Abstract
Panicle architecture and grain size are two important agronomic traits which determine grain yield directly in rice. In the present study, a mutant named ltbsg1 (longer top branch and shorter grain 1) was isolated from the cultivar “Zhenong 34” (Oryza sativa L. ssp. indica) by ethyl methane sulfonate (EMS) mutagenesis. The target gene was studied through phenotype observation, genetic analysis, map-based cloning and functional analysis. The histocytological analysis indicated that the elongated top branch and shorter grain of mutant ltbsg1 were caused from the defects of cell elongation. The ltbsg1 gene in mutant revealed a single nucleotide substitution (G-A) in the exon 2 of LOC_Os10g25780, causing an amino acid variation (Glycine-Arginine) in the FAD (Flavin-adenine dinucleotide)-binding domain of delta (24)-sterol reductase, which was involved in the brassinosteroid (BR) biosynthesis. LTBSG1 was constitutively expressed and the protein was widely localized in chloroplast, nucleus and cytomembrane. The ltbsg1 seedlings had a lower endogenous BR level and could be restored to the phenotype of wild type by exogenous BR. The LTBSG1 knock-out lines showed similar phenotype defects as mutant ltbsg1, which confirmed that LTBSG1 was responsible for top branch elongation and grain size reduction. Furthermore, LTBSG1 along with other BR-related genes were feedback-regulated due to their obvious altered expression in mutant ltbsg1. This study demonstrated that LTBSG1 could play a new role in regulating panicle and grain development by BR biosynthetic pathway.
Keywords: rice, LTBSG1, longer top branch, shorter grain, brassinosteroid (BR)
1. Introduction
Rice (Oryza sativa L.) is one of the most important cereal crops, which feeds over half of the world’s population. With the rapid increase of population, the world is facing more serious food security challenges. The improving of crop productivity is, hence, becoming ever more urgent and important in the rice breeding. The panicle number per plant, the grain number per panicle and the grain weight were the three main factors which determines the rice grain yield directly [1]. The panicle number per plant is largely influenced by the tillering ability; the grain number per panicle is closely related to the panicle length, the branch number, the branch length, the grain density and fertility; while the grain weight is mainly dependent on the size of grain referring to a combination of the grain length, the grain width and the thickness [2,3]. Thus, both panicle architecture and grain size are important agronomic traits directly affecting the grain yield, which have been the ongoing major purposes of crop improvement programs. In recent decades, the panicle architecture genes including FZP, DEP1, LP, FUWA and the grain size genes including GW2, GL3.1, TGW6, GL7, qSW5/GW5 and GW7 have been characterized consecutively [4,5,6,7,8,9,10,11,12,13,14]. However, the molecular mechanisms underlying the regulations of panicle architecture and grain size remained unclear, especially for the genes controlling the panicle morphology and grain size.
The existing data suggested that the panicle architecture and the grain size were not only controlled by genetic factors, which determined the states of inflorescence and spikelet meristem and regulated the cell division but also affected by the external environment (temperature, light, humidity) and hormone signals such as auxin (IAA), cytokinin (CK), gibberellin (GA) and brassinosteroids (BRs). BRs, a class of plant steroidal hormones, were involved in multiple development progresses in plants, such as organ elongation, vascular differentiation, light signaling, seed germination, reproductive development, biotic and abiotic stress response [15,16,17]. Brassinolide (BL) was the most bioactive compound among the BRs. BR biosynthesis and signaling pathways are complex regulatory processes involving multiple genes. Utilizing many mutants, the BR pathway has been transparently studied in dicotyledons Arabidopsis. BRs are perceived by a plasma membrane-localized BRI1 (BR INSENSITIVE 1)-BAK1 (BRI1-ASSOCIATED RECEPTOR KINASE 1) receptor complex and terminate in the nucleus [18,19,20,21], then a phosphorylation-dephosphorylation cascade involving the GSK3 (GLYCOGEN SYNTHASE KINASE 3)-like kinase BIN2 (BR INSENSITIVE 1), two downstream plant-specific transcription factors BRZ1 (BRASSINAZOLE-RESISTANT 1) and BES1 (BRI1-EMS-SUPPRESSOR 1) [17,22,23,24]. The dephosphorylated forms of BES1 and BZR1 in the nucleus could bind to the promoter of thousands of target genes and finally activate or repress their transcription [25,26]. However, the BR pathway in rice, a model plant of monocots, was less understood. Until now, there are only some BR related genes that have been identified from previous studies, which included BR biosynthetic genes BRD1, D2, BRD2, D11, CPD1, CPD2 and OsDWARF4 [27,28,29,30,31,32], as well as BR signaling gene D61/OsBRI1, OsBZR1, 14-3-3, BU1, OsBAK1 and DLT [33,34,35,36,37].
In most cases, BR-deficient or -insensitive mutants of rice showed dwarf phenotype, tortuous leaf blades or erect leaf, delayed flowering, reduced fertility and grain length [27,28,29,30,33,35]. In many studies, dwarfism and erect leaf were the most common phenotypes among all of the BR-related mutants because of the inhibitions of cell proliferation and cell elongation, which indicated that BR could regulate the organ size by affecting the cell cycle [38,39,40]. On the other hand, the studies conducted on the panicle and the grain development mediated by BR were not so much, and the genetic and molecular mechanisms remained unclear. The genes BRD1, D2 and D61 were directly acted on the BR biosynthesis and the signaling pathways, and their loss-of-function mutants were identified as the dwarf mutants with the phenotypes of short panicle and grain, but the genetic and regulated mechanism of panicle development mediated by these genes was not studied further [27,28,33]. In addition, some genes such as XIAO, SG1, SMG1 and GL2 controlling the grain size by cell proliferation and elongation were reported in influencing the BR responses and were speculated in involving the signaling and homeostasis of BR [41,42,43,44]. Therefore, the mechanism of BR regulating the panicle and the grain development needs a more in-depth research. Recently, a mutant cpb1 (clustered primary branch 1) was identified as a new allele of DWARF11 (D11), which encoded a cytochrome P450 protein and regulated the panicle architecture and grain size by BR biosynthesis pathway. In addition, using the panicle-specific promoter to drive CPB1/D11 could increase the grain size and the yield in rice [30,45]. As mentioned above, much evidence has revealed that the proper genetic manipulation of BR related genes involved in the biosynthesis and signaling pathways could remarkably enhance the grain yield in plant [46,47].
Here, a BR-deficient mutant ltbsg1 showing longer top branch and shorter grain was identified in present study. The target gene LTBSG1 was located on chromosome 10 finally. The sequencing result showed that a nucleotide substitution (G-A) occurred at the exon 2 of LOC_Os10g25780, which caused the amino acid variation (Glycine-Arginine) in the FAD-binding domain where was comparatively conserved among different plants. The LTBSG1 knock-out lines also showed similar defective phenotypes as mutant ltbsg1, which confirmed that LTBSG1 was responsible for top branch elongation and grain size reduction. LTBSG1 was a new allele of BRD2 which encoded a delta (24)-sterol reductase and could catalyze the conversion of 24-methylenecholesterol (24-MC) to campesterol (CR) in the early stage of BR biosynthesis [29]. The panicle architecture and the grain size, as two important agronomic traits, are closely associated with the rice grain yield. In view of many reported BR-related genes were focused on the dwarfism or the leaf angle, and the new phenotype of top branch elongation in mutant ltbsg1 was rare in rice mutants and even on BR-related mutants. Thus, this study will provide some new thoughts on the role of gene LTBSG1, which could regulate the yield traits such as panicle and grain development by BR pathway.
2. Materials and Methods
2.1. Plant Materials
The mutant ltbsg1 with longer top branch and shorter grain phenotype, was isolated from an indica cultivar Zhenong 34 through ethylmethane sulfonate (EMS) treatment. After multiple generations of continuous self-crossing, this mutated phenotype could be stably inherited. All the plant materials were grown in the paddy field of Zhejiang University in Hangzhou (120°19′ E, 30°26′ N), China.
2.2. Measurement of Agronomic Traits
The main panicles of ten plants at mature stage were randomly selected for measuring the agronomic traits of WT and ltbsg1 including the plant height (cm), the branch length (cm), the branch number (No.) and the seed setting rate (%). After air-dried, ten fully developed grains were measured for the grain length (mm) and the width (mm) by using the vernier caliper. The 1000-grain weight (g) was estimated from the weight of 200 grains and repeated three times.
2.3. Histocytological Analysis
To analyze the paraffin section, the fresh samples were fixed in FAA (3.7% formaldehyde, 50% ethanol and 5.0% glacial acetic acid) overnight at 4 °C, then dehydrated using a graded ethanol series (50%, 70%, 85% and 95% for 90 min each and 100% for 60 min) and finally embedded in paraffin. The embedded tissue sections (10 μm) were de-paraffined with xylene, and then stained with safranin and fast green. Finally, digital images were collected by the light microscopy (LEICA DMI4000, Wetzlar, Germany).
For scanning electron microscopy (SEM), fresh samples were fixed in 2.5% glutaraldehyde solution for at least 2 h, and then washed with a sodium phosphate buffer (0.1 M, pH 7.2) three times. The samples were fixed in 1% osmic acid for 1.5 h, and then dehydrated through an ethanol series. After incubated in the ethanol-isoamyl acetate (1:1, v/v) and isoamyl acetate, in turn, the samples were dried, mounted and coated with gold. Finally, they were observed and photographed by the scanning electronic microscope (HITACHI TM-1000, Tokyo, Japan).
2.4. BR Sensitivity Test
The BR sensitivity test of the seedlings and the lamina joint (between leaf sheath and blade) were performed as described previously [40]. To detect the BR sensitivity of mutant ltbsg1, 24-EBL (24-Epibrassinolide) (Sigma Aldrich, Saint Louis, MO, USA), a type of active BR, was selected in this study. Firstly, a 0.010 M mother solution was made by dissolving 25 mg 24-EBL solid powder in 2.5 mL of 95% (v/v) ethanol completely. In addition, then we added this mother solution of 0 μL, 0.1 μL, 1 μL, 10 μL and 100 μL into liquid culture solution (1 L) to prepare the test solution of 0.000 µM, 0.001 µM, 0.010 µM, 0.100 µM and 1.000 µM 24-EBL, respectively. The sterilized seeds of WT and mutant ltbsg1 were grown in the liquid culture solution with different doses of 24-EBL in a constant growth chamber (28 °C, 16 h/light and 8 h/dark). Then the seedlings at the three-leaf stage were photographed. In the lamina joint bending assay, the detached lamina joints were photographed after incubation for 7 days under the dark condition.
2.5. The Determination of Endogenous Brassinolide
Whole plants of WT and mutant ltbsg1 without any BR treatment were harvested at the 8th week after germination for measuring the endogenous brassinolide level. The methods of extraction and quantification of brassinolide were in reference to that described by Huo et al. [48]. The brassinolide, a standard in this experiment, was purchased from Sigma Aldrich with a purity of 90%.
The fresh samples were grinded to powder in liquid nitrogen with a mortar and pestle. 2 g powder was extracted in 10 mL of 80% methanol (precooled at 4 °C) for 2 h at 4 °C. Then centrifugation at 10,000 r/min, 4 °C for 5 min, and the supernatant was transferred to the Bond Elut prepacked column (Aglient, Santa Clara, CA, USA) and eluted with 3 mL of methanol. Using methanol diluted the eluent obtained in the last step by 50% and transferred it to the strata-X cartridge (33 mm, 3 mL) (Phenomenex), eluting with 3 mL of methanol. After dried by the pressure blowing concentrator, the dry matter was dissolved with 200 μL of methanol. The solution was filtered with a 0.22 μm filter, and it stored at −20 °C until HPLC-MS/MS (High performance liquid chromatography-tandem mass spectrometry) analysis.
The HPLC separation was performed based on the system of Aglient1290 High Performance Liquid Chromatography (Aglient, Santa Clara, CA, USA), and the analytical column was a ZORBAX SB-C18 Reversed-Phase Column (2.1 × 150, 3.5 μm). The flow rate of mobile phase was set at 0.35 mL/min with column temperature of 35 °C and injection volume of 5 μL. The separation was performed by gradient elution using mobile phase (A) (0.1% formic acid) and the aqueous organic phase (B) (Methanol). The gradient elution program was employed during the separation process (Solution A: 80% in 0–2 min, 80–95% in 2–3.5 min, 95% in 3.5–6 min, 95–80% in 6–6.1 min and 80% in 6.1–10 min). The HPLC system was coupled to a SCIEX-6500Q trap (MS/MS), (SCIEX, Redwood City, CA, USA) in the positive mode. Data was acquired by the multiple reaction monitoring (MRM) mode. The capillary voltage was set at 5500 V and the rate of nebulization gas was set as 800 L/h at 550 °C. The ion source temperature was set at 60 °C. The protonated molecule was chosen as the precursor ion and the most intensive product ion was selected for the quantification. The selected quantification ion plus another specific product ion were chosen for the confirmation. MRM mass spectrometric parameters of the two analytes were summarized in Table 1.
Table 1.
The reaction monitoring conditions for protonated brassinolide ([M + H]+).
| Analyte | Polarity | Precursor Ion (m/z) | Product Ion (m/z) | Cone Voltage (V) | Collision Energy (V) |
|---|---|---|---|---|---|
| brassinolide | + | 481.6 | 445.3/315.3 | 48 | 29/57 |
m: The detected ion mass. z: The charge on the detected ion. m/z: The mass-to-charge ratio.
2.6. Genetic Analysis and Map-Based Cloning of ltbsg1
The segregation ratio of ltbsg1 phenotype and WT plants in the F2 population from the crossing ltbsg1 and Zhenong 34 plants was analyzed by the Microsoft Excel.
To map the gene ltbsg1, F2 population was constructed by the crossing from ltbsg1 with Zhenongda 104 (Oryza sativa L. ssp. japonica) plants. Totally 729 individuals with clear ltbsg1 phenotype from the F2 population were selected for the gene mapping by using the mapping method performed as described by Zhang et al. [49]. The polymorphic primers were designed by DNASTAR and Primer premier 5.0 software based on the sequence differences between indica (Oryza sativa L.) and japonica (Oryza sativa L.). To investigate the functional annotations of genes in the candidate region, databases of RGAP (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/) and RiceData (China Rice Data Center, http://www.ricedata.cn/gene/) were used, thereby sequencing the candidate genes of WT and ltbsg1 to confirm the mutation site. The gene structure and sequence were referred to http://ensembl.gramene.org/Oryza_indica/Info/Index. The primes for the gene mapping are listed in Table S1A.
2.7. Generation of Knock-Out Transgenic Plants
To construct the knock-out vector of LTBSG1, the pYLCRISPR/Cas9Pubi-H system was used in this study [50]. The sequence ATGGATTCTGGTGATCTTTG close to the start codon of LTBSG1 was selected as the target of single guide RNA (sgRNA). The overlapping PCR was used to introduce the target sequences into sgRNA expression cassettes, and then the sgRNA expression cassette was cloned into the pYLCRISPR/Cas9Pubi-H binary vector by golden gate cloning strategy. Finally, the vector was introduced into Nipponbare (Oryza sativa L. ssp. japonica) callus by Agrobacterium-mediated transformation using EHA105 strain. To identify mutation by CRISPR (Clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated 9) editing, the primers flanking the designed target site of genomic DNA from transgenic plants were used for the PCR amplification and sequencing. Primers used for vector construction are listed in Table S1B. In this experiment, totally eight positive lines by CRISPR/Cas9 editing were obtained. Three homozygous lines with different mutations (Cas-k1, Cas-k2 and Cas-k3) and Nipponbare were used for analysis. The data for expression level of LTBSG1 in each knock-out line was derived from three biological replications. Ten plants from each line were selected for the measurements of agronomic traits and the details followed the way of the WT and mutant ltbsg1.
2.8. β-Glucuronidase Assay
β-glucuronidase (GUS) assay in transgenic plants was performed as previously described by Jefferson et al. [51]. To generate the promoter: GUS vector, a 2262-bp fragment upstream from the ATG codon of LTBSG1 was amplified. Primers used for vector construct are listed in Table S1B. After the amplified fragment ligated into the binary vector pCAMBIA1301 upstream of the GUS reporter gene, this vector was introduced into Nipponbare callus by Agrobacterium-mediated transformation using EHA105 strain. The tested tissues were immersed in a solution (1 mM 5-bromo-4-chloro 3-indolyl-glucuronic acid, 100 mM sodium phosphate (pH 7.0), 0.1 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide and 0.1% Triton X-100). Then the samples were vacuumed for half an hour and incubated at 37 °C, for 24 h. After removal chlorophyll from by 70% ethanol, digital images were taken by light microscopy (LEICA DMI4000, Wetzlar, Germany).
2.9. Subcellular Localization of LTBSG1
For subcellular localization, the full-length coding sequence of LTBSG1 without the stop codon was amplified (Primers are listed in Table S1B) and ligated into the empty GFP (Green Fluorescent Protein) vector. Then the empty vector and the fusion constructs GFP-LTBSG1 vectors were introduced into the rice protoplasts as described by He et al. [52]. The fluorescence was detected by laser confocal microscopy (ZEISS LSM 700, Jena, Germany).
2.10. RNA Isolation and qRT-PCR Analysis
Total RNA was extracted using Trizol reagent as the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). PrimeScript RT reagent Kit with gDNA Eraser and SYBR Premix Ex Taq II were used for qRT-PCR (Quantitative Real Time-PCR) as described by instruction of Takara Company (Tokyo, Japan). Primers for qRT-PCR were listed in the Table S1C. The expression levels were analyzed using a Real-Time System of Roche LightCycler® 96 (Basel, Switzerland) with rice OsActin as an internal control. The reaction solution (20 μL) contained 0.8 μL of each primer (10 μM), 10 μL 2 × SYBR Premix Ex Taq II, 1.6 μL cDNA and 6.8 μL ddH2O. The three-step protocol was performed in this qRT-PCR: activation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 55 °C for 20 s, and extending at 72 °C for 10 s. Values of expression levels represent the means ± standard deviation (SD) of three biological replicates (n = 3). * p < 0.05; ** p < 0.01.
2.11. Alignment Analysis and Phylogenic Analysis
All the protein sequences were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/BLAST/), using the LTBSG1 full-length protein sequence as a query against the nonredundant protein database. Amino acid sequence alignment was performed by the ClustalW [53]. The phylogenetic tree was constructed using the MEGA 6.0 [54] with the neighbor-joining method by 1000 bootstrap replicates.
3. Results
3.1. Phenotype Characterization of Mutant ltbsg1
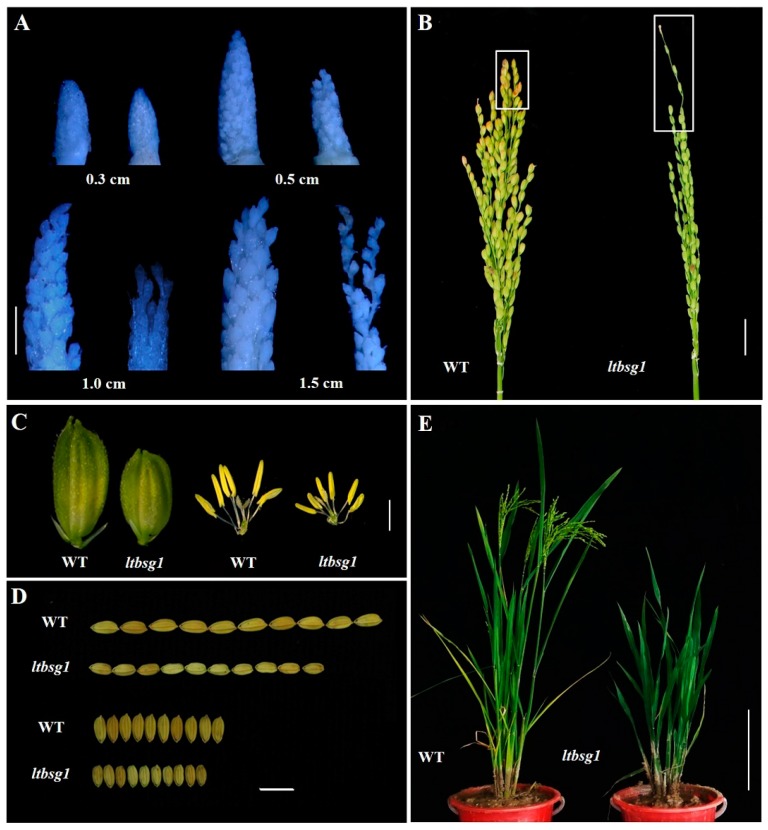
The phenotype observation showed that mutant ltbsg1 displayed the new phenotype of longer top branch in panicle and shorter grain (Figure 1A–D; Table 2). In addition, ltbsg1 exhibited severe dwarfism, erect lamina joint and late mature phenotype (Figure 1E; Figure S1; Table 2). In the early stage of panicle development, the panicle of ltbsg1 was a little smaller compared with WT. When the panicle length of ltbsg1 reached to 0.5 cm, the top branch of ltbsg1 started to protract and it was dramatically elongated later compared with that of WT (Figure 1A). At grain-filling stage, the top branch of ltbsg1 was much longer than that of WT, whereas the other branches were relatively short. The primary and secondary branches of ltbsg1 were much less than those of WT (Figure 1B; Table 2). Furthermore, the spikelet and floret of ltbsg1 were obviously smaller than those of WT (Figure 1C). The mature grain of mutant ltbsg1 also displayed significantly reduced length and width, which led to a much decreased 1000-grain weight (16.63 g) (Figure 1D; Table 2). The pollen vitality of ltbsg1 was much lower than that of WT, while the seed setting rate was just 8.42% correspondingly (Figure S2; Table 2).
Figure 1.
Phenotype characterization of wild type (WT) and mutant ltbsg1 (longer top branch and shorter grain 1). (A) The young panicles of different length in WT and mutant ltbsg1. Bar = 2 mm. Left: WT; Right: ltbsg1; (B) The panicles of WT and mutant ltbsg1 at grain-filling stage. The white box indicated the top branch of panicle. Bar = 2 cm; (C) The spikelets and flowers of WT and mutant ltbsg1. Bar = 2 mm; (D) The grains length and width of WT and mutant ltbsg1. Bar = 1 cm; (E) Plants of WT and mutant ltbsg1 at grain-filling stage. Bar = 20 cm.
Table 2.
Agronomic traits of WT and mutant ltbsg1.
| Traits | WT | ltbsg1 |
|---|---|---|
| Plant height (cm) | 82.90 ± 1.30 | 54.41 ± 1.37 ** |
| Top branch length (cm) | 3.55 ± 0.21 | 5.63 ± 0.19 ** |
| Primary branch length (cm) 1 | 7.60 ± 0.46 | 5.80 ± 0.35 ** |
| Secondary branch length (cm) | 2.13 ± 0.16 | 1.60 ± 0.21 ** |
| No. of primary branches | 12.67 ± 0.82 | 7.50 ± 1.05 ** |
| No. of secondary branches | 20.83 ± 1.17 | 1.83 ± 0.75 ** |
| Grain length (mm) | 8.12 ± 0.03 | 5.93 ± 0.02 ** |
| Grain width (mm) | 3.17 ± 0.02 | 2.83 ± 0.01 * |
| 1000-grain weight (g) | 25.83 ± 0.12 | 16.63 ± 0.03 ** |
| No. of grains per panicle | 190.90 ± 3.51 | 84.70 ± 3.83 ** |
| Seed setting rate (%) | 90.95 ± 1.38 | 8.42 ± 1.10 ** |
1 The primary branch length represented the average length of all primary branches without the top branch. Values represent the means ± SD (n = 10). * p < 0.05; ** p < 0.01.
3.2. LTBSG1 Affected the Top Branch and the Grain Size by Regulating the Cell Elongation
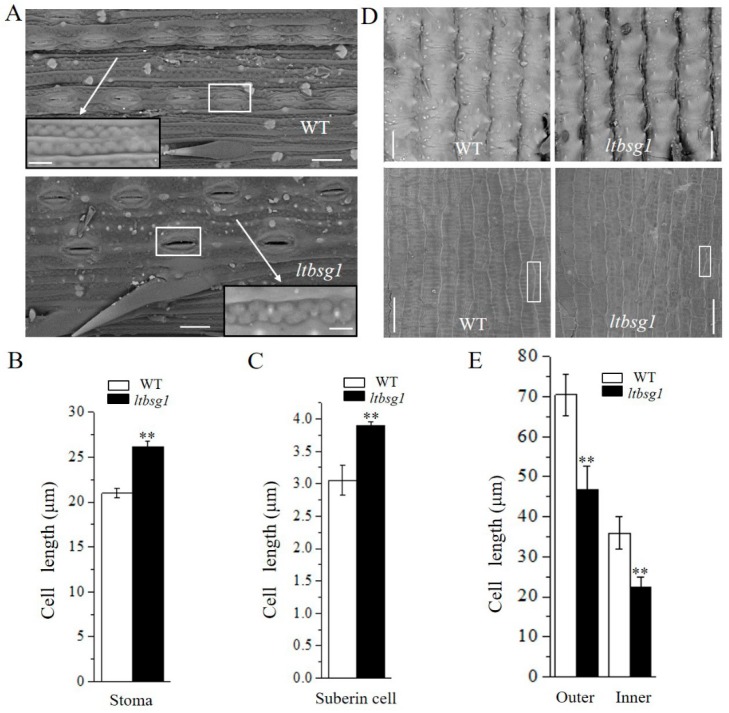
To clarify whether cell proliferation or cell elongation affecting the lengths of branch and grain in ltbsg1, assays of scanning electron microscopes were performed in the present experiment. The lengths of stomata and suberin cell of mutant ltbsg1 were significantly longer in top branch than those of WT, indicating that the cell elongation was exacerbated in ltbsg1 (Figure 2A–C). However, the cell lengths of both outer and inner epidermal surfaces of spikelet were much shorter in mutant ltbsg1 compared to those in WT, which conferred the reduced grain length of mutant (Figure 2D,E).
Figure 2.
The effect of LTBSG1 on top branch and grain development. (A) The scanning electron microscopic analysis of the top branch surfaces of WT and mutant ltbsg1. Bars = 20 μm. The white frames indicated the stoma and the white arrows indicated the magnified views of suberin cell. Bars = 5 μm; (B) The length of stoma in top branch surfaces of WT and mutant ltbsg1; (C) The length of suberin cell in top branch surfaces of WT and mutant ltbsg1; (D) The scanning electron microscopic analysis of spikelet hulls of WT and mutant ltbsg1. The outer surface and inner surface were showed up panel (Bars = 50 μm) and bottom panel (Bars = 30 μm), respectively; (E) The cell length of outer and inner surfaces of spikelet hulls in WT and mutant ltbsg1. Values represent the means ± SD (n = 10). ** p < 0.01.
3.3. Map-Based Cloning of the ltbsg1 Gene
The genetic analysis showed that the panicle morphology of F1 plants was normal as that of wild type, and it produced a phenotypic separation between wild type (normal panicle) and ltbsg1 phenotype (deformed panicle with longer top branch and shorter grain) in the F2 population (ltbsg1/Zhenong 34). In addition, the segregation ratio of wild type to ltbsg1 phenotype accorded with a Mendel model of 3:1 (, n = 408). This result indicated that the phenotype of longer top branch and shorter grain in ltbsg1 was controlled by a single recessive nuclear gene.
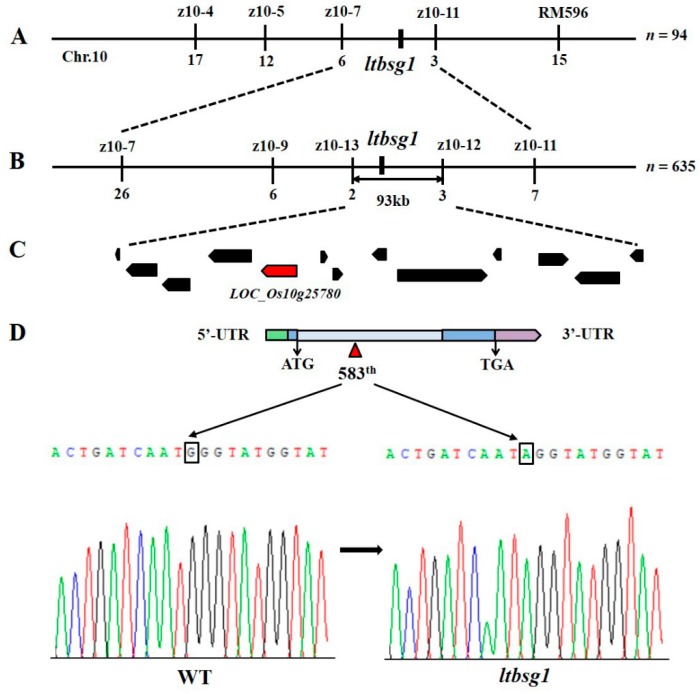
Map-based cloning was used for locating the ltbsg1 gene based on the F2 population (ltbsg1/Zhenongda104). LTBSG1 was primarily mapped on the long arm of chromosome 10 between InDel (Insertion-Deletion) markers z10-7 and z10-11 (Figure 3A). To fine-map the gene, another 635 recessive individuals and several polymorphic markers were used, and the target locus was narrowed down to a 93-kb interval between markers z10-13 and z10-12 (Figure 3B). A total of 13 putative genes were distributed in this region according to database of RGAP and the genes function annotations of them are listed in Table S2. Among these genes, one locus LOC_Os10g25780 was annotated as FAD-linked oxidoreductase protein and it was allelic to BRD2, which was involved in BR biosynthesis (Figure 3C). After comparing the genomic sequences of these genes in this region between WT and ltbsg1, only one base substitution (G-A) at 583th base in the exon 2 of LOC_Os10g25780 was detected, which caused an amino acid substitution Glycine-Arginine (G-R, GGG-AGG) on the FAD-binding domain (Figure 3D and Figure 4). Thus, these results suggested that LOC_Os10g25780 was the candidate gene, which was responsible for longer top branch and shorter grain.
Figure 3.
Map-based cloning and confirmation of the gene ltbsg1. (A) Primary mapping of ltbsg1; (B) The fine mapping of gene ltbsg1; (C) The candidate genes in the target region; (D) The gene structure and the mutation site confirmed by sequencing analysis of LOC_Os10g25780 in WT and mutant ltbsg1. The black box indicated the single nucleotide mutation G (WT) to A (ltbsg1) at the 583th base in the exon 2. UTR: Untranslated Regions; Chr.: Chromosome.
Figure 4.
ClustalW alignment of the orthologs of LTBSG1. The orthologs of LTBSG1 from different plants were Zea mays (Accession Number NP_001105560.1), Sorghum bicolor (XP_021306085.1), Arabidopsis thaliana (NP_188616.1), Gossypium hirsutum (XP_016697581.1) and Brassica napus (XP_013721305.1), respectively. The red line region represented the FAD-binding domain (110-237 amino acids) and the amino acid substitution (G-R) was indicated by the yellow frame.
3.4. Analysis of LTBSG1 Orthologous Proteins
The LTBSG1 gene encoded a delta (24)-sterol reductase and played an important role in BR biosynthesis by catalyzing the conversion of 24-methylenecholesterol (24-MC) to campesterol (CR) [29]. LTBSG1 contained a FAD-binding domain. To identify the conservation of LTBSG1 protein sequences, another five orthologs of LTBSG1 from Zea mays, Sorghum bicolor, Arabidopsis thaliana, Gossypium hirsutum and Brassica napus were compared. They shared high similarity in the FAD-binding domain and the amino acid (G) where the substitution occurred was unified among the six plants (Figure 4). This result indicated that this domain sequence was highly conserved and played a very important role in maintaining the function of LTBSG1. As shown in phylogenetic analysis of 20 representative proteins, compared with the dicots, the LTBSG1 protein was more closely related to the monocots Oryza sativa japonica, Oryza brachyantha, Setaria italic, Zea mays and Sorghum bicolor (Figure S3).
3.5. Subcellular Localization of LTBSG1
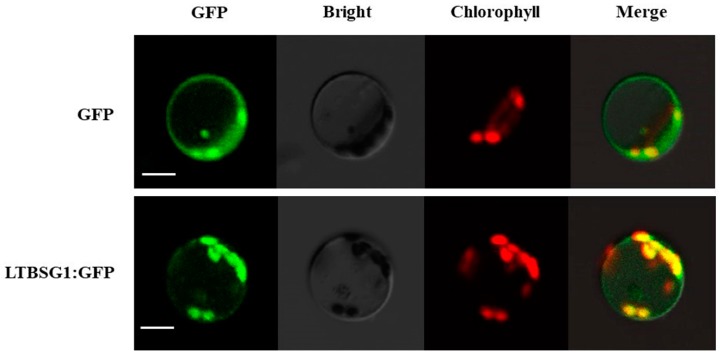
To confirm the subcellular localization of LTBSG1, the LTBSG1-GFP fusion vector was transiently expressed in the rice protoplasts. LTBSG1-GFP signals in the rice protoplasts were distributed almost in whole cell, which was strongly expressed in chloroplast and slightly in nucleus, cytomembrane (Figure 5). Therefore, the widely localization of LTBSG1 implied that it might be involved in multiple functional pathways in plant development.
Figure 5.
Subcellular localization of LTBSG1-GFP in rice protoplasts. Free GFP (Green Fluorescent Protein) (up panel) and LTBSG1-GFP (bottom panel) vectors were introduced into rice protoplasts. The horizontal four columns referred to GFP fluorescence images, bright images, chlorophyll spontaneous light images and their merged images for the same cells, respectively. Bars = 50 μm.
3.6. Expression Pattern of LTBSG1
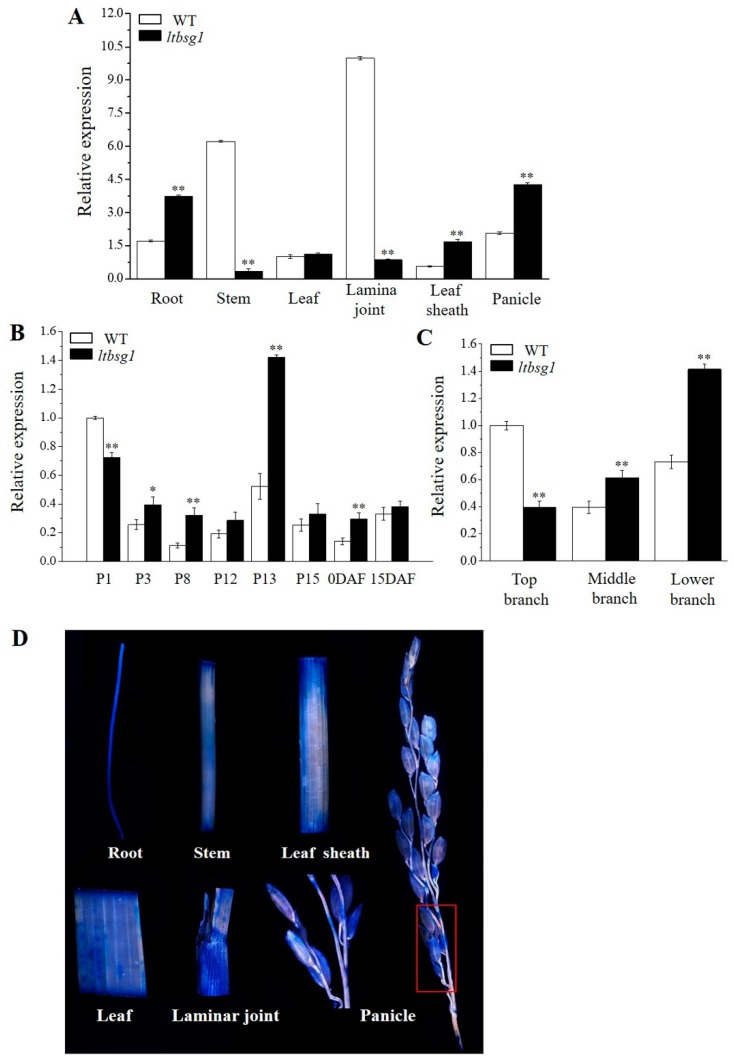
The expression profile of LTBSG1 at the heading stage showed that it could be expressed in all the tested organizations and was highly expressed in root, stem, lamina joint and panicle in WT. The expression levels of LTBSG1 in stem and lamina joint were much down-regulated in mutant ltbsg1 and up-regulated in root, leaf sheath and panicle, but the difference in leaf was not significant between WT and mutant ltbsg1 (Figure 6A). As the morphological states of branch and spikelet could be determined before the grain-filling stage, panicles in the length of 1 cm (P1) to 15 cm (P15), flowering day (0 DAF) and 15 days after fertilization (15 DAF) were selected for determining the expression levels of LTBSG1. LTBSG1 transcripts in both WT and ltbsg1 accumulated more in P1 and then decreased much in later several stages, and it reached the strongest level of the expression in P13 and then became lower again until the 15 DAF. Obviously, the expression levels of LTBSG1 in mutant ltbsg1 except for P1 were higher than those in WT during panicle development although they were not significant for P12 and P15 (Figure 6B). Moreover, the expression distinction of LTBSG1 in different branches of one panicle was also observed. The expression level of LTBSG1 in top branch of WT was significantly higher than those in the middle and lower branches, while that in top branch of mutant ltbsg1 was expressed at the lowest level (Figure 6C). These results revealed the expression variance of LTBSG1 in top branch between mutant and WT, which might be the reason for the elongation of the top branch in mutant. In agreement with the above gene expression profile in Figure 5A, LTBSG1-GUS could be expressed in all of the examined tissues at heading stage especially for root, lamina joint, spikelet and branch of the panicle (Figure 6D).
Figure 6.
Expression pattern analysis of LTBSG1. (A) Temporal and spatial patterns of LTBSG1 gene in WT and mutant ltbsg1 at heading stage; (B) LTBSG1 gene expression in panicles of 1 cm (P1) to 15 cm (P15), flowering day (0DAF) and 15 days after fertilization (15DAF); (C) The expression of LTBSG1 in different branches of young panicle in WT and mutant ltbsg1; (D) β-glucuronidase (GUS) assay staining analysis of the LTBSG1 promoter in different tissues. The red box in panicle was magnified. The rice OsActin gene was used as an internal control. Values represent the means ± SD of three biological replicates (n = 3). * p < 0.05; ** p < 0.01.
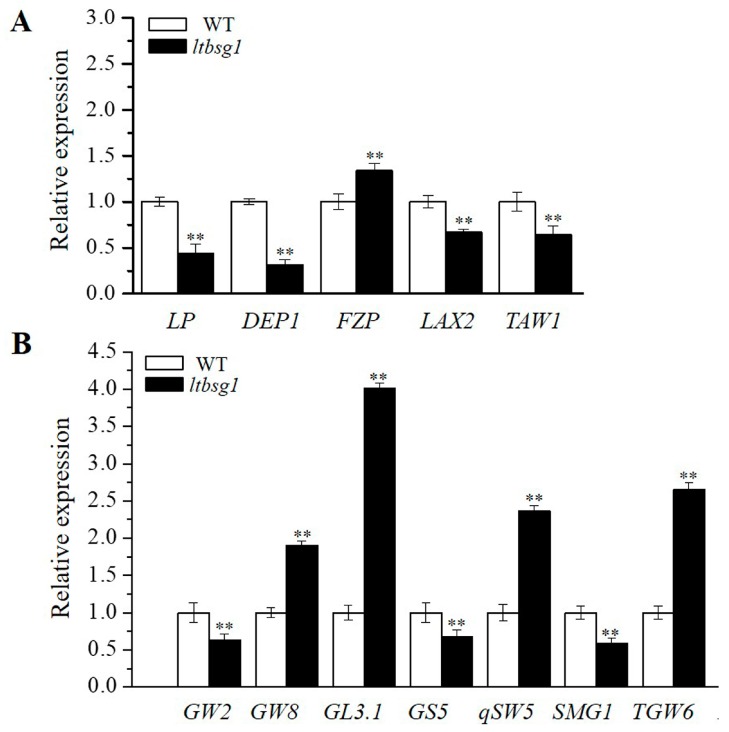
3.7. The Expression Analysis of Panicle Architecture and Grain Size Related Genes
To identify the role of LTBSG1 in the regulatory pathway about panicle and grain development, the expression levels of genes associated with the panicle architecture and the grain size were examined correspondingly. The results showed that the expressions of LP, DEP1, LAX2 and TAW1 were much down-regulated in the mutant ltbsg1, whereas FZP was higher increased compared with those in WT (Figure 7A). As well, the expression levels of genes related to the grain size revealed that the expressions of GW8, GL3.1, qSW5 and TGW6 but not GW2, GS5 and SMG1 were significantly increased in mutant ltbsg1 (Figure 7B). These results suggested that LTBSG1 might regulate the panicle and the grain development relying on the panicle and the grain regulation pathway by affecting these related genes’ expression.
Figure 7.
The expression of genes related to the panicle and the grain development in WT and mutant ltbsg1. (A) The expression of genes related to the panicle development in young panicles of WT and mutant ltbsg1; (B) The expression of genes related to the grain size in young panicles of WT and mutant ltbsg1. The rice OsActin gene was used as an internal control. Values represent the means ± SD of three biological replicates (n = 3). ** p < 0.01.
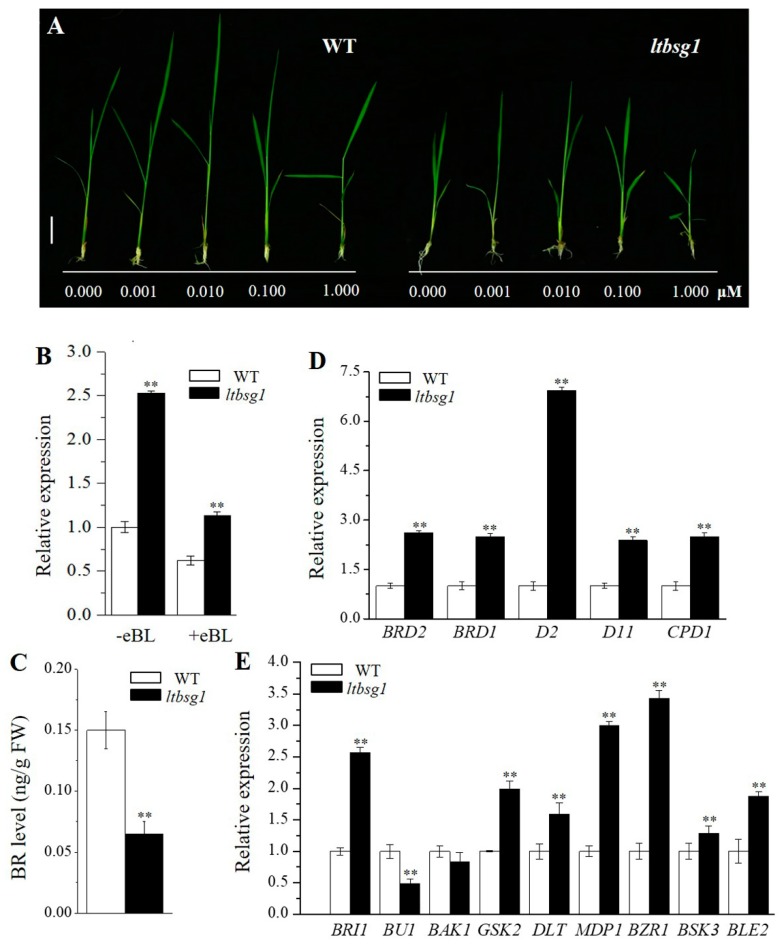
3.8. ltbsg1 Was a BR-Deficient Mutant and LTBSG1 Was Feedback-Regulated by 24-EBL
It is well known that the altered plant height and lamina joint bending were the typical responses to BR in rice for which were highly sensitive to BR. Those were also considered as the proofs to determine whether mutant was BR-deficient or BR-insensitive [28]. To test the BR sensitivity of mutant ltbsg1, different doses of 24-EBL, a bioactive BR compound, were used at the seedling stage. The mutant ltbsg1 showed the highly response to 24-EBL-like WT in both seedling height and lamina joint bending, thus, it was considered to be a BR-deficient mutant. The heights of seedlings for both WT and were increased with the increasing 24-EBL dose and it reached the highest at 0.010 µM, then suppressed by 24-EBL of 0.100 µM and 1.000 µM gradually (Figure 8A). It indicated that the plant height was dose-dependent on BR. However, the lamina joint bending was positive promoted with the dose increasing. The bending of lamina joint in mutant ltbsg1 at 0.000 µM 24-EBL was almost invisible, but it could bend more greatly as the 24-EBL dose increased, even the leaf angle was near 90° in both WT and mutant ltbsg1 at 1.000 µM (Figure 8A). In terms of the response to 24-EBL, 0.010–0.100 µM might be the optimal concentration region for recovering the phenotype of mutant. Given that the expressions of BR biosynthesis genes, such as BRD1, D2 and OsDWARF4 were feedback regulated by the end product of the BR biosynthesis pathway [27,28,32], the effect of 24-EBL on LTBSG1 expression was studied here. Without 24-EBL, the LTBSG1 expression level was higher in mutant ltbsg1 seedlings compared with that in WT. In addition, it was dramatically reduced in WT or mutant plants after treated with 24-EBL, implying that the expression of LTBSG1 was feedback-regulated by 24-EBL (Figure 8B). Correspondingly, the level of endogenous brassinolide in mutant ltbsg1 was reduced to one-half of that in WT, confirming that the mutant ltbsg1 was deficient in the biosynthesis of BR (Figure 8C).
Figure 8.
Rescue of the phenotype of mutant ltbsg1 by 24-EBL and the feedback regulation of BR-related genes. (A) Phenotypic observation of WT (left) and mutant ltbsg1 (right) at three-leaf stage treated with different doses of 24-EBL (0.000 to 1.000 μM). Bar = 1.5 cm; (B) The expression of LTBSG1 in leaf treated with 0.01 μM 24-EBL; (C) Quantification of endogenous brassinolide contents of WT and mutant ltbsg1; (D) The expression levels of genes related to BR biosynthesis in young panicles of WT and mutant ltbsg1; (E) The expression levels of genes related to BR signaling in young panicles of WT and mutant ltbsg1. The rice OsActin gene was used as an internal control. Values represent the means ± SD of three biological replicates (n = 3). ** p < 0.01.
In addition, the expression levels of BR biosynthesis genes BRD1, D2, D11 and CPD1 in young panicle of mutant ltbsg1 were much higher than those in WT (Figure 8D). The significantly higher expression of gene D2 among these biosynthetic genes indicated it was the most responsive to BR defect. This result indicated the feedback regulations of BR biosynthetic genes in young panicle. Meanwhile, BR signaling genes including BRI1, GSK2, DLT, MDP1, BZR1, BSK3 and BLE2 except for BU1 and BAK1 had the increased expression levels in mutant ltbsg1 (Figure 8E). The responses of BR signaling genes in mutant ltbsg1 revealed that the BR signaling genes were required in the feedback regulation of BR biosynthetic genes.
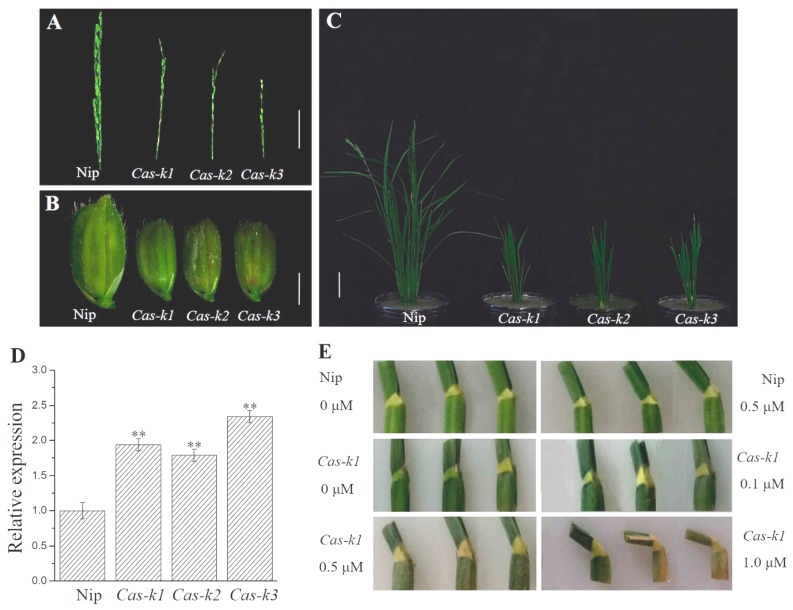
3.9. The Confirmation of LTBSG1 Gene Function
Since the indica material Zhenong34 was difficult to regenerate in plants, Nipponbare was used for producing knock-out transgenic lines of LTBSG1 by CRISPR/Cas9 strategy, to confirm the inactivation of LTBSG1 contributed to the elongation of top branch and the reduction of grain size in mutant ltbsg1. All the eight positive lines showed similar defects on the panicle development as the mutant ltbsg1. Among them, three independent homozygous lines Cas-k1, Cas-k2 and Cas-k3 accompanied with nucleotide deletions at 173th bp, 174th bp and 175th bp of exon 2 respectively were selected for phenotypes analysis (Figure S4). They exhibited obvious elongation of top branches on the panicle (Figure 9A, Table S3). Meanwhile, the grains were much shorter with the decreased length and width compared to those of Nipponbare (Figure 9B, Table S3). The expression level of LTBSG1 in young panicles of each line was much increased than that in Nipponbare, which was consistent with the expression trend in the mutant ltbsg1 (Figure 9C). Beyond that, the plants of knock-out lines displayed severe dwarfism and the lamina joints did not show any bending and leaf blades were upright, which led to compact plant architecture in the knock-out plants (Figure 9D, Table S3). In addition, the agronomic traits of each line also showed serious defects such as the reduced grain number per panicle and seed setting rate (Table S3), which were also consistent with those of mutant ltbsg1. The BR sensitivity test revealed that the lamina joints of knock-out plants was hyper-sensitive to 24-EBL and it could bend more obviously with the increasing BR dose (Figure 9E). Therefore, we concluded that the gene LTBSG1 was responsible for the longer top branch and shorter grain of mutant ltbsg1 as well as the knock-out plants.
Figure 9.
Phenotype analysis of three knock-out lines Cas-k1, Cas-k2 and Cas-k3. (A) The morphological analysis of panicles of knock-out lines. Bar = 5 cm; (B) The morphological analysis of spikelets in knock-out lines. Bar = 2 mm; (C) Plant phenotype of knock-out lines. Bar = 10 cm; (D) The expression levels of LTBSG1 in the young panicles of knock-out lines; (E) Sensitivity test of lamina joints to 24-EBL in knock-out line cas-k1. Values represent the means ± SD of three biological replicates (n = 3). ** p < 0.01.
4. Discussion
The panicle architecture and grain size are important agronomic traits, which are closely related to grain production. As many mutants and genes have been identified, people are more aware of the mechanisms of panicle and grain development now. In general, rice plant showed normal panicle with the short top branch. However, the mutant ltbsg1 in the present experiment displayed longer top branch, which was rare in previous studies. Meanwhile, the grains of mutant were much shorter than those of WT. The genetic analysis indicated that this phenotype was controlled by a single recessive gene. Through map-based cloning, the locus of LOC_Os10g25780 in mutant ltbsg1 revealed a single nucleotide substitution (G-A) in exon 2, causing an amino acid variation (G-R) on the FAD-binding domain. The gene LTBSG1 was allelic to BRD2, which encoded delta (24)-sterol reductase functioning in the BR biosynthesis pathway by catalyzing the conversion of 24-methylenecholesterol to campesterol [29]. Interestingly, the gene LTBSG1 was identified to be a new allele of BRD2, for the mutation site and phenotype were different from the previous two mutants brd2 and lhdd10, even though all the three mutants showed the typical BR-deficient phenotype such as severe dwarf, erect lamina joint, late mature and low fertility [29,55]. More importantly, the new phenotype of top branch elongation in mutant ltbsg1 was rare on BR-related mutants. Besides the longer top branch, the internode pattern among these three mutants was also obviously different. The mutant ltbsg1 exhibited similar inhibition in internode length as brd2, which was in japonica background and had a premature stop codon on exon 2. Compared to WT, the first two internodes of ltbsg1 from the base were so short that could not be distinguished easily, and the upper three internodes were also reduced a lot, which was caused by the inhibition of cell elongation (Figure S1). However, the allele lhdd10 with indica background had a single nucleotide substitution in exon 2 and showed a very different type. The first two internodes from the base of lhdd10 plant were almost not changed, only the upper three internodes were shortened. Given the above, the gene LTBSG1 showing different effects on plant development might due to the different genetic background and the mutation way.
Many genes related to biosynthesis and signal transduction of BR were reported recently and had effect on plant height, panicle architecture and grain size such as GW5, XIAO, SMG1, GL2, CPB1/D11 and GS6/DLT, which also influenced the BR responses and the expression of BR-related genes [13,41,42,43,44,45,56]. Previous studies suggested that BR regulated the organ size by cell elongation and cell division in rice [27,41,57]. The organs of BR-related mutants were usually short and small, but the regulation mechanism was still not clear. Here, mutant ltbsg1 exhibited the defects in cell elongation in different organ including short stem and grain. On the other hand, it showed longer top branch due to the obvious cell elongation were (Figure 2; Figure S1). This contrary phenomenon was also occurred on previous BR related mutants d2 and d61 [28,33]. They not only showed the different elongation of internodes such as mutant ltbsg1, but also produced the over-elongation of panicle neck internode. The gene BRI1/D61 was expressed higher in the uppermost and fourth internodes, and it allowed these internodes to respond to BR by inducing elongation. The expression levels of LTBSG1 were also varied significantly among different organs, suggesting that their response to BR was also different. This might be the reason for different elongation patterns in mutant ltbsg1.
A recent study suggested that the BRs could promote the pollen and grain development in rice by enhancing the expression of Carbon Starved Anther (CSA), which could directly trigger the expression of sugar partitioning and metabolic genes [58]. OsSPL16 (GW8), an SBP-domain transcription factor related to the grain width, could directly bound to the GW7 promoter and repressed its expression to further regulate the grain size [14]. In the present study, the gene LTBSG1 might act on the panicle and grain development relying on the related genes regulatory network. In support of this suggestion, the expression levels of most panicle and grain related genes were alerted in mutant ltbsg1 (Figure 7). The expression level of DEP1 was repressed in mutant ltbsg1, which regulated the panicle size negatively, indicating its negative relationship with LTBSG1. The increased expression levels of qSW5 and TGW6 (two negative regulators of grain size) and the decreased expression levels of GS5 and SMG1 (two positive regulators of grain size) in mutant ltbsg1 might indicate that these genes also contributed to the shorter grain of mutant ltbsg1 [10,12,43,59].
The mutants defective in BR biosynthesis pathway were usually sensitive to exogenous BR, and the defects could be rescued when given exogenous BR [29,30]. However, the mutants involved in BR signaling transduction could not perceive exogenous BR and the signal would not be passed on normally [33,35,37]. The sensitivity test to exogenous BR was also considered as the basis to determine whether it was a BR-deficient mutant or not. As mutant brd2, ltbsg1 was a BR-deficient mutant given that it was highly sensitive to 24-EBL and contained less endogenous brassinolide, especially the phenotype of mutant ltbsg1 could be recovered by a moderate dose of 24-EBL (Figure 8A,C). It was common that the biosynthesis of plant hormones was controlled by the level of the end products in a feedback manner through regulating the biosynthetic genes. Such feedback mechanism could maintain the endogenous hormone homeostasis. The phenomenon has been demonstrated on BR and other hormones, such as ethylene and GA [60,61,62]. Previous studies have revealed that expressions of genes BRD1, D2 and D11, which encoded key enzymes involved in BR biosynthesis, were feedback regulated by bioactive BR [27,28,30]. Here, LTBSG1 showed a similar feedback manner to the above genes, and it was negatively regulated by BR level. LTBSG1 had a higher expression in the BR-deficient mutant ltbsg1 than that of WT, and its expression was down-regulated after treated with 24-EBL (Figure 8B). Moreover, the other BR biosynthetic genes tested in this study such as BRD1, D2, D11 and CPD1 were found to have up-regulated expressions in the mutant ltbsg1 (Figure 8D), which were consistent with the feedback regulation of these genes in BR-deficient mutants. The significantly higher expression of gene D2 among these biosynthetic genes indicated it was the most responsive to BR defect to maintain the BR homeostasis. This feedback regulation was also found in Arabidopsis cytochrome P450 genes DWF4/CYP90B1, CPD/CYP90A1, DWARF/CYP85A1 and CYP90D1, which were downregulate by BL [63,64]. These findings indicated that the negative feedback regulation of BR biosynthetic genes was common in both dicot and monocot plants.
The BR signaling pathway was considered to be intact in the BR-deficient mutants. In view of the increased expression levels of LTBSG1 and other BR biosynthetic genes in the BR non-treated mutant ltbsg1 than those in WT, the feedback regulation of BR biosynthetic genes might be regulated through the BR signaling pathway. In addition, the expression result of BR signaling genes revealed the altered expressions between mutant and WT (Figure 8E), which indicated that their responses to the defect of LTBSG1 was changed. Thus, they were required for feedback regulation of BR biosynthesis genes. BRI1 encoded a receptor kinase and could perceive the BR signal on cell surface [33]. BRI1 was higher expressed in mutant ltbsg1 compared with WT, we proposed that, thus, the highly sensitivity of mutant ltbsg1 to 24-EBL might be caused by the accumulation of the BR receptor kinase. This similar result was also observed in the BR-deficient mutants d2 and d11. In the study of Arabidopsis, the BZR1 protein, a transcription factor in BR signaling pathway, could mediate the feedback inhibition of the BR biosynthetic genes such as BRD1, CPD and DWF4 [34]. The significantly higher expression of BZR1 in mutant ltbsg1 also confirming its role in BR response. Therefore, we proposed that this negative feedback regulation of the BR biosynthetic genes was regulated through BR-signaling molecules, such as the BRI1 and BZR1 proteins. The pathway of BR signaling introduction was a complicated network regulation, which involved in many kinds of receptor kinases, a series of phosphorylation and dephosphorylation reactions and transcription factor transmitting the signal to the target genes downstream [65,66]. Therefore, it needs more studies and efforts to uncover the roles of these genes they worked in the BR pathways.
In addition, the knock-out lines of LTBSG1 displayed typical development defects of BR related mutants such as ltbsg1, brd1, d2 and d11, which could also be restored by 24-EBL. Moreover, the top branch elongation and shorter grains were clearly identified in the knock-out plants (Figure 9). This work further confirmed the important role of LTBSG1 in regulating panicle and grain development by BR biosynthesis pathway.
In this study, besides the new phenotype of longer top branch and shorter grain, the mutant showed the pleiotropic phenotypes such as dwarfism, erect lamina joint, poor fertility and late maturity, which was consistent with the phenotypes of BR related mutants [17,40]. This indicated that the BR biosynthesis pathway or the BR hormone itself played important roles in plant various development processes. To gain insight into the function of LTBSG1, the gene ontology enrichment analysis of differentially expressed genes (DEGs) in biological process was performed. The resulted revealed many DEGs involved in the different biosynthetic process, metabolic process and catabolic process. Except that, there were also some DEGs which were enriched in the process of biotic and abiotic stresses responses such as defense response to bacterium, fungus and oxidative stress (Table S4). Therefore, these results further confirmed the multiple roles of LTBSG1. The mechanism of plant growth development regulated by BR is a complicated process, which requires more research to clarify in the future.
5. Conclusions
In conclusion, LTBSG1 was a new allele of BRD2, which was responsible for the longer top branch and shorter grain by involving in the brassinosteroid biosynthetic pathway.
Acknowledgments
We thank Yaoguang Liu for providing the CRISPR/Cas9 system and Liang Wu for the technical assistance in vector construction.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/6/292/s1, Figure S1: Analysis of internode pattern and lamina joints of WT and ltbsg1, Figure S2: The detection of pollen vitality between WT and ltbsg1, Figure S3: Phylogenetic tree of LTBSG1 with 20 homologous proteins in different plants, Figure S4: The confirmation of mutation sites of LTBSG1 three knock-out lines by sequencing, Table S1: Primer list of this study, Table S2: Predicated function analysis of candidate genes of ltbsg1, Table S3: Agronomic traits of Nipponbare (Nip) and three knock-out lines, Table S4: Gene ontology enrichment analysis for part of differentially expressed genes involved in biological process.
Author Contributions
Conceptualization, C.S.; Data curation, C.Y. and D.A.; Formal analysis, R.Q., D.Z. and X.J.; Funding acquisition, C.S.; Investigation, R.Q., D.Z., C.Y. and M.A.; Methodology, X.J.; Project administration, C.S.; Resources, C.S.; Software, D.A. and M.A.; Supervision, C.S.; Validation, R.Q.; Visualization, R.Q., D.Z. and C.Y.; Writing—original draft, R.Q.; Writing—review and editing, R.Q., D.Z., C.Y., D.A., M.A., X.J. and C.S.
Funding
This work was supported by Science and Technology Office of Zhejiang Province grant number 2016C32085 and 2016C02050-6.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Singh V.K., Ellur R.K., Singh A.K., Nagarajan M., Singh B.D., Singh N.K. Effect of qGN4.1 QTL for grain number per panicle in genetic backgrounds of twelve different mega varieties of rice. Rice. 2018;11:8. doi: 10.1186/s12284-017-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang E., Wang J., Zhu X.D., Hao W., Wang L.Y., Li Q., Zhang L.X., He W., Lu B.R., Lin H.X., et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- 3.Xing Y., Zhang Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130:3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- 5.Huang X.Z., Qian Q., Liu Z.B., Sun H.Y., He S.Y., Luo D., Xia G.M., Chu C.C., Li J.Y., Fu X.D. Natural variation at the dep1 locus enhances grain yield in rice. Nat. Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 6.Li M., Tang D., Wang K.J., Wu X.R., Lu L.L., Yu H.X., Gu M.H., Yan C.J., Cheng Z.K. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol. J. 2011;9:1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen J., Gao H., Zheng X.M., Jin M., Weng J.F., Ma J., Ren Y., Zhou K., Wang Q., Wang J., et al. An evolutionarily conserved gene, FUWA, plays a role in determining panicle architecture, grain shape and grain weight in rice. Plant J. 2015;83:427–438. doi: 10.1111/tpj.12895. [DOI] [PubMed] [Google Scholar]
- 8.Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. A QTL for rice grain width and weight encodes a previously unknown Ring-type E3 ubiquitin ligase. Nat. Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 9.Qi P., Lin Y.S., Song X.J., Shen J.B., Huang W., Shan J.X., Zhu M.Z., Jiang L., Gao J.P., Lin H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishimaru K., Hirotsu N., Madoka Y., Murakami N., Hara N., Onodera H., Kashiwagi T., Ujiie K., Shimizu B., Onishi A., et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y.X., Xiong G.S., Hu J., Jiang L., Yu H., Xu J., Fang Y.X., Zeng L.J., Xu E.B., Xu J., et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015;47:944. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- 12.Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 13.Liu J.F., Chen J., Zheng X.M., Wu F.Q., Lin Q.B., Heng Y.Q., Tian P., Cheng Z.J., Yu X.W., Zhou K.N., et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants. 2017;3:17043. doi: 10.1038/nplants.2017.43. [DOI] [PubMed] [Google Scholar]
- 14.Wang S.K., Li S., Liu Q., Wu K., Zhang J.Q., Wang S.S., Wang Y., Chen X.B., Zhang Y., Gao C.X., et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015;47:949. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- 15.Krishna P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003;22:289–297. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- 16.Vriet C., Russinova E., Reuzeau C. Boosting crop yields with plant steroids. Plant Cell. 2012;24:842–857. doi: 10.1105/tpc.111.094912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J.Y., Sae-Seaw J., Wang Z.Y. Brassinosteroid signalling. Development. 2013;140:1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Nam K.H., Vafeados D., Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cano-Delgado A., Yin Y.H., Yu C., Vafeados D., Mora-Garcia S., Cheng J.C., Nam K.H., Li J.M., Chory J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 20.Belkhadir Y., Chory J. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science. 2006;314:1410–1411. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- 21.Gou X.P., Yin H.J., He K., Du J.B., Yi J., Xu S.B., Lin H.H., Clouse S.D., Li J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8:e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Y.H., Wang Z.Y., Mora-Garcia S., Li J.M., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 23.Yin Y.H., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 24.Wang W.F., Bai M.Y., Wang Z.Y. The brassinosteroid signaling network—A paradigm of signal integration. Curr. Opin. Plant Biol. 2014;21:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X.F., Li L., Zola J., Aluru M., Ye H.X., Foudree A., Guo H.Q., Anderson S., Aluru S., Liu P., et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 26.Guo H.Q., Li L., Aluru M., Aluru S., Yin Y.H. Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 2013;16:545–553. doi: 10.1016/j.pbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Hong Z., Ueguchi-Tanaka M., Shimizu-Sato S., Inukai Y., Fujioka S., Shimada Y., Takatsuto S., Agetsuma M., Yoshida S., Watanabe Y., et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002;32:495–508. doi: 10.1046/j.1365-313X.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- 28.Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., Yoshida S., Ashikari M., Kitano H., Matsuoka M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell. 2003;15:2900–2910. doi: 10.1105/tpc.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong Z., Ueguchi-Tanaka M., Fujioka S., Takatsuto S., Yoshida S., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17:2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17:776–790. doi: 10.1105/tpc.104.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto T., Matsuoka M. Characterization of constitutive photomorphogenesis and dwarfism homologs in rice (Oryza sativa L.) J. Plant Growth Regul. 2006;25:245–251. doi: 10.1007/s00344-006-0041-6. [DOI] [Google Scholar]
- 32.Sakamoto T., Morinaka Y., Ohnishi T., Sunohara H., Fujioka S., Ueguchi-Tanaka M., Mizutani M., Sakata K., Takatsuto S., Yoshida S., et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006;24:105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- 33.Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka A., Nakagawa H., Tomita C., Shimatani Z., Ohtake M., Nomura T., Jiang C.J., Dubouzet J.G., Kikuchi S., Sekimoto H., et al. BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009;151:669–680. doi: 10.1104/pp.109.140806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D., Wang L., Wang M., Xu Y.Y., Luo W., Liu Y.J., Xu Z.H., Li J., Chong K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 2009;7:791–806. doi: 10.1111/j.1467-7652.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- 37.Tong H.N., Liu L.C., Jin Y., Du L., Yin Y.H., Qian Q., Zhu L.H., Chu C.C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell. 2012;24:2562–2577. doi: 10.1105/tpc.112.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura A., Fujioka S., Sunohara H., Kamiya N., Hong Z., Inukai Y., Miura K., Takatsuto S., Yoshida S., Ueguchi-Tanaka M., et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006;140:580–590. doi: 10.1104/pp.105.072330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S.N., Wang S.K., Xu Y.X., Yu C.L., Shen C.J., Qian Q., Geisler M., Jiang D.A., Qi Y.H. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015;38:638–654. doi: 10.1111/pce.12397. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto T., Yamada K., Iwasaki I., Yoshizawa Y., Oh K. New compounds induce brassinosteroid deficient-like phenotypes in rice. Plants. 2013;2:521–529. doi: 10.3390/plants2030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y.H., Bao L., Jeong S.Y., Kim S.K., Xu C.G., Li X.H., Zhang Q.F. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 2012;70:398–408. doi: 10.1111/j.1365-313X.2011.04877.x. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa H., Tanaka A., Tanabata T., Ohtake M., Fujioka S., Nakamura H., Ichikawa H., Mori M. SHORT GRAIN1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 2012;158:1208–1219. doi: 10.1104/pp.111.187567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan P.G., Rao Y.C., Zeng D.L., Yang Y.L., Xu R., Zhang B.L., Dong G.J., Qian Q., Li Y.H. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 2014;77:547–557. doi: 10.1111/tpj.12405. [DOI] [PubMed] [Google Scholar]
- 44.Che R.H., Tong H.N., Shi B.H., Liu Y.Q., Fang S.R., Liu D.P., Xiao Y.H., Hu B., Liu L.C., Wang H.R., et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants. 2016;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 45.Wu Y.Z., Fu Y.C., Zhao S.S., Gu P., Zhu Z.F., Sun C.Q., Tan L.B. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 2016;14:377–386. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choe S., Fujioka S., Noguchi T., Takatsuto S., Yoshida S., Feldmann K.A. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- 47.Wu C.Y., Trieu A., Radhakrishnan P., Kwok S.F., Harris S., Zhang K., Wang J.L., Wan J.M., Zhai H.Q., Takatsuto S., et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20:2130–2145. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huo F., Wang X., Han Y., Bai Y., Zhang W., Yuan H., Liu H. A new derivatization approach for the rapid and sensitive analysis of brassinosteroids by using ultra high performance liquid chromatography-electrospray ionization triple quadrupole mass spectrometry. Talanta. 2012;99:420–425. doi: 10.1016/j.talanta.2012.05.073. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q.F., Shen B.Z., Dai X.K., Mei M.H., Maroof M.A.S., Li Z.B. Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male-sterility in rice. Proc. Natl. Acad. Sci. USA. 1994;91:8675–8679. doi: 10.1073/pnas.91.18.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma X.L., Zhang Q.Y., Zhu Q.L., Liu W., Chen Y., Qiu R., Wang B., Yang Z.F., Li H.Y., Lin Y.R., et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Jefferson R.A., Kavanagh T.A., Bevan M.W. Beta-Glucuronidase (GUS) as a sensitive and versatile gene fusion marker in plants. J. Cell Biochem. 1987;6:57. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He F., Chen S., Ning Y., Wang G.L. Rice (Oryza sativa) protoplast isolation and its application for transient expression analysis. Curr. Protoc. Plant Biol. 2016;1:373–383. doi: 10.1002/cppb.20026. [DOI] [PubMed] [Google Scholar]
- 53.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evolut. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., Feng Z.M., Zhou C.L., Ren Y.K., Mou C.L., Wu T., Yang C.Y., Liu S.J., Jiang L., Wan J.M. Brassinosteroid (BR) biosynthetic gene lhdd10 controls late heading and plant height in rice (Oryza sativa L.) Plant Cell Rep. 2016;35:357–368. doi: 10.1007/s00299-015-1889-3. [DOI] [PubMed] [Google Scholar]
- 56.Sun L.J., Li X.J., Fu Y.C., Zhu Z.F., Tan L.B., Liu F.X., Sun X.Y., Sun X.W., Sun C.Q. GS6, a member of the gras gene family, negatively regulates grain size in rice. J. Integr. Plant Biol. 2013;55:938–949. doi: 10.1111/jipb.12062. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez-Garcia M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-Garcia S., Russinova E., Cano-Delgado A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- 58.Zhu X., Liang W., Cui X., Chen M., Yin C., Luo Z., Zhu J., Lucas W.J., Wang Z., Zhang D. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 2015;82:570–581. doi: 10.1111/tpj.12820. [DOI] [PubMed] [Google Scholar]
- 59.Li Y.B., Fan C.C., Xing Y.Z., Jiang Y.H., Luo L.J., Sun L., Shao D., Xu C.J., Li X.H., Xiao J.H., et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011;43:12661269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 60.Mathur J., Molnar G., Fujioka S., Takatsuto S., Sakurai A., Yokota T., Adam G., Voigt B., Nagy F., Maas C., et al. Transcription of the Arabidopsis cpd gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313X.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- 61.Yang S.F., Hoffman N.E. Ethylene biosynthesis and its regulation in higher-plants. Annu. Rev. Plant Phys. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 62.Oikawa T., Koshioka M., Kojima K., Yoshida H., Kawata M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 2004;55:687–700. doi: 10.1007/s11103-004-1692-y. [DOI] [PubMed] [Google Scholar]
- 63.Goda H., Shimada Y., Asami T., Fujioka S., Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bancos S., Nomura T., Sato T., Molnar G., Bishop G.J., Koncz C., Yokota T., Nagy F., Szekeres M. Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002;130:504–513. doi: 10.1104/pp.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Xu Y.Y., Ma Q.B., Li D., Xu Z.H., Chong K. Heterotrimeric g protein α subunit is involved in rice brassinosteroid response. Cell Res. 2006;16:916–922. doi: 10.1038/sj.cr.7310111. [DOI] [PubMed] [Google Scholar]
- 66.Vert G., Nemhauser J.L., Geldner N., Hong F.X., Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.