGF-1 and the IGF-1 receptor (IGF-1R) are implicated in cardiac aging and longevity.68 In the present study, atrogin-1 Tg+ hearts have enhanced FoxO1/3a transcription factor activities (Figure 6B), which would predictably inhibit IGF-1 signaling, as previously described in atrogin-1 Tg+.31 The observed decreases in cardiac mass (Figure 1E) likely reflect an enhanced age-related loss of cardiac muscle (sarcopenia) mediated by these effects. The significant increase in atrogin-1 Tg+ LV mass index normalized to tibia length (Table 1) and cardiomyocyte cross-sectional area (Figure 2B) suggests some compensatory increases in LV muscle despite the overall loss of muscle mass. IGF-1 signaling has important roles in cardiac aging.68 Deletion of IGF-1R has been shown to delay the development of senescence-associated pathologies.69 Specifically, aging is associated with the induction of IGF-1R in hearts, resulting in age-associated cardiac hypertrophy.69 Deletion of the IGF-1R resulted in reduced hypertrophy, and fibrosis was reduced in aged IGF-1R−/− mice compared with aged wild-type controls.69 Furthermore, IGF-1R−/− hearts had an attenuation of age-related proinflammatory cytokines (IL-1α, IL-1β, IL-6, and receptor activator of nuclear factor κ-B ligand).69 In cultured cardiomyocytes, IGF-1 induction of senescence evidence has been reported, evidenced by increased β-galactosidase staining (inhibited by phosphoinositide 3-kinase inhibitor).69 Despite these potential benefits of inhibiting IGF-1 signaling in the atrogin-1 Tg+ hearts, IGF-1 receptor haploinsufficiency can contribute to age-dependent development of metabolic syndrome in skeletal muscle, so it should be considered as a system-wide target to aging-associated changes in striated muscle.70
The role of atrogin-1 in regulating autophagy was recently discovered, with atrogin-1−/− mouse heart developing cardiomyopathy and premature death, including significantly increased fibrosis.17 In these studies, in vivo pulsing of stable isotope labeling amino acids in cell culture proteomics revealed increases in the CHMP2B, which is part of the endosomal sorting complex required for autophagy.17 Atrogin-1−/− mice with increased CHMP2B protein had impaired autophagy, accumulation of aggregates, and activation of the unfolded protein response, which increased progressively with age.17 In the current study, we identified that atrogin-1 Tg+ hearts have significant increases in CHMP2B protein in 18-month–old mouse hearts (Figure 6C), which may indicate that the enhanced degradation expected with increased atrogin-1 is counteracted by other regulatory mechanisms. The increased protein levels may be because of increased transcription, and not enhanced degradation, because multiple transcription factors have binding sites in the CHMP2B gene promoter, including AREB6, POU3F1, USF1, USF2, Sox9, and POU3F2 (GeneCards identification: GC03P087277). Although there is limited information on any links between autophagy and fibrosis, the present study may be the first to demonstrate a link between the increased atrogin-1 Tg+ mediated CHMP2B protein levels with the observed decrease in collagen accumulation. In support of this notion, liver fibrosis was suppressed by the direct inhibition of Beclin1-mediated autophagy.71
Cardiomyocyte proliferation and growth during development is regulated by FoxO1/FoxO3 transcription factors.72 However, myocyte-specific transgenic expression of FoxO1 during heart development leads to decreased myocyte proliferation and lethality.72 In contrast, dominant negative FoxO1 in cardiomyocytes leads to increased myocyte proliferation.72 In the present study, we identified a significant increase in atrogin-1 Tg+ nuclei density compared with wild-type hearts (Figure 2, E and F) without alterations in fibroblasts (Figure 3). Because the α-major histocompatibility complex expression predominates after birth, at which stage myocytes are binucleate,73, 74, 75 the effects of increasing atrogin-1 with age may alter cardiomyocyte number in an unpredictable manner.
Acknowledgments
We thank Dr. William Claycomb for gifting the HL-1 cells used in this study and for guidance in detailing their care and use, Dawud Hilliard (University of North Carolina Lineberger Center Animal Histopathology Laboratory) for preparation and staining of the histologic specimens, and Bentley Midkiff (University of North Carolina Translational Pathology Laboratory) for assistance in digitally scanning slides with Aperio VERSA Digital Pathology Scanner and for the histologic nuclei quantification analysis performed.
Footnotes
Supported by the Jefferson-Pilot Corporation Fellowship in Academic Medicine (M.S.W.), the NIH National Heart, Lung, and Blood Institute grant R01HL104129 (M.S.W.), the Leducq Foundation grant 11CVD04 (M.S.W. and C.P.), and the China Scholarship Council State Scholarship Fund (Z.Q.).
R.M. and T.L.P. contributed equally to this work.
Disclosure: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.04.007.
Supplemental Data
Supplemental Figure S1.
Immunofluorescence (IF) analysis of vimentin in atrogin-1 Tg+ histologic sections. A: Atrogin-1 fibroblast areas were accessed for colocalized vimentin-positive cells by immunohistochemistry in four quadrants (Q1–Q4) as oriented to the right and left ventricles (RV and LV). B: Representative vimentin (green) and DAPI (blue) counterstaining of atrogin-1 Tg+ and wild-type histologic sections in four quadrants (Q1–Q4). Original magnification, ×20 (B).
Supplemental Figure S2.
Increasing atrogin-1 and its effect on steady-state glucocorticoid receptor (GR) levels in HL-1 cells by immunoblot (IB) analysis. A: Representative immunoblot of atrogin-1, GR, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in cardiomyocyte-derived HL-1 cells. B: Densitometric analysis of atrogin-1 protein levels with increasing levels of adenovirus expressing atrogin-1 (Ad-Atrogin-1) multiplicity of infection (MOI). C: Densitometric analysis of GR protein levels. Data are expressed as means ± SEM (B and C). n = 3 biological replicates per group. AU, arbitrary unit.
Supplemental Figure S3.
Analysis of glucocorticoid receptor–regulated genes (and proteins) in atrogin-1 Tg+ hearts at 18 months of age by real-time quantitative PCR. A–E: Relative quantity of dystrophin (Dmd; A), Kruppel-like factor 15 (Klf15; B), ryanodine receptor 2 (Ryr2; C), nuclear receptor subfamily 3 group C member 1 (Nr3c1; D), and prostaglandin D2 synthase (Ptgds; E) mRNA. A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM. n = 4 biological replicates per group.
Protein expression in 18-month–old atrogin-1 Tg+ hearts. Immunoblot (IB; A) and densitometric (B) analysis of atrogin-1 (C), tissue inhibitor of metalloproteases-1 (TIMP-1; D), TIMP-2 (E), and TIMP-3 (F) in atrogin-1 Tg+ and age-matched wild-type hearts. A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM (B, D, E, and F). n = 4 biological replicates per group (). AU, arbitrary unit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Supplemental Figure S5.
Protein turnover in adult and aged atrogin-1 Tg+ hearts. Immunoblot (IB) and densitometric analysis of puromycin (protein turnover) in adult 6-month–old (A) and aged 16-month–old (B) atrogin-1 Tg+ and litter-matched wild-type hearts. A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM. n = 4 (A, adult mice: wild type); n = 5 (A, adult mice: atrogin-1 Tg+); n = 2 (B, aged mice: wild type and atrogin-1 Tg+).
Supplemental Figure S6.
Serum chemistry in atrogin-1 Tg+ mice. Analysis of serum alanine aminotransferase (ALT; A), alkaline phosphatase (ALP; B), blood urine nitrogen (BUN; C), and creatinine (D). A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM. n = 3 (wild type); n = 8 (atrogin-1 Tg+).
References
- 1.Steenman M., Lande G. Cardiac aging and heart disease in humans. Biophys Rev. 2017;9:131–137. doi: 10.1007/s12551-017-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu B., Daimon M. Cardiac aging phenomenon and its clinical features by echocardiography. J Echocardiogr. 2016;14:139–145. doi: 10.1007/s12574-016-0292-6. [DOI] [PubMed] [Google Scholar]
- 3.Barger J.L., Kayo T., Vann J.M., Arias E.B., Wang J., Hacker T.A., Wang Y., Raederstorff D., Morrow J.D., Leeuwenburgh C., Allison D.B., Saupe K.W., Cartee G.D., Weindruch R., Prolla T.A. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns M., Gondro C., Tellam R.L., Radley-Crabb H.G., Grounds M.D., Shavlakadze T. Molecular analyses provide insight into mechanisms underlying sarcopenia and myofibre denervation in old skeletal muscles of mice. Int J Biochem Cell Biol. 2014;53:174–185. doi: 10.1016/j.biocel.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Rivas D.A., Lessard S.J., Rice N.P., Lustgarten M.S., So K., Goodyear L.J., Parnell L.D., Fielding R.A. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014;28:4133–4147. doi: 10.1096/fj.14-254490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang S.H., Lee H.A., Kim M., Lee E., Sohn U.D., Kim I. Forkhead box O3 plays a role in skeletal muscle atrophy through expression of E3 ubiquitin ligases Murf-1 and Atrogin-1 in Cushing's syndrome. Am J Physiol Endocrinol Metab. 2017;312:E495–E507. doi: 10.1152/ajpendo.00389.2016. [DOI] [PubMed] [Google Scholar]
- 7.Willis M.S., Schisler J.C., Patterson C. Appetite for destruction: E3 ubiquitin-ligase protection in cardiac disease. Future Cardiol. 2008;4:65–75. doi: 10.2217/14796678.4.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodine S.C., Baehr L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab. 2014;307:E469–E484. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altun M., Besche H.C., Overkleeft H.S., Piccirillo R., Edelmann M.J., Kessler B.M., Goldberg A.L., Ulfhake B. Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J Biol Chem. 2010;285:39597–39608. doi: 10.1074/jbc.M110.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertaggia E., Coletto L., Sandri M. Posttranslational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol. 2012;302:C587–C596. doi: 10.1152/ajpcell.00142.2011. [DOI] [PubMed] [Google Scholar]
- 12.Pomies P., Blaquiere M., Maury J., Mercier J., Gouzi F., Hayot M. Involvement of the FoxO1/MuRF1/Atrogin-1 signaling pathway in the oxidative stress-induced atrophy of cultured chronic obstructive pulmonary disease myotubes. PLoS One. 2016;11:e0160092. doi: 10.1371/journal.pone.0160092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavazis A.N., Smuder A.J., Powers S.K. Effects of short-term endurance exercise training on acute doxorubicin-induced FoxO transcription in cardiac and skeletal muscle. J Appl Physiol (1985) 2014;117:223–230. doi: 10.1152/japplphysiol.00210.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paula-Gomes S., Goncalves D.A., Baviera A.M., Zanon N.M., Navegantes L.C., Kettelhut I.C. Insulin suppresses atrophy- and autophagy-related genes in heart tissue and cardiomyocytes through AKT/FOXO signaling. Horm Metab Res. 2013;45:849–855. doi: 10.1055/s-0033-1347209. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds T.H., Merrell E., Cinquino N., Gaugler M., Ng L. Disassociation of insulin action and Akt/FOXO signaling in skeletal muscle of older Akt-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2012;303:R1186–R1194. doi: 10.1152/ajpregu.00358.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng B., Ohkawa S., Li H., Roberts-Wilson T.K., Price S.R. FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. FASEB J. 2010;24:2660–2669. doi: 10.1096/fj.09-151480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaglia T., Milan G., Ruhs A., Franzoso M., Bertaggia E., Pianca N., Carpi A., Carullo P., Pesce P., Sacerdoti D., Sarais C., Catalucci D., Kruger M., Mongillo M., Sandri M. Atrogin-1 deficiency promotes cardiomyopathy and premature death via impaired autophagy. J Clin Invest. 2014;124:2410–2424. doi: 10.1172/JCI66339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel E.D., Doenst T. Mitochondrial adaptations to physiological vs. pathological cardiac hypertrophy. Cardiovasc Res. 2011;90:234–242. doi: 10.1093/cvr/cvr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamilpa R., Ibarra J., de Castro Bras L.E., Ramirez T.A., Nguyen N., Halade G.V., Zhang J., Dai Q., Dayah T., Chiao Y.A., Lowell W., Ahuja S.S., D'Armiento J., Jin Y.F., Lindsey M.L. Transgenic overexpression of matrix metalloproteinase-9 in macrophages attenuates the inflammatory response and improves left ventricular function post-myocardial infarction. J Mol Cell Cardiol. 2012;53:599–608. doi: 10.1016/j.yjmcc.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sternlicht M.D., Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meschiari C.A., Ero O.K., Pan H., Finkel T., Lindsey M.L. The impact of aging on cardiac extracellular matrix. Geroscience. 2017;39:7–18. doi: 10.1007/s11357-017-9959-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu T., Narang N., Chen P., Yu B., Knapp M., Janardanan J., Blair J., Liao J.K. Fibroblast deletion of ROCK2 attenuates cardiac hypertrophy, fibrosis, and diastolic dysfunction. JCI Insight. 2017 doi: 10.1172/jci.insight.93187. [Epub ahead of print] doi: 10.1172/jci.insight.93187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al Darazi F., Zhao W., Zhao T., Sun Y., Marion T.N., Ahokas R.A., Bhattacharya S.K., Gerling I.C., Weber K.T. Small dedifferentiated cardiomyocytes bordering on microdomains of fibrosis: evidence for reverse remodeling with assisted recovery. J Cardiovasc Pharmacol. 2014;64:237–246. doi: 10.1097/FJC.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H.H., Kedar V., Zhang C., McDonough H., Arya R., Wang D.Z., Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana M.T., Parry T.L., He J., Yates C.C., Sidorova T.N., Murray K.T., Bain J.R., Newgard C.B., Muehlbauer M.J., Eaton S.C., Hishiya A., Takayama S., Willis M.S. Cardiomyocyte-specific human Bcl2-associated anthanogene 3 P209L expression induces mitochondrial fragmentation, Bcl2-associated anthanogene 3 haploinsufficiency, and activates p38 signaling. Am J Pathol. 2016;186:1989–2007. doi: 10.1016/j.ajpath.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willis M.S., Wadosky K.M., Rodriguez J.E., Schisler J.C., Lockyer P., Hilliard E.G., Glass D.J., Patterson C. Muscle ring finger 1 and muscle ring finger 2 are necessary but functionally redundant during developmental cardiac growth and regulate E2F1-mediated gene expression in vivo. Cell Biochem Funct. 2014;32:39–50. doi: 10.1002/cbf.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feist P., Hummon A.B. Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int J Mol Sci. 2015;16:3537–3563. doi: 10.3390/ijms16023537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risling T.E., Caulkett N.A., Florence D. Open-drop anesthesia for small laboratory animals. Can Vet J. 2012;53:299–302. [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman C.A., Hornberger T.A. Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc Sport Sci Rev. 2013;41:107–115. doi: 10.1097/JES.0b013e3182798a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wadosky K.M., Rodriguez J.E., Hite R.L., Min J.N., Walton B.L., Willis M.S. Muscle RING finger-1 attenuates IGF-I-dependent cardiomyocyte hypertrophy by inhibiting JNK signaling. Am J Physiol Endocrinol Metab. 2014;306:E723–E739. doi: 10.1152/ajpendo.00326.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H.H., Willis M.S., Lockyer P., Miller N., McDonough H., Glass D.J., Patterson C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jazbutyte V., Fiedler J., Kneitz S., Galuppo P., Just A., Holzmann A., Bauersachs J., Thum T. MicroRNA-22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordr) 2013;35:747–762. doi: 10.1007/s11357-012-9407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collier P., Watson C.J., van Es M.H., Phelan D., McGorrian C., Tolan M., Ledwidge M.T., McDonald K.M., Baugh J.A. Getting to the heart of cardiac remodeling: how collagen subtypes may contribute to phenotype. J Mol Cell Cardiol. 2012;52:148–153. doi: 10.1016/j.yjmcc.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Naugle J.E., Olson E.R., Zhang X., Mase S.E., Pilati C.F., Maron M.B., Folkesson H.G., Horne W.I., Doane K.J., Meszaros J.G. Type VI collagen induces cardiac myofibroblast differentiation: implications for postinfarction remodeling. Am J Physiol Heart Circ Physiol. 2006;290:H323–H330. doi: 10.1152/ajpheart.00321.2005. [DOI] [PubMed] [Google Scholar]
- 35.Fitzgerald J., Rich C., Zhou F.H., Hansen U. Three novel collagen VI chains, alpha4(VI), alpha5(VI), and alpha6(VI) J Biol Chem. 2008;283:20170–20180. doi: 10.1074/jbc.M710139200. [DOI] [PubMed] [Google Scholar]
- 36.Reiter E., Jiang Q., Christen S. Anti-inflammatory properties of alpha- and gamma-tocopherol. Mol Aspects Med. 2007;28:668–691. doi: 10.1016/j.mam.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oakley R.H., Ren R., Cruz-Topete D., Bird G.S., Myers P.H., Boyle M.C., Schneider M.D., Willis M.S., Cidlowski J.A. Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proc Natl Acad Sci U S A. 2013;110:17035–17040. doi: 10.1073/pnas.1302546110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren R., Oakley R.H., Cruz-Topete D., Cidlowski J.A. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology. 2012;153:5346–5360. doi: 10.1210/en.2012-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace A.D., Cidlowski J.A. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem. 2001;276:42714–42721. doi: 10.1074/jbc.M106033200. [DOI] [PubMed] [Google Scholar]
- 40.Tintignac L.A., Lagirand J., Batonnet S., Sirri V., Leibovitch M.P., Leibovitch S.A. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem. 2005;280:2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- 41.Lagirand-Cantaloube J., Cornille K., Csibi A., Batonnet-Pichon S., Leibovitch M.P., Leibovitch S.A. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLoS One. 2009;4:e4973. doi: 10.1371/journal.pone.0004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mearini G., Gedicke C., Schlossarek S., Witt C.C., Kramer E., Cao P., Gomes M.D., Lecker S.H., Labeit S., Willis M.S., Eschenhagen T., Carrier L. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc Res. 2010;85:357–366. doi: 10.1093/cvr/cvp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stitt T.N., Drujan D., Clarke B.A., Panaro F., Timofeyva Y., Kline W.O., Gonzalez M., Yancopoulos G.D., Glass D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 44.Sacheck J.M., Ohtsuka A., McLary S.C., Goldberg A.L. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–E601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 45.Willis M.S., Schisler J.C., Li L., Rodriguez J.E., Hilliard E.G., Charles P.C., Patterson C. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ Res. 2009;105:80–88. doi: 10.1161/CIRCRESAHA.109.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattox T.A., Young M.E., Rubel C.E., Spaniel C., Rodriguez J.E., Grevengoed T.J., Gautel M., Xu Z., Anderson E.J., Willis M.S. MuRF1 activity is present in cardiac mitochondria and regulates reactive oxygen species production in vivo. J Bioenerg Biomembr. 2014;46:173–187. doi: 10.1007/s10863-014-9549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Yacoub N., Shaheen R., Awad S.M., Kunhi M., Dzimiri N., Nguyen H.C., Xiong Y., Al-Buraiki J., Al-Habeeb W., Alkuraya F.S., Poizat C. FBXO32, encoding a member of the SCF complex, is mutated in dilated cardiomyopathy. Genome Biol. 2016;17:2. doi: 10.1186/s13059-015-0861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagase H., Visse R., Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Brew K., Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toba H., Cannon P.L., Yabluchanskiy A., Iyer R.P., D'Armiento J., Lindsey M.L. Transgenic overexpression of macrophage matrix metalloproteinase-9 exacerbates age-related cardiac hypertrophy, vessel rarefaction, inflammation, and fibrosis. Am J Physiol Heart Circ Physiol. 2017;312:H375–H383. doi: 10.1152/ajpheart.00633.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harty M.W., Huddleston H.M., Papa E.F., Puthawala T., Tracy A.P., Ramm G.A., Gehring S., Gregory S.H., Tracy T.F., Jr. Repair after cholestatic liver injury correlates with neutrophil infiltration and matrix metalloproteinase 8 activity. Surgery. 2005;138:313–320. doi: 10.1016/j.surg.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Sun X., Chen W.D., Wang Y.D. DAF-16/FOXO transcription factor in aging and longevity. Front Pharmacol. 2017;8:548. doi: 10.3389/fphar.2017.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lapierre L.R., Kumsta C., Sandri M., Ballabio A., Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–880. doi: 10.1080/15548627.2015.1034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McElwee J., Bubb K., Thomas J.H. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 55.Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 56.Dong M.Q., Venable J.D., Au N., Xu T., Park S.K., Cociorva D., Johnson J.R., Dillin A., Yates J.R., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 57.Schuster E., McElwee J.J., Tullet J.M., Doonan R., Matthijssens F., Reece-Hoyes J.S., Hope I.A., Vanfleteren J.R., Thornton J.M., Gems D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y.H., Zhang G.G. Towards understanding the lifespan extension by reduced insulin signaling: bioinformatics analysis of DAF-16/FOXO direct targets in Caenorhabditis elegans. Oncotarget. 2016;7:19185–19192. doi: 10.18632/oncotarget.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang H., Regan K.M., Wang F., Wang D., Smith D.I., van Deursen J.M., Tindall D.J. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato S., Ding J., Pisck E., Jhala U.S., Du K. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J Biol Chem. 2008;283:35464–35473. doi: 10.1074/jbc.M801011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J.Y., Zong C.S., Xia W., Yamaguchi H., Ding Q., Xie X., Lang J.Y., Lai C.C., Chang C.J., Huang W.C., Huang H., Kuo H.P., Lee D.F., Li L.Y., Lien H.C., Cheng X., Chang K.J., Hsiao C.D., Tsai F.J., Tsai C.H., Sahin A.A., Muller W.J., Mills G.B., Yu D., Hortobagyi G.N., Hung M.C. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li F., Xie P., Fan Y., Zhang H., Zheng L., Gu D., Patterson C., Li H. C terminus of Hsc70-interacting protein promotes smooth muscle cell proliferation and survival through ubiquitin-mediated degradation of FoxO1. J Biol Chem. 2009;284:20090–20098. doi: 10.1074/jbc.M109.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu M.C., Lee D.F., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Kobayashi R., Hung M.C. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 64.Takahashi Y., Daitoku H., Hirota K., Tamiya H., Yokoyama A., Kako K., Nagashima Y., Nakamura A., Shimada T., Watanabe S., Yamagata K., Yasuda K., Ishii N., Fukamizu A. Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF-16. Cell Metab. 2011;13:505–516. doi: 10.1016/j.cmet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 65.Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., Fukamizu A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Blice-Baum A.C., Zambon A.C., Kaushik G., Viswanathan M.C., Engler A.J., Bodmer R., Cammarato A. Modest overexpression of FOXO maintains cardiac proteostasis and ameliorates age-associated functional decline. Aging Cell. 2017;16:93–103. doi: 10.1111/acel.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adamo M.L., Farrar R.P. Resistance training, and IGF involvement in the maintenance of muscle mass during the aging process. Ageing Res Rev. 2006;5:310–331. doi: 10.1016/j.arr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Lee W.S., Kim J. Insulin-like growth factor-1 signaling in cardiac aging. Biochim Biophys Acta. 2017;1864:1931–1938. doi: 10.1016/j.bbadis.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 69.Ock S., Lee W.S., Ahn J., Kim H.M., Kang H., Kim H.S., Jo D., Abel E.D., Lee T.J., Kim J. Deletion of IGF-1 receptors in cardiomyocytes attenuates cardiac aging in male mice. Endocrinology. 2016;157:336–345. doi: 10.1210/en.2015-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thakur S., Garg N., Zhang N., Hussey S.E., Musi N., Adamo M.L. IGF-1 receptor haploinsufficiency leads to age-dependent development of metabolic syndrome. Biochem Biophys Res Commun. 2017;486:937–944. doi: 10.1016/j.bbrc.2017.03.129. [DOI] [PubMed] [Google Scholar]
- 71.Chen J., Yu Y., Li S., Liu Y., Zhou S., Cao S., Yin J., Li G. MicroRNA-30a ameliorates hepatic fibrosis by inhibiting Beclin1-mediated autophagy. J Cell Mol Med. 2017;21:3679–3692. doi: 10.1111/jcmm.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Evans-Anderson H.J., Alfieri C.M., Yutzey K.E. Regulation of cardiomyocyte proliferation and myocardial growth during development by FOXO transcription factors. Circ Res. 2008;102:686–694. doi: 10.1161/CIRCRESAHA.107.163428. [DOI] [PubMed] [Google Scholar]
- 73.Soonpaa M.H., Field L.J. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 74.Soonpaa M.H., Kim K.K., Pajak L., Franklin M., Field L.J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 75.Weiss A., Leinwand L.A. The mammalian myosin heavy chain gene family. Annu Rev Cell Dev Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein expression in 18-month–old atrogin-1 Tg+ hearts. Immunoblot (IB; A) and densitometric (B) analysis of atrogin-1 (C), tissue inhibitor of metalloproteases-1 (TIMP-1; D), TIMP-2 (E), and TIMP-3 (F) in atrogin-1 Tg+ and age-matched wild-type hearts. A two-tailed t-test was performed to determine significance between groups. Data are expressed as means ± SEM (B, D, E, and F). n = 4 biological replicates per group (). AU, arbitrary unit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.