Abstract
We have identified Klp2p, a new kinesin-like protein (KLP) of the KAR3 subfamily in fission yeast. The motor domain of this protein is 61% identical and 71% similar to Pkl1p, another fission yeast KAR3 protein, yet the two enzymes are different in behavior and function. Pkl1p is nuclear throughout the cell cycle, whereas Klp2p is cytoplasmic during interphase. During mitosis Klp2p enters the nucleus where it forms about six chromatin-associated dots. In metaphase-arrested cells these migrate back and forth across the nucleus. During early anaphase they segregate with the chromosomes into two sets of about three, fade, and are replaced by other dots that form on the spindle interzone. Neither klp2+ nor pkl1+ is essential, and the double deletion is also wild type for both vegetative and sexual reproduction. Each deletion rescues different alleles of cut7ts, a KLP that contributes to spindle formation and elongation. When either or both deletions are combined with a dynein deletion, vegetative growth is normal, but sexual reproduction fails: klp2Δ,dhc1-d1 in karyogamy, pkl1Δ,dhc1-d1 in multiple phases of meiosis, and the triple deletion in both. Deletion of Klp2p elongates a metaphase-arrested spindle, but pkl1Δ shortens it. The anaphase spindle of klp2Δ becomes longer than the cell, leading it to curl around the cell's ends. Apparently, Klp2p promotes spindle disassembly and contributes to the behavior of mitotic chromosomes.
INTRODUCTION
The accurate and timely segregation of chromosomes is an essential aspect of mitotic cell division. Such chromosome motion is dependent on the activity of the mitotic microtubules (MTs) and requires several MT-dependent force-generating mechanisms. Experiments in diverse cells have demonstrated that kinesin-like proteins (KLPs) are important for mitotic spindle formation and the organized segregation of chromosomes (reviewed in Hoyt et al., 1997; Endow, 1999; Sharp et al., 2000b). Both plus- and minus-end–directed KLPs contribute to mitotic movements, and in some organisms cytoplasmic dynein is also involved (reviewed in Saunders et al., 1995; Merdes et al., 1996; Starr et al., 1998; O'Connell and Wang, 2000; Hildebrandt and Hoyt, 2000). Motors can also affect tubulin dynamics, for example, KAR3 acts to destabilize MT minus-ends (Endow et al., 1994) and XKCM1 effects general MT disassembly (Walczak et al.,1996). Indeed, mitosis seems to depend on both the coordination of multiple motor enzymes (Sharp et al., 2000a) and the factors that control tubulin polymerization and depolymerization (Inoue, 1997). For example, spindle forces generated by plus-end–directed KLPs of the BimC subfamily and various minus-end–directed motors coordinate the separation of centrosomes during prophase (Sharp et al., 2000b). In Aspergillus nidulans, the bimC4ts allele is rescued by deletion of the KAR3 homologue KlpA (O'Connell et al., 1993). In Drosophila, plus-end–directed Klp61F and minus-end–directed dynein are thought to move the poles apart, whereas minus-end ncd pulls the poles together to achieve a metaphase spindle (Sharp et al., 1999, 2000a). In mammals, plus-end–directed Eg5 is balanced by two minus-end motors, the KAR3 homologue HSET (Mountain et al., 1999) and the dynein–dynactin complex (Gaglio et al., 1996).
To understand the complexity of mitosis one wants experimental systems that permit detailed, rigorous and informative study. We have selected the fission yeast Schizosaccharomyces pombe for our research on chromosome motion because of its suitability for molecular and genetic work (Moreno et al., 1991) and its useful mitotic cytology (Hagan and Hyams, 1988; Ding et al., 1993, 1997; Hagan, 1998; Nabeshima et al., 1998). It carries only three comparatively large chromosomes, each of which contains a centromere that includes many tens of kilobases (kb) of DNA (Fishel et al., 1988; Chikashige et al., 1989). In this way it is more similar than budding yeast to other eukaryotic cells (reviewed in Clarke, 1990), and it constitutes an attractive model for the study of chromosome–MT interactions.
Two MT-dependent motor enzymes that may be important for fission yeast mitosis have already been described. Cut7p is a KLP of the BimC family that is essential for spindle formation in prophase and spindle elongation in anaphase B (Hagan and Yanagida, 1992). Pkl1p is a KLP of the KAR3 family that is not essential but localizes to the nucleus during interphase and to the spindle throughout mitosis; further, deletion of pkl1+ is known to rescue some alleles of cut7ts (Pidoux et al., 1996). S. pombe expresses one dynein heavy chain (Yamamoto et al., 1999), and although this motor is essential for nuclear migration during the sexual phase of the life cycle, it plays no detectable role in mitosis. Klp3p (Brazer et al., 2000) and Klp4p, also known as Tea2p (Browning et al., 2000) are cytoplasmic motors in S. pombe that have no detectable role in mitosis, nor are their cytoplasmic functions essential. Given the plethora of motors in other organisms and the fact that several organisms have more than one KLP from the KAR3 subfamily (reviewed in Endow, 1999), we have sought additional KLPs in fission yeast that might function together with Cut7p and Pkl1p in mitosis.
A PCR screen was used to search for KAR3 homologues in S. pombe. With this screen we rediscovered Pkl1p and found a new member of the KAR3 family, Klp2p. This motor, too, is not essential for growth, but its genetic interactions and localization at different times in the cell cycle implicate it both in the establishment and maintenance of the bipolar spindle and as a kinetochore-associated KLP that promotes MT shortening. Furthermore, Klp2p, together with Dhc1p, is essential for karyogamy in the formation of zygotes.
MATERIALS AND METHODS
Strains and Cell Culture
Strains were constructed and maintained as detailed in Moreno et al. (1991). Cultures were grown in rich medium containing yeast extract plus supplements (YES) or in Edinburgh minimal medium (EMM; Moreno et al., 1991) supplemented as appropriate. To compare the growth of different klp mutant strains by plating assays, we placed 4 × 107 cells of each strain in the first well of a row in a 96-well plate and serially diluted them: 1:10, 1:10, 1:5, 1:5, 1:5. Cells were then stamped in triplicate onto YES plates or YES plates containing 10 μg/ml thiabendazole (TBZ), an MT-depolymerizing drug, and incubated at appropriate temperatures until significant growth occurred for at least one strain. The wild-type strain used for comparison was ade6-M216,leu1-32,h+ (PN 69), and mutant strains were isogenic to the wild-type strain with respect to these nutritional markers. The growth of pkl1Δ and klp2Δ strains was examined at 20, 25, 29, 32, and 36°C. Restrictive temperature for cut7tsstrains was 36°C, a temperature at which pkl1Δ by itself shows no increased sensitivity to TBZ. The restrictive temperature for cut11ts was 29°C. All plating assays were repeated at least twice, with the use of cell cultures grown on different days. Cells were prepared for analysis by flow cytometry essentially as described (Sazer and Sherwood, 1990), except that a buffer of 0.2 M Tris-HCl, pH 7.5, 20 mM EDTA was used instead of the 50 mM Na citrate solution.
The success of zygotic meioses for various crosses was assessed in two ways. First, the number of spores per ascus was determined by counting in a hemacytometer. Second, spore viability was determined by tetrad analysis. Crosses performed at 29°C (the standard temperature) were allowed to progress 2 d. Crosses performed at 25 or 20°C were allowed to progress 3 or 7 d, respectively. Tetrads were dissected on YES plates and incubated for 2–3 d at 32°C to determine spore viability. All tetrad counts were repeated two to four times, and 200–400 spores were examined per count. To compare the success of azygotic meiosis in wild type with those in klp2Δ/klp2Δ; dhc1-d1/dhc1-d1 diploids, the number of spores per ascus and spore viability were determined as described for zygotic crosses. Observations on wild-type diploids were carried out twice, and the observations of mutant diploids three times.
PCR Strategy for Identifying KLPs from the Kar3 Subfamily
Degenerate primers encoding conserved portions of the kinesin motor domain were used to amplify genomic DNA prepared as described in Moreno et al. (1991). The 5′ primer was 5′-ATHTTYGCNTAYGGNCARWC-3′, which encodes IFAYGQT, and the 3′ primer was 5′- GCGCGAATTCNTCRTTRTADATYTC-3′, which encodes EIYND/E (where H is A,C,T; N is A,C,T,G; R is G,A; W is A,T and Y is C,T). The latter primer was optimized to match known KLPs with motors located at their C terminus, including Kar3p (Saccharomyces cerevisiae) and KlpAp (A. nidulans). PCR amplifications were performed on ca. 10 ng genomic DNA as follows: 94°C 3 min; add Taq polymerase; one cycle of 94°C for 60 s, 30°C 90 s, 60 s ramp to 72°C; 5 cycles of 92°C 60 s, 47°C 90 s, 60 s ramp to 72°C, 72°C 60 s; 35 cycles of 94°C 60 s, 50°C 90 s, 60 s ramp to 72°C, 72°C 60 s; and then 72°C 15 min. PCR products of ≥180 nt were selected for further analysis. Vectors with inserts were prepared and sequenced according to standard methods (Sambrook et al., 1989). Fragments of two novel KLP genes were identified in this screen and named klp1+ and klp2+.
Cloning and Mapping of pkl1+ and klp2+
Standard molecular methods (Sambrook et al., 1989) were used to clone and characterize the corresponding genes, except when noted otherwise. An S. pombe genomic DNA library (generously provided by Dr. A. Carr; Barbet et al., 1992) was screened with the two PCR fragments, essentially as described by Woods (1984). Two unique genomic clones, ca. 6.9 and 7.3 kb, were isolated. Sequence from both ends of each insert showed that they encoded full-length copies of klp1+and klp2+, respectively. klp1+ was identical to pkl1+, which has already been described (Pidoux et al., 1996), so this name will be used henceforth. pkl1+and klp2+ were mapped to the left arm of chromosome I by probing a filter containing ordered arrays of cosmids (Hoheisel et al., 1993), with the use of the original PCR products and with the kind assistance of Dr. Elmar Maier (Imperial Cancer Research Fund, Genome Analysis Laboratory, London).
Molecular Characterization of klp2+
An apparent 5′-intron in klp2+ (see Figure 1A) was confirmed by amplifying the corresponding region from a cDNA library (generous gift from C. Norbury, Imperial Cancer Research Fund, London, United Kingdom) with the use of the 5′ primer 5′- GGAAGAAGAAGGACATA-3′ and the 3′ primer 5′- AAGAACTCGAGGACTGA-3′ and then sequencing the resulting PCR product directly, with the use of automated sequencing. Total RNA was isolated from S. pombe cells according to Moreno et al. (1991), and polyA+ fractions were selected with the use of oligo(dT) cellulose columns (GIBCO BRL, Rockville MD) according to the manufacturer's directions. Northern blots were performed with the use of formaldehyde-agarose gels with 1 μg of RNA per lane, probed with the 32P-labeled open reading frame (ORF) of klp2+, which was amplified by PCR with the use of the 5′ primer 5′-ATGTCGACAGAAGAAGAAGGACATAAAAGTTTA-3′ and the 3′ primer 5′-GAAGATCTTCATTTTGTGACTTTGCGTGCTGT-3′. A message of ca. 3.5 kb was detected, consistent with a single transcript from klp2+ (our unpublished observations).
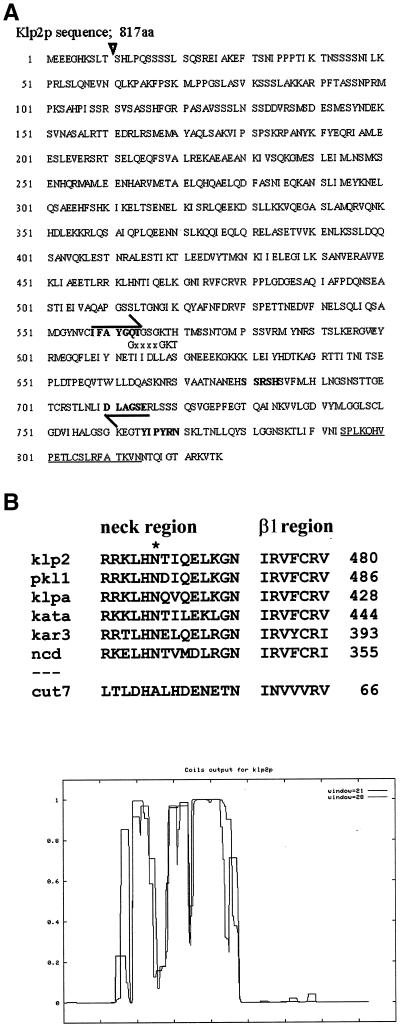
Figure 1.
(A) The DNA sequence of klp2+ encodes a polypeptide of 817 amino acids that contains motifs characteristic of KLPs (bold sequences). The sequences used to design degenerate primers for PCR are indicated by arrows. The position of an intron is shown by a ▿, a putative MT-binding domain is underlined, and the “P-loop” for nucleotide binding is indicated by its consensus sequence, GxxxGKT. (B) klp2+ and pkl1+ both contain a sequence that is conserved among members of the KAR3 family (O'Connell et al., 1993). The sequence has been determined to include a neck region that contains an N residue (*) important for motor directionality and the β1 region of the motor with which it may interact (Endow and Higuchi, 2000). KlpA is from A. nidulans; Kata is from A. thaliana; and ncd is from D. melanogaster. The corresponding part of Cut7p, a fission yeast KLP of the BimC family that lacks the neck region, is shown for comparison. (C) The p that different parts of Klp2p form coiled-coils was predicted by COILS (Lupas, 1996).
Computer-aided Sequence Analysis
Direct sequence comparisons were made with the Bestfit program from the Wisconsin Package from the Genetics Computer Group (GCG; Madison, WI). Comparisons of the motor heads were from the Kar3 consensus through the 3′ ends of the peptides. Sequences compared included A. nidulans KlpA (O'Connell et al., 1993), S. cerevisiae KAR3 (Meluh and Rose, 1990), and Drosophila melanogaster ncd (McDonald et al., 1990). Coiled-coil predictions were made with the use of the COILS program at http://www.ch.embnet.org/software/COILS_form.html (Lupas, 1996). The Kar3p consensus regions of several motors (O'Connell et al., 1993) were aligned by the PILEUP program available from GCG.
Disruption of klp2+
A complete disruption of the pkl1+gene has been made with the use of the genomic klp1+ clone described above (Pidoux et al., 1996). A complete replacement of the klp2+ by ura4+ was made by a single-step gene replacement protocol (Rothstein, 1991), with the use of a cassette containing two genomic regions flanking the either side of the klp2+ gene and ura4+ as the selectable marker. A 2900-nt PstI-HindIII fragment corresponding to the sequence −2900 to −68 nt upstream of the klp2+ ORF was subcloned into the PstI-HindIII site of the bacterial cloning vector pSPORT1 (GIBCO BRL), which had been modified to carry the ura4+ gene (pSPORT1-URA4; West et al., 1998). A 1.3-kb BglII-HindIII fragment corresponding to the sequence +83 nt to +1043 nt downstream of the klp2+ORF was blunt-ended and subcloned into the KpnI site of pSPORT1-URA4 with the aid of KpnI linkers. A subclone, pKLP2KO, containing the fragment of the correct orientation was identified by restriction analysis. The cassette was excised from pKLP2KO and used to transform a ura4− diploid strain (ade6-M210/ade6-M216,leu1-32/leu1-32,ura4-D18/ura4-D18,h+/h−) by the PLATE method (Elble, 1992). Diploid transformants with a ura4+ phenotype were identified by plating to selective media and sporulated by nitrogen starvation. Homologous integrants were identified by colony PCR (Troxell's protocol, available at http://pingu.salk.edu/∼Forsburg/pcr.html) and confirmed by Southern blot analysis. Colony PCR with a 5′ primer (5′-GAGTTTTGAATAACGAC-3′) to the C terminus of klp2+ and a 3′ primer (5′-CATTGGTGTTGGAACAG-3′) to ura4+ was used in addition to the ura4+ marker to follow the klp2Δ in crosses.
Construction of the klp2+-pk-GFP Homologous Integrant
klp2+ was tagged at its C terminus with three tandem repeats of the pk1 tag (Craven et al., 1998) followed in frame by green fluorescent protein (GFP; S65T allele; Heim and Tsien, 1996). The ura4+ gene was placed downstream of the tags to serve as a marker for the strain. The strain was made by first constructing a vector that was not gene specific, pCS2pkSu, which carried simply the pk1 epitopes, GFP, and the SV40 polyadenylation signal followed by the ura4+ gene. This tagging cassette was made specific for integration in frame at the C terminus of klp2+ by a one-step PCR amplification that used long primers designed to add 85 bp of sequence identical to the locus of integration on either end of the tagging cassette. The 5′ primer contains the final 85 bp of klp2+ (with the exception of the stop codon), a flexible linker (6-Gly), and the first 15 bp of the 3-pk1 tag (5′-ACATTATGCAGTTTGAGATTTGCAACAAAGGTAAAT-AATACTCAAATTGGCACAGCACGCAAAGTCACAAAATCCG-GAGGAGGTGGAGGAGAGCTCATGGGTATTCCTAAT-3′). The 3′ primer contains 25 bp encoding the 3′ end of ura4+ and 85 bp of a region 560 bp downstream from the klp2+locus (5′-AAAATATACAGTGGGTAGTAGAATTTCTAATATTTTTATTATCAAGAGA-GAATTAAAGGAACTGCCAAGTTTGAGAAAAAAAAAATAGCTTCATTAAGAGAAAGTCTTTGCTGATATGCCTT-3′). The resulting ca. 3100-nt PCR product was used to transform a ura-D18 strain. Homologous integration was confirmed by Southern blotting. We confirmed that the ORF was preserved from klp2+ through the GFP gene by PCR amplifying the corresponding regions and directly sequencing the resulting PCR products. The klp2+-pk-GFP strain was also identified by a colony PCR assay with the use of a 5′ primer near the end of the klp2+gene (5′- TACTCATTTCCTCTTCTTGA-3′) and a 3′ primer to the end of the GFP gene in the tag (5′-TACTCATTTCCTCTTCTTGA-3′).
Fluorescence Microscopy
For measurements of metaphase spindle length, cells in early- to mid-log phase were prepared for immunofluorescence by aldehyde fixation (Hagan and Hyams, 1988). Tubulin was stained with a mouse mAb against α-tubulin (kindness of Margaret Fuller, Stanford University, Stanford, CA) and visualized with rhodamine-conjugated goat anti-mouse secondary antibodies (Jackson Laboratories, Bar Harbor, ME). DNA was stained with 4,6-diamino-2-phenylindole dihydrochloride (DAPI; Sigma, St. Louis, MO) as suggested by Moreno et al. (1991). Various Klp deletions were crossed into nuc2-663ts, and each double mutant was grown at 32°C to increase its mitotic index. Metaphase spindle lengths were measured in mitotic-arrested cells stained with antitubulin and DAPI, with the use of cells judged to be in metaphase by three criteria: 1) that spindle length did not exceed the diameter of an interphase nucleus, 2) that the spindle lay very close to one focal plane, and 3) that chromatin, as imaged by DAPI, was tightly compacted on the spindle. Cells were viewed with a Zeiss fluorescence microscope, with the use of an Empix charge-coupled device camera and the Metaphorph software for image capture, processing and measurement (Universal Imaging, West Chester, PA). Final images were exported to Adobe Photoshop (San Jose, CA) for figure preparation. To examine nuclear movement in crosses between klp2Δ,dhc1-d1 strains, cut11::GFP (West et al., 1998) was used as a marker for the nuclear envelope.
For live cell microscopy, cells were grown to midlog phase in YES or EMM+S at 25°C. To stain their DNA, cells were transferred to YES, pH 7.5, containing 3 μg/ml Hoescht 33342 and incubated at 25°C for 15–30 min (West et al., 2001). To visualize MTs klp2Δ cells were transformed with a plasmid (pDQ105; Ding et al., 1998) expressing α-tubulin-GFP. These cells were cultured in EMM containing 15 nM thiamin to minimize expression from the plasmid (Maundrell, 1990). Klp2p was imaged by GFP fluorescence from the tagged strain described above, in which klp2p-GFP expression is driven by the klp2+promoter. Cells were mounted on glass coverslips, and microscopy was performed at room temperature with a Zeiss Axiophot2 fluorescence microscope with the use of a Xenon arc lamp, filter cubes from Chroma Technologies (Brattlebourgh, VT), and either a 100× Neofluar lens (n.a. = 1.3), for single time point observations or a 100× Plan-APO lens (n.a. = 1.4) for time course observations. Image sets were collected on a Cooke SensiCam CCD camera, and Slidebook software (3I Inc., Denver, CO) was used for image capture and processing. For each series of observations, 4–10 focal planes were imaged at 400-nm intervals in the Z-axis, with the use of 0.2- to 2-s exposures, depending on image brightness. These images were subsequently deconvolved with the use of a “no neighbors” algorithm, and a two-dimensional projection was made by using the value of the brightest pixel at each position in X and Y through the complete Z-series. Images were taken every 10–200 s, depending on the rate of photobleaching and the duration of the process under study and then transferred as TIFF files to Adobe Photoshop for final compilation.
RESULTS
Identification and Characterization of the klp2+ Gene
We used genomic DNA and PCR with degenerate primers to amplify and clone sequences that might correspond to genes encoding KLPs in S. pombe. Our primers encoded highly conserved sequences from the motor domain of kinesins (Figure 1A, bold-faced sequences marked with arrows), and the resulting PCR products identified two distinct kinesin like genes in a fission yeast genomic library (Barbet et al., 1992). One was identified by sequence as pkl1+ (Pidoux et al., 1996); the other is shown as its predicted protein product in Figure 1A. This gene encodes a KLP by the criterion that its predicted amino acid sequence includes all the motifs characteristic of this protein superfamily (Goldstein, 1991; Moore and Endow, 1996; Endow, 1999). Given the order of its discovery in S. pombe, we have called the gene klp2+,pkl1+ is located on chromosome I, NotI-F, between the probes 13e2 and 7f6; klp2+is located on chromosome I, NotI-D, next to the probes 57bI and 57a9 on cosmid clone 31E3c. klp2+ has been identified on chromosome I, cosmid c664, and given protein_id number CAB65811.1 by the S. pombe Genome Sequencing Project at the Sanger Center (http://www.sanger. ac.uk/Projects/S_pombe/).
The motor domain of klp2+ lies near the C terminus of the predicted polypeptide, and analyses of sequence similarity, with the use of the BLAST and PILEUP algorithms, suggest that this protein is a member of the KAR3 subfamily (Table 1, Figure 1B). Both Klp2p and Pkl1p are predicted to contain conserved residues in the neck region and in the β1 region of the motor core (Figure 1B) that are important for minus-end–directed movement along a MT (Endow and Higuchi, 2000). Klp2p shares an additional structural feature with KLPs from this family in containing a domain with high likelihood of forming an α-helical coiled-coil (Figure 1C). We conclude that fission yeast contains at least two members of the KAR3 family, unlike S. cerevisiae, which has one (Meluh and Rose, 1990).
Table 1.
Bestfit comparisons (% similarity/% identity) of selected members of the KAR3 subfamily
| Pklp1 | Klp2p | |
|---|---|---|
| Entire peptides compared | ||
| S. pombe Pkl1p | — | 48%/39% |
| S. pombe Klp2p | 48%/39% | — |
| A. nidulans KlpA | 53/45 | 54/46 |
| S. cerevisiae KAR3 | 56/42 | 54/43 |
| D. melanogaster NCD | 53/43 | 49/41 |
| Pkl1p | Klp2p | |
|---|---|---|
| Head domains compared | ||
| S. pombe Pkl1p | — | 71/61 |
| S. pombe Klp2p | 71/61 | — |
| A. nidulans KlpA | 66/58 | 70/63 |
| S. cerevisiae KAR3 | 66/55 | 64/55 |
| D. melanogaster NCD | 59/49 | 57/48 |
Neither Klp2p nor Pkl1p Is Essential
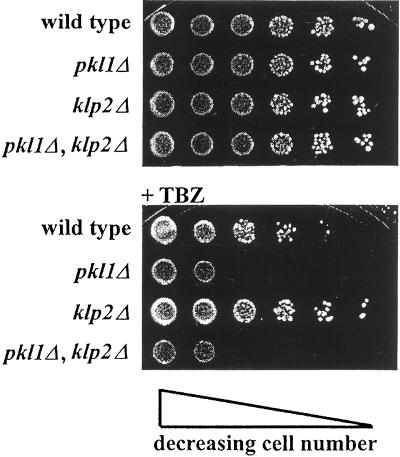
Previous work has shown that Pkl1p is not essential for either vegetative or sexual reproduction of S. pombe, so we were curious to know whether the deletion of either klp2+ or of both these members of the KAR3 family would have an effect on cell behavior. Neither pkl1+ nor klp2+ is essential, because the vegetative growths of both the singly and the doubly deleted strains are indistinguishable from wild type (Figures 2A and 3; see also Figure 6). This result has been confirmed at temperatures ranging from 20 to 36°C, the customary range of growth for this organism. Because a variety of KLPs have been shown to influence microtubule stability, and because MT depolymerizing agents could enhance or suppress these effects, we tested the effect of the MT poison thiabendazole (TBZ) on the viability of strains deleted for the KAR3 motors. The klp2Δ strain is more resistant than wild type to TBZ, whereas the pkl1Δ or the double deletion is less resistant than the wild-type strain. The two KAR3 motors can thus be distinguished in that the absence of Klp2p makes MTs more stable, whereas the absence of Pkl1p makes them more labile.
Figure 2.
The deletion of klp2+ and/or pkl1+ has no effect on cell growth, as seen by plating serial dilutions of each strain. At 32°C, klp2Δ is slightly more resistant than wild type to 10 μg/ml TBZ, whereas pkl1Δ and pkl1Δ,klp2Δ are more sensitive. (See Figures 3 and 6 for growth of these strains at 36, 25, and 29°C.)
Figure 6.
Serial dilutions of pkl1Δ and/or klp2Δ in either a wild-type or a cut11-1 background, grown at temperatures that are permissive for growth of all of these mutants. pkl1Δ, and this nuclear envelope component show a synthetic phenotype.
Three Temperature-sensitive Alleles of the Mitotic Motor, Cut7p, Are Rescued in an Allele-specific Manner by klp2Δ,pkl1Δ and Thiabendazole
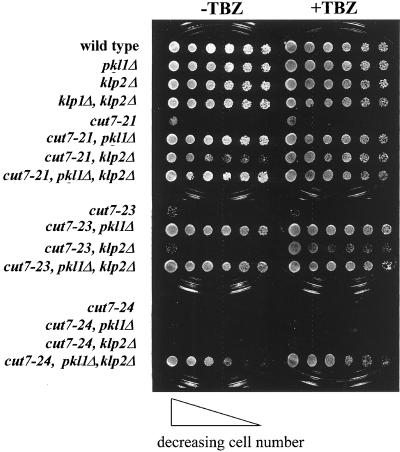
Although deletions of pkl1+and klp2+ have no obvious effect on mitotic growth rates, they interact distinctively with mutants of cut7+ (Hagan and Yanagida, 1992), suggesting that all three of these motors play some role in mitosis (Figure 3 and also Pidoux et al., 1996). Deletion of either C-terminal motor suppresses the temperature sensitive (ts) phenotype of cut7-21; either pkl1Δ alone or klp2Δ plus TBZ suppresses cut7-23, and only the double KLP deletion suppresses cut7-24, with or without TBZ. These allele-specific phenotypes support the hypothesis that both members of the KAR3 family play a role in mitosis of fission yeast but that their functions differ.
Figure 3.
The deletion of klp2+ and/or pkl1+ suppresses cut7ts in an allele-specific manner, as shown by plating serial dilutions of the various strains and incubating them at the restrictive temperature (36°C). Note that pkl1Δ does not show hypersensitivity to TBZ at 36°C.
Deletion of pkl1+ and/or klp2+ and the Dynein Heavy-Chain Gene, dhc1+, Results in Meiotic Defects
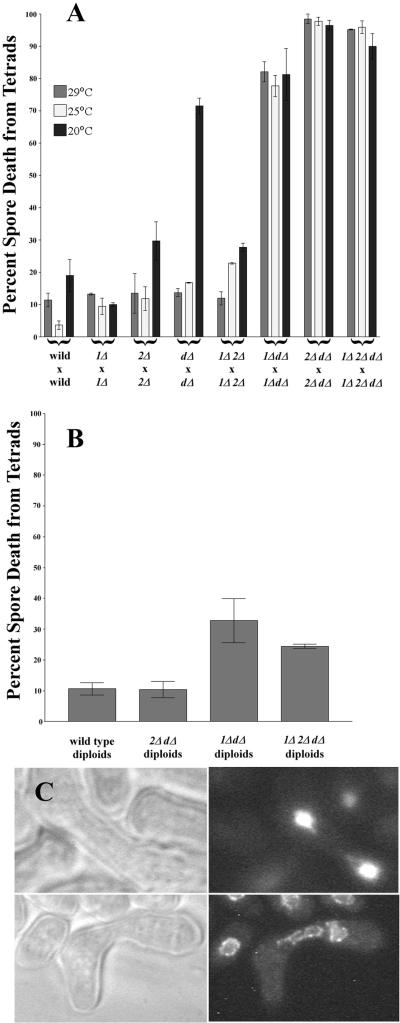
The viability of S. pombe upon the deletion of two putative minus-end–directed motors suggested that some additional motor(s) might be present to perform comparable functions in their absence. One likely candidate is cytoplasmic dynein heavy chain, dhc1+. Because a deletion for the dynein heavy chain dhc1+ has been described, dhc1-d, (Yamamoto et al., 1999), we were able to compare the behaviors of strains that were deleted for each of these motors and for all possible combinations. All of the double deletions and the triple deletion grew vegetatively at rates that were indistinguishable from wild type (our unpublished observations), but all of the double deletions that lack dhc1+ showed meiotic abnormalities (Figure 4A, Table 2). The rates of spore death at 25 or 29°C are modest in crosses between wild-type strains and in homozygous crosses between strains lacking either pkl1+, klp2+, or dhc1+. We did, however, note a cold sensitivity resulting in decreased spore viability in dhc1-dl × dhc1-d1 crosses (Figure 4A). At all temperatures tested, the crosses of klp2Δ,dhc1-d1 with itself, pkl1Δ,dhc1-d1 with itself, and the triple delete with itself all produced almost no viable spores (Figure 4A), suggesting a severe defect in some aspect of sexual reproduction. These defects were accompanied by deviations from the wild-type condition of four spores per ascus (Table 2). In particular, 44% of the asci from klp2Δ,dhc1-d1 × klp2Δ,dhc1-d1 crosses and 52% from crosses of the triple delete strain contained more than four spores per ascus, suggesting that meiosis had proceeded in the absence of karyogamy to result in catastrophic rates of spore death (Figure 4A).
Figure 4.
(A) Crosses between wild-type strains or crosses of either klp2Δ with itself, pkl1Δ with itself or dhc1-d1 with itself result in low levels of spore death, except the latter cross at 20°C. Homozygous crosses of klp2Δ,dhc1-d1, of pkl1Δ,dhc1-d1, or of the triple deletion, on the other hand, result in catastrophic levels of spore death. (B) The percent spore death resulting from sporulation of wild-type azygotic diploids and of azygotic diploids that were homozygous klp2Δ,dhc1-d1 were indistinguishable. Other azygotic dipoids show more spore death, but the percentage is still reduced, relative to the zygotic crosses. (C) A typical early zygote, resulting from klp2Δ,dhc1-d1 × klp2Δ,dhc1-d1. The two unfused nuclei have a shape that is characteristic of a late zygotic nucleus in wild-type cells during the processes of “horsetailing” (Ding et al., 1998).
Table 2.
Number of spores per zygotic ascus at 29°C
| Strains Crossed | % Asci with <4 spores | % Asci with 4 spores | % Asci with 5+ spores | n |
|---|---|---|---|---|
| Wild × wild | 5.5 | 94.0 | 0.5 | 830 |
| dhc-d1 × dhc-d1 | 34.7 | 64.3 | 1.0 | 793 |
| pkl1Δ × pkl1Δ | 13.2 | 86.6 | 0.2 | 514 |
| klp2Δ × klp2Δ | 3.5 | 95.0 | 1.5 | 529 |
| 1Δ2Δ × 1Δ2Δ | 7.0 | 90.0 | 3.0 | 534 |
| dhc-d1,1Δ × dhc-d1,1Δ | 50.0 | 39.1 | 10.9 | 952 |
| dhc-d1,2Δ × dhc-d1,2Δ | 30.0 | 26.0 | 44.0 | 806 |
| dhc-d1,1Δ2Δ × dhc-d1,1Δ2Δ | 25.3 | 22.6 | 52.1 | 447 |
We were able to recover diploid cells from all of these homozygous crosses, and they grew vegetatively at wild-type rates. When the diploid cells were plated on malt extract plates to induce sporulation, the viabilities of spores from these azygotic meioses were much more similar to wild type than were the zygotic crosses (Figure 4B). This effect is most pronounced for the klp2Δ,dhc1-d1 diploid, where azygotic spore viability is indistinguishable from wild type, suggesting that these two genes work together to effect karyogamy but play no essential role later in the meiotic process. This inference is corroborated by a reduction in the percentage of azygotic asci with greater than four spores to near wild type levels (Table 3). Also, microscopy of zygotes formed early in the cross of klp2Δ,dhc1-d1 with itself, with the use of either DAPI to visualize the nuclei or strains carrying Cut11p-GFP to mark the nuclear envelopes (West et al., 1998) revealed that zygotes with a single nucleus were rare in this cross. Most contained two nuclei whose morphology resembled that of diploid nuclei during the “horse-tailing” motions of meiotic prophase (Figure 4C; Chikashige et al., 1994). This is in contrast to the nuclear morphology when dhc1-d1 is crossed with itself. In these cells, karyogamy generally occurred producing a single round nucleus that failed to undergo horse-tailing. Sporulation of the pkl1Δ,dhc1-d1 diploid, on the other hand, still led to significant spore death (Figure 4B), suggesting that these two motors participate in the events of meiosis as well as playing some role in karyogamy. We interpret these observations to mean that karyogamy in fission yeast is particularly dependent on cooperation between Klp2p and Dhc1p with some contribution from Pkl1p and that coordination of Pkl1p with Dhc1p is more important in later meiotic events.
Table 3.
Number of spores per azygotic ascus at 29°C
| Strains sporulated | % Asci with <4 spores | % Asci with 4 spores | % Asci with >4 spores | n |
|---|---|---|---|---|
| Wild-type diploid | 5.0 | 93.5 | 1.5 | 572 |
| dhc-d1/dhc-d1;1Δ/1Δ | 15.9 | 83.5 | 0.6 | 702 |
| dhc-d1/dhc-d1;2Δ/2Δ | 9.25 | 85.75 | 5.0 | 559 |
| dhc-d1/dhc-d1;1Δ/1Δ;2Δ/2Δ | 19.0 | 81.0 | 0.0 | 816 |
Genetic Interactions between the KAR3 Motors and Temperature-sensitive Alleles of the Mitotic Genes, cut11+ and cut12+
Cut12p, also known as Skf1p, is a structural component of the fission yeast spindle pole body whose function is essential for mitotic progression (Bridge et al., 1998). Both pkl1Δ and klp2Δ show a synthetic phenotype with a temperature-sensitive allele of this gene, cut12-1 (Figure 5). At 32°C, a temperature that is permissive for the growth of cut12-1, either double mutant (cut12-1,pkl1Δ, or cut12-1,klp2Δ) shows poor growth and a disastrously inaccurate segregation of chromosomes, as visualized by flow cytometry (Figure 5). The arrows indicate the amount of DNA characteristic of a normal diploid cell and the distributions reveal the accumulation of numerous aneuploid cells. A triple mutant (cut12-1,pkl1Δ,klp2Δ) accumulates some cells with the haploid amount of DNA and others with almost no DNA at all, suggesting severe chromosome instability.
Figure 5.
The numbers of cells containing various amounts of DNA, as seen by flow cytometry. Deletion of klp2+ and/or pkl1+ has no effect in a wild-type background at 32°C, but in a cut12-1 background at the same temperature, either deletion leads to aneuploidy. The double deletion accumulates some cells with a haploid amount of DNA and others with almost no DNA.
Functional Cut11p is required at the onset of mitosis to allow the cell's spindle pole body to enter and attach to an opening that forms in the nuclear envelope; it is required for the formation of a normal mitotic spindle (West et al., 1998). In a cut11-1 background, pkl1Δ shows defective growth at the semipermissive temperature of 29°C, at which temperature cut11-1 and cut11-1,klp2Δ grow like wild type (Figure 6). The triple mutant (cut11-1,pkl1Δ,klp2Δ) displays an intermediate phenotype, indicating a partial rescue of pkl1Δ by klp2Δ. The same results (our unpublished observations) were obtained with cut11–2, an allele with a distinguishable phenotype (West et al., 1998). These observations support the ideas that motors of the KAR3 family are important for normal mitosis in fission yeast, but that the functions of these motors are different.
Localization of Klp2p in Fission Yeast
We constructed a plasmid in which the 3′ end of klp2+ was linked in frame with DNA encoding both the Pk1 epitope (Craven et al., 1998) and the GFP (Chalfie et al., 1994; Figure 7A). This element was homologously integrated at the klp2+ locus such that the chimeric protein was expressed under the control of the klp2+ promoter. The resulting fluorescence in vivo was dim, suggesting that the normal expression level of this protein is low; but, with the sensitive optical system described in MATERIALS AND METHODS, we were able to record the distribution of signal and follow its redistribution with time.
Figure 7.
Localizations of Klp2p at different times in the cell cycle. (A) A diagram showing the organization of the pk-gfp tag that was integrated at the klp2+ locus to assure uniform levels of tagged protein expression driven by the klp2+ promoter. (B) Localization of Klp2p-GFP in logarithmically growing, interphase cells. Linear arrays of small spots are seen in the cytoplasm. (C) The Klp2p-GFP spots move along straight lines. The time after the beginning of the series is given in seconds for each exposure. The cell on the right lacks distributed dots and shows only one cluster of GFP staining. (D) Cells expressing Klp2p-GFP and vitally stained with Hoechst 33342. The positions of GFP (green) and DNA (purple) show that Klp2p-GFP localizes to the nucleus in cells with condensed or condensing chromatin. In early mitosis Klp2p-GFP localizes to several distinct spots within the nucleus. (E) In later mitosis, as indicated by the presence of two pools of DNA stain, a few spots are found located either in each nucleus or in a line between the nuclei. Bar, 4 μm.
Interphase cells contain numerous dots of Klp2-GFP, which are distributed in linear arrays throughout the cytoplasm (Figure 7B). This arrangement is particularly evident in cells that are unusually long (Figure 7C). Here arrows indicate the positions of the same dots at successive times, with images taken at the intervals shown. Some of the dots move along the line defined by their linear arrangement. It seems likely that these dots are associated with interphase MTs. Figure 7C also shows a cell that we interpret as mitotic (cell on the right side). It lacks the several distributed dots, but now mobile dots are visible in a cluster near the cell's middle, presumably the nucleus.
A nuclear localization of Klp2p-GFP during mitosis has been confirmed by counterstaining live cells with the vital DNA dye, Hoechst's 33342 (Chikashige et al., 1994). As the cells enter mitosis, the cytoplasmic dots gradually disappear, although an occasional cytoplasmic dot can be seen in a cell with condensed DNA. In general, cytoplasmic staining during mitosis is weak, and dots of GFP are now found in the nucleus. In early mitosis, when the cell's DNA appears as a single object, the dots are distributed either as a row across the nucleus or as a cluster (Figure 7D). The number of dots is difficult to count with certainty, given their proximity, but as many as six have been identified in several cells. In later mitosis, as defined by the presence of two masses of DNA staining, there are dots in each daughter nucleus; now the largest number of dots per nucleus is three (Figure 7E). Late in anaphase, as indicated by the distance separating the daughter nuclei, the dots are positioned along a line between the DNA masses (Figure 7E). Colocalization of this staining with Cut11p-GFP, a marker for the nuclear envelope, suggests that at this stage the motor protein still lies within the nucleus, situated on the isthmus that persists between daughter nuclei as they separate in late anaphase (our unpublished observations; Hagan, 1998). This behavior is reminiscent of CENP-E, a kinetochore protein of vertebrates (Yen et al., 1992; Brown et al., 1996).
Because a kinetochore localization would be of interest for a putative minus-end–directed KLP, we have explored this possibility by following Klp2p-GFP localization in live cells that carry a ts allele of one component of the anaphase-promoting complex, nuc2-633 (Hirano et al., 1988). In these metaphase-arrested cells, every nucleus contains either dots or lines of Klp2p-GFP staining (Figure 8A). Images of the same cells taken at successive times show that the distribution of stain changes within the nucleus (Figure 8B), forming one cluster, two clusters, or a strand of stain, as kinetochores in metaphase-arrested cells are known to do (Goshima et al., 1999). It therefore seems likely that Klp2-GFP is localized at or near the kinetochores of fission yeast.
Figure 8.
Klp2p-GFP forms dots that oscillate across the nucleus in metaphase-arrested cells. (A) nuc2-633,klp2+-pk-gfp cells were arrested at 36oC for 3 h and stained with Hoechst before visualization. Klp2p-GFP is found along a line that runs across the nucleus, frequently concentrated at or near the ends of the line in unequal masses. (B) A time course of Klp2p-GFP localization in nuc2-633ts cells at 36°C reveals that the majority of the fluorescence coalesces and disperses along a diameter of this metaphase-arrested nucleus.
Motors of the KAR3 Subfamily Affect Spindle Length in Fission Yeast
In budding yeast, Kar3p helps to define the length of the metaphase spindle (Saunders et al., 1997b). We have used nuc2-633 32°C to obtain a large population of mitotic cells, particularly cells in metaphase (Figure 9). The length of the spindle was measured in hundreds of such cells for each genotype, and their lengths were compared (Table 4). The absence of Pkl1p leads to spindles of significantly reduced length, whereas the absence of Klp2p induces spindles to elongate. Spindles in the double deletion are indistinguishable from wild type, showing that pkl1Δ is not epistatic to klp2Δ by this assay. These results suggest that although the two fission yeast motors of the KAR3 family are similar in the primary structure of their motor domains, they are likely to function quite differently in mitosis.
Figure 9.
Spindle morphology in a fission yeast cell, strain nuc2-633, grown at the semirestrictive temperature of 32°C. MTs have been stained with antitubulin and DNA with DAPI. A single mass of compacted chromatin lies roughly at the spindle equator, and the length of the well-defined spindle is easy to measure. Bar, 4 μm.
Table 4.
The effects on metaphase spindle length of deleting pkl1+ and/or klp2+
| Strain | Mean spindle length (μm) | SD | SEM | Valid N | p |
|---|---|---|---|---|---|
| nuc2-663ts | 2.03 | 0.313 | 0.021 | 229 | — |
| nuc2-663ts,pkl1Δ | 1.81 | 0.281 | 0.017 | 285 | 0.000000 |
| nuc2-663ts,klp2Δ | 2.13 | 0.349 | 0.021 | 286 | 0.001724 |
| nuc2-663ts,pkl1,klp2Δ | 2.01 | 0.332 | 0.021 | 250 | 0.359041 |
In these preparations we also noticed that the number of spindles with detectable astral MTs showed a systematic variation with genotype. The absence of Pkl1p increased the fraction of spindles with asters relative to wild type, with or without Klp2p, whereas the lack of Klp2p had no obvious effect (Table 5).
Table 5.
The effects on percentage of spindles with astral microtubules of deleting pkl1+ or klp2+
| Strain | No. of spindles with astral MTs | No. sampled | % Spindles with astral MTs |
|---|---|---|---|
| nuc2-663ts | 36 | 928 | 4 |
| nuc2-663ts,pkl1Δ | 96 | 593 | 16 |
| nuc2-663ts,klp2Δ | 45 | 801 | 6 |
| nuc2-663ts,pkl1,klp2Δ | 48 | 312 | 15 |
The effects of Klp2p deletion on spindle length can also be seen in otherwise wild-type S. pombe during anaphase B. Figure 10A shows a time sequence from a klp2Δ cell that is expressing α-tubulin-GFP (Ding et al., 1998). The observed hyper-extension of the late anaphase spindle was seen in all anaphase cells examined by this method (n = 16). Thirty percent of these showed an additional mitotic problem: the interzone spindle broke or accumulated additional MT bundles that are not normally seen (Figure 10B). These results show that Klp2p is important for the controlled elongation of the anaphase spindle and its appropriate disassembly at the completion of mitosis.
Figure 10.
klp2Δ cells display spindle defects in late anaphase. Cells deleted for Klp2p were transformed with the plasmid pDQ105, which expresses α-tubulin-GFP. (A) The localizations of DNA (blue) and tubulin (green), determined as a function of time in minutes, reveal a failure of the spindle to disassemble in late anaphase and a concomitant hyper-extension of the distance between daughter nuclei. (B) Approximately 30% of the cells deleted for Klp2p also display a spindle breakage phenotype (arrowheads).
DISCUSSION
We have identified a second KLP of the KAR3 family in fission yeast and have shown that these two structurally similar motor enzymes play distinct roles for this organism in vivo. The previously described Pkl1p is confined to the nucleus, whereas the newly identified Klp2p is cytoplasmic during interphase and nuclear during mitosis. The localization of Klp2p to about six dots in the early mitotic nucleus, combined with the facts that these dots coalesce and separate in a metaphase-arrested nucleus and that they segregate into two sets of about three when anaphase proceeds, suggest that Klp2p is kinetochore-associated during early cell division. This protein then migrates from the chromatin masses to an intranuclear line that runs between the two late-anaphase chromatin masses, suggesting that it leaves the kinetochores in anaphase and moves to the spindle interzone, a behavior characteristic of other kinetochore motor enzymes (Yen et al., 1992; Brown et al., 1996). The increased length of metaphase-arrested spindles in cells that lack Klp2p suggests that this protein normally shortens the spindle, perhaps by pulling kinetochore-associated MTs toward the kinetochores. The increased length of late anaphase spindles in klp2Δ, compared with wild-type spindles, suggests that Klp2p has a MT-disassembling action.
The general similarity in predicted amino acid sequence between the motor domains of Klp2p and Kar3p, together with the position of their motor domains at the proteins' C-termini and the details of their sequence identity in the region between motor and stalk, implies that Klp2p has properties in common with Kar3p, for example, the ability to move toward the minus end of a MT and to induce MT shortening (Endow et al., 1994). These properties for Klp2p remain to be tested, but the localization of the protein in vivo, together with the details of its deletion phenotype, are consistent with both of these properties. Localization at kinetochores, minus-end–directed motility, and the ability to depolymerize MTs would all help to explain the elongation of metaphase arrested spindles in the klp2Δ strain, whereas the protein's localization to the wild-type spindle midzone during late anaphase, coupled with the same functional properties, would explain the hyper-extension of klp2Δ mutant spindles at the end of mitosis.
A kinetochore function has been proposed for Kar3p in S. cerevisiae, based on the ability of this motor to bind to the protein complex, CBF3, which associates with an essential element of the budding yeast centromere (Middleton and Carbon, 1994). Localizations of Kar3p in vivo have, however, always suggested an association with the spindle pole body, rather than with the chromosomes (Saunders et al., 1997a). It must be said, however, kinetochores in budding yeast may spend much of the cell cycle in close proximity to the SPB. Moreover, detecting a kinetochore-specific localization during early mitosis, with only a few molecules of Kar3p at each of the 32 kinetochores, would be a formidable technical challenge. These loci are spread quite widely in the nucleoplasm (Winey et al., 1995; He et al., 2000), so the antigen is not concentrated as it may later be on the spindle pole body, making it far harder to see. Thus, although there is little evidence to support this model, we cannot conclude on current evidence that Kar3p is not kinetochore-associated in budding yeast.
Kar3p in budding yeast binds with two distinct companion polypeptides, Cik1p (Page et al., 1994) and Vik1p (Manning et al., 1999). These associated proteins bind with Kar3p and help to localize it to different places (Vik1p to the poles and Cik1p to the spindle and to “nuclear patches”), and they seem to induce it to perform different functions. A comparable associating protein has not yet been found for either Klp2p or Pkl1p, but the fact that fission yeast has two motor heavy chains may provide a functional diversity similar to that conferred on Kar3p by its two accompanying proteins. It will be interesting to seek a companion protein(s) for Pkl1p and Klp2p and to determine its (their) functions.
Phenotypic comparisons among these KAR3 family members are instructive, but they do not yet define the function of each motor complex. For example, in S. cerevisiae Vik1p localizes with Kar3p to spindle pole bodies, as does Pkl1p in S. pombe; in addition, cik1Δ displays increased MT length in mitosis similar to klp2Δ. In contrast, pkl1Δ increases the sensitivity of fission yeast to a MT poison, whereas cik1Δ has the same effect on budding yeast (Figure 2; Page et al., 1994). Moreover, klp2Δ in S. pombe and cik1Δ in S. cerevisiae provide resistance to MT poisons (Figure 2; Manning et al., 1999). In addition, the deletion of VIK1 partially rescues the cik1Δ phenotype, similar to the effect seen when Klp2 is deleted in a pkl1Δ,cut11-1 background in S. pombe. In addition, there are overlapping roles of these motors in S. pombe; although vik1Δ rescues ts alleles of the BimC class motors in budding yeast and cik1Δ does not, in fission yeast both members of this family rescue certain alleles of the corresponding BimC motor (Figure 3). Furthermore, Cik1p is essential for karyogamy in budding yeast, but Vik1p is not, whereas in fission yeast both Pkl1p and Klp2p appear to cooperate with cytoplasmic dynein in this process, albeit Klp2p plays the greater role. Thus, the parallels between the two yeasts are not strict, and further work will be required to sort out just what each motor does in these two microorganisms.
Other motors in fission yeast may function coordinately with Klp2p and Pkl1p. Klp5p and Klp6p are KLPs of the KIP3 subfamily that promote both the disassembly of MTs and the alignment of metaphase chromosomes (R.R. West, T. Malmstrom, C.L. Troxell, and J.R. McIntosh, unpublished observations). A KLP that is likely to be a chromokinesin has been identified by the S. pombe genome sequencing project, and dynein may perform some as yet unidentified role in mitosis. There are also nonmotor proteins that contribute to spindle morphogenesis and chromosome alignment. These include the kinetochore proteins Mis12p and Mis6p, because loss-of-function alleles of the corresponding genes show a large increase in spindle length (Goshima et al., 1999). The establishment of a metaphase spindle requires a restraining activity from the MT-associated protein, Dis1p and the related Mtc1p; loss of function of Dis1p leads to a precocious anaphase-like elongation of the spindle (Nabeshima et al., 1998; Nakaseko et al., 2001). In addition, Pkl1p interacts with γ-tubulin, a protein found at the spindle pole body that is essential for MT nucleation (Paluh et al., 2000). It appears, therefore, that there are many proteins that help to regulate the structure and function of the mitotic apparatus. We surmise that the experimental flexibility of an organism like fission yeast will make it a good model system in which to work out some of these complex interactions. Further studies of this organism should elucidate just what each of the motors does in mitosis and how their mechanochemical action is related to the control of MT polymerization and depolymerization.
ACKNOWLEDGMENTS
We thank Heidi Browning, Katya Grishchuk, Paula Grissom, and other members of the McIntosh Lab as well as Susan Forsburg, Shelly Jones, Pam Meluh, and Shelly Sazer for helpful discussions. Yu Ming Han operated the Boulder Automated DNA Sequencing Facility. The work was supported in part by National Science Foundation Postdoctoral Fellowships to C.L.T. and M.A.S. and by GM33787 to J.R.M., who is a Research Professor of the American Cancer Society.
REFERENCES
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Brazer SC, Williams HP, Chappell TG, Cande WZ. A fission yeast kinesin affects Golgi membrane recycling. Yeast. 2000;16:149–166. doi: 10.1002/(SICI)1097-0061(20000130)16:2<149::AID-YEA514>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Morphew M, Bartlett R, Hagan IM. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Wood KW, Cleveland DW. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J Cell Sci. 1996;109(Pt 5):961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
- Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol. 2000;151:15–27. doi: 10.1083/jcb.151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;263:390–393. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Clarke L. Centromeres of budding and fission yeasts. Trends Genet. 1990;6:150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–152. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Endow SA. Microtubule motors in spindle and chromosome motility. Eur J Biochem. 1999;262:12–18. doi: 10.1046/j.1432-1327.1999.00339.x. [DOI] [PubMed] [Google Scholar]
- Endow SA, Higuchi H. A mutant of the motor protein kinesin that moves in both directions on microtubules [see comments] Nature. 2000;406:913–916. doi: 10.1038/35022617. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. The kinesin superfamily: tails of functional redundancy. Trends Cell Biol. 1991;1:93–98. doi: 10.1016/0962-8924(91)90036-9. [DOI] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–75. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths, and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Hirano T, Hiraoka Y, Yanagida M. A temperature-sensitive mutation of the Schizosaccharomyces pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Hoheisel JD, Maier E, Mott R, McCarthy L, Grigoriev AV, Schalkwyk LC, Nizetic D, Francis F, Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Hyman AA, Bahler M. Motor proteins of the eukaryotic cytoskeleton. Proc Natl Acad Sci USA. 1997;94:12747–12748. doi: 10.1073/pnas.94.24.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S. The role of microtubule assembly dynamics in mitotic force generation and functional organization of living cells. J Struct Biol. 1997;118:87–93. doi: 10.1006/jsbi.1996.3839. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- McDonald HB, Steward RJ, Goldstein LSB. The kinesin-like NCD protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion [published erratum appears in Cell 1990 May 4;61(3):548] Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Middleton K, Carbon J. KAR3-encoded kinesin is a minus-end-directed motor that functions with centromere binding proteins (CBF3) on an in vitro yeast kinetochore. Proc Natl Acad Sci USA. 1994;91:7212–7216. doi: 10.1073/pnas.91.15.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Nakatani K, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Sequencing and characterization of the kinesin-related genes katB and katC of Arabidopsis thaliana. Plant Mol Biol. 1994;25:865–876. doi: 10.1007/BF00028881. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Identification of a gene family (kat) encoding kinesin-like proteins in Arabidopsis thaliana and the characterization of secondary structure of KatA. Mol Gen Genet. 1993;238:362–368. doi: 10.1007/BF00291995. [DOI] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast. Microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11(8):537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MJ, Meluh PB, Rose MD, Morris NR. Suppression of bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans. J Cell Biol. 1993;120:153–162. doi: 10.1083/jcb.120.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux AL, Cande WZ. A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p [In Process Citation] Mol Biol Cell. 2000;11:1225–39. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, LeDizet M, Cande WZ. Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol Biol Cell. 1996;7:1639–1655. doi: 10.1091/mbc.7.10.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997a;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997b;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000b;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR. cut11(+): a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, R.R., Malmstrom, T., Troxell, C.L., and McIntosh, J.R. (2001). Kinesins klp5+ and klp6+ footer microtubule disassembly and metaphase chromosome alignment in fission yeast. Mol. Biol. Cell 12, (in press). [DOI] [PMC free article] [PubMed]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. Oligonucleotide screening of cDNA libraries. Focus. 1984;6:1–3. [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis [see comments] Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]