Abstract
Whole genome sequencing of actinomycetes has uncovered a new immense realm of microbial chemistry and biology. Most biosynthetic gene clusters present in genomes were found to remain “silent” under standard cultivation conditions. Some small molecules—chemical elicitors—can be used to induce the biosynthesis of antibiotics in actinobacteria and to expand the chemical diversity of secondary metabolites. Here, we outline a brief account of the basic principles of the search for regulators of this type and their application.
Keywords: actinomycetes, antibiotic biosynthesis, silent biosynthetic pathways, γ-butyrolactones, HiTES, translation inhibitors
1. Introduction
Since the discovery of streptomycin by Selman A. Waksman, actinomycetes have become one of the most fruitful sources of new antibiotics. Most antibiotic classes in current clinical use were discovered during the “golden era”, 1940–1960s, by phenotypic screening of soil microorganisms. Moreover, since the 1960s, a significant number of approved drugs has been designed using chemical modifications of natural scaffolds. Due to an “innovation gap” in this area, society is now facing an emerging threat of microbial drug resistance. An urgent need for new effective antimicrobials has become an important social and political issue [1,2].
On the other hand, achievements in genome sequencing of actinomycetes has revealed a difference between their potential and observed biosynthetic gene expression. Biosynthetic gene clusters (BGCs) are several generally contiguous genes encoding enzymes responsible for a stepwise assembly of complex bioactive molecules. According to data from the majority of published genomes, most BGCs remain “silent” under standard cultivation conditions. These silent, or cryptic, BGCs represent a potential source of new scaffolds for the discovery of novel antimicrobials [3,4,5]. Several techniques for activation of silent BGCs have been developed in recent decades, e.g., direct identification of BGCs and their expression in heterologous hosts [6,7,8,9,10], and systematic alteration of cultivation parameters (the “one strain—many compounds” (OSMAC) approach) [11,12,13]. These strategies are extremely powerful although still remaining laborious and resource-intensive, especially for large (>40 kb) BGCs. Further techniques comprise co-cultivation [14,15], ribosome engineering [16,17,18], and the use of chemical elicitors—compounds that induce the synthesis of antibiotics in actinomycetes [14,19,20,21]. The last approach is accounted and discussed succinctly in this review, covering literature up to December 2017. Here, we focus on small organic molecules capable in nanomolar to micromolar minimum effective concentrations to induce biosynthesis of secondary metabolite in actinomycetes.
2. γ-Butyrolactones (GBL) and Related (auto)Regulators
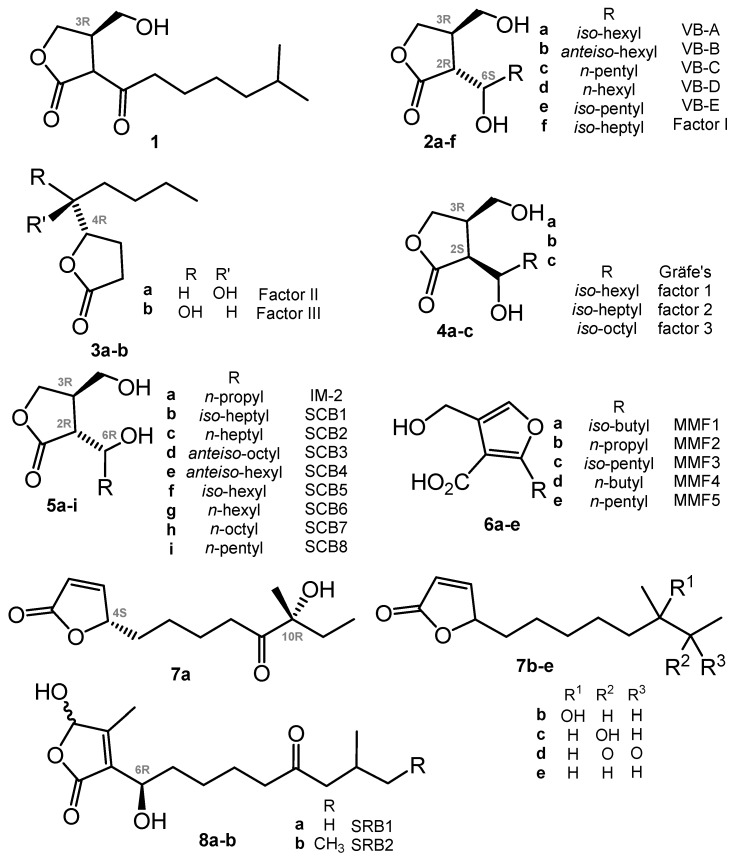
Historically, A-factor (1, Chart 1) (A for “autoregulation”) was the first compound that revolutionized our views of the secondary metabolism and development cycle of actinomycetes. This γ-butyrolactone (GBL) derivative was discovered by Prof. Khokhlov and co-workers in 1967 [22].
Chart 1.
GBL and closely related regulators: butenolides (avenolide, S. rochei butenolides or SRB) and furans (methylenomycin furans or MMF).
A-factor acts as a pleiotropic regulator: it binds to the A-factor receptor protein (ArpA) and causes dissociation of this suppressor from DNA. This triggers the transcription of the adpA gene encoding the transcription activator AdpA, which in turn induces morphological differentiation, spore formation, and biosynthesis of secondary metabolites [23,24].
In further decades, many other closely related autoregulators were discovered, e.g., Streptomyces virginiae butanolides (VBs A-E, 2a–e) [25,26,27], Factor I (2f) [28], Factors II and III (3a,b) [29], Gräfe’s factors (4a–c) from S. bikiniensis and S. cyaneofuscatus [30], IM-2 (5a) [31,32], SCB1 (5b) [33,34], SCB2,3 (5c,d) [35] and SCB4–8 (5e–i) [36], methylenomycin furans (MMFs, 6) [37], avenolide (7a) from S. avermitilis [38], related compounds (7b–e) from S. albus [39], and two S. rochei butenolides (SRBs, 8a,b) [40]. The stereoconfiguration of avenolide analogues 7b–e reported very recently has not been assigned [39]. All these compounds have a high degree of similarity in their chemical nature (butanolides, butenolides, or furans), biosynthesis pathways, and in mechanisms of action on actinomycetes. The transduction of the chemical signal starts from binding with a specific receptor protein (like ArpA in the A-factor signaling cascade) belonging to the TetR family of transcriptional regulators; the binding of compounds probably causes conformational changes, thus inducing the relocation of the DNA-binding domain and DNA release [23].
TetR transcriptional repressors are well represented and widespread in bacteria [41,42]. The recent study on the distribution of the GBL (IM-2, VB)-type and butenolide (avenolide)-type signaling showed these hormones to be common in actinomycetes. Production of regulators was screened using S. lavendulae FRI-5 (ΔfarX), S. virginiae (ΔbarX), and S. avermitilis (Δaco) strains with disrupted essential biosynthetic genes for the corresponding hormone-like molecules. Twenty percent of the investigated strains produced 5a (IM-2), another 20% produced VBs, and 24% of actinomycetes showed avenolide activity [43].
Thus, the potential for various applications of A-factor-like regulators is quite high: they induce biosynthesis of antibiotics of different classes, e.g., aminoglycosides (streptomycin), streptogramins (virginamycin), peptide ionophores (valinomycin) and others, including clinically significant antibiotics. For more detail see ref. [24].
3. Other Types of (auto)Regulators
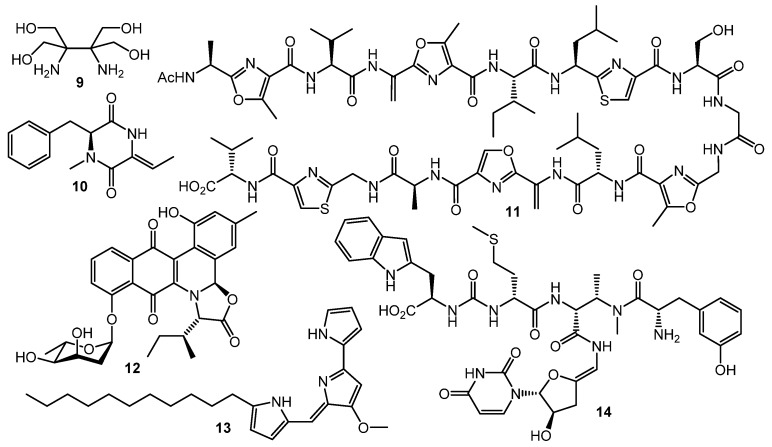
However, GBLs and related compounds are not the only type of regulators found in actinomycetes. Hitherto, a few different types of autoregulators were reported (Chart 2), e.g., the pi-factor from S. natalensis (9) [44] and l-N-methylphenylalanyl-dehydrobutyrine diketopiperazine (10) from S. globisporus [45]. These molecules have no structural similarity to GBL-type compounds and their mechanisms of action remain unclear [46,47].
Chart 2.
Other (non-GBL) types of regulators.
Goadsporin (11) [48,49], an active 19-aa peptide found in the culture broth of Streptomyces sp. TP-A0584, shows extremely broad elicitation activity on actinobacteria. Induction of sporulation and growth inhibition were observed in about 80% of Streptomyces sp. and Micromonospora sp. isolates after goadsporin treatment [50]. Possible mechanism of the goadsporin action on susceptible cells was postulated by Prof. H. Onaka and co-workers. Bioinformatics analysis of godI—self-resistance gene from the goadsporin BGC—showed a high degree of homology with ffh encoding the Ffh protein. The Ffh and 4.5S RNA form the signal recognition particles (SRPs) in prokaryotic cells. The SPRs recognize the signal peptide of membrane or secretory proteins and facilitate their translocation to the cytoplasmic membrane. Binding of Ffh protein with goadsporin causes the upregulation of protein translocation and maturation, growth inhibition, and alternation of secondary metabolism [51].
It was suggested that goadsporin and its analogs [51,52] could be useful in crop protection: S. scabies, which is known to cause potato scab, is effectively inhibited by the low concentrations of goadsporin.
Two “atypical” transcriptional regulators were discovered in S. venezuelae: JadR1 activates the transcription of jadomycin B (12) biosynthetic genes while repressing its own gene. Jadomycin B binds to the N-terminal receiver domain of JadR1, causing dissociation of JadR1 from target promoters. High expression of the biosynthetic enzyme genes leads to accumulation of the product in a concentration sufficient to inhibit the initial activation. Therefore, jadomycin acts as an autoregulator of its own synthesis. Similarly, the RedZ protein regulating production of undecylprodigiosine (13) is modulated by the end product. These findings demonstrate that end-product-mediated control of antibiotic biosynthesis may be widespread in nature, offering seminal basis for interspecies signaling and interaction [53,54]. Production of another class of antibiotics, uridyl peptides sansanmycins (sansanmycin A, 14), active against multidrug-resistant Mycobacterium tuberculosis, is also controlled at the transcriptional level by the end-product-mediated mechanism [55].
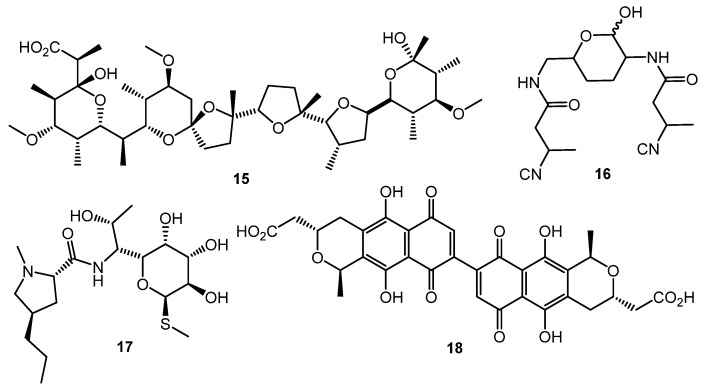
Polyether antibiotics at sub-inhibitory concentrations can elicit the synthesis of secondary metabolites. Promomycin (15, Chart 3) and closely related compounds (salinomycin, monensin, and nigericin) promoted the synthesis of cryptic isonitrile antibiotic SF2768 (16) by S. griseorubiginosus [56,57]. This was the first example of isolation of a novel original compound using the chemical elicitation approach. However, the action of polyether antibiotics has not yet been studied on the molecular level.
Chart 3.
Antibiotic-antibiotic induction; elicitors and products.
Lincomycin (17) at one tenth of its MIC increased the expression of the pathway-specific regulatory gene actII-ORF4 in the enzyme gene cluster producing the blue-pigmented antibiotic actinorhodin (18) in S. lividans, thus resulting in actinorhodin overproduction [58]. Chloramphenicol activated biosynthetic genes at the transcriptional level and increased the amino acid pool 1.5- to 6-fold, enhancing the production of non-ribosomal peptide antibiotics [59]. Other ribosome-targeted antibiotics, especially thiosteptone and spectinomycin, at sub-inhibitory concentrations were also capable of altering colony morphology and antibiotics (13 and 18) production in S. coelicolor M145 [60].
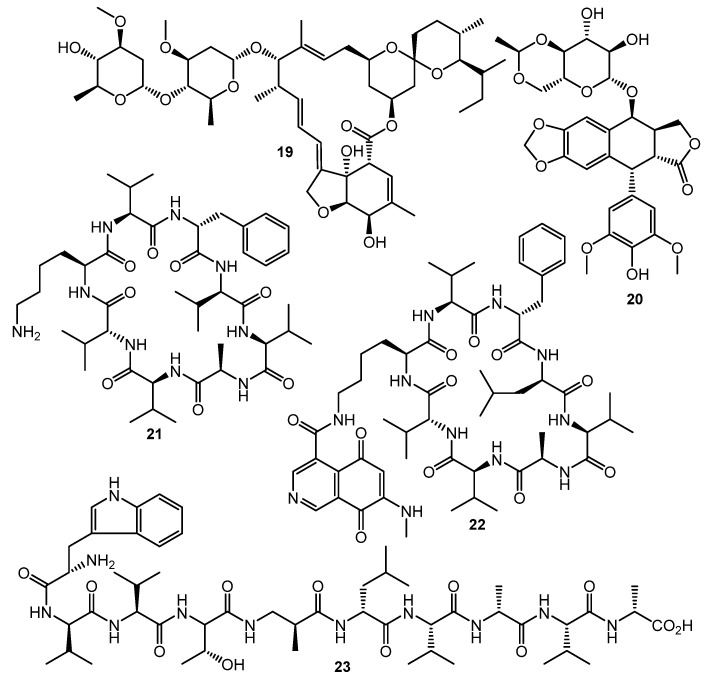
The HiTES (High-Throughput Elicitor Screening) approach [61,62,63] was developed to discover small molecule elicitors of silent biosynthetic gene clusters and novel secondary metabolites using a simple fluorescent assay format. Three copies of eGFP (enhanced green fluorescent protein encoding gene) inserted in the S. albus J1074 genome both at a neutral site (attB) and inside the biosynthetic cluster of interest (sur, surugamide BGC). The difference in fluorescence between attB::Psur-eGFPx3 (the Psur promoter region (~260 bp) upstream of surE) and surE::eGFPx3 strains after testing compound treatment was used as an indicator of BGC’s activation. Screening of a commercially-available natural products library (ca. 500 compounds) identified 6 potential elicitors, among these ivermectin b1a (19, Chart 4) and etoposide (20) had the maximum surugamide BGC activation effect and markedly changed the secondary metabolome profile of S. albus J1074—14 novel small molecules were isolated and characterized using these two inducers, e.g., surugamide J (21), albucyclone F (22), and surugamide F2 (23) [63].
Chart 4.
Natural products eliciting surugamide biosynthesis in S. albus J1074 (detected by HiTES).
4. Metabolism Remodeling and Epigenetic Control
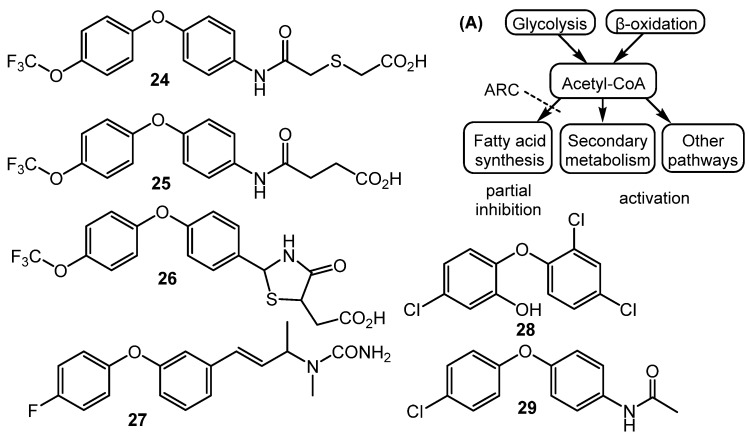
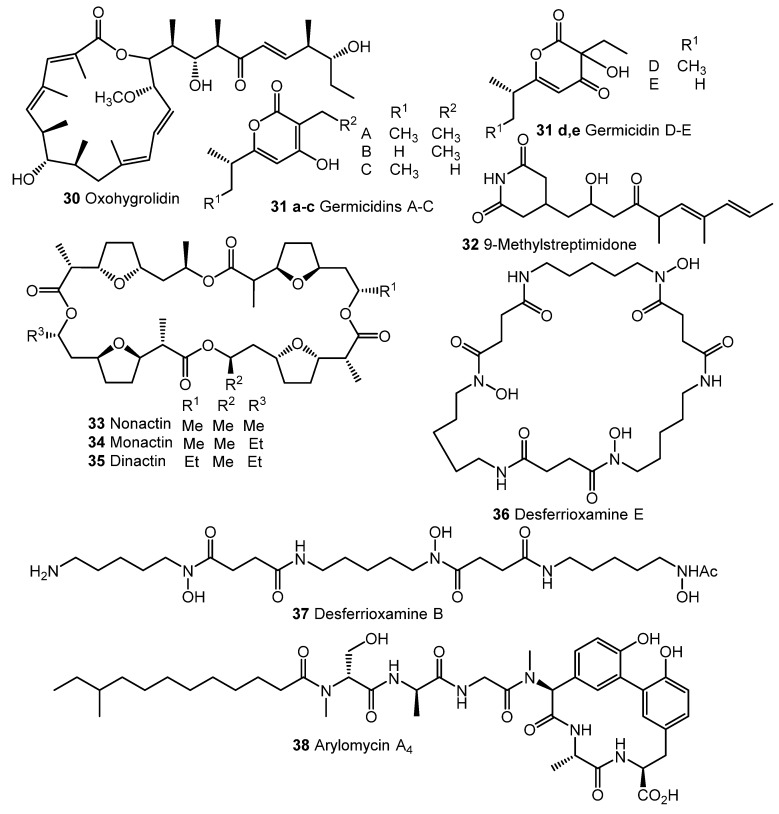
Fully synthetic chemicals also can be utilized as elicitors. ARC2 (24, ARC from “Antibiotic-Remodeling-Compound”) and similar diphenyl ethers (ARC3–5, 25–27) (Chart 5) have become the first inducer examples discovered by systematic screening of 30,569 small molecules [64,65]. Taking into account the structural similarity of ARC compounds with triclosan (28), a well-known biocide and inhibitor of fatty acid biosynthesis, the mechanism of ARCs’ action (Chart 5A) has been proposed. Generally, ARCs act as metabolism remodeling compounds, changing the balance between primary and secondary metabolic pathways. The mode of action was confirmed by the experimental data: ARCs changed fatty acid pool by inhibition of key biosynthetic enzymes like FabI. Remarkably, triclosan also can stimulate polyketide antibiotic synthesis at sub-inhibitory concentrations [59]. Further investigation of the different ARCs’ effects on the streptomycetes biochemistry revealed the targets for different molecules (even structurally close) as not being the same; however, all of them influenced fatty acid metabolism [66]. Chlorinated analog Cl-ARC (29) was used to enhance the expression of cryptic biosynthetic genes, and the observed difference between growth on control and elicitor-containing media was studied using a metabolomics approach. More than 100 induced secondary metabolites were detected, including rare antibiotics [67]. Several identified structures are represented in Chart 6: oxohygrolidin (30), germicidins A–E (31a–e) 9-methylstreptimidone (32), nactins (33, 34, 35), desferrioxamines B,E (36, 37), and arylomycin A4 (38).
Chart 5.
Synthetic diphenyl ethers (ARCs) as elicitors and their mode of action (A).
Chart 6.
Cl-ARC-induced biosynthesis: examples of identified antibiotics [67].
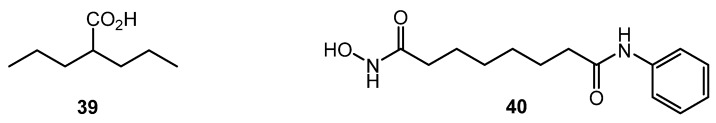
Despite the significant difference in structural DNA organization between eukaryotes and actinomycetes, inhibitors of histone deacetylase (HDAC) like valproic acid (39) and suberanilohydroxamic acid (SAHA, 40) (Chart 7) showed effect on antibiotic production by several Streptomyces strains, especially on nutrient-poor media. The finding was explained by the homology between several S. coelicolor enzymes (SCO0452, SCO6464, SCO3330) and human HDAC enzymes [68]. Epigenetic modification could be used for the control of antibiotic biosynthesis; the approach, however, is not well studied for actinobacteria.
Chart 7.
Histone deacetylase (HDAC) inhibitors.
5. Conclusions
Activation of antibiotics’ biosynthesis in actinomycetes by using small-molecule elicitors seems to be a useful technique for drug discovery. The approach allows identification of new metabolite scaffolds and enhances synthesis of low yield secondary metabolites. Moreover, antibiotic-antibiotic induction relationships are believed to be the basis of chemical signaling between different actinobacterial strains and are, therefore, of interest for chemical ecology. Elicitation is an obvious cost-effective approach in biotechnology for increasing antibiotic yields and reducing fermentation time.
Acknowledgments
This work was supported in part by RFBR (project No. 18-33-00044) and by the budget funding of the topic 006 “Isolation and physico-phemical characterization of microbially produced antibiotic compounds” at the Gause Institute of New Antibiotics.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. [(accessed on 27 February 2017)]; Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/
- 2.The National Strategy to Prevent the Antimicrobial Resistance Spreading (2017–2030) [(accessed on 3 October 2017)]; Available online: http://government.ru/docs/29477/
- 3.Nett M., Ikeda H., Moore B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 5.Genilloud O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017;34:1203–1232. doi: 10.1039/C7NP00026J. [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka K., Reynolds K.A., Kersten R.D., Ryan K.S., Gonzalez D.J., Nizet V., Dorrestein P.C., Moore B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nah H.-J., Pyeon H.-R., Kang S.-H., Choi S.-S., Kim E.-S. Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front. Microbiol. 2017;8:394. doi: 10.3389/fmicb.2017.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberti F., Khairudin K., Venegas E.R., Davies J.A., Hayes P.M., Willis C.L., Bailey A.M., Foster G.D. Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives. Nat. Commun. 2017;8:1831. doi: 10.1038/s41467-017-01659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y., Zhang L., Zhou Z., Yan X. Diversity of gene clusters for polyketide and nonribosomal peptide biosynthesis revealed by metagenomics analysis of the Yellow Sea sediment. Front. Microbiol. 2018;9:295. doi: 10.3389/fmicb.2018.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suroto D.A., Kitani S., Arai M., Ikeda H., Nihira T. Characterization of the biosynthetic gene cluster for cryptic phthoxazolin A in Streptomyces avermitilis. PLoS ONE. 2018;13:e0190973. doi: 10.1371/journal.pone.0190973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode H.B., Bethe B., Höfs R., Zeeck A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Reen F.J., Romano S., Dobson A.D.W., O’Gara F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs. 2015;13:4754–4783. doi: 10.3390/md13084754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M., Grkovic T., Liu X., Han J., Zhang L., Quinn R.J. A systems approach using OSMAC, Log P and NMR fingerprinting: An approach to novelty. Synth. Syst. Biotechnol. 2017;2:276–286. doi: 10.1016/j.synbio.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelmohsen U.R., Grkovic T., Balasubramanian S., Kamel M.S., Quinn R.J., Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015;33:798–811. doi: 10.1016/j.biotechadv.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Van der Meij A., Worsley S.F., Hutchings M.I., van Wezel G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017;41:392–416. doi: 10.1093/femsre/fux005. [DOI] [PubMed] [Google Scholar]
- 16.Wang G., Hosaka T., Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl. Environ. Microbiol. 2008;74:2834–2840. doi: 10.1128/AEM.02800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tojo S., Tanaka Y., Ochi K. Activation of antibiotic production in Bacillus spp. by cumulative drug resistance mutations. Antimicrob. Agents Chemother. 2015;59:7799–7804. doi: 10.1128/AAC.01932-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochi K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017;70:25–40. doi: 10.1038/ja.2016.82. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H., Sandiford S.K., van Wezel G.P. Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol. 2014;41:371–386. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 20.Okada B.K., Seyedsayamdost M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017;41:19–33. doi: 10.1093/femsre/fuw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarins-Tutt J.S., Barberi T.T., Gao H., Mearns-Spragg A., Zhang L., Newman D.J., Goss R.J.M. Prospecting for new bacterial metabolites: A glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 2016;33:54–72. doi: 10.1039/C5NP00111K. [DOI] [PubMed] [Google Scholar]
- 22.Khokhlov A.S., Tovarova I.I., Borisova L.N., Pliner S.A., Shevchenko L.N., Kornitskaia E.I., Ivkina N.S., Rapoport I.A. The A-factor, responsible for streptomycin biosynthesis by mutant strains of Actinomyces streptomycini. Dokl. Akad. Nauk SSSR. 1967;177:232–235. [PubMed] [Google Scholar]
- 23.Niu G., Chater K.F., Tian Y., Zhang J., Tan H. Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 2016;40:554–573. doi: 10.1093/femsre/fuw012. [DOI] [PubMed] [Google Scholar]
- 24.Efremenkova O.V. A-factor-like autoregulators. Russ. J. Bioorg. Chem. 2016;42:457–472. doi: 10.1134/S1068162016050058. [DOI] [Google Scholar]
- 25.Yanagimoto M., Terui G. Physiological studies on staphylomycin production II, formation of a substance effective in inducing staphylomycin production. J. Ferment. Technol. 1971;49:611–618. [Google Scholar]
- 26.Yanagimoto M., Yamada Y., Terui G. Extraction and purification of inducing material produced in staphylomycin fermentation. Hakko Kogaku Kaishi. 1979;57:6–14. [Google Scholar]
- 27.Yamada Y., Sugamura K., Kondo K., Yanagimoto M., Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J. Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 28.Gräfe U., Schade W., Eritt I., Fleck W.F., Radics L. A new inducer of anthracycline biosynthesis from Streptomyces viridochromogenes. J. Antibiot. 1982;35:1722–1723. doi: 10.7164/antibiotics.35.1722. [DOI] [PubMed] [Google Scholar]
- 29.Gräfe U., Reinhardt G., Schade W., Krebs D., Eritt I., Fleck W.F., Heinrich E. Isolation and structure of novel autoregulators from Streptomyces griseus. J. Antibiot. 1982;35:609–614. doi: 10.7164/antibiotics.35.609. [DOI] [PubMed] [Google Scholar]
- 30.Gräfe U., Reinhardt G., Schade W., Eritt I., Fleck W.F., Radics L. Interspecific inducers of cytodifferentiation and anthracycline biosynthesis from Streptomyces bikinensis and S. cyaneofuscatus. Biotechnol. Lett. 1983;5:591–596. doi: 10.1007/BF00130838. [DOI] [Google Scholar]
- 31.Sato K., Nihira T., Sakuda S., Yanagimoto M., Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J. Ferment. Bioeng. 1989;68:170–173. doi: 10.1016/0922-338X(89)90131-1. [DOI] [Google Scholar]
- 32.Hashimoto K., Nihira T., Sakuda S., Yamada Y. IM-2, a butyrolactone autoregulator, induces production of several nucleoside antibiotics in Streptomyces sp. FRI-5. J. Ferment. Bioeng. 1992;73:449–455. doi: 10.1016/0922-338X(92)90136-I. [DOI] [Google Scholar]
- 33.Takano E., Nihira T., Hara Y., Jones J.J., Gershater C.J.L., Yamada Y., Bibb M. Purification and structural determination of SCB1, a γ-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2) J. Biol. Chem. 2000;275:11010–11016. doi: 10.1074/jbc.275.15.11010. [DOI] [PubMed] [Google Scholar]
- 34.Takano E., Chakraburtty R., Nihira T., Yamada Y., Bibb M. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2) Mol. Microbiol. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 35.Hsiao N.-H., Nakayama S., Merlo M.E., de Vries M., Bunet R., Kitani S., Nihira T., Takano E. Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem. Biol. 2009;16:951–960. doi: 10.1016/j.chembiol.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Sidda J.D., Poon V., Song L., Wang W., Yang K., Corre C. Overproduction and identification of butyrolactones SCB1–8 in the antibiotic production superhost Streptomyces M1152. Org. Biomol. Chem. 2016;14:6390–6393. doi: 10.1039/C6OB00840B. [DOI] [PubMed] [Google Scholar]
- 37.Corre C., Song L., O’Rourke S., Chater K.F., Challis G.L. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. USA. 2008;105:17510–17515. doi: 10.1073/pnas.0805530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitani S., Miyamoto K.T., Takamatsu S., Herawati E., Iguchi H., Nishitomi K., Uchida M., Nagamitsu T., Omura S., Ikeda H., et al. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA. 2011;108:16410–16415. doi: 10.1073/pnas.1113908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen T.B., Kitani S., Shimma S., Nihira T. Butenolides from Streptomyces albus J1074 act as external signals to stimulate avermectin production in Streptomyces avermitilis. Appl. Environ. Microbiol. 2018;84:e02791-17. doi: 10.1128/AEM.02791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arakawa K., Tsuda N., Taniguchi A., Kinashi H. The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. ChemBioChem. 2012;13:1447–1457. doi: 10.1002/cbic.201200149. [DOI] [PubMed] [Google Scholar]
- 41.Ramos J.L., Martínez-Bueno M., Molina-Henares A.J., Terán W., Watanabe K., Zhang X., Gallegos M.T., Brennan R., Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuthbertson L., Nodwell J.R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 2013;77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thao N.B., Kitani S., Nitta H., Tomioka T., Nihira T. Discovering potential Streptomyces hormone producers by using disruptants of essential biosynthetic genes as indicator strains. J. Antibiot. 2017;70:1004–1008. doi: 10.1038/ja.2017.85. [DOI] [PubMed] [Google Scholar]
- 44.Recio E., Colinas A., Rumbero A., Aparicio J.F., Martin J.F. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 2004;279:41586–41593. doi: 10.1074/jbc.M402340200. [DOI] [PubMed] [Google Scholar]
- 45.Matselyukh B., Mohammadipanah F., Laatsch H., Rohr J., Efremenkova O., Khilya V. N-methylphenylalanyl-dehydrobutyrine diketopiperazine, an A-factor mimic that restores antibiotic biosynthesis and morphogenesis in Streptomyces globisporus 1912-B2 and Streptomyces griseus 1439. J. Antibiot. 2015;68:9–14. doi: 10.1038/ja.2014.86. [DOI] [PubMed] [Google Scholar]
- 46.Recio E. Glycerol, ethylene glycol and propanediol elicit pimaricin biosynthesis in the PI-factor-defective strain Streptomyces natalensis npi287 and increase polyene production in several wild-type actinomycetes. Microbiology. 2006;152:3147–3156. doi: 10.1099/mic.0.28953-0. [DOI] [PubMed] [Google Scholar]
- 47.Vicente C.M., Santos-Aberturas J., Guerra S.M., Payero T.D., Martín J.F., Aparicio J.F. PimT, an amino acid exporter controls polyene production via secretion of the quorum sensing pimaricin-inducer PI-factor in Streptomyces natalensis. Microb. Cell Fact. 2009;8:8–33. doi: 10.1186/1475-2859-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onaka H., Tabata H., Igarashi Y., Sato Y., Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J. Antibiot. 2001;54:1036–1044. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- 49.Igarashi Y., Kan Y., Fujii K., Fujita T., Harada K.-I., Naoki H., Tabata H., Onaka H., Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. II. Structure determination. J. Antibiot. 2001;54:1045–1053. doi: 10.7164/antibiotics.54.1045. [DOI] [PubMed] [Google Scholar]
- 50.Onaka H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J. Antibiot. 2017;70:865–870. doi: 10.1038/ja.2017.51. [DOI] [PubMed] [Google Scholar]
- 51.Onaka H., Nakaho M., Hayashi K., Igarashi Y., Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151:3923–3933. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- 52.Ozaki T., Yamashita K., Goto Y., Shimomura M., Hayashi S., Asamizu S., Sugai Y., Ikeda H., Suga H., Onaka H. Dissection of goadsporin biosynthesis by in vitro reconstitution leading to designer analogues expressed in vivo. Nat. Commun. 2017;8:14207. doi: 10.1038/ncomms14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., Yang K., Tan H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. USA. 2009;106:8617–8622. doi: 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W., Ji J., Li X., Wang J., Li S., Pan G., Fan K., Yang K. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA. 2014;111:5688–5693. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q., Wang L., Xie Y., Wang S., Chen R., Hong B. SsaA, a member of a novel class of transcriptional regulators, controls sansanmycin production in Streptomyces sp. strain SS through a feedback mechanism. J. Bacteriol. 2013;195:2232–2243. doi: 10.1128/JB.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amano S., Morota T., Kano Y., Narita H., Hashidzume T., Yamamoto S., Mizutani K., Sakuda S., Furihata K., Takano-Shiratori H., et al. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot. 2010;63:486–491. doi: 10.1038/ja.2010.68. [DOI] [PubMed] [Google Scholar]
- 57.Amano S., Sakurai T., Endo K., Takano H., Beppu T., Furihata K., Sakuda S., Ueda K. A cryptic antibiotic triggered by monensin. J. Antibiot. 2011;64:703. doi: 10.1038/ja.2011.69. [DOI] [PubMed] [Google Scholar]
- 58.Imai Y., Sato S., Tanaka Y., Ochi K., Hosaka T. Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl. Environ. Microbiol. 2015;81:3869–3879. doi: 10.1128/AEM.04214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka Y., Izawa M., Hiraga Y., Misaki Y., Watanabe T., Ochi K. Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs. Appl. Microbiol. Biotechnol. 2017;101:4417–4431. doi: 10.1007/s00253-017-8216-6. [DOI] [PubMed] [Google Scholar]
- 60.Wang H., Zhao G., Ding X. Morphology engineering of Streptomyces coelicolor M145 by sub-inhibitory concentrations of antibiotics. Sci. Rep. 2017;7:13226. doi: 10.1038/s41598-017-13493-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seyedsayamdost M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. USA. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okada B.K., Wu Y., Mao D., Bushin L.B., Seyedsayamdost M.R. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem. Biol. 2016;11:2124–2130. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu F., Nazari B., Moon K., Bushin L.B., Seyedsayamdost M.R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 2017;139:9203–9212. doi: 10.1021/jacs.7b02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Craney A., Ozimok C., Pimentel-Elardo S.M., Capretta A., Nodwell J.R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012;19:1020–1027. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 65.Ochi K., Okamoto S. A magic bullet for antibiotic discovery. Chem. Biol. 2012;19:932–934. doi: 10.1016/j.chembiol.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 66.Ahmed S., Craney A., Pimentel-Elardo S.M., Nodwell J.R. A synthetic, species-specific activator of secondary metabolism and sporulation in Streptomyces coelicolor. ChemBioChem. 2013;14:83–91. doi: 10.1002/cbic.201200619. [DOI] [PubMed] [Google Scholar]
- 67.Pimentel-Elardo S.M., Sørensen D., Ho L., Ziko M., Bueler S.A., Lu S., Tao J., Moser A., Lee R., Agard D., et al. Activity-independent discovery of secondary metabolites using chemical elicitation and cheminformatic inference. ACS Chem. Biol. 2015;10:2616–2623. doi: 10.1021/acschembio.5b00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore J.M., Bradshaw E., Seipke R.F., Hutchings M.I., McArthur M. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Meth. Enzymol. 2012;517:367–385. doi: 10.1016/B978-0-12-404634-4.00018-8. [DOI] [PubMed] [Google Scholar]