Publisher's Note: There is a Blood Commentary on this article in this issue.
Key Points
hsa-mir183/EGR1/E2F1 is a novel and critical factor for CML SPC survival.
E2F1 plays a pivotal role in regulating CML SPC proliferation status.
Abstract
Chronic myeloid leukemia (CML) stem/progenitor cells (SPCs) express a transcriptional program characteristic of proliferation, yet can achieve and maintain quiescence. Understanding the mechanisms by which leukemic SPCs maintain quiescence will help to clarify how they persist during long-term targeted treatment. We have identified a novel BCR-ABL1 protein kinase–dependent pathway mediated by the upregulation of hsa-mir183, the downregulation of its direct target early growth response 1 (EGR1), and, as a consequence, upregulation of E2F1. We show here that inhibition of hsa-mir183 reduced proliferation and impaired colony formation of CML SPCs. Downstream of this, inhibition of E2F1 also reduced proliferation of CML SPCs, leading to p53-mediated apoptosis. In addition, we demonstrate that E2F1 plays a pivotal role in regulating CML SPC proliferation status. Thus, for the first time, we highlight the mechanism of hsa-mir183/EGR1–mediated E2F1 regulation and demonstrate this axis as a novel, critical factor for CML SPC survival, offering new insights into leukemic stem cell eradication.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disease of hemopoietic stem cell (HSC) origin resulting from the chromosomal translocation t(9;22)(q34;q11) that gives rise to the fusion gene, BCR-ABL1. ABL tyrosine kinase inhibitors (TKIs) lead to long-term remission in the majority of CML patients, but ∼50% of cases relapse when treatment is discontinued, as TKIs are unable to fully eradicate quiescent stem/progenitor cells (SPCs),1-9 despite being able to inhibit BCR-ABL signaling in these cells.6,9,10 Investigations have demonstrated that TKI-resistant SPCs in patients show reduced levels of BCR-ABL1 expression as compared with baseline, and exhibit a more primitive, quiescent transcriptional signature that becomes predominant over time in response to TKI therapy.11-14 It is not currently known whether this quiescent signature is driven by CML SPC intrinsic signaling by the microenvironment, or by a combination of both.
One possible candidate for cell-intrinsic regulation of the quiescent CML phenotype is the transcription factor E2F1, which regulates cell proliferation by activating genes important for G1–S-phase progression.15 In mice, deletion of E2f1 resulted in increased T-cell numbers,16,17 whereas combined loss of E2f1/2/3 affected mature hemopoietic cell proliferation18,19 and survival of the myeloid lineage,16,20 yet no effect was demonstrated on HSC function.21,22 In CML, E2F3 was shown to be important for disease initiation23 and silencing of E2F1 in K562 cells or CD34+ cells led to activation of PP2A and BCR-ABL1 suppression.24 In mouse fibroblasts, triple inactivation of E2f1/2/3 led to p53 activation and cell-cycle arrest,25 whereas deletion of p53 restored E2F1 transcriptional activity.26 In recent work, we have demonstrated that p53 acts as a key signaling hub to maintain survival of CML SPCs.27
As BCR-ABL had previously been shown to modulate microRNA (miRNA) levels,28 to gain insight into how CML SPCs maintain quiescence, we investigated novel and publicly deposited messenger RNA (mRNA)/microRNA transcriptomic data sets derived from primitive human CML SPCs.27,29-34 Here, we show that in CML SPCs, the cancer-related miRNA hsa-mir183 is highly expressed in a BCR-ABL1–dependent manner and hypothesize that this miRNA deregulates specific SPC-intrinsic mechanisms. Interestingly, it has been reported that hsa-mir183 targets early growth response 1 (EGR1), a member of the immediate early response transcription factor family that regulates proliferation and mobilization of healthy SPCs. The genetic network regulated by EGR1 responsible for stem cell division and migration has not been entirely elucidated. Interestingly, EGR1 loss has been shown to promote development of BCR-ABL1–mediated leukemia, whereas constitutive Egr1 is able to mitigate leukemia conferred by deregulated E2F1 leukemic cells.35-37 Furthermore, it has recently been shown that genetic deletion of Egr1 accelerates BCR-ABL1–driven CML.38 Here, we identify a novel CML-specific pathway in which BCR-ABL1 protein kinase regulates hsa-mir183–mediated inhibition of EGR1 leading to upregulation of E2F1. In addition, we investigate the role that hsa-mir183/EGR1–mediated E2F1 expression plays in priming proliferation in CML SPCs.
Methods
Cell isolation and culture
Fresh leukapheresis or peripheral blood (PB) samples were obtained from patients (informed consent) with chronic-phase (CP) CML at diagnosis or non-CML donors (defined as healthy). The in vitro studies with patient material were approved by the West of Scotland Research Ethics Committee 4, National Health Service (NHS) Greater Glasgow and Clyde (15-WS-0077). Samples were enriched for CD34+, Pyronin Y− (PY) Hoechst− (Ho), CD34+38+, and CD34+38− populations as described.4 Dual-fluorescence in situ hybridization (D-FISH) was performed as previously described.39 Where indicated, cells were treated with imatinib, dasatinib, nilotinib (Selleckchem), or colcemid. Colony-forming assays (CFCs) in methylcellulose medium (H4434, M3434; StemCell Technologies) were performed as described.40
RNA extraction and Q-PCR
RNA extraction was performed using the RNeasy Mini kit (Qiagen); complementary DNA (cDNA) was synthesized using the High-Capacity cDNA RT kit (Applied Biosystems), direct 1-step quantitative polymerase chain reaction (Q-PCR; Invitrogen), the Cells-to-CT kit for miRNA (Ambion), or the Power SYBR Green Cells-to-Ct kit (Ambion/Lifetech). Q-PCR was performed on the ABI7900 (Applied Biosystems) or Fluidigm platforms (Fluidigm Corporation).
Microarray data analysis
The CML vs healthy microarray data were obtained from ArrayExpress (E-MTAB-2508; Affymetrix Human Genome U133A) and describe gene expression in quiescent CML/healthy CD34+PY−Ho− cells. Differential expression was calculated using rank products (RPs; false discovery rate [FDR] calculated using 1000 permutations).41
For the TKI treatment data, cells were treated for 8 hours (CD34+38−) or 7 days (CD34+) with 5 μM imatinib, 150 nM dasatinib, or 5 μM nilotinib (no growth factors). cDNA from viable cells (7 days only) was hybridized to Affymetrix Human Gene 1.0 ST arrays (E-MTAB-2594) and normalized using robust multiarray average.42 Differentially expressed genes were identified using a paired-sample Limma analysis43 with pooled TKI treatments and a significance threshold of FDR ≤ 0.05 (Benjamini-Hochberg multiple testing correction was applied).44
For miRNA data, cDNA was hybridized to a miRNA chip based on Sanger miRBase Release 14 (LC Sciences) and normalized using a locally weighted regression (LOWESS) method on the background-subtracted data. Raw data are publicly available via ArrayExpress (accession E-MTAB-3220) and normalized data are provided in supplemental Table 1 (available on the Blood Web site). The resulting miRNA data were analyzed using Limma.43
Enrichment analysis
A PANTHER enrichment test (release 20141219) was used to identify enrichment of Gene Ontology (GO) biological process terms (release 20150111) in the list of genes identified as differentially expressed in CML vs healthy cells using RPs (supplemental Table 2). The Bonferroni correction was applied to the P values to account for multiple testing. The Gene Set Enrichment Analysis (GSEA) was carried out using GSEA (2-2.2.2) as obtained from the Broad Institute (gsea2-2.2.2.jar; http://software.broadinstitute.org/gsea/index.jsp)45; q values were calculated using 10 000 permutations of the phenotype label. The hypergeometric distribution was used to calculate enrichment statistics.
Comparison of distributions
One-sided Kolmogorov-Smirnov tests (using ks.test in the base R stats package) were carried out to identify positive shifts in distribution for a gene set, as compared with background. Ten thousand random subsamplings of the transcriptomic data were used to generate the expected null distribution for the Kolmogorov-Smirnov statistic for calculation of FDRs.
Western blotting
Western blotting was performed as described elsewhere.46 Antibodies used were c-ABL1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and tubulin (Cell Signaling); E2F1 (Upstate); p53-Do-1 (Santa Cruz Biotechnology).
Fluorescence-activated cell sorting, flow cytometry, and imaging analysis
Cells were stained with 7-aminoactinomycin D (7-AAD; Becton Dickinson), Zombie Aqua (Biolegend), the CellTrace Violet Cell Proliferation kit (Invitrogen), and 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) according to the manufacturer’s instructions. For intracellular analysis, cells were fixed and permeabilized using Fix and Perm (Merck Chemicals Ltd) or the Fixation/Permeabilization Solution kit (Becton Dickinson). Primary antibodies were phospho-p53-Ser15, p27 (R&D Systems), p21 (Santa Cruz Biotechnology), phospho-AKT-T308, phospho-STAT5-Y694, BCL2, Ki-67, Annexin V, and CD34 and CD38 (Becton Dickinson). Lin−Sca-1+c-Kit+ (LSK) cells were isolated as previously described.27
Retinoblastoma (Rb) phosphorylation was measured with the Cellomic Phospho-Rb activation kit (ThermoScientific) according to the manufacturer’s instructions and analyzed by the Operetta High Content Imaging System (Operetta; PerkinElmer UK).
Gene knockdown
E2F1 (short hairpin [sh]-E2F1, sh-E2F1-1, and sh-E2F1-2) and scrambled shRNAs were subcloned into the pLKO.1 GFP vector. Lentiviral infection was carried out as described elsewhere.9 A custom Mirzip lentiviral system (PGK promoter) and mirzip scrambled vector (Cambridge Bioscience Ltd) were used. E2F1-small interfering RNA (siRNA), -siRNA1, -siRNA2; p53-siRNA, -siRNA1; EGR1-siRNA; scrambled siRNA; scrambled siRNA1 (100 nM) were obtained from Ambion.
Luciferase assay
EGR1 cDNA was amplified from human mononuclear cells. The 3′untranslated region (UTR) sequence containing the hsa-mir183–binding site or a mutated binding site were cloned into the pmirGLO vector (Dual Glo Luciferase Assay System; Promega). KCL22 cells were cotransfected with pmirGLO vector containing a wild-type (WT) or mutated (MUT) EGR1 sequence, and 50 mM hsa-mir183 mimic or scrambled control (Integrated DNA Technologies) by electroporation. Luminescence was measured after 48 hours using a Glomax 20/20 luminometer (Promega).
Mice and BM transplantation
E2f1−/− mice were obtained from The Jackson Laboratory. Bone marrow (BM) transplants were performed by tail-vein injection into lethally irradiated (2 doses of 4.25 Gy) SJL C57/B6 CD45.1 recipients. One thousand LSK cells were transplanted from E2f1+/+ and E2f1−/− donors (CD45.2+) alongside 2 × 105 unfractionated BM support cells from SJL CD45.1 mice and kept on Baytril antibiotic for 2 weeks. Peripheral blood was analyzed at 4, 8, 12, and 16 weeks after transplantation, and BM was analyzed at 16 weeks posttransplant. Populations analyzed were LSK, long-term hemopoietic stem cells, short-term hemopoietic stem cells, multipotent progenitor 1, multipotent progenitor 2, granulocyte/macrophage progenitors, megakaryocyte/erythrocyte progenitors, common myeloid progenitors, myeloid cells, and B cells.
For secondary transplantations, 2000 CD45.2+ LSK cells from primary recipients were transplanted with 2 × 105 CD45.1+ support cells into CD45.1+ irradiated recipient mice.
Monoclonal antibodies against CD45.1 and CD45.2 were used. For C-kit+ (CD117+) cell isolation, BM cells were passed through MACS separation columns (Miltenyi Biotec) using the manufacturer’s instructions. C-kit–enriched cells were isolated using a FACSAria cell sorter (Becton Dickinson).
Statistics
Statistical analyses were performed using the Student t test. A threshold of P < .05 was defined as statistically significant (*). Levels of P < .01 (**), and P < .001 (***) were taken to be highly statistically significant.
Study approval
All animal experiments were carried out according to UK Home Office regulations.
Results
CML SPCs are predominantly quiescent despite expressing active BCR-ABL1
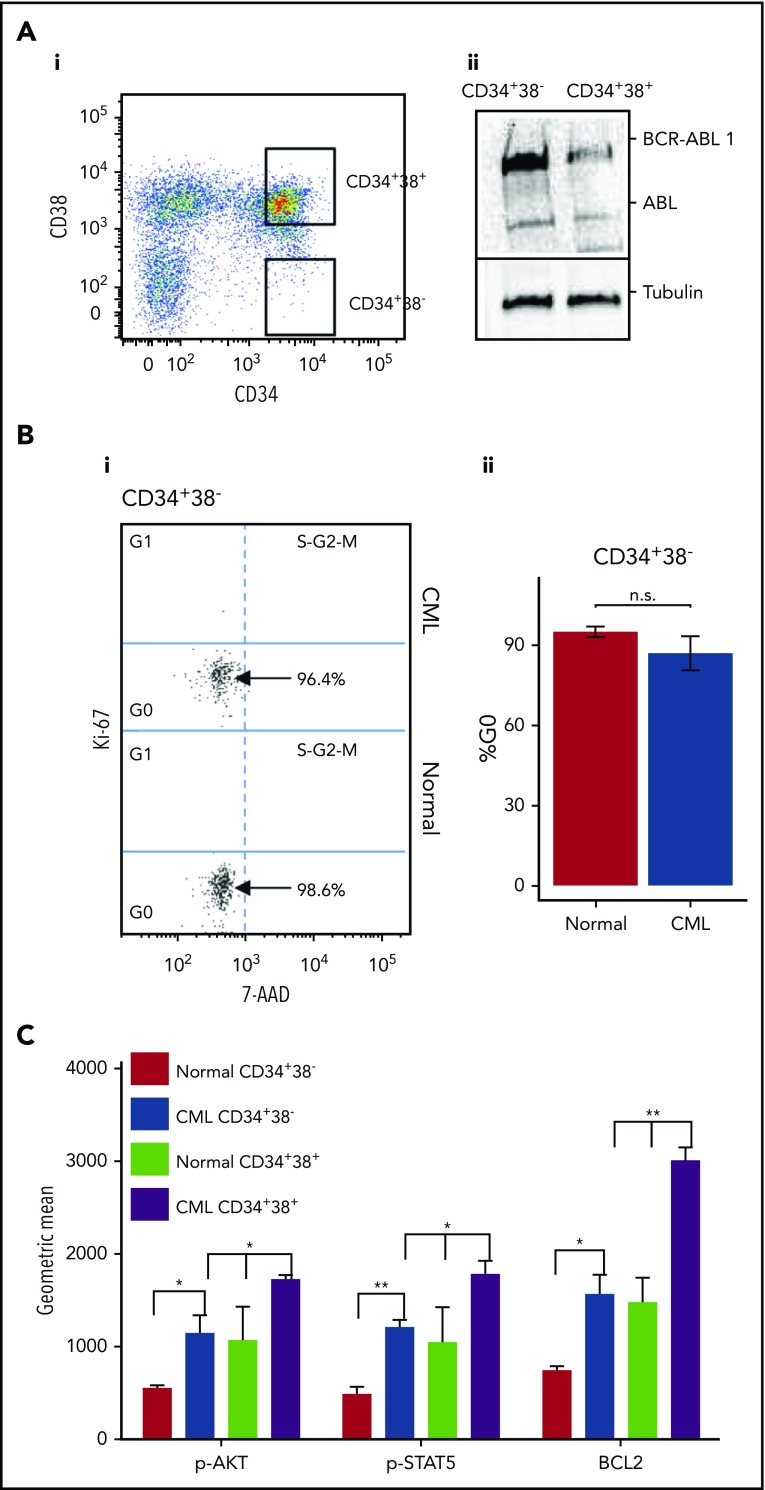
BCR-ABL1 confers a proliferative advantage to primitive CML cells.32 To investigate cell-cycle status, SPCs (CD34+38−) and more mature cells (CD34+38+) were isolated from primary healthy and CML CP samples (Figure 1Ai). As previously reported, CML SPCs expressed high levels of BCR-ABL1 (Figure 1Aii) when compared with more mature cells prior to treatment.40,47,48 Despite this, CML SPCs showed no significant difference in the percentage of quiescent cells (Figure 1B; replicates shown in supplemental Figure 1A). Similarly, mRNA levels of the cell-cycle inhibitors p21, p27, p57, and protein levels of p21 and p27 did not differ between healthy and CML SPCs (supplemental Figure 1B-C). The levels of phospho-AKT, phospho-STAT5, and BCL2, proteins downstream of BCR-ABL1, were higher in CML than healthy SPCs (Figure 1C), showing that, despite being quiescent at the functional level, BCR-ABL1 kinase activity and transcriptional machinery are active, suggesting that CML SPCs are “primed to proliferate,” as previously reported.49 Consistent with the published literature,6,9,10 BCR-ABL1 activity is affected by treatment with TKIs in SPCs, as evidenced by a decrease in the levels of its downstream targets phospho-STAT5 and phospho-AKT (supplemental Figure 1D). Although CML CD34+38− SPCs showed lower levels of BCL2 and less phosphorylation of AKT and STAT5 compared with mature CML CD34+38+ cells, these levels were still higher than in the equivalent population of healthy cells (Figure 1C).
Figure 1.
CML SPCs are predominantly quiescent. (Ai) Gating strategy for healthy and CML SPCs enriched for CD34+38− and CD34+38+ cells. (ii) Level of BCR-ABL1 and ABL measured in CD34+38− and CD34+38+ cells. (Bi) Representative dot plots showing the cell-cycle phases in healthy and CML SPCs measured by Ki-67/7-AAD staining. N = 1 representative sample shown. (ii) Percentage of healthy (n = 4) and CML (n = 4) SPCs in G0 cell-cycle phase. (C) Phosphorylation of AKT and STAT5 and level of BCL2 measured by fluorescence-activated cell sorting (FACS) in CML CD34+38− and CD34+38+cells. (*P < .05; **P < .01). n.s., not significant; p-AKT, phospho-AKT; p-STAT5, phospho-STAT5.
BCR-ABL1 regulates hsa-mir183 and EGR1 in CML SPCs
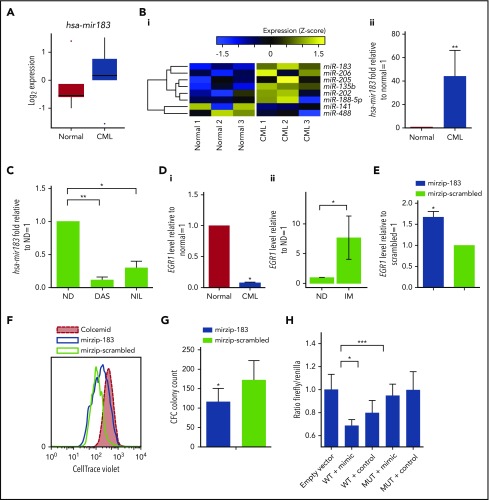
BCR-ABL1 modulates the expression of miRNAs,28 therefore, to elaborate networks that may regulate SPCs quiescence, we performed global miRNA expression profiling in healthy and CML SPCs. Several miRNAs were differentially expressed, including hsa-mir183, which was upregulated in CML (Figure 2A). Validation by OncoMir array confirmed that hsa-mir183 was significantly upregulated by 38-fold in CML vs healthy SPCs (Figure 2B). The upregulation of hsa-mir183 was BCR-ABL1 kinase dependent as its expression decreased upon treatment with TKIs (Figure 2C). Published evidence,35,36 together with computational target prediction by the miRWalk database,50 indicated that hsa-mir183 targets and downregulates EGR1, which plays a role in healthy SPC proliferation and development of BCR-ABL1–mediated leukemia. As predicted, EGR1 mRNA levels were significantly lower in CML vs healthy SPCs (Figure 2Di). This effect appeared to be BCR-ABL dependent as primary CML CD34+ cells demonstrated a significant increase in EGR1 expression following long-term exposure to TKI (P = .01; Figure 2Dii). To further confirm that the regulation of EGR1 was hsa-mir183 dependent, hsa-mir183 expression was knocked down in CML SPCs using a green fluorescent protein (GFP)-lentiviral-based anti-miRNA. GFP+ SPCs were analyzed for the level of the EGR1 transcripts, the expression of which was rescued by hsa-mir183 knockdown (Figure 2E). In addition, we found that SPCs with hsa-mir183 knockdown proliferated less than control cells (Figure 2F) and generated fewer colonies in CFC assays (Figure 2G).
Figure 2.
CML SPCs show regulation of the hsa-mir183/EGR1 axis. (A) Genome-wide miRNA expression for healthy and CML SPCs (N = 5 biological replicates). (Bi) Heatmap for the OncoMir MiRNA Q-PCR Array in SPCs showing statistically significant regulation of miRNAs between healthy and CML SPCs. (ii) Q-PCR for hsa-mir183 regulation in CML vs healthy SPCs. (C) CML SPCs treated for 24 hours with dasatinib (DAS; 150 nM) and nilotinib (NIL; 1 µM) and analysis by Q-PCR for hsa-mir183. (Di) mRNA level of the hsa-mir183 target gene EGR1 in healthy and CML CD34+38− cells. (ii) CML CD34+ cells treated for 7 days with imatinib (IM; 5 µM) and live cells analyzed by Q-PCR for EGR1 expression (N = 6). (E) hsa-mir183 knockdown GFP+ CML SPCs sorted and analyzed for EGR1 mRNA level by Q-PCR. Mir zip-scrambled vector was used as negative control. (F) Representative plot for cell divisions analyzed in hsa-mir183 knockdown GFP+ CML SPCs using Cell Trace Violet staining. Colcemid treatment used to visualize undivided cells. (G) CFC analysis carried out in hsa-mir183 knockdown CML SPCs. (H) Luciferase assay showing binding between of hsa-mir183 and EGR1 in KCL22 cells. Cells transfected with oligos containing the EGR1 3′UTR (along with the binding site for hsa-mir183, WT) or a mutant version (MUT) together with hsa-mir183 mimic or scrambled negative control. Each experiment had N = 3 biological replicates; *P < .05; **P < .01; *** P < .001. ND, no drug.
To prove direct binding between hsa-mir183 and EGR1 in CML cells, we generated oligos complementary to the EGR1 3′UTR (WT) encompassing the binding site for hsa-mir183 (supplemental Figure 1F). As a control, a mutated version of the binding site was designed by inserting a BamHI restriction site. Both oligos were transfected into CML KCL22 cells, together with either 50 nM hsa-mir183 mimic or a negative control. After 48 hours, cell lysates showed a significant decrease in luciferase signal in the cells containing the WT EGR1 sequence and the hsa-mir183 mimic; however, there was no effect in the cells containing the EGR1 MUT sequence (Figure 2H), confirming that hsa-mir183 binds directly to the predicted sequence within EGR1 to prevent its transcription. In summary, these data support a model where BCR-ABL1–induced hsa-mir183 leads to a decrease of EGR1.
E2F1-signaling pathway is deregulated in CML SPCs
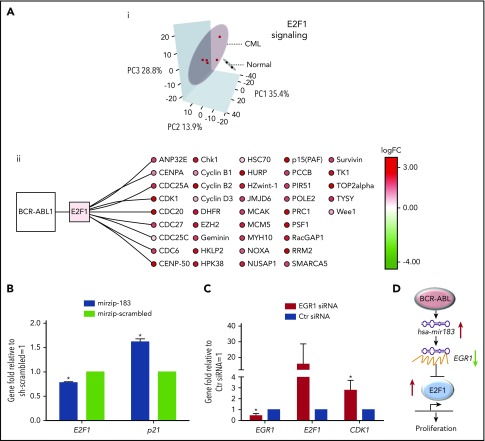
To investigate how this hsa-mir183/EGR1 axis relates to transcriptional control of the cell cycle, specifically in relation to E2F1, we have performed in silico and in vitro assays. Comparison of previously published transcriptomic data from quiescent (G0) healthy and CML cells, defined as CD34+ PY−Ho− (ArrayExpress accession E-MTAB-2508)32 by principal component analysis (PCA), demonstrates clear separation of CML and healthy samples using E2F1 target gene expression data (Figure 3Ai; targets extracted from the MetaCoreKB; supplemental Table 3). Overall, these E2F1 targets exhibit a statistically significant positive shift (ie, toward upregulation) with respect to differential expression when comparing primary quiescent CML SPCs to quiescent healthy SPCs32 (supplemental Figure 2Ai; D = 0.18, P = 2.06 × 10−36, Kolmogorov-Smirnov test). Random resampling of these data demonstrated that this difference is unlikely to occur by chance (supplemental Figure 2Aii; FDR < 0.0001, 10 000 iterations). This statistically significant positive shift for E2F1 targets was also observed in a second, complementary data set of primary CML and healthy quiescent cells (Gene Expression Omnibus [GEO] accession GSE24739)29 (supplemental Figure 2Bi; D = 0.11, P = 8.90 × 10−13, Kolmogorov-Smirnov test); here, again, a difference of the same magnitude or greater is very unlikely to occur by chance (supplemental Figure 2Bii; FDR < 0.0001, 10 000 iterations).
Figure 3.
CML SPCs show deregulation of E2F1-signaling networks. (Ai) Analysis of mRNA screen for CD34+PY−Ho− healthy (N = 2 biological replicates) and CML (N = 5 biological replicates) cells. PCA reduced the high-dimensional data set to 3 dimensions using E2F1 targets as obtained from MetaCoreKB (targets listed in supplemental Table 3). Each sample is represented as a dot; CML and healthy samples are colored red and black, respectively. Three-dimensional (3D) ellipses are drawn around each set of samples (CML or healthy) using the mean and covariance of that set, to represent each sample type’s 95% confidence region. Axes labels indicate the percentage of variability accounted for by each principal component (PC). (ii) The network of BCR-ABL1, E2F1, and E2F1 targets, significantly deregulated in CML is shown. Node color indicates transcriptional deregulation in CML vs healthy G0 cells (ArrayExpress accession E-MTAB-2508), with red/green indicating up/downregulation, respectively; color intensity indicates the extent of the deregulation (as indicated by the color bar). (B) Levels of E2F1 and p21 mRNA measured by Q-PCR in hsa-mir183 knockdown CML SPCs. (C) Levels of EGR1, E2F1, and CDK1 mRNA measured by Q-PCR after knocking down EGR1 by siRNA in CML SPCs. (D) Schematic summary of the hsa-mir183/EGR1–mediated regulation of E2F1 (each experiment had N = 3 biological replicates;*P < .05). Ctr, control; logFC, log fold change.
Cell-cycle–related GO51 terms were significantly overrepresented in the list of differentially expressed genes32,41 (supplemental Table 2). E2F1 target genes identified as significantly differentially upregulated in the first CML vs healthy G0 transcriptional data set (ArrayExpress accession E-MTAB-2508) are summarized in Figure 3Aii; these same genes were shown to be significantly upregulated in the second CML/healthy G0 data set by GSEA45 (supplemental Figure 2C; NES = 1.40, P = .05). Interestingly, although E2F1, E2F2, and E2F3 share a high degree of functional redundancy, E2F2 and E2F3 were not differentially expressed between healthy and CML cells (supplemental Figure 2D). Taken together, these in silico results suggest that E2F1 transcriptional targets are upregulated in quiescent CML SPCs as compared with quiescent healthy SPCs.
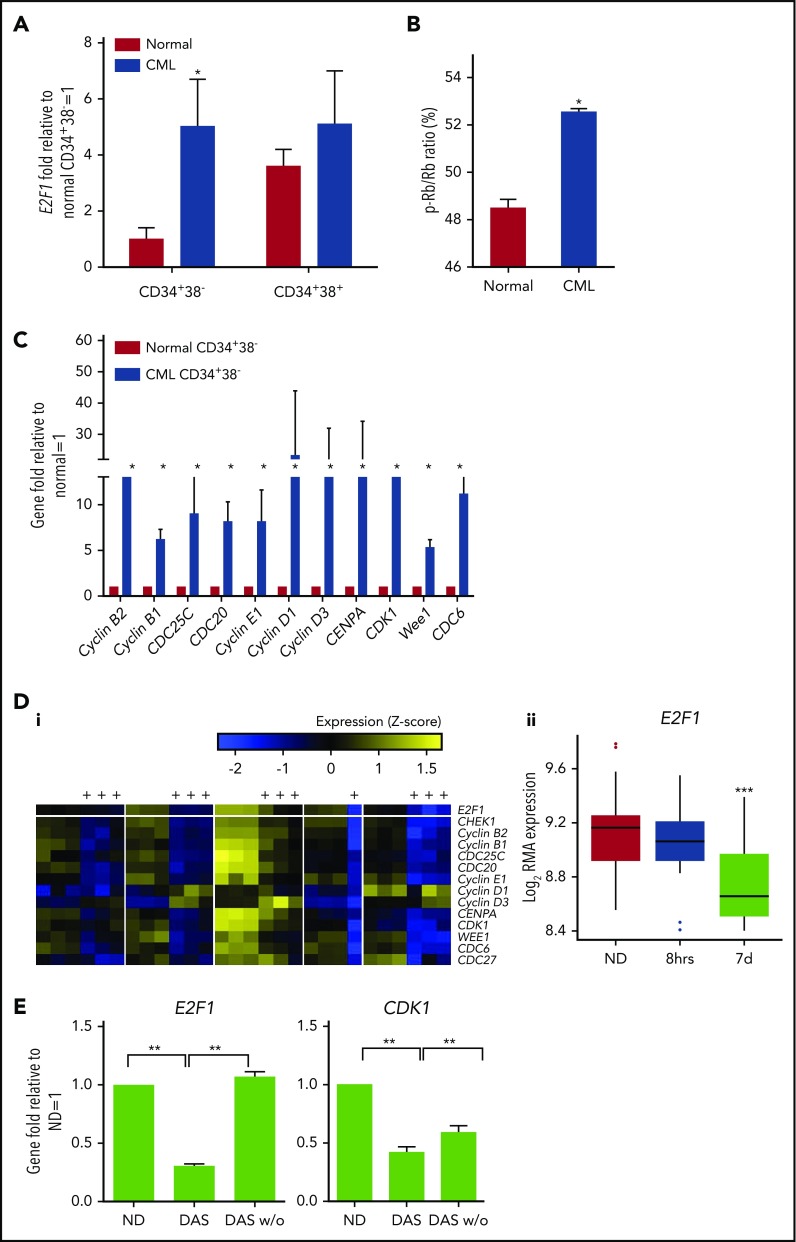
To validate these in silico findings, we next performed in vitro functional experiments. Upregulation of E2F1 mRNA in CML SPCs was confirmed by Q-PCR (Figure 4A). As expected, given that Rb protein phosphorylation (ie, Rb inactivation) often coincides with E2F1 upregulation,52 phospho-Rb (inactive) was high in CML SPCs (Figure 4B; raw data provided in supplemental Table 4). The higher activity of E2F1 in CML SPCs, in terms of cell-cycle regulation, was confirmed by expression of key downstream genes and additional E2F1 targets (Figure 4C). Similar to the bulk SPC data (Figure 4A-C), CML and healthy G0 SPCs (CD34+38−PY−Ho) showed upregulation of E2F1 and its downstream genes (supplemental Figure 3A-C).
Figure 4.
E2F1 regulation in CML SPCs by BCR-ABL1. (A) Nuclear levels of E2F1 mRNA measured in healthy and CML CD34+38− and CD34+38+ cells by Q-PCR. (B) Phosphorylation level of Rb measured in healthy and CML SPCs by a high-content screening-based platform. (C) mRNA levels of E2F1 downstream genes measured in healthy and CML CD34+38− and CD34+38+ cells by Q-PCR. (Di) Heatmap showing regulation of E2F1 and its signaling (yellow, upregulation; blue, downregulation, see color bar) in CML SPCs after treatment with TKIs (values for imatinib, dasatinib, and nilotinib pooled together) for 7 days (treated columns indicated by “+”). (ii) E2F1 response to TKI treatment of 8 hours and 7 days. (E) E2F1 and CDK1 mRNA levels from CML SPCs treated with dasatinib (DAS, 150 nM) or washed out and cultured for a further 3 days (DAS w/o). ●, Outliers as calculated using the Tukey method.24 Each experiment had N = 3 biological replicates. *P < .05; **P < .01; ***P < .001. RMA, robust multiarray average.
To determine whether hsa-mir183/EGR1 were involved in E2F1 regulation of the cell cycle, we again knocked down hsa-mir183 and observed a decrease in E2F1 and an increase in p21 mRNA levels, suggesting that the upregulation of E2F1 was mediated by hsa-mir183, presumably through EGR1 (Figure 3B). To confirm this, we knocked down EGR1 using siRNA, which indeed caused an increase in E2F1 and CDK1 mRNA levels compared with control (Figure 3C).
These data therefore support a model whereby BCR-ABL1–induced hsa-mir183 expression leads to a decrease of EGR1 expression, which in turn results in the upregulation of E2F1 activity in CML SPCs (Figure 3D).
E2F1 regulation is dependent on BCR-ABL1 kinase activity
To investigate whether E2F1 regulation in CML SPCs was BCR-ABL1 kinase dependent as found for hsa-mir183 (Figure 2), we compared transcriptional data for SPCs plus or minus TKIs (8 hours) and CD34+ cells plus or minus TKI (7 days) (Figure 4D). BCR-ABL1+ (confirmed by D-FISH) cells were treated, sorted again for viable cells, and analyzed using Affymetrix Human Gene 1.0 ST chips. Analysis revealed a limited effect on gene expression at 8 hours (49 differentially expressed genes, FDR = 0.05), but an extensive effect (n ≈ 20 000, FDR = 0.05) at 7 days; expression of E2F1 (Figure 4D) and most of its downstream targets (supplemental Figure 4) was significantly decreased. To determine whether the BCR-ABL1–driven regulation of E2F1 in the SPCs of patients was sensitive to TKI therapy in vivo, we interrogated transcriptomic data from CML CD34+ cells harvested from 6 patients, before and at 7 days after imatinib treatment (GEO accession GSE12211).30 We observed that significantly more of the E2F1 targets were downregulated by TKIs in vivo than expected by chance (29 of 54; P = 2.16 × 10−24). To confirm that these effects were the result of BCR-ABL1 kinase inhibition rather than an enrichment for resistant leukemic cells, CML SPCs were treated for 7 days with dasatinib, washed, and cultured for a further 3 days without drug; levels of E2F1 and CDK1 were restored following washout (Figure 4E). Similar behavior was seen in other representative E2F1 target genes (supplemental Figure 5A). Taken together, these data suggest that E2F1 regulation in CML SPCs is BCR-ABL1 kinase dependent. Interestingly, E2F1 target genes were similarly deregulated in primary CD34+ CML vs healthy PB samples, for both TKI-responders (TKI-Rs) and TKI-nonresponders (TKI-NRs) (GEO accession GSE14671)33: random sampling demonstrated that E2F1 targets were significantly more correlated across TKI-Rs and TKI-NRs than we would expect by chance (r = 0.70; q < 0.001) (supplemental Figure 5B). The same result was found for aggressive and indolent CML samples (ArrayExpress accession E-MIMR-17) (r = 0.73; q < 0.001) (supplemental Figure 5C).34 Together, these analyses indicate that the E2F1-dependent deregulation observed in our unselected CML samples is also present in TKI-NRs and aggressive CML phenotypes, and that targeting this deregulation may have wide clinical scope.
E2F1 is dispensable for healthy SPC survival
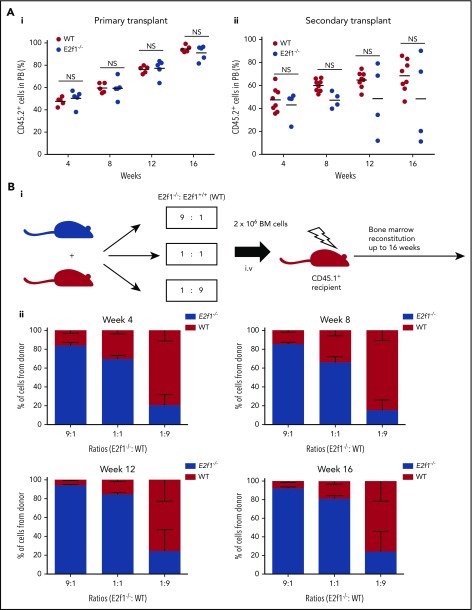
Before considering the hsa-mir-183/EGR1/E2F1 axis for therapeutic targeting in CML SPCs, we first wished to confirm that targeting E2F1 would not be detrimental to healthy SPC homeostasis using E2f1−/− mice.17 The reconstitution capacity of healthy SPCs lacking E2f1 was tested by transplanting E2f1−/− and WT CD45.2+ LSK cells into lethally irradiated syngeneic CD45.1+ recipients. Cells of both genotypes contributed equally to long-term hemopoiesis following primary and secondary transplantation (Figure 5A). Full characterization of the BM of primary and secondary transplanted mice showed minimal differences in the reconstitution potential of WT and E2f1−/− LSK cells in terms of long- and short-term stem cells, various progenitor populations, and mature myeloid and B cells (supplemental Figure 6A-B). Finally, E2f1−/− (CD45.2+) and E2f1 WT (CD45.1+) BM cells were transplanted in ratios of 9:1, 1:1, and 1:9 into lethally irradiated syngeneic CD45.1+ recipient mice to test long-term reconstitution primary ability of mutant vs WT cells. Analysis of PB at 4, 8, 12, and 16 weeks posttransplant showed that E2f1−/− cells exhibited comparable reconstitution ability to WT cells (Figure 5B). Together, these data suggest that loss of E2F1 has no detrimental effect on healthy SPC maintenance and function.
Figure 5.
Healthy E2f1−/−SPCs cells retain functionality. (A) E2f1−/− or WT BM LSK cells transplanted into lethally irradiated CD45.1+ WT recipient mice (n = 5) for long-term reconstitution (i). Sixteen weeks posttransplantation, CD45.2+ LSK WT or E2f1−/− cells purified from harvested BM and transplanted into lethally irradiated CD45.1+ WT secondary recipient mice (N = 4/5) (ii). The engraftment ability of E2f1+/+ WT (black) and E2f1−/− (gray) LSK cells was assessed by the percentage of CD45.2+/CD45.1− cells in PB at weeks 4, 8, 12, and 16 post primary and secondary transplantation. (Bi) Schematic representation of experimental design. E2f1−/− (CD45.2+) and E2f1+/+ WT BM cells (CD45.1+) were transplanted in ratios of 9:1, 1:1, and 1:9 into lethally irradiated (7Gy) CD45.1+ WT recipient mice (N = 5) for long-term reconstitution. (ii) Percentages of CD45.2+ E2f1−/− donor vs recipient cells (CD45.1+) at weeks 4, 8, 12, and 16 posttransplant. NS, not significant with P > .05.
E2F1 is required for CML SPC survival in vitro
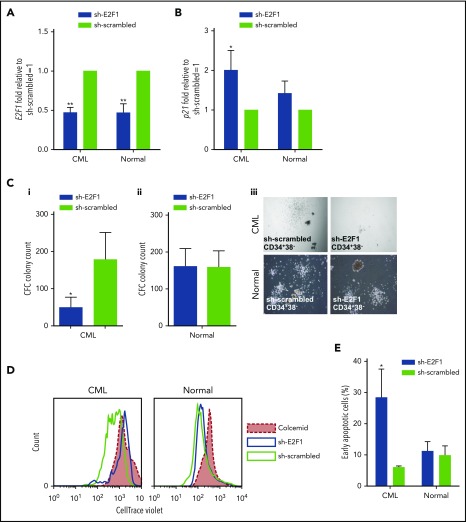
We next asked whether increased activity of E2F1 in CML SPCs led to an increased dependency on E2F1 for survival. E2F1 was knocked down in healthy and CML SPCs using a GFP-lentiviral shRNA. The GFP+CD34+38− cells showed a significant decrease in E2F1 expression in both healthy and CML cells, and an increase in p21 mRNA in CML (Figure 6A-B). In CFC assays, the number of colonies derived from CML, but not healthy, SPCs was significantly decreased, suggesting that E2F1 is required for CML SPC colony-forming potential (Figure 6C). Furthermore, knockdown of E2F1 inhibited proliferation of CML SPCs to a similar extent as the colcemid control (Figure 6D). E2F1 depletion also induced a significant increase in cell death in CML, but not in healthy SPCs, as indicated by an increased percentage of Annexin V+ cells (Figure 6E). These data suggest that E2F1 is required for CML SPC survival and proliferation in vitro. The efficacy of the knockdown system and the subsequent effects in primary CML cells was corroborated using different sets of shRNAs (supplemental Figure 7A-B).
Figure 6.
E2F1 knockdown induces a block in proliferation and increased cell death in CML SPCs. mRNA levels of (A) E2F1 and (B) p21 by Q-PCR in E2F1 knockdown GFP+ healthy and CML SPCs. Transfection with scrambled vector was used as negative control. (C) CFC analysis to measure colony-forming ability following E2F1 knockdown in (i) CML and (ii) healthy SPCs. (iii) Representative images for CFC from healthy and CML SPCs upon E2F1 knockdown (phase-contrast images, ×10 magnification). (D) Representative histogram of cell divisions by CellTrace Violet staining in E2F1 knockdown healthy and CML SPCs. Colcemid treatment was used to visualize undivided cells. (E) Percentage of early apoptosis indicated by Annexin V+/7-AAD− cells. Each experiment had N = 3 biological replicates. *P < .05; **P < .01.
p53 is known to mediate cell-cycle arrest and apoptosis in primary mouse fibroblasts with triple inactivation of E2f1/2/3.25 Given this and the increase in p21 mRNA levels we observed following E2F1 knockdown, we investigated whether the cell-cycle arrest and apoptosis upon depletion of E2F1 in CML SPCs was mediated by p53 activation. CML SPCs were transfected with an E2F1-specific siRNA or control sequence. Although the level of E2F1 was decreased, the level of p53 protein was unchanged (supplemental Figure 8Ai). However, an increase in serine 15 (Ser15) phosphorylation on p53, a crucial site for the induction of cell-cycle arrest,53 was observed in SPCs transduced with E2F1 siRNA compared with control (supplemental Figure 8Aii), suggesting that a decrease in E2F1 resulted in activation of p53. The efficacy of the knockdown system in primary CD34+ CML cells was confirmed using different sets of siRNA (supplemental Figure 8B).
To determine whether the changes in apoptosis in E2F1 knockdown CML SPCs were the direct result of increased p53 activity, we performed knockdowns of E2F1 and p53, separately and in combination, with the double knockdown resulting in reduced E2F1 on a background of low p53 (expression of Puma and CDK1 were measured as representative indicators of p53 and E2F1 activity, respectively) (supplemental Figure 8C). p53 activity was higher upon E2F1 knockdown, whereas E2F1 activity was higher upon p53 knockdown. In the combination, no changes were seen in either the p53 or E2F1 downstream targets. Three days after transfection, apoptosis levels were significantly higher in the E2F1 knockdown, whereas there was no difference in the combination relative to control (supplemental Figure 8D). These data imply that the apoptosis and inhibition of proliferation arising from E2F1 knockdown in CML SPCs is mediated by p53.
Discussion
The oncogenic role of the E2F family members has previously been reported.52,54-56 E2F3 has been shown to play a role in controlling BCR-ABL1–driven leukemogenesis in vitro and in vivo,23 whereas in vitro studies have indicated that E2F1 regulates the cell cycle of BCR-ABL1+ cell lines.57,58 Here, we have used primary patient-derived cells to demonstrate that E2F1 is required for the survival of CML but not healthy SPCs in vitro, and that E2F1 is regulated in CML SPCs via the BCR-ABL1/hsa-mir183/EGR1 axis. In CML SPCs, E2F1 was the effector of a novel signaling pathway mediated by upregulation of hsa-mir183 and inhibition of its direct target EGR1. Indeed, recent evidence demonstrates that leukemia development is accelerated in the absence of EGR1 in a CML mouse model.38 E2F1 upregulation was BCR-ABL1 kinase dependent and its inhibition led to a decrease in colony-forming potential, cell-cycle arrest, and induction of p53-mediated cell death, suggesting the dependency of leukemic SPCs on E2F1.
Our data provide mechanistic insight into how CML SPC death induced by E2F1 inhibition may be mediated by p53. We have previously shown that the apoptotic activity of p53 is downregulated in leukemic stem cells27 and here we have demonstrated that its activity is increased upon inhibition of E2F1 in CML SPCs (by showing that increased cell death in E2F1 knockdown CML SPCs was associated with posttranscriptional modification of Ser15 on p53). To investigate the functional link between E2F1 and p53 in CML SPCs, we hypothesized that cell death due to E2F1 knockdown might be rescued by simultaneous knockdown of p53. Although these data suggest that knockdown of p53 may be sufficient to ameliorate cell death of E2F1-deficient CML SPCs, further work will be required to definitively prove the relevance of this mechanism in CML.
Despite E2F1 upregulation being BCR-ABL dependent, recent work in single CML SPCs has demonstrated that E2F1 signaling and proliferation-associated gene expression are active in a subpopulation of BCR-ABL+ cells that persist following TKI treatment.14 This suggests that therapeutic intervention (via therapeutic targeting of E2F1 or the other components of our model) could address a pressing unmet clinical need for CML patients.
Of relevance to designing a novel therapy for CML, our work indicated that healthy SPCs were not affected by inhibition of the E2F1 pathway, suggesting a potential therapeutic window and CML specificity. Although our studies suggest that inhibition of E2F1 itself may represent a promising therapeutic target, transcription factors are notoriously challenging in drug development, and alternative routes to target the BCR-ABL1/hsa-mir183/EGR1/E2F1 axis may prove to be more tractable. Future investigations will explore the therapeutic potential of the BCR-ABL1/hsa-mir183/EGR1/E2F1 axis in CML.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank all of the patients and healthy donors who have contributed to this work by providing samples of PB and BM; the authors thank K. Dunn, E. K. Allan, J. Cassels, L. McGarry, D. Vetrie, A. Michie, and the Beatson Institute for Cancer Research Animal Unit for their support. The authors thank K. Ryan for providing support and reagents and E. Gómez-Castañeda for providing cDNA for in vitro validation. The authors thank the Glasgow Experimental Cancer Medicine Centre for provision of patient material.
F.P., L.E.M.H., M.A.-O., A.D.W., and T.L.H. were supported by Cancer Research UK (C11074/A11008). F.P. was supported by the Elimination of Leukaemia Fund (F217). L.P. and M.M.S. were supported by the Kay Kendall Leukaemia Fund (KKL690). A.S. was supported by the Biotechnology and Biological Sciences Research Council (BB/F016050/1). S.A.A., M.T.S., and A.D.W. were supported by Bloodwise (14005, 13035, 08071, and 08004). L.J. is supported by an MRC DTP Precision Medicine Studentship (MR/K500847/1) and by the Scottish Cancer Foundation. C.J.C. is supported by Bloodwise (Specialist Programme 14033). Fluorescence-activated cell sorting (FACS) was supported by the Kay Kendall Leukaemia Fund (KKL501) and The Howat Foundation. The Glasgow Experimental Cancer Medicine Centre was supported by Cancer Research UK and by the Chief Scientist's Office, Scotland and the SPIRIT Trials Management Group. L.E.M.H. is a Leuka John Goldman fellow (Leuka2017/JGF/0003) and Kay Kendall Leukaemia Fund Intermediate fellow (KKL1148). G.V.H. is a Kay Kendall Leukaemia Fund Intermediate fellow (KKL698). K.R.K. is a Cancer Research UK Senior Cancer Research Fellow.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.P. and L.E.M.H. designed and performed research, analyzed data, and wrote the manuscript; L.P., M.M.S., L.J., C.J.C., and A.S. performed experiments and reviewed the manuscript; M.T.S., S.A.A., A.H., G.V.H., M.A.-O., R.B., G.L., K.R.K., and A.D.W. provided material, interpreted data, and reviewed the manuscript; and T.L.H. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for F.P. is The Beatson Institute for Cancer Research, Glasgow, United Kingdom.
Tessa L. Holyoake, an inspirational, generous, and enthusiastic clinician and scientist, died on 30 August 2017.
Correspondence: Lisa E. M. Hopcroft, Paul O’Gorman Leukaemia Research Centre, Gartnavel General Hospital, 1053 Great Western Rd, Glasgow G12 0YN, United Kingdom; e-mail: lisa.hopcroft@glasgow.ac.uk.
REFERENCES
- 1.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561-566. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia R, Holtz M, Niu N, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101(12):4701-4707. [DOI] [PubMed] [Google Scholar]
- 3.Mahon FX, Réa D, Guilhot J, et al. ; Intergroupe Français des Leucémies Myéloïdes Chroniques. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029-1035. [DOI] [PubMed] [Google Scholar]
- 4.Graham SM, Jørgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319-325. [DOI] [PubMed] [Google Scholar]
- 5.Chen IM, Harvey RC, Mullighan CG, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119(15):3512-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121(1):396-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomel JC, Bonnet ML, Sorel N, et al. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. 2011;118(13):3657-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94(6):2056-2064. [PubMed] [Google Scholar]
- 9.Hamilton A, Helgason GV, Schemionek M, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119(6):1501-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallipoli P, Cook A, Rhodes S, et al. JAK2/STAT5 inhibition by nilotinib with ruxolitinib contributes to the elimination of CML CD34+ cells in vitro and in vivo. Blood. 2014;124(9):1492-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomel JC, Sorel N, Guilhot J, Guilhot F, Turhan AG. BCR-ABL expression in leukemic progenitors and primitive stem cells of patients with chronic myeloid leukemia. Blood. 2012;119(12):2964-2965, author reply 2965-2966. [DOI] [PubMed] [Google Scholar]
- 12.Burchert A, Neubauer A, Hochhaus A. Response: too much BCR-ABL to live on, but too little BCR-ABL to die on? Blood. 2012;119(12):2965-2966. [Google Scholar]
- 13.Warfvinge R, Geironson L, Sommarin MNE, et al. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood. 2017;129(17):2384-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giustacchini A, Thongjuea S, Barkas N, et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017;23(6):692-702. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12(15):2245-2262. [DOI] [PubMed] [Google Scholar]
- 16.Trikha P, Sharma N, Opavsky R, et al. E2f1-3 are critical for myeloid development. J Biol Chem. 2011;286(6):4783-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Field SJ, Tsai FY, Kuo F, et al. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85(4):549-561. [DOI] [PubMed] [Google Scholar]
- 18.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3(1):11-20. [DOI] [PubMed] [Google Scholar]
- 19.Bhardwaj G, Murdoch B, Wu D, et al. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol. 2001;2(2):172-180. [DOI] [PubMed] [Google Scholar]
- 20.Allen LF, Sebolt-Leopold J, Meyer MB. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK). Semin Oncol. 2003;30(5 suppl 16):105-116. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, Bremner R. Division and apoptosis of E2f-deficient retinal progenitors. Nature. 2009;462(7275):925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong JL, Wenzel PL, Sáenz-Robles MT, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462(7275):930-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eiring AM, Neviani P, Santhanam R, et al. Identification of novel posttranscriptional targets of the BCR/ABL oncoprotein by ribonomics: requirement of E2F3 for BCR/ABL leukemogenesis. Blood. 2008;111(2):816-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucas CM, Harris RJ, Holcroft AK, et al. Second generation tyrosine kinase inhibitors prevent disease progression in high-risk (high CIP2A) chronic myeloid leukaemia patients. Leukemia. 2015;29(7):1514-1523. [DOI] [PubMed] [Google Scholar]
- 25.Timmers C, Sharma N, Opavsky R, et al. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27(1):65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma N, Timmers C, Trikha P, Saavedra HI, Obery A, Leone G. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281(47):36124-36131. [DOI] [PubMed] [Google Scholar]
- 27.Abraham SA, Hopcroft LE, Carrick E, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534(7607):341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venturini L, Battmer K, Castoldi M, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399-4405. [DOI] [PubMed] [Google Scholar]
- 29.Affer M, Dao S, Liu C, et al. Gene expression differences between enriched normal and chronic myelogenous leukemia quiescent stem/progenitor cells and correlations with biologicssal abnormalities. J Oncol. 2011;2011:798592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruennert D, Czibere A, Bruns I, et al. Early in vivo changes of the transcriptome in Philadelphia chromosome-positive CD34+ cells from patients with chronic myelogenous leukaemia following imatinib therapy. Leukemia. 2009;23(5):983-985. [DOI] [PubMed] [Google Scholar]
- 31.Cramer-Morales K, Nieborowska-Skorska M, Scheibner K, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122(7):1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham SM, Vass JK, Holyoake TL, Graham GJ. Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells. 2007;25(12):3111-3120. [DOI] [PubMed] [Google Scholar]
- 33.McWeeney SK, Pemberton LC, Loriaux MM, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010;115(2):315-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yong AS, Szydlo RM, Goldman JM, Apperley JF, Melo JV. Molecular profiling of CD34+ cells identifies low expression of CD7, along with high expression of proteinase 3 or elastase, as predictors of longer survival in patients with CML. Blood. 2006;107(1):205-212. [DOI] [PubMed] [Google Scholar]
- 35.Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2(4):380-391. [DOI] [PubMed] [Google Scholar]
- 36.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70(23):9570-9580. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs JD, Liebermann DA, Hoffman B. Egr-1 abrogates the E2F-1 block in terminal myeloid differentiation and suppresses leukemia. Oncogene. 2008;27(1):98-106. [DOI] [PubMed] [Google Scholar]
- 38.Maifrede S, Magimaidas A, Sha X, Mukherjee K, Liebermann DA, Hoffman B. Loss of Egr1, a human del5q gene, accelerates BCR-ABL driven chronic myelogenous leukemia. Oncotarget. 2017;8(41):69281-69294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellicano F, Copland M, Jorgensen HG, Mountford J, Leber B, Holyoake TL. BMS-214662 induces mitochondrial apoptosis in chronic myeloid leukemia (CML) stem/progenitor cells, including CD34+38- cells, through activation of protein kinase Cbeta. Blood. 2009;114(19):4186-4196. [DOI] [PubMed] [Google Scholar]
- 40.Copland M, Hamilton A, Elrick LJ, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107(11):4532-4539. [DOI] [PubMed] [Google Scholar]
- 41.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1-3):83-92. [DOI] [PubMed] [Google Scholar]
- 42.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249-264. [DOI] [PubMed] [Google Scholar]
- 43.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289-300. [Google Scholar]
- 45.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellicano F, Simara P, Sinclair A, et al. The MEK inhibitor PD184352 enhances BMS-214662-induced apoptosis in CD34+ CML stem/progenitor cells. Leukemia. 2011;25(7):1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang X, Saw KM, Eaves A, Eaves C. Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst. 2007;99(9):680-693. [DOI] [PubMed] [Google Scholar]
- 48.Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657-667. [DOI] [PubMed] [Google Scholar]
- 49.Neviani P, Harb JG, Oaks JJ, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest. 2013;123(10):4144-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839-847. [DOI] [PubMed] [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, et al. ; The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94(14):7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banin S, Moyal L, Shieh S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674-1677. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Timmers C, Maiti B, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414(6862):457-462. [DOI] [PubMed] [Google Scholar]
- 55.Foster CS, Falconer A, Dodson AR, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23(35):5871-5879. [DOI] [PubMed] [Google Scholar]
- 56.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Birchenall-Roberts MC, Yoo YD, Bertolette DC III, et al. The p120-v-Abl protein interacts with E2F-1 and regulates E2F-1 transcriptional activity. J Biol Chem. 1997;272(14):8905-8911. [DOI] [PubMed] [Google Scholar]
- 58.Stewart MJ, Litz-Jackson S, Burgess GS, Williamson EA, Leibowitz DS, Boswell HS. Role for E2F1 in p210 BCR-ABL downstream regulation of c-myc transcription initiation. Studies in murine myeloid cells. Leukemia. 1995;9(9):1499-1507. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.