Abstract
Previous studies suggest that signal transducer and activator of transcription 3 (STAT3) and CCAAT/enhancer binding protein beta (C/EBPβ) play an essential role in ovarian granulosa cells (GCs) for mammalian follicular development. Several C/EBPβ putative binding sites were previously predicted on the STAT3 promoter in mammals. However, the molecular regulation of C/EBPβ on STAT3 and their effects on cell proliferation and apoptosis remain virtually unexplored in GCs. Using porcine GCs as a model, the 5′-deletion, luciferase report assay, mutation, chromatin immunoprecipitation, Annexin-V/PI staining and EdU assays were applied to investigate the molecular mechanism for C/EBPβ regulating the expression of STAT3 and their effects on the cell proliferation and apoptosis ability. We found that over and interfering with the expression of C/EBPβ significantly increased and decreased the messenger RNA (mRNA) and protein levels of STAT3, respectively. The dual luciferase reporter assay showed that C/EBPβ directly bound at −1397/−1387 of STAT3 to positively regulate the mRNA and protein expressions of STAT3. Both C/EBPβ and STAT3 were observed to inhibit cell apoptosis and promote cell proliferation. Furthermore, C/EBPβ might enhance the antiapoptotic and pro-proliferative effects of STAT3. These results would be of great insight in further exploring the molecular mechanism of C/EBPβ and STAT3 on the function of GCs and the development of ovarian follicles in mammals.
Keywords: ovarian granulosa cells, C/EBPβ, STAT3, cell proliferation, cell apoptosis
1. Introduction
Ovarian dysfunction causes the reproductive failure and infertility in female mammals. The main functions of ovaries are to produce mature oocytes that can propagate the species [1,2] and synthetize the steroids that support secondary sexual characteristics [3,4]. It was widely thought that granulosa cells (GCs) supported the development and maturation of follicles though the complex interactions [5,6], and that the growth and proliferation of GCs play critical roles in the biological processes of recruitment, selection, atresia, ovulation and luteolysis of follicles [7,8]. Previous studies have found that steroids, growth factors, and cytokine factors secreted by GCs are essential for the survival and growth of follicles [9,10], and moreover, the high apoptosis of GCs can impair folliculogenesis and result in the increased follicular atresia [11,12].
In mammals, the signal transducer and activator of transcription 3 (STAT3) protein has been suggested to be involved in folliculogenesis [13,14]. It has been reported that STAT3 expressed highly in porcine GCs [15], and porcine complementary DNA (cDNA) of STAT3 is 93% and 90% homologous to humans and mice, respectively [15]. In mares, mRNA of STAT3 expresses higher in adult ovaries than in fetal ovaries [13]. In chickens, phosphorylated STAT3 is activated by the epidermal growth factor [15], which is known to decrease the P450scc and follicle-stimulating hormone receptor mRNA abundance to regulate the biological functions of GCs [16]. In mouse, reduced expression of STAT3 enhances the early apoptosis rate of mGCs [17], and moreover, specifically deleted STAT3 in ovarian GCs can impair fertility with significantly fewer litters and smaller litter size [18]. However, the cell function of STAT3 is seldom investigated in porcine GCs.
Much evidence has suggested that CCAAT/enhancer binding protein beta (C/EBPβ) plays an essential role in female reproduction [19,20]. In mice and rats, C/EBPβ mRNA is specifically and rapidly induced by luteinizing hormone in GCs [20,21]. A targeted deletion of C/EBPβ results in reproductive defects in female mice [20]. Moreover, the GC-specific C/EBPβ knockout causes subfertility with the absence of corpus luteums in 70% and the reduction expression of Ptgs2, Star, and Cyp11a1 in mice [19]. These results supported the proposed and essential role of C/EBPβ in GCs for mammalian folliculogenesis. However, the functions of C/EBPβ on cell apoptosis and proliferation remained virtually unexplored for ovarian GCs.
Previous studies report that C/EBPβ has a cis-acting element in the promoter PGS-2 in rat GCs [21], and moreover, we found several C/EBPβ potential binding sites were predicted on the STAT3 promoter of humans, mice, and pigs (see Materials and Methods). We hypothesized that C/EBPβ might play a cis-acting regulatory role in transcription of STAT3 and thus regulate the function of GCs in mammals. In this study, using porcine GCs as a model, the molecular mechanism regarding the regulation between C/EBPβ and STAT3 was first identified, and then its biological functions were explored for cell apoptosis and proliferation.
2. Materials and Methods
2.1. Ethics Approval
All experiments in the present study were performed in accordance with the guidelines of the Animal Care and Use Committee of South China Agricultural University Guangzhou, China (approval number: SCAU#2013-10).
2.2. Prediction of Putative C/EBPβ Binding Sites on the Promoter of STAT3
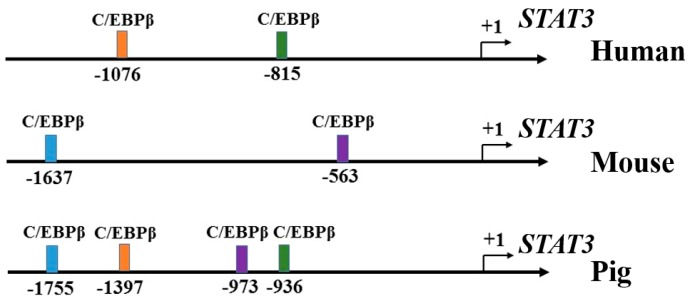
The promoter sequences of STAT3 (upstream 2 kb) were download from NCBI for human [22], mouse [23] and pig [24]. TFBIND [25], Biobase [26], Jaspar [27] and Research [28] were used to predict the putative binding location of C/EBPβ. The putative binding sites of C/EBPβ concurrently predicted by all of those four tools were used for further analysis. The putative binding sites of C/EBPβ on the promoter of STAT3 in humans, mice, and pigs are shown in Figure 1.
Figure 1.
Putative binding sites of CCAAT/enhancer binding protein beta (C/EBPβ) on the promoter of Signal transducer and activator of transcription 3 (STAT3) in humans, mice, and pigs.
2.3. Construction of STAT3 5′ Deletion and Luciferase Assay
The genomic DNA of porcine ovary tissues was used as a template. PCR was performed using PrimerSTAR® (TaKaRa, Dalian, Liaoning, China) high fidelity enzyme to obtain the STAT3 promoter of 2575 bp. Primers are presented in Table 1. Then PCR products were purified by gelatinization and the addition of “A” tail to combine with pMD-18T, which were transformed into competent cells DH5α, inoculated on ampicillin-containing lysogeny broth (LB) plates at 37 °C for overnight. Further, monoclonal bacteria was added from platelets to ampicillin of LB medium, and incubated overnight at 37 °C shaker. The bacteria were collected by centrifugation and the plasmids were extracted. The correct plasmid for sequencing was named T-STAT3. Then we used T-STAT3 as a template and designed five different upstream primers to amplify deletion fragments. The longest deletion fragment was named P1. The location of each deletion fragment of STAT3 was P1 (−2199/+375), P2 (−1532/+375), P3 (−1035/+375), P4 (−587/+375) and P5 (−167/+375). We used the same method to obtain plasmids of each deletion fragment containing MIuI and XhoI cleavage sites (Table 1). Simultaneously, we cloned each deletion fragment into the eukaryotic expression vector pGL3-Basic, which digested with MIuI and XhoI restriction endonuclease. According to Promega’s dual luciferase reporter assay kit (Promega, Madison, WI, USA) and previous study [29], we use the BioTek Synergy 2 multifunctional microplate reader (BioTek, Winooski, VT, USA) for fluorescence detection. The ratio of the expression of firefly luciferase to renilla luciferase was the target fragment activity.
Table 1.
Primers used in the present study.
| Name | Sequence * | Product (bp) | Accession Number |
|---|---|---|---|
| P1 (−2199/+375) | F: CGACGCGT TCCTCAACCCACCAAGAAAG | 2575 | NM_001044580 |
| R: CCCTCGAG CTCCCGGTCTCTTCGTATCC | |||
| P2 (−1532/+375) | F: CGACGCGT CTCCAAGTCATTGATTTTCT | 1908 | NM_001044580 |
| R: ditto | |||
| P3 (−1035/+375) | F: CGACGCGT TACTAAACAAACACAATAAA | 1410 | NM_001044580 |
| R: ditto | |||
| P4 (−587/+375) | F: CGACGCGT CTGAGGTTCAAAGCAGGCGG | 963 | NM_001044580 |
| R: ditto | |||
| P5 (−167/+375) | F: CGACGCGT CTCTCCTCATTGGCAAGTGG | 543 | NM_001044580 |
| R: ditto | |||
| qRT-PCR-STAT3 | F: GGGCTTTATCAGTAAGGAGA R: GGAATGTCAGGGTAGAGGTA |
276 | NM_001044580 |
| qRT-PCR -C/EBPβ | F: CGGACTGCAAGCGGAAGGAGGA R: GGCTGGACGACGAGGATGTGGA |
153 | NM_001199889 |
| qRT-PCR-GAPDH | F: TCCCGCCAACATCAAAT R: CACGCCCATCACAAACAT |
201 | NM_001206359 |
| ChIP-STAT3 | F: ATAGCTATCCTTGGGGAGG R: AAGGGCCTGTTATCTCAC |
150 | NM_001044580 |
* The underlined is enzyme-cutting sites. The gray part is base protection.
2.4. Culture of Porcine GCs In Vitro
Porcine ovarian GCs were cultured as previously described [30]. Briefly, porcine ovaries from prepubertal gilts were collected from a local slaughterhouse in Guangzhou, and transferred to our laboratory in phosphate-buffered saline (PBS) containing penicillin (100 IU/mL) and streptomycin (100 μg/mL) (Invitrogen, Shanghai, China) at a storage temperature of >30 °C. Subsequently, 3–5 mm follicles were punctured for GC collection using a 1-mL syringe, and the isolated GCs were washed twice with PBS preheated to 37 °C. The cells were seeded into 25-cm2 flasks and cultured at 37 °C under 5% CO2 in DMEM (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 IU/mL penicillin, and 100 μg/mL streptomycin.
2.5. Chromatin Immunoprecipitation Assay
GCs were cross-linked until the cell density reached 70%. Then we discarded the original culture medium in the flask and the cells were sequentially treated with formaldehyde, glycine, PBS-Halt Cocktail and centrifuged to collect the cell pellet. The chromatin immunoprecipitation (ChIP) assay (Thermofisher, Rockford, IL, USA) was carried out according to the previous ChIP protocol [31]. ChIP primer for −1397/−1387 of STAT3 is presented in Table 1 and was used to detect the binding of STAT3 and H3. After immunoprecipitation, the C/EBPβ binding site was identified by PCR amplification. Total fragmented DNA was used as input. DNA Marker was 100 bp.
2.6. Real-Time Quantitative PCR Analysis
At least three wells per group were collected for extraction of total RNA. Total RNA was extracted using TRIzol reagent (TaKaRa, Tokyo, Japan) and then reverse-transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) for mRNAs. The relative expression levels of mRNAs were quantified using Maxima SYBR Green qRT-PCR Master Mix (2×) (Thermo Scientific, Waltham, MA, USA) and THUNDERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan) in a LightCycler Real-Time PCR system. The expression levels of GAPDH mRNAs were used as endogenous controls, and the fold changes of STAT3 and C/EBPβ were calculated using the 2−ΔΔct method. The primer sequences are listed in Table 1.
2.7. Cell Proliferation and Apoptosis Assay
Cell proliferation assays were performed using Cell-Light Edu Apollo 567 In Vitro Kit (RiboBio Co., Ltd., Guangzhou, Guangdong, China). GCs were seeded into 48-well plates at one day prior to transfection. When the cells reached 30–50% coverage of one well, pcDNA3.1-C/EBPβ, pcDNA3.1-STAT3, pcDNA3.1-Control, C/EBPβ-siRNA, STAT3-siRNA or siRNA-NC were transfected into the cells at different final concentrations for 48 h. The specific steps are: the Edu solution was diluted 1:1000 with cell culture media to prepare 50 μM Edu medium, add 100 μL of 50 μM Edu media to each well for 2 h, discard the culture medium, add 100 μL of cell fixing solution (80% acetone) to each well for 30 min at room temperature, wash twice with PBS, add 100 μL of penetrant (0.5% TritonX-100 in PBS) to permeabilize the cells and rinse once with PBS, add 100 μL of 1 × Apollo Staining Solution and incubate for 30 min at room temperature in the dark, discard staining solution and add 100 μL DAPI per well incubate for 30 min at room temperature in the dark, then add PBS to take pictures under microscope.
Cell apoptosis assays were performed using an Annexin V-FITC Apoptosis Detection Kit (BioVision, Milpitas, CA, USA) according to the manufacturer’s instructions. Briefly, GCs (1–5 × 105 cells/well) were cultured in triplicate in 6-well plates at one day prior to transfection. When the cells reached 30–50% coverage of one well, pcDNA3.1-C/EBPβ, pcDNA3.1-STAT3, pcDNA3.1-Control, C/EBPβ-siRNA, STAT3-siRNA or siRNA-NC were transfected into the cells at different final concentrations for 48 h. The cells were then harvested, washed twice with ice-cold PBS, and resuspended in 500 μL of binding buffer. Next, 1.25 μL of Annexin V-FITC was added in the dark for 15 min at room temperature, then 1000× g centrifugation for 5 min at room temperature to remove the supernatant. The cells were gently resuspended with 0.5 mL precooling 1 × solution, and 10 μL of PI (propidium iodide; 50 μg/mL) were added. Last, the cells were analyzed in a flow cytometer (Becton Dickinson Co., San Jose, CA, USA) using the FITC signal detector (FL1) and phycoerythrin emission signal detector (FL2). All experiments were performed at least three times. Cells in the lower right quadrant are annexin-positive/PI-negative early apoptotic cells. The cells in the upper right quadrant are annexin-positive/PI-positive late apoptotic cells.
2.8. Western Blot Analysis
The cells were harvested and analyzed for their expression levels of total STAT3 using an anti-STAT3 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The molecular weight of STAT3 was 91 kDa. Protein concentrations were determined using a BCA Protein Assay Kit (Vigorous Bio-technology Beijing Co., Ltd., Beijing, China), and equal amounts of protein were separated by SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. The membranes were blocked with 5% nonfat milk in PBS containing a percentage of Tween-20 for 1 h, and then incubated with a primary antibody against hamartin (1:1000; Biorbyt, San Francisco, CA, USA) overnight at 4 °C. An anti-GAPDH antibody (1:3000; Sigma, St. Louis, MO, USA) was used as an internal control. After incubation with secondary antibodies for 1 h at room temperature, antibody-bound protein bands were visualized using an ECL-PLUS Kit (Amersham Biosciences, Piscataway, NJ, USA). The gray scale values of the bands were calculated using ImageJ software, which was free downloaded from NIH. The relative protein expression level of STAT3 was normalized by β-Actin values. At least three replicates were conducted for each group.
2.9. Data Analysis
All experiments were repeated at least three times independently. Data were expressed as means ± standard deviation (SD) of repeated experiments. Statistical analyses were carried out using R software. The significance of differences in means between two groups was analyzed by using Student’s t-test (two-tailed). * indicates p < 0.05; ** indicates p < 0.01.
3. Results
3.1. C/EBPβ Promotes the mRNA and Protein Level of STAT3
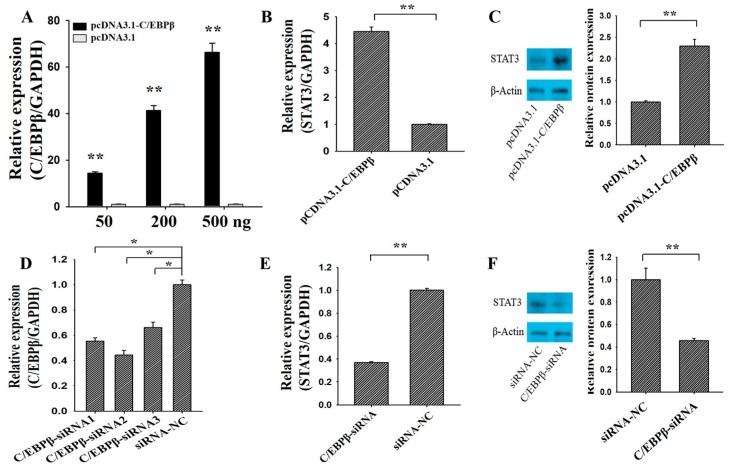
The overexpression plasmid and small interfering RNA (siRNA) of C/EBPβ were first built to explore the effects of C/EBPβ on the expression of STAT3 (Figure 2). We found that the mRNA expression of C/EBPβ was increasing along with the concentration of pcDNA3.1-C/EBPβ (Figure 2A), and the 200 ng of pcDNA3.1-C/EBPβ plasmid was selected and used for further analysis. Compared to the control group, overexpression of C/EBPβ significantly up-regulated the mRNA (Figure 2B, p < 0.01) and protein (Figure 2C, p < 0.05) levels of STAT3. Three C/EBPβ-specific small interfering RNA (siRNA) (C/EBPβ-siRNA1, C/EBPβ-siRNA2 and C/EBPβ-siRNA3) and a negative control (siRNA-NC) were transfected into GCs to evaluate the inhibition efficiency for C/EBPβ (Figure 2D). As shown in Figure 2D, C/EBPβ-siRNA2 exhibited the best inhibition efficiency, and thus C/EBPβ-siRNA2 was selected for the knockdown of C/EBPβ in GCs. Compared with the control group, interfering with the expression of C/EBPβ significantly down-regulated the mRNA (Figure 2E, p < 0.01) and protein (Figure 2F, p < 0.05) levels of STAT3. These observations indicated that C/EBPβ could positively regulate the mRNA and protein amounts of STAT3 in porcine GCs.
Figure 2.
C/EBPβ promotes the mRNA and protein levels of STAT3. (A) Relative expression of C/EBPβ against the different concentrations of pcDNA3.1-C/EBPβ plasmid; the mRNA (B) and protein (C) expression of STAT3 was stimulated by pcDNA3.1-C/EBPβ; (D) relative expression of C/EBPβ knockdown by three siRNAs; the mRNA (E) and protein (F) expression of STAT3 depressed by C/EBPβ-siRNA. ** indicates p < 0.01; * indicates p < 0.05. Data were represented as means ± SD. siRNA: small interfering RNA; siRNA-NC: a siRNA negative control.
3.2. C/EBPβ Binding at −1397/−1387 Region of STAT3
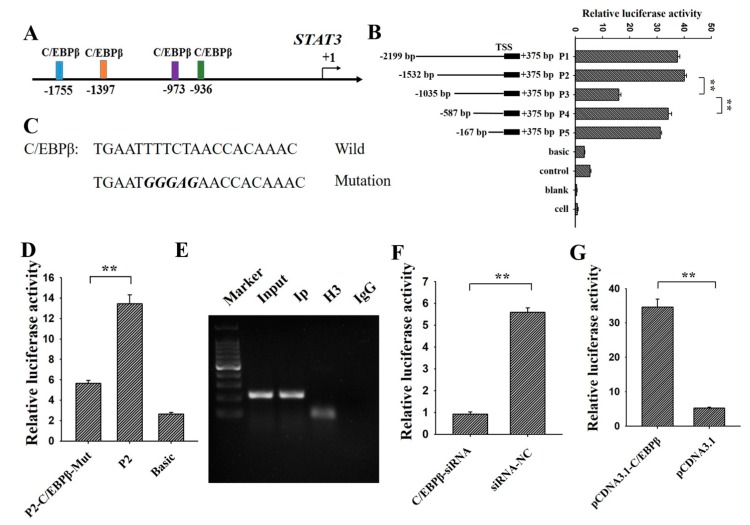
Four putative binding sites of C/EBPβ were predicted on the promoter of STAT3 in pigs (Figure 3A), suggesting STAT3 might be a direct target of C/EBPβ. To investigate the molecular mechanism of C/EBPβ regulating the expression of STAT3, the 5′-deletions and gene reporter assay were constructed for STAT3 (Figure 3B). Compared with P1, the relative luciferase activity of P2 did not showed significant changes with the deletion of the first putative binding site (−1755/−1747). Deletion of the second potential binding site (−1397/−1387) significantly reduced the relative luciferase activity of P2 (Figure 3B), which is accordance with the results of Figure 2. However, compared with P3, deletion of the third (−973/−964) and forth binding site (−936/−925) could significantly increase the relative luciferase activity (Figure 3B), which didn’t correspond with the results of Figure 2. Therefore, −1397/−1387 might be the exact binding site of C/EBPβ and was selected for further analysis.
Figure 3.
C/EBPβ binding at −1397/−1387 region of STAT3. (A) Predictions of potential C/EBPβ binding sites at the promoter of STAT3; (B) relative luciferase activity of STAT3 promoter after the 5′ deletion of the potential C/EBPβ binding sites; (C) mutation of the potential C/EBPβ binding site (−1397/−1387); (D) relative luciferase activity of P2 after the mutation of −1397/−1387; (E) further confirmation of C/EBPβ binding at −1397/−1387 by ChIP. DNA Marker was 100 bp. Relative luciferase activity of P2 after the knockdown and over-expression of C/EBPβ by C/EBPβ-siRNA (F) and pcDNA3.1-C/EBPβ (G), respectively. ** indicates p < 0.01; * indicates p < 0.05. Data were represented as means ± SD.
To validate STAT3 as a target of C/EBPβ, this potential site of P2 was mutated and then were cloned into the dual-luciferase reporter (Figure 3C). We found that mutation significantly reduced the relative luciferase activity of P2 (Figure 3D). Moreover, ChIP further confirmed that C/EBPβ bound at (−1397/−1387) in porcine GCs (Figure 3E), and siRNA-C/EBPβ significantly decreased the relative luciferase of P2 (Figure 3F, p < 0.01). Moreover, pcDNA3.1-C/EBPβ significantly increased the relative luciferase of P2 (Figure 3G, p < 0.01). These results suggested that C/EBPβ directly bound at −1397/−1387 of STAT3 to positively regulate the transcription of STAT3 in porcine GCs.
3.3. STAT3 Inhibits Apoptosis and Promotes Proliferation of Ovarian GCs
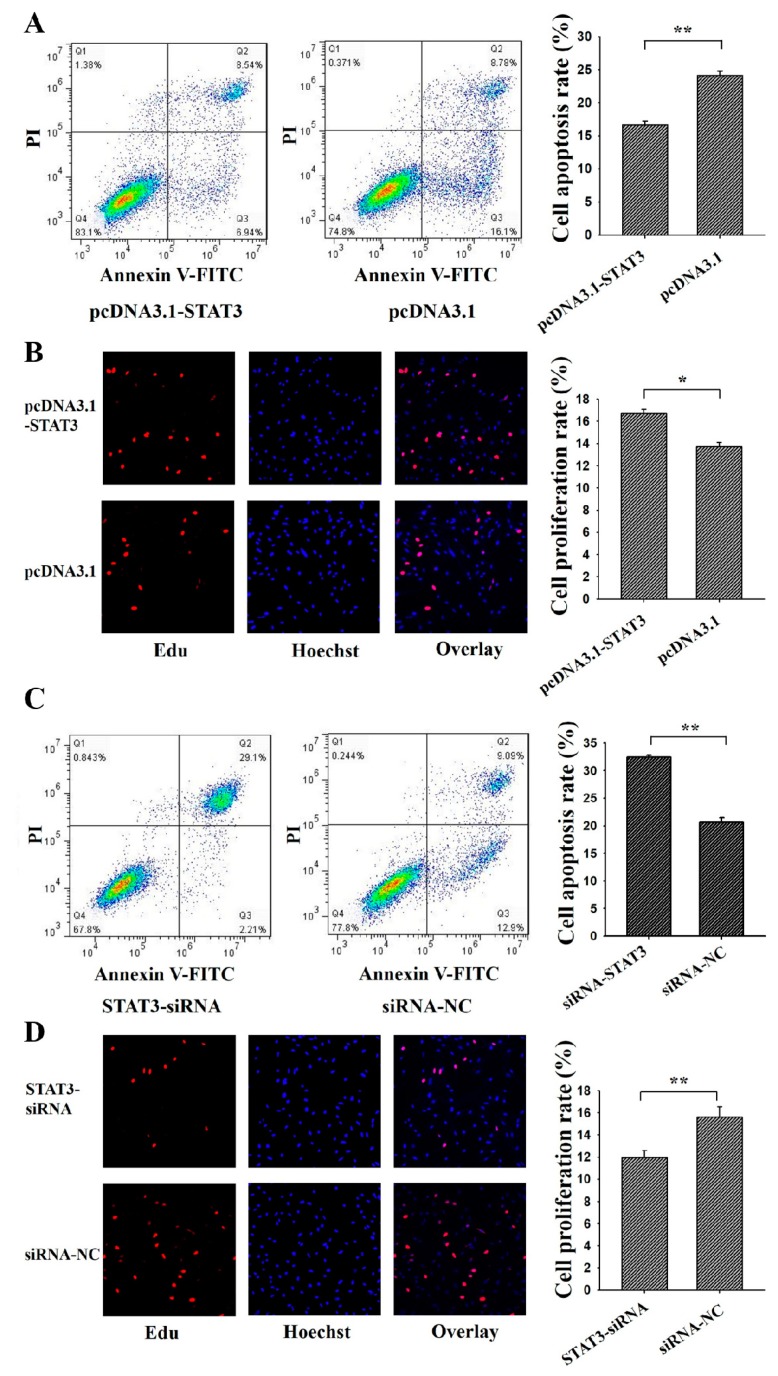
To determine the cellular function of STAT3 on the apoptosis and proliferation, pcDNA3.1-STAT3 or STAT3-siRNA was transfected into porcine GCs (Figure 4). Annexin V-FITC flow cytometry and Edu staining were used to analysis the cell apoptosis and proliferation, respectively. Results showed that the apoptosis rate of GCs in the pcDNA3.1-STAT3 group was significantly lower than the control group (Figure 4A), and the proliferation rate in the pcDNA3.1-STAT3 group was significantly higher than that in the control group (Figure 4B). Moreover, the apoptosis rate of GCs in STAT3-siRNA was significantly higher than that in the control group (Figure 4C), and the proliferation rate in STAT3-siRNA group was significantly lower than that in the control group (Figure 4D). These results suggested that STAT3 might inhibit apoptosis and promote proliferation of ovarian GCs.
Figure 4.
STAT3 regulates cell apoptosis and proliferation of ovarian granulosa cells (GCs). PcDNA3.1-STAT3 decreased cell apoptosis (A) and increased cell proliferation (B) in GCs as assessed by Annexin V-FITC/PI and Edu staining, respectively. SiRNA-STAT3 increased cell apoptosis (C,D) decreased cell proliferation in porcine GCs as assessed by Annexin V-FITC/PI and Edu staining, respectively. For Annexin V-FITC/P analysis, cells in the lower right quadrant were annexin-positive/PI-negative early apoptotic cells. The cells in the upper right quadrant were annexin-positive/PI-positive late apoptotic cells. The percentage of cells undergoing early and late apoptosis were presented in the relating barplot. ** indicates p < 0.01; * indicates p < 0.05. Data were represented as means ± SD.
3.4. C/EBPβ Inhibits Apoptosis and Promotes Proliferation of Ovarian GCs
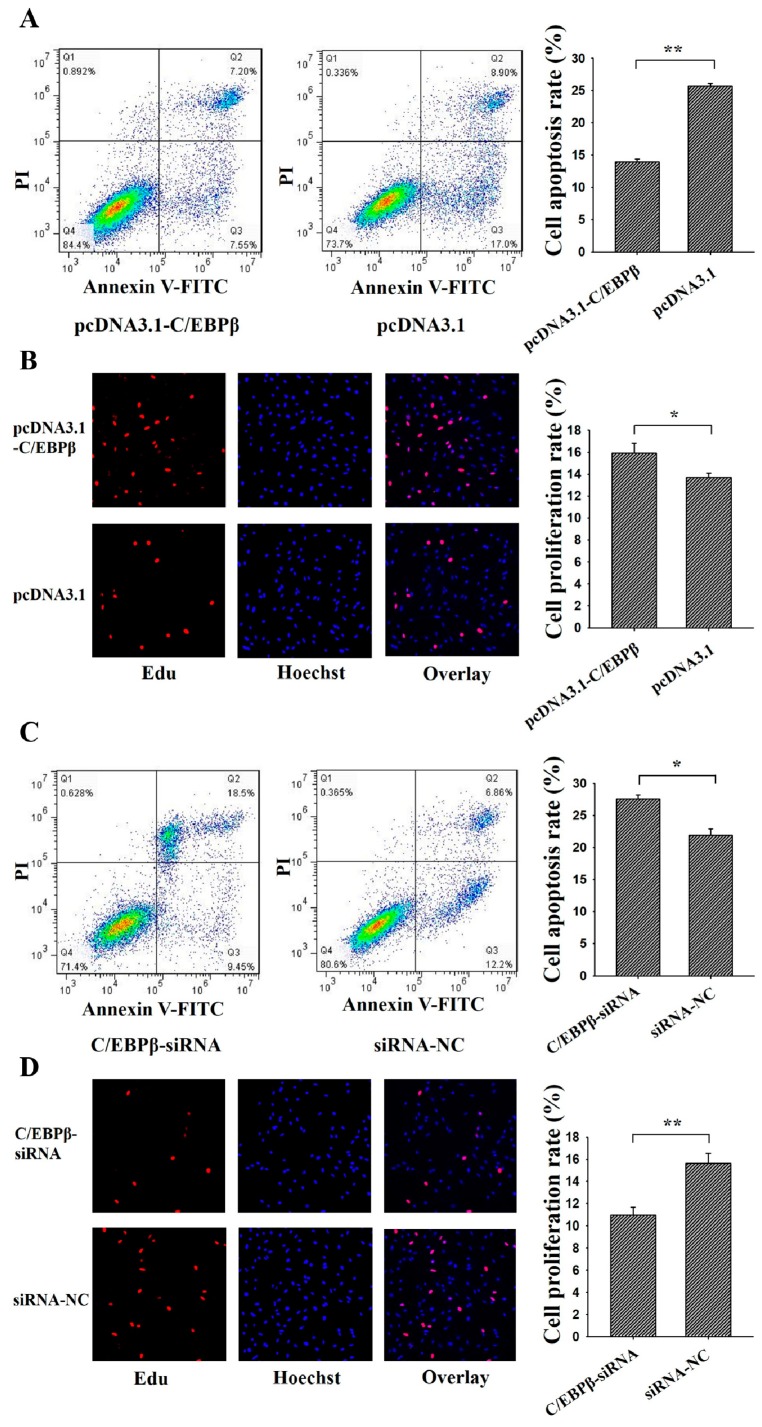
To investigate the function of C/EBPβ on cell apoptosis and cell proliferation, pcDNA3.1-C/EBPβ or C/EBPβ-siRNA was transfected into porcine GCs (Figure 5). Results showed that the apoptosis rate of GCs in pcDNA3.1-C/EBPβ group was significantly lower than that in the control group (Figure 5A), and the proliferation rate in the pcDNA3.1-C/EBPβ group was significantly higher than that in the control group (Figure 5B). Furthermore, the apoptosis rate of GCs in the C/EBPβ-siRNA group was significantly higher than that in the control group (Figure 5C), and the proliferation rate in the C/EBPβ-siRNA group was significantly lower than that in the control group (Figure 5D). These results demonstrated that C/EBPβ might inhibit cell apoptosis and promote cell proliferation for porcine GCs.
Figure 5.
C/EBPβ regulates the apoptosis and proliferation of ovarian GCs. PcDNA3.1-C/EBPβ decreased cell apoptosis (A) and increased cell proliferation (B) in granulosa cells as assessed by Annexin V-FITC/PI and Edu staining, respectively. SiRNA-C/EBPβ increased cell apoptosis (C,D) decreased cell proliferation in granulosa cells as assessed by Annexin V-FITC/PI and Edu staining, respectively. For Annexin V-FITC/P analysis, cells in the lower right quadrant were annexin-positive/PI-negative early apoptotic cells. The cells in the upper right quadrant were annexin-positive/PI-positive late apoptotic cells. The percentage of cells undergoing early and late apoptosis were presented in the relating barplot. ** indicates p < 0.01; * indicates p < 0.05. Data were represented as means ± SD.
3.5. C/EBPβ Enhanced the Antiapoptotic and Pro-Proliferation Effects of STAT3 in Ovarian GCs
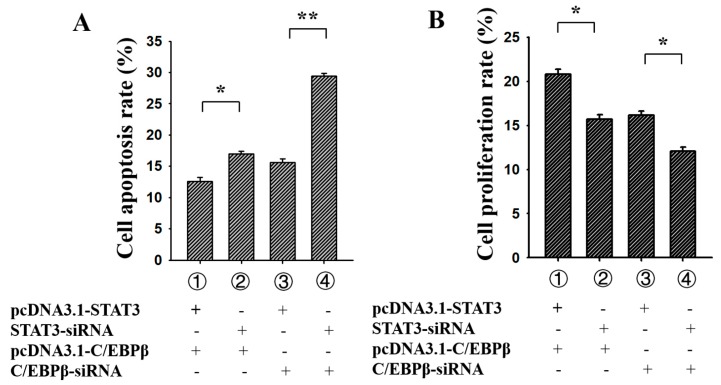
PcDNA3.1-C/EBPβ, C/EBPβ-siRNA, pcDNA3.1-STAT3 and STAT3-siRNA were co-transfected into porcine GCs to analyze the effect of C/EBPβ on the function of STAT3, respectively (Figure 6). For the apoptosis rate of porcine GCs (Figure 6A), group 1 (pcDNA3.1-C/EBPβ + pcDNA3.1-STAT3) was significantly lower than group 2 (pcDNA3.1-C/EBPβ + STAT3-siRNA), and group 3 (pcDNA3.1-STAT3 + C/EBPβ-siRNA) was significantly lower than group 4 (STAT3-siRNA + C/EBPβ-siRNA). These results indicated that C/EBPβ might enhance the antiapoptotic effect of STAT3 in ovarian GCs. For the proliferation rate of porcine GCs (Figure 6B), group 1 (pcDNA3.1-C/EBPβ + pcDNA3.1-STAT3) was significantly higher than group 2 (pcDNA3.1-C/EBPβ + STAT3-siRNA), while group 3 (pcDNA3.1-STAT3 + C/EBPβ-siRNA) was significantly higher than group 4 (STAT3-siRNA + C/EBPβ-siRNA). These results suggested that C/EBPβ might enhance pro-proliferation effects of STAT3 in ovarian GCs.
Figure 6.
C/EBPβ enhanced the antiapoptotic and pro-proliferation effects of STAT3 in porcine GCs. (A) C/EBPβ enhanced the antiapoptotic effects of STAT3; (B) C/EBPβ enhanced the pro-proliferation effects of STAT3. The percentage of cells undergoing early and late apoptosis were presented. ** indicates p < 0.01; * indicates p < 0.05. Data were represented as means ± SD.
4. Discussion
It is widely known that the regular cell apoptosis and proliferation of GCs supported the development of follicles in mammals [5,6]. In mice, GC-specific STAT3 knockout could impair the fertility with a significant reduction in litters and litter sizes [18], but oocyte-specific STAT3 knockout had no effect on fertility [18]. Moreover, the GC-specific C/EBPβ knockout also resulted in the subfertility with a reduction of corpus luteums in 70% [19]. These results suggested that STAT3 and C/EBPβ had exhibited an essential role in folliculogenesis and female reproduction. Previous studies recommended that C/EBPβ might display a cis-regulatory role in mammalian GCs [21,32]. We previously found there were several potential C/EBPβ sites on the promoter of STAT3 in humans, mice, and pigs (Figure 1), and furthermore, we found that mRNA expression of STAT3 had significantly increased or decreased along with the overexpression or depression of C/EBPβ (Figure 2). This observation indicated that C/EBPβ was likely to play a positive regulatory role for STAT3 in porcine GCs.
In pigs, four putative C/EBPβ binding sites were predicted at the promoter of STAT3 (Figure 3A), suggesting that STAT3 might be a direct target of C/EBPβ. Based on the dual luciferase reporter system, the deletion of −1397/−1387 binding sites resulted in a significant reduction of the relative luciferase activity. This observation was in accordance with results shown in Figure 2. Furthermore, after mutation of this putative binding site, the relative luciferase activity showed a significant reduction compared to that of P2 (Figure 3C,D). Then C/EBPβ was further confirmed to bind at −1397/−1387 of STAT3 by ChIP (Figure 3E). These resulted demonstrated that C/EBPβ directly targeted at STAT3 to positively regulate the expression of STAT3 in porcine GCs.
Previous studies have found that lower levels of phosphorylated STAT3 in GCs may be related to ovarian leptin resistance and decreased fertilization in polycystic ovarian syndrome women [33]. STAT3 is required to regulate the expression of luteinizting hormone receptor mRNA during the cell differentiation of GCs in mice [34]. In bovine GCs, protein expression of STAT3 appears to be stronger in the ovulatory follicles as compared to the small follicles and dominant follicles, and these results mean that STAT3 in GCs may be associated with the follicular growth [35], and phosphorylated STAT3 levels are dramatically higher in GCs of subordinate follicles than those in dominant follicles in bovines [36]. Furthermore, it is recently proposed that the stronger amount of pSTAT3 in small follicles is associated with GCs death and follicular atresia [37]. These observations suggested that STAT3 got involved in the functions of GCs and follicular development. Most importantly, in human GCs, promoting ovarian GCs apoptosis and affecting the steroidogenesis could suppress the activation of STAT3 [38], and reduced expression of STAT3 could enhance the early apoptosis rate of mouse GCs [17]. These results indicated STAT3 might induce the apoptosis of mammalian GCs. In the present study, we found that STAT3 inhibited cell apoptosis and promotes cell proliferation in porcine GCs (Figure 4), and these results further identified the biological functions of STAT3 on cell apoptosis and cell proliferation in mammals.
C/EBPβ is essential for ovulation and luteinization [39,40]. Previous studies report that C/EBPα and C/EBPβ double-mutant could change expressions of Prlr, Abcb1b1, Plxnd1 and Cyp19a1 in mouse GCs, and moreover, putative C/EBPβ binding sites have been identified in the promoters of these genes [39]. C/EBPβ knockdown could alter Star, Cyp11a1, and Hsd3b2 expressions in granulosa tumour-derived KGN cells [41] and result in a reduction of Runx2 mRNA levels in periovulatory GC of mice and thus might impact cell differentiation of GCs to become luteal cells [42]. These observations suggested C/EBPβ was an important transcript factor in mammalian GCs. Moreover, by binding at the promoter, C/EBPβ was likely to be essentially required for Star, which was a vital accessory protein required for biosynthesis of steroid hormones from cholesterol, in response to hormones in rat [32], mouse [43], human [44], and porcine [45] GCs. In the present study, we found that C/EBPβ inhibited cell apoptosis and promoted cell proliferation in porcine GCs (Figure 5). We also found C/EBPβ enhanced the antiapoptotic and pro-proliferation effects of STAT3 in porcine GCs (Figure 6). These results further certified and confirmed that STAT3 was a target of C/EBPβ in mammalian GCs. Collectively, in porcine ovarian GCs, C/EBPβ targeted at −1397/−1387 region to promote the expression of STAT3, and moreover, C/EBPβ might enhance the antiapoptotic and pro-proliferative effects of STAT3. These present works could provide usefully biological information for further studies on the molecular mechanism of C/EBPβ and STAT3 in ovarian follicular development.
Acknowledgments
This work was supported by the earmarked fund for China Agriculture Research System (CARS-35), the Basic Work of Science and Technology Project (2014FY120800) and Guangdong Sailing Program (2014YT02H042).
Author Contributions
Conceived and designed the experiments: X.Y., X.Z. and J.L. Prepared biological samples: Y.H., Y.Z. and A.Z. Wrote the paper: X.Y., X.Z. and J.L. Revised the paper: J.L., Z.Z., H.Z. and A.Z. All authors reviewed and approved the final manuscript.
Funding
This research was funded by the earmarked fund for China Agriculture Research System (CARS-35), the Basic Work of Science and Technology Project (2014FY120800) and Guangdong Sailing Program (2014YT02H042).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Shirasawa H., Terada Y. In vitro maturation of human immature oocytes for fertility preservation and research material. Reprod. Med. Biol. 2017;16:258–267. doi: 10.1002/rmb2.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiwari M., Prasad S., Tripathi A., Pandey A.N., Ali I., Singh A.K., Shrivastav T.G., Chaube S.K. Apoptosis in mammalian oocytes: A review. Apoptosis. 2015;20:1019–1025. doi: 10.1007/s10495-015-1136-y. [DOI] [PubMed] [Google Scholar]
- 3.Svechnikov K., Soder O. Ontogeny of gonadal sex steroids. Best Pract. Res. Clin. Endocrinol. Metab. 2008;22:95–106. doi: 10.1016/j.beem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Newman D.M., Jones P.L., Ingram B.A. Age-related changes in ovarian characteristics, plasma sex steroids and fertility during pubertal development in captive female murray cod Maccullochella peelii peelii. Comp. Biochem. Phys. 2008;150:444–451. doi: 10.1016/j.cbpa.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Rooke J.A., Ewen M., Mackie K., Staines M.E., McEvoy T.G., Sinclair K.D. Effect of ammonium chloride on the growth and metabolism of bovine ovarian granulosa cells and the development of ovine oocytes matured in the presence of bovine granulosa cells previously exposed to ammonium chloride. Anim. Reprod. Sci. 2004;84:53–71. doi: 10.1016/j.anireprosci.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Kubo N., Cayo-Colca I.S., Miyano T. Effect of estradiol-17β during in vitro growth culture on the growth, maturation, cumulus expansion and development of porcine oocytes from early antral follicles. Anim. Sci. J. 2015;86:251–259. doi: 10.1111/asj.12283. [DOI] [PubMed] [Google Scholar]
- 7.McGee E.A., Hsueh A.J. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000;21:200–214. doi: 10.1210/er.21.2.200. [DOI] [PubMed] [Google Scholar]
- 8.Hunter M.G. Oocyte maturation and ovum quality in pigs. Rev. Reprod. 2000;5:122–130. doi: 10.1530/ror.0.0050122. [DOI] [PubMed] [Google Scholar]
- 9.De Cesaro M.P., Dos Santos J.T., Ferst J.G., Nobrega J.E., Jr., Rosa P., Rovani M.T., Ilha G.F., Bohrer R.C., Ferreira R., Gasperin B.G., et al. Natriuretic peptide system regulation in granulosa cells during follicle deviation and ovulation in cattle. Reprod. Domest. Anim. 2018;53:710–717. doi: 10.1111/rda.13161. [DOI] [PubMed] [Google Scholar]
- 10.Skowronski M.T., Mlotkowska P., Tanski D., Lepiarczyk E., Oklinski M.K., Nielsen S., Skowronska A. Pituitary gonadotropins, prolactin and growth hormone differentially regulate AQP1 expression in the porcine ovarian follicular cells. Int. J. Mol. Sci. 2018;19:5. doi: 10.3390/ijms19010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda F., Inoue N., Manabe N., Ohkura S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 12.Hamm M.L., Bhat G.K., Thompson W.E., Mann D.R. Folliculogenesis is impaired and granulosa cell apoptosis is increased in leptin-deficient mice. Biol. Reprod. 2004;71:66–72. doi: 10.1095/biolreprod.104.027292. [DOI] [PubMed] [Google Scholar]
- 13.Hall S.E., Upton R.M.O., McLaughlin E.A., Sutherland J.M. Phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) follicular signalling is conserved in the mare ovary. Reprod. Fertil. Dev. 2018;30:624–633. doi: 10.1071/RD17024. [DOI] [PubMed] [Google Scholar]
- 14.Kim H., Nakajima T., Hayashi S., Chambon P., Watanabe H., Iguchi T., Sato T. Effects of diethylstilbestrol on programmed oocyte death and induction of polyovular follicles in neonatal mouse ovaries. Biol. Reprod. 2009;81:1002–1009. doi: 10.1095/biolreprod.108.070599. [DOI] [PubMed] [Google Scholar]
- 15.Wen L., Craig J., Dyce P.W., Li J. Cloning of porcine signal transducer and activator of transcription 3 cDNA and its expression in reproductive tissues. Reproduction. 2006;132:511–518. doi: 10.1530/rep.1.01055. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez A.G., Bahr J.M. Role of FSH and epidermal growth factor (EGF) in the initiation of steroidogenesis in granulosa cells associated with follicular selection in chicken ovaries. Reproduction. 2003;125:683–691. doi: 10.1530/rep.0.1250683. [DOI] [PubMed] [Google Scholar]
- 17.Wang C., Li D., Zhang S., Xing Y., Gao Y., Wu J. MicroRNA-125a-5p induces mouse granulosa cell apoptosis by targeting signal transducer and activator of transcription 3. Menopause. 2016;23:100–107. doi: 10.1097/GME.0000000000000507. [DOI] [PubMed] [Google Scholar]
- 18.Robker R.L., Watson L.N., Robertson S.A., Dunning K.R., McLaughlin E.A., Russell D.L. Identification of sites of STAT3 action in the female reproductive tract through conditional gene deletion. PLoS ONE. 2014;9:e101182. doi: 10.1371/journal.pone.0101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan H.Y., Liu Z., Shimada M., Sterneck E., Johnson P.F., Hedrick S.M., Richards J.S. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324:938–941. doi: 10.1126/science.1171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sterneck E., Tessarollo L., Johnson P.F. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirois J., Richards J.S. Transcriptional regulation of the rat prostaglandin endoperoxide synthase 2 gene in granulosa cells. Evidence for the role of a cis-acting C/EBP beta promoter element. J. Biol. Chem. 1993;268:21931–21938. [PubMed] [Google Scholar]
- 22.Signal Transducer and Activator of Transcription 3 Homo sapiens (Human) [(accessed on 28 May 2018)]; Available online: https://www.ncbi.nlm.nih.gov/gene/6774.
- 23.Signal Transducer and Activator of Transcription 3Mus musculus (House Mouse) [(accessed on 28 May 2018)]; Available online: https://www.ncbi.nlm.nih.gov/gene/20848.
- 24.Signal Transducer and Activator of Transcription 3 Sus scrofa (Pig) [(accessed on 28 May 2018)]; Available online: https://www.ncbi.nlm.nih.gov/gene/733648.
- 25.Tsunoda T., Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 26.Biobase. [(accessed on 28 May 2018)]; Available online: http://gene-regulation.com/pub/programs/alibaba2/index.html.
- 27.Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R., et al. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Research. [(accessed on 28 May 2018)]; Available online: http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3.
- 29.Lai T.C., Li H.F., Li Y.S., Hung P.Y., Shyu M.K., Hu M.C. Proximal GATA-binding sites are essential for human HSD3B1 gene transcription in the placenta. Sci. Rep. 2017;7:4271. doi: 10.1038/s41598-017-04133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Du X., Zhou J., Pan Z., Liu H., Li Q. MicroRNA-26b functions as a proapoptotic factor in porcine follicular Granulosa cells by targeting Sma-and Mad-related protein 4. Biol. Reprod. 2014;91:146. doi: 10.1095/biolreprod.114.122788. [DOI] [PubMed] [Google Scholar]
- 31.Chen L.J., Feng L.J., Wang X., Du J., Chen Y., Yang W., Zhou C.Y., Cheng L., Shen Y.J., Fang S.Y., et al. Mesencephalic astrocyte-derived neurotrophic factor is involved in inflammation by negatively regulating the NF-kB pathway. Sci. Rep. 2015;5:8133. doi: 10.1038/srep08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman E., Eimerl S., Orly J. CCAAT enhancer-binding protein beta and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells. J. Biol. Chem. 1999;274:17987–17996. doi: 10.1074/jbc.274.25.17987. [DOI] [PubMed] [Google Scholar]
- 33.Li M.G., Ding G.L., Chen X.J., Lu X.P., Dong L.J., Dong M.Y., Yang X.F., Lu X.E., Huang H.F. Association of serum and follicular fluid leptin concentrations with granulosa cell phosphorylated signal transducer and activator of transcription 3 expression in fertile patients with polycystic ovarian syndrome. J. Clin. Endocrinol. Metab. 2007;92:4771–4776. doi: 10.1210/jc.2007-0978. [DOI] [PubMed] [Google Scholar]
- 34.Imai F., Kishi H., Nakao K., Nishimura T., Minegishi T. IL-6 up-regulates the expression of rat LH receptors during granulosa cell differentiation. Endocrinology. 2014;155:1436–1444. doi: 10.1210/en.2013-1821. [DOI] [PubMed] [Google Scholar]
- 35.Ndiaye K., Castonguay A., Benoit G., Silversides D.W., Lussier J.G. Differential regulation of Janus kinase 3 (JAK3) in bovine preovulatory follicles and identification of JAK3 interacting proteins in granulosa cells. J. Ovarian Res. 2016;9:71. doi: 10.1186/s13048-016-0280-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasperin B.G., Rovani M.T., Ferreira R., Ilha G.F., Bordignon V., Goncalves P.B., Duggavathi R. Functional status of STAT3 and MAPK3/1 signaling pathways in granulosa cells during bovine follicular deviation. Theriogenology. 2015;83:353–359. doi: 10.1016/j.theriogenology.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Ilha G.F., Rovani M.T., Gasperin B.G., Antoniazzi A.Q., Goncalves P.B., Bordignon V., Duggavathi R. Lack of FSH support enhances LIF-STAT3 signaling in granulosa cells of atretic follicles in cattle. Reproduction. 2015;150:395–403. doi: 10.1530/REP-15-0026. [DOI] [PubMed] [Google Scholar]
- 38.Ding X., Kou X., Zhang Y., Zhang X., Cheng G., Jia T. Leptin siRNA promotes ovarian granulosa cell apoptosis and affects steroidogenesis by increasing NPY2 receptor expression. Gene. 2017;633:28–34. doi: 10.1016/j.gene.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 39.Fan H.Y., Liu Z.L., Johnson P.F., Richards J.S. CCAAT/enhancer-binding proteins (C/EBP)-alpha and -beta are essential for ovulation, luteinization, and the expression of key target genes. Mol. Endocrinol. 2011;25:253–268. doi: 10.1210/me.2010-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren Y.A., Liu Z.L., Mullany L.K., Fan C.M., Richards J.S. Growth arrest specific-1 (GAS1) is a C/EBP target gene that functions in ovulation and corpus luteum formation in mice. Biol. Reprod. 2016;94 doi: 10.1095/biolreprod.115.133058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizutani T., Ju Y.F., Imamichi Y., Osaki T., Yazawa T., Kawabe S., Ishikane S., Matsumura T., Kanno M., Kamiki Y., et al. C/EBPβ (CCAAT/enhancer-binding protein β) mediates progesterone production through transcriptional regulation in co-operation with SF-1 (steroidogenic factor-1) Biochem. J. 2014;460:459–471. doi: 10.1042/BJ20131522. [DOI] [PubMed] [Google Scholar]
- 42.Park E.S., Lind A.K., Dahm-Kahler P., Brannstrom M., Carletti M.Z., Christenson L.K., Curry T.E., Jr., Jo M. RUNX2 transcription factor regulates gene expression in luteinizing granulosa cells of rat ovaries. Mol. Endocrinol. 2010;24:846–858. doi: 10.1210/me.2009-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Son D.S., Terranova P.F., Roby K.F. Interaction of adenosine 3′,5′-cyclic monophosphate and tumor necrosis factor-alpha on serum amyloid A3 expression in mouse granulosa cells: dependence on CCAAT-enhancing binding protein-beta isoform. Endocrinology. 2010;151:3407–3419. doi: 10.1210/en.2009-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christenson L.K., Johnson P.F., McAllister J.M., Strauss J.F. CCAAT/enhancer-binding proteins regulate expression of the human steroidogenic acute regulatory protein (StAR) gene. J. Biol. Chem. 1999;274:26591–26598. doi: 10.1074/jbc.274.37.26591. [DOI] [PubMed] [Google Scholar]
- 45.LaVoie H.A., Singh D., Hui Y.Y. Concerted regulation of the porcine steroidogenic acute regulatory protein gene promoter activity by follicle-stimulating hormone and insulin-like growth factor I in granulosa cells involves GATA-4 and CCAAT/enhancer binding protein beta. Endocrinology. 2004;145:3122–3134. doi: 10.1210/en.2003-1719. [DOI] [PubMed] [Google Scholar]