Abstract
Corynebacterium ulcerans is an emerging pathogen, which is increasingly recognized as an etiological agent of diphtheria, but can also evoke ulcers of the skin and systemic infections in humans. Besides man, the bacteria can colonize a wide variety of different animals, including cattle and pet animals, which might serve as a reservoir for human infections. In this study, surface-located proteins and the exoproteome of two Corynebacterium ulcerans strains were analyzed, since these may have key roles in the interaction of the pathogen with host cells. Strain 809 was isolated from a fatal case of human respiratory tract infection, while strain BR-AD22 was isolated from a nasal swap of an asymptomatic dog. While a very similar pattern of virulence factors was observed in the culture supernatant and surface protein fractions of the two strains, proteome analyses revealed a higher stability of 809 cells compared to strain BR-AD22. During exponential growth, 17% of encoded proteins of strain 809 were detectable in the medium, while 38% of the predicted proteins encoded by the BR-AD22 chromosome were found. Furthermore, the data indicate differential expression of phospholipase D and a cell wall-associated hydrolase, since these were only detected in strain BR-AD22.
Keywords: diphtheria, exoproteome, genomics, host-pathogen interaction, surface proteome, virulence factors
1. Introduction
Corynebacterium ulcerans is a pathogenic member of the genus Corynebacterium, which is part of the family Corynebacteriaceae, the order Actinomycetales, and the phylum Actinobacteria [1]. C. ulcerans was first described by Gilbert and Stewart, who isolated the bacteria from the throat of a patient with respiratory diphtheria-like illness [2]. In fact, when lysogenized by a tox gene-carrying corynephage, C. ulcerans can—similar to Corynebacterium diphtheria—produce diphtheria toxin and evoke the typical symptoms of respiratory diphtheria (for a recent review on corynophages, see Reference [3]).
C. ulcerans is an emerging pathogen [4,5,6,7,8], and during the past decade diphtheria-like infections with toxigenic C. ulcerans have outnumbered those caused by toxigenic C. diphtheriae in many industrialized countries [9]. Moreover, during the last years, other human infections associated with C. ulcerans appear to be increasing in various countries, and can most often be ascribed to zoonotic transmission (for an example, see References [10,11]; for a review, see Reference [7]). The range of mammals that may serve as a reservoir for human infections is extremely broad. C. ulcerans was isolated from cattle, goats, pigs, wild boars, dogs, cats, ground squirrels, otters, camels, monkeys, orcas, and water rats (for a review, see Reference [7]).
In 2011, two C. ulcerans strains from the metropolitan area of Rio de Janeiro, Brazil, were sequenced [12]—BR-AD22, isolated from a nasal swab of an asymptomatic dog, and 809, isolated from a bronchoalveolar lavage sample of an 80-year-old woman with fatal pulmonary infection [13,14]. Based on these genome sequences and comparative genomics approaches, a number of putative virulence factors were annotated. Unfortunately, functional analyses are scarce, and up to now, a limited number of data concerning adhesion and invasion of epithelial cells, interaction with macrophages, fibrinogen, fibronectin and collagen binding, antimicrobial profiles, and arthritogenic potential of isolates were published [15,16,17,18]. To get deeper insights into C. ulcerans physiology and pathology, proteome analyses were carried out. Similar studies focusing on different corynebacteria, e.g., C. diphtheriae, Corynebacterium jeikeium, and Corynebacterium pseudotuberculosis strains have been previously published [19,20,21,22,23,24,25,26]. The study presented here describes proteome analyses of C. ulcerans strains 809 and BR-AD22, focusing on surface-located and medium-secreted proteins of the two strains, since these proteins may represent key components of pathogen-host interaction, and may influence pathogenicity as well as immune response.
2. Materials and Methods
2.1. Strains and Growth Conditions
C. ulcerans 809 and BR-AD22 were grown in heart infusion (HI) medium (25 g HI broth; Becton, Dickinson, ND, USA). For solid media, 15 g/L Bacto Agar (Oxoid, Basinstoke, UK) was added. Incubation of liquid cultures of C. ulcerans was carried out at 30 °C under shaking in baffled flasks. Overnight cultures were used to inoculate fresh media to an OD600 of approximately 0.1, bacteria were incubated at 30 °C until the exponential growth phase was reached (OD600 approximately 2), and cultures were further processed as described below.
2.2. Isolation of Extracellular Proteins Secreted into the Medium
For preparation of extracellular proteins, the cells were separated by centrifugation (25 min, 5000× g, 4 °C) and Complete EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany) was added to the supernatant to avoid proteolysis. To prevent contamination of extracellular proteins by cells or cell debris, the supernatant was centrifuged again (1 h, 7000× g, 4 °C) and subsequently filtered using 0.2 µm pore-size filters (Minisart, Sartorius, Göttingen, Germany) [27]. Proteins were precipitated under constant stirring by dropwise addition of trichloroacetic acid (10% w/v) and incubation at 4 °C overnight. Precipitated proteins were harvested by centrifugation (1 h, 5500× g, 4 °C). Pellets were washed three times with acetone, incubated for 5 min on ice and centrifuged (10 min, 5500× g, 4 °C). This step was repeated twice with 80% acetone and once with 100% acetone. The proteins were air-dried under sterile conditions and resuspended in 250 µL dehydration buffer (8 M urea, 50 mM Tris, 20 mM DTT, 1% sodium desoxycholate).
2.3. In-Solution Tryptic Digest
A total of 1.5 µg of proteins (see Section 2.2) were transferred to 10 kDa Hydrosart® membrane filters (Sartorius Stedim biotech, Göttingen, Germany) for modified filter-aided sample preparation (FASP) [28]. Proteins were reduced with 250 µL of reduction buffer containing 8 M urea, 100 mM triethylammonium bicarbonate buffer (TEAB, Sigma-Aldrich, Munich, Germany), and 20 mM dithiothreitol (DTT) for 30 min at room temperature. Alkylation of sulfhydryl groups was carried out in the same buffer containing 40 mM chloroacetamide, instead of DTT for 30 min. Proteins were washed with 250 µL of 1 M urea in 50 mM TEAB and trypsinized with 1 µg of sequencing-grade trypsin (Promega, Mannheim, Germany) in 1 M urea in 50 mM TEAB for 18 h at 37 °C. Peptides were extracted by centrifugation and desalted on C18 stage tips. Prior to nanoLC-MS/MS analysis, tryptic peptides were dried under vacuum and resuspended in 0.1% trifluoroacetic acid (TFA).
2.4. NanoLC-MS/MS Analysis
Mass spectrometric analyses by nanoLC-MS/MS were carried out as described [27]. In short, resulting peptides (approx. 1 µg) were loaded on a nanoflow Ultimate 3000 HPLC (Dionex, Sunnyvale, CA, USA) for separation on EASY-Spray column (Thermo Fisher Scientific; C18 with 2 μm particle size, 50 cm × 75 μm), with a flow rate of 200 nL/min by increasing acetonitrile concentrations over 120 min. All samples were analyzed on an Orbitrap Fusion (Thermo Fisher Scientific, Waltham, MA, USA) with the previously described MS/MS settings [27]. The mass spectrometer was operating with 2000 V spray voltage, 300–2000 (m/z) scan range, a maximum injection time of 50 ms, and an AGC target of 400,000 for the first stage of mass analysis t (MS1). The most intense ions were selected for collision-induced dissociation with collision energy of 35%, a maximum injection time of 250 ms, and an AGC target of 100 for the second stage of mass analysis (MS2).
Raw data files were processed against C. ulcerans databases 809 (UniProt UP 000008886, rel.1/16, 2180 sequences) and BR-AD22 (UniProt UP000008887, rel. 2/16, 2335 sequences), using Proteome Discoverer 2.0 (Thermo Scientific, Waltham, MA, USA). Theoretical masses were generated by trypsin with a maximum of two missed cleavages for full-tryptic and semi-tryptic peptides, and their product ions were compared to the measured spectra with the following parameters: carbamidomethyl modification was set as fixed and oxidation of methionine residues and carbamidomethylated lysine residues were set as an optional modification. Mass tolerance was set to 10 ppm for survey scans and 0.5 Da for fragment mass measurements. Peptide charges of 2–7 were allowed. Only resulting peptides with false discovery rate (FDR) below 1% were regarded as identified.
2.5. Isolation of Cell Surface Proteins by Tryptic Shaving
The preparation of surface proteins was based on a previously published protocol [29]. Harvested cells were resuspended in PBS and washed three times in this buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH (HCl) 7.4). Cells were treated with 25 U sequencing-grade trypsin (Promega, Mannheim, Germany) in 500 µL PBS buffer, for 15 min at 37 °C. Subsequently, bacteria were separated by centrifugation (30 min, 4 °C, 13,000× g), and the supernatant was filtered using 0.2 µm pore-size filters (Minisart, Sartorius, Göttingen, Germany). Mass spectrometry was carried out as described.
2.6. Zymography
To test for protease activity, the protein fractions were analyzed using casein gels for zymography [30]. The samples were prepared in a non-reducing loading buffer (125 mM Tris, 20% glycerol, 6% SDS, 0.02% bromophenol blue) and incubated for 25 min at room temperature. A 10% polyacrylamide gel containing 0.1% casein was used for electrophoresis. After electrophoresis, SDS was removed by incubation for 30 min in 2.5% Triton X-100. The gel was equilibrated for 30 min and subsequently incubated for 12 h at 37 °C in digestion buffer (50 mM Tris, 200 mM NaCl, 5 mM CaCl2 × 2 H2O, 240 mM Brij35 (pH 7.4)). Staining was carried out by incubation for 5 h in 40% methanol, 7% acetic acid, 0.5% Coomassie brilliant Blue R. Gels were destained by 15 min incubation in 40% methanol, 7% acetic acid.
2.7. Inverse CAMP Test
For detection of phospholipase D activity, an inverse CAMP (Christie–Atkins–Munch-Petersen) test was carried out [31]. For this purpose, Staphylococcus aureus ATCC 29213 was streaked-out as a vertical line and C. ulcerans BR-AD22 and 809 as parallel horizontal lines on Columbia Blood Agar plates (Oxoid, Basingstoke, UK), and bacteria were incubated for two days at 37 °C.
3. Results
3.1. Analysis of Proteins in Extracellular and Surface Fraction
When supernatants of C. ulcerans strains 809 and BR-AD22 cultures grown to exponential phase were analyzed, strain-dependent differences in the number of proteins were observed. In strain 809, 221 proteins were identified in three biological replicates, and an additional 144 proteins in less than three replicates, leading to a total number of 365 identified proteins. In contrast, 517 proteins were identified in three biological replicates, and an additional 369 in less than three replicates, leading to a total number of 886 proteins for strain BR-AD22. 134 proteins were identified in culture supernatants of both strains (for supporting information, see Tables S1 and S2). The high number of proteins observed in BR-AD22 supernatants, 38% of the predicted proteins encoded by its chromosomal DNA [12], indicated a considerable lysis of bacterial cells occurring under standard growth conditions in shaking flasks. In contrast, strain 809 was less prone to cell damage. In this case, only 17% of the predicted proteins [12] were detected.
Surface-located proteins were released by trypsin treatment of cells. When corresponding fractions of C. ulcerans 809 and BR-AD22 were analyzed after trypsin-shaving, isolation of the supernatant, and subsequent mass spectrometry, again, fractions of strain 809 revealed less proteins compared to BR-AD22. For 809, a total 528 proteins was identified in the shaving fraction, with 305 protein present in triplicates and 223 in less than three samples. For strain BR-AD22, 1025 proteins were identified, with 577 proteins found in triplicates and 448 in less than three samples.
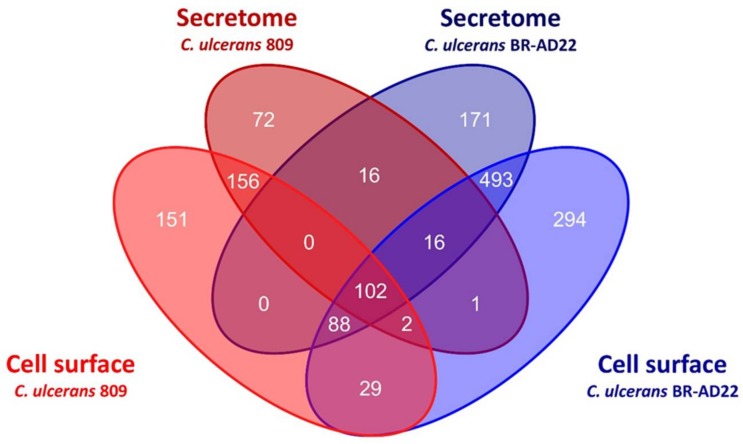
From the 102 proteins found in all samples, 23 were attributed to ribosome function, 13 were annotated as uncharacterized proteins, and 15 as putative secreted proteins. Only two were annotated in the databases as extracellular (Figure 1, Table 1). In summary, the annotation of the two C. ulcerans genomes used is rather preliminary today, and comprises a high number of hypothetical and uncharacterized proteins. Nevertheless, the data obtained here show that proteome analysis may be helpful for a basic characterization of protein expression and localization.
Figure 1.
Overview of proteomic data from C. ulcerans 809 and BR-AD22. Venn diagram mapping the overlapping proteins in culture supernatant and cell surface fraction.
Table 1.
List of proteins identified in all fractions of C. ulcerans 809 and BR-AD22. Accession numbers, annotated functions, and cellular localization, as indicated in the different data bases, are given.
| Accession No. | Description | Cellular Localisation |
|---|---|---|
| G0CTA8; embl-cds:AEG81534 | 50S ribosomal protein L35 | ribosome |
| G0CTV7; embl-cds:AEG81724 | ABC-type transport system involved in Fe-S cluster assembly ATP-binding protein | membrane |
| G0CU06; embl-cds:AEG81772 | Elongation factor EF-P | cytoplasm |
| G0CU14; embl-cds:AEG81780 | Putative secreted protein | |
| G0CU20; embl-cds:AEG80548 | Uncharacterized protein | |
| G0CU44; embl-cds:AEG81799 | Putative secreted protein | |
| G0CU89; embl-cds:AEG81844 | DtxR-family transcription regulator | cytoplasm |
| G0CU95; embl-cds:AEG81850 | Alkyl hydroperoxide reductase | cytoplasm |
| G0CUC6; embl-cds:AEG80578 | Uncharacterized protein | |
| G0CUG4; embl-cds:AEG80613 | Uncharacterized protein | |
| G0CUM1; embl-cds:AEG81896 | Polyribonucleotide nucleotidyltransferase | cytoplasm |
| G0CUR3; embl-cds:AEG81937 | Elongation factor Ts | cytoplasm |
| G0CUW8; embl-cds:AEG80687 | Putative secreted protein | |
| G0CV31; embl-cds:AEG81974 | Uncharacterized protein | |
| G0CV41; embl-cds:AEG81983 | Putative secreted protein | |
| G0CV58; embl-cds:AEG82000 | Putative secreted protein | |
| G0CV74; embl-cds:AEG82016 | Putative secreted protein | |
| G0CV86; embl-cds:AEG80721 | Pyridoxal 5′-phosphate synthase subunit PdxS | cytoplasm |
| G0CVA1; embl-cds:AEG80736 | Nucleoid-associated protein CULC22_00193 | cytoplasm |
| G0CVC7; embl-cds:AEG80762 | Aspartokinase | cytoplasm |
| G0CVK8; embl-cds:AEG82067 | Branched-chain-amino-acid aminotransferase | cytoplasm |
| G0CVR2; embl-cds:AEG80808 | Cold shock-like protein A | cytoplasm |
| G0CVS1; embl-cds:AEG80815 | Putative secreted protein | |
| G0CVT0; embl-cds:AEG80823 | Dihydrolipoyl dehydrogenase | cytoplasm |
| G0CVT9; embl-cds:AEG80832 | Putative secreted protein | |
| G0CVU4; embl-cds:AEG80837 | Uncharacterized protein | |
| G0CVU8; embl-cds:AEG80841 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | cytoplasm |
| G0CVX3; embl-cds:AEG80866 | Thiol-disulfide isomerase/thioredoxin | cytoplasm |
| G0CW00; embl-cds:AEG82124 | Glutamine--fructose-6-phosphate aminotransferase (isomerizing) | cytoplasm |
| G0CW81; embl-cds:AEG80892 | 50S ribosomal protein L11 | ribosome |
| G0CW82; embl-cds:AEG80893 | 50S ribosomal protein L1 | ribosome |
| G0CW87; embl-cds:AEG80898 | 50S ribosomal protein L7/L12 | ribosome |
| G0CW95; embl-cds:AEG80906 | DNA-directed RNA polymerase subunit beta | cytoplasm |
| G0CWB3; embl-cds:AEG80924 | Elongation factor G | cytoplasm |
| G0CWB4; embl-cds:AEG80925 | Elongation factor Tu | cytoplasm |
| G0CWC3; embl-cds:AEG80934 | 30S ribosomal protein S10 | ribosome |
| G0CWC4; embl-cds:AEG80935 | 50S ribosomal protein L3 | ribosome |
| G0CWC5; embl-cds:AEG80936 | 50S ribosomal protein L4 | ribosome |
| G0CWC6; embl-cds:AEG80937 | 50S ribosomal protein L23 | ribosome |
| G0CWC7; embl-cds:AEG80938 | 50S ribosomal protein L2 | ribosome |
| G0CWC9; embl-cds:AEG80940 | 50S ribosomal protein L22 | ribosome |
| G0CWD0; embl-cds:AEG80941 | 30S ribosomal protein S3 | ribosome |
| G0CWD1; embl-cds:AEG80942 | 50S ribosomal protein L16 | ribosome |
| G0CWD8; embl-cds:AEG80949 | 50S ribosomal protein L14 | ribosome |
| G0CWE7; embl-cds:AEG80958 | 50S ribosomal protein L18 | ribosome |
| G0CWL5; embl-cds:AEG82198 | 50S ribosomal protein L27 | ribosome |
| G0CWL6; embl-cds:AEG82199 | 50S ribosomal protein L21 | ribosome |
| G0CWM9; embl-cds:AEG82211 | ATP-dependent Clp protease proteolytic subunit | cytoplasm |
| G0CWN2; embl-cds:AEG80960 | 50S ribosomal protein L30 | ribosome |
| G0CWN3; embl-cds:AEG80961 | 50S ribosomal protein L15 | ribosome |
| G0CWN5; embl-cds:AEG80963 | Maltotriose-binding protein | membrane |
| G0CWQ1; embl-cds:AEG80980 | DNA-directed RNA polymerase subunit alpha | cytoplasm |
| G0CWS7; embl-cds:AEG81005 | 10 kDa chaperonin | cytoplasm |
| G0CWU3; embl-cds:AEG81019 | Putative secreted protein | |
| G0CX97; embl-cds:AEG81093 | LytR-family transcription regulator | cytoplasm |
| G0CXC9; embl-cds:AEG81124 | Putative secreted protein | |
| G0CXG7; embl-cds:AEG82307 | Succinyl-CoA:Coenzyme A transferase | redox chain |
| G0CXJ0; embl-cds:AEG82330 | Phosphoribosylformylglycinamidine synthase subunit PurS | cytoplasm |
| G0CXK8; embl-cds:AEG81127 | Uncharacterized protein | |
| G0CXM1; embl-cds:AEG81140 | Uncharacterized protein | |
| G0CXP5; embl-cds:AEG81163 | Carbonic anhydrase | cytoplasm |
| G0CXQ6; embl-cds:AEG81174 | Resuscitation-promoting factor | extracellular |
| G0CXQ9; embl-cds:AEG81178 | Putative secreted protein | |
| G0CXR6; embl-cds:AEG81185 | Citrate synthase | cytoplasm |
| G0CXR9; embl-cds:AEG81188 | Superoxide dismutase [Cu-Zn] | cell envelope |
| G0CY06; embl-cds:AEG82362 | Putative secreted protein | |
| G0CY28; embl-cds:AEG81208 | Uncharacterized protein | |
| G0CY42; embl-cds:AEG81222 | Uncharacterized protein | |
| G0CY56; embl-cds:AEG81236 | 50S ribosomal protein L28 | ribosome |
| G0CY58; embl-cds:AEG81238 | 50S ribosomal protein L32 | ribosome |
| G0CY59; embl-cds:AEG81239 | Two-component system transcriptional regulatory protein | cytoplasm |
| G0CYA4; embl-cds:AEG81281 | 50S ribosomal protein L25 | ribosome |
| G0CYH1; embl-cds:AEG82442 | Purine phosphoribosyltransferase | cytoplasm |
| G0CYH2; embl-cds:AEG82443 | Putative membrane protein | membrane |
| G0CYK0; embl-cds:AEG81292 | Enolase | extracellular; cytoplasm; cell surface |
| G0CYK9; embl-cds:AEG81301 | Transcription elongation factor GreA | cytoplasm |
| G0CYM4; embl-cds:AEG81317 | Fumarate hydratase class II | cytoplasm |
| G0CYQ3; embl-cds:AEG81345 | Putative secreted protein | |
| G0CYQ7; embl-cds:AEG81349 | Uncharacterized protein | |
| G0CYS8; embl-cds:AEG82468 | Fructose-bisphosphate aldolase | cytoplasm |
| G0CYV5; embl-cds:AEG82498 | Alcohol dehydrogenase | cytoplasm |
| G0CYV6; embl-cds:AEG82499 | Aldehyde dehydrogenase | cytoplasm |
| G0CYW2; embl-cds:AEG82505 | Chaperone protein DnaK | cytoplasm |
| G0CYY1; embl-cds:AEG82523 | Urease accessory protein UreG | cytoplasm |
| G0CYY4; embl-cds:AEG82526 | Urease subunit alpha | cytoplasm |
| G0CYY5; embl-cds:AEG82527 | Urease subunit beta | cytoplasm |
| G0CZ22; embl-cds:AEG81380 | 2-oxoglutarate dehydrogenase E1 component | membrane |
| G0CZ53; embl-cds:AEG81411 | Uncharacterized protein | |
| G0CZ62; embl-cds:AEG81420 | Peptide chain release factor 1 | cytoplasm |
| G0CZ71; embl-cds:AEG81429 | ATP synthase subunit alpha | membrane |
| G0CZ73; embl-cds:AEG81431 | ATP synthase subunit beta | membrane |
| G0CZ86; embl-cds:AEG81444 | Electron transfer flavoprotein beta subunit | redox chain |
| G0CZ94; embl-cds:AEG82552 | Phosphoenolpyruvate carboxykinase (GTP) | cytoplasm |
| G0CZA6; embl-cds:AEG82564 | Putative secreted protein | |
| G0CZA9; embl-cds:AEG82567 | Trehalose corynomycolyl transferase | cytoplasm |
| G0CZB9; embl-cds:AEG82576 | UDP-galactopyranose mutase | cytoplasm |
| G0CZC6; embl-cds:AEG82583 | Serine-tRNA ligase | cytoplasm |
| G0CZC8; embl-cds:AEG82585 | Putative secreted protein | |
| G0CZG8; embl-cds:AEG82621 | Uncharacterized protein | |
| G0CZP6; embl-cds:AEG81523 | 30S ribosomal protein S1 | ribosome |
| G0CZQ1; embl-cds:AEG81528 | Uncharacterized protein | cytoplasm |
| G0CZR5; embl-cds:AEG82638 | 30S ribosomal protein S6 | ribosome |
3.2. Identification of Putative Virulence Factors
A number of putative virulence factors was annotated in a comparative genome sequencing study of C. ulcerans 809 and BR-AD22 [12]. When the results of mass spectrometric fingerprint analysis were monitored for these proteins, almost all virulence factors annotated in the genome sequencing project were observed (Table 2). From the annotated virulence factors, only the pilus subunits comprised LPxTG surface anchoring motifs.
Table 2.
Identification of annotated virulence factors in the exoproteome and surface-located proteome of C. ulcerans. Localization of the observed proteins from strains 809 and BR-AD22 as well as presence of an LPxTG motif for cell wall anchoring is indicated.
| Name (NCBI) | Gene | Uniprot Identifier | Localization of Protein | LPxTG Motif | ||||
|---|---|---|---|---|---|---|---|---|
| Supernatant | Cell Surface | |||||||
| 809 | BR-AD22 | 809 | BR-AD22 | 809 | BR-AD22 | |||
| Putative ribosome binding protein | rbp | AEG80717 | - | - | - | - | - | none |
| Corynebacterial protease CP40 precursor | cpp | AEG82501 | AEG84830 | + | + | - | - | none |
| Phospholipase D | pld | AEG80581 | AEG82759 | - | + | - | - | none |
| Surface-anchored protein, fimbrial subunit | spaF | AEG82476 | AEG84808 | + | + | + | + | LPKTG |
| Surface-anchored protein, fimbrial subunit | spaE | AEG82477 | AEG84809 | - | - | - | - | LPLTG |
| Surface-anchored protein, fimbrial subunit | spaD | AEG82479 | AEG84811 | + | + | - | - | LPMTG |
| Surface-anchored protein, fimbrial subunit | spaC | AEG82506 | AEG84835 | + | + | + | + | LPLTG |
| Surface-anchored protein, fimbrial subunit | spaB | AEG82507 | AEG84836 | + | + | - | - | LARTG |
| Resuscitation-promoting factor interacting protein | rpfI | AEG81666 | AEG83858 | + | + | + | + | none |
| Cell wall-associated hydrolase | cwlH | AEG82053 | AEG84247 | - | + | - | + | none |
| Sialidase precursor | nanH | AEG80974 | AEG83155 | + | + | + | + | none |
| Venom serine protease KN13 | vsp1 | AEG81049 | AEG83233 | + | + | + | + | none |
| Venom serine protease 2A | vsp2 | AEG82491 | - | - | - | - | - | none |
| Trypsin-like serine protease | tspA | AEG82376 | AEG84712 | + | + | - | - | none |
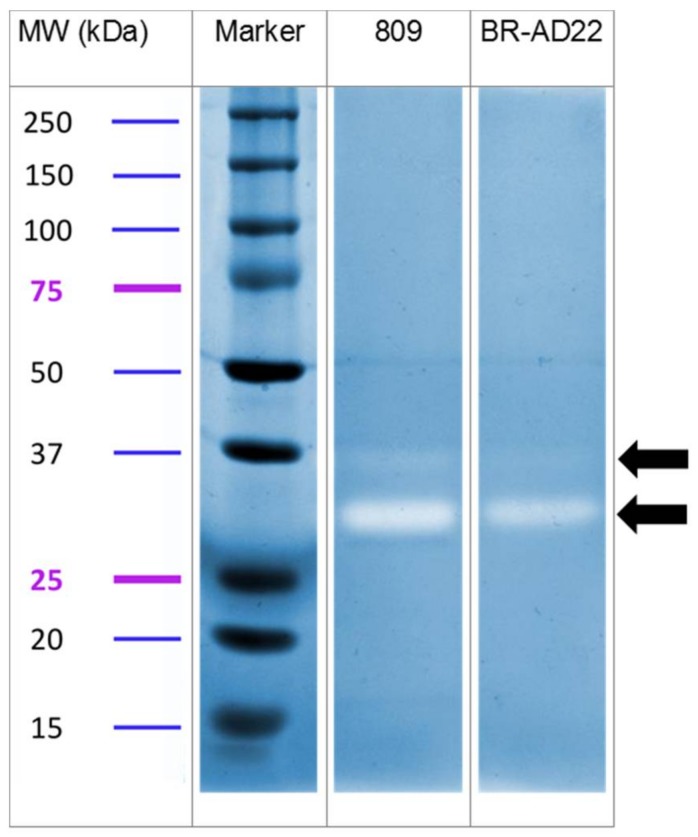
From the 14 annotated virulence proteins, only three were absent in the surface protein fraction and supernatant, namely, a putative ribosome binding protein with homology to Shiga-like toxin, the surface-anchored pilus protein SpaE, and the venom serine protease 2A. In contrast, three other secreted proteases were identified in this study. In fact, protease activity was clearly detectable in samples of supernatants using casein-containing gels. In each fraction, a minor band with an apparent molecular weight of 36 kDa, and a major band with weight 30 kDa, were observed (Figure 2), which fits with the calculated mass of the secreted proteases found. This result indicates a role of proteolysis in pathogenicity of C. ulcerans, and, in fact, tissue destruction is often observed in C. ulcerans infections.
Figure 2.
Casein zymography of C. ulcerans culture supernatants. Samples were separated using non-reducing, casein-containing gels. After renaturation and incubation in reaction buffer, the gels were stained with Coomassie brilliant blue. Protease activity results in clear bands, which are indicated by arrows. Apparent molecular weights of marker proteins (Dual Colour, Biorad, Munich, Germany) are indicated.
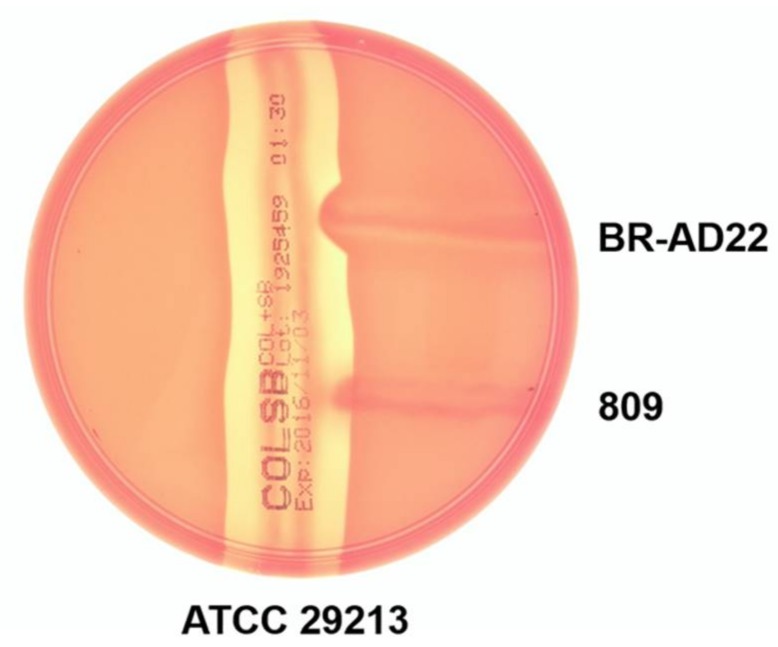
The results obtained also provide insight into the possible regulation of virulence factors. Although encoded in the genome of the two strains, phospholipase D (PLD) was only found in supernatants of BR-AD22. To verify this result, an inverse CAMP test was carried out. In this assay, hemolysis of sheep erythrocytes by S. aureus strain ATCC 29213 is inhibited when phospholipase activity alters the surface of the erythrocytes. In fact, only strain BR-AD22 influences the CAMP reaction of S. aureus, supporting expression of PLD in BR-AD22, and its absence in strain 809 (Figure 3).
Figure 3.
Reverse CAMP test. Hemolysis by S. aureus strain ATCC 29213, which is indicated by a halo around the vertical streak-out, is inhibited by strain BR-AD22, while strain 809 had no negative effect on hemolysis.
4. Discussion
Proteome analyses of C. ulcerans strains grown in vitro indicated that the human isolate 809 is more stable and stress-resistant than strain BR-AD22, which was isolated from an asymptomatic dog. The fact that a considerably high number of intracellular proteins is found at all is rather astonishing, since corynebacteria are typically very robust, due to their complex cell wall structure [32,33,34]. Furthermore, significantly less proteins were found in the supernatant of other pathogenic corynebacteria in previous studies [19,20,21,22,23,24,25,26].
Besides a number of proteins with annotated function, many functionally non-annotated and hypothetical proteins were detected, supporting the idea that proteome analyses may be helpful for a basic characterization of protein expression and localization, even in the case of a very basic genome annotation.
The study presented here was initiated in order to achieve an overview of surface-anchored and secreted proteins, which may have key functions in host interaction. While general expression patterns of putative virulence factors were rather similar for the two investigated strains, one of the main virulence factors of C. ulcerans, phospholipase D, was not found in strain 809. In this strain, the ribosome binding protein, exclusively found in its genome sequence, and a cell wall-associated hydrolase were not expressed. The lack of these proteins was unexpected, since strain 809 was isolated from a fatal case of human infection and was expected to be quite pathogenic. These observations highlight that the virulence of C. ulcerans, and especially the regulation of its pathogenicity determinants, is hardly understood, and more basic research is necessary to study this emerging human pathogen in order to predict the outcome of future infections. Further proteome studies might be helpful in this respect, especially in combination with other -omics analyses of host-pathogen interactions [35].
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft in frame of SFB796 (project Z1). The inverse CAMP assay was kindly carried out by Isabel Aly (Erlangen, Germany).
Supplementary Materials
The following are available online at http://www.mdpi.com/2227-7382/6/2/18/s1, Table S1: List of proteins identified as secretome proteins of C. ulcerans 809 and BR-AD22, Table S2: List of proteins identified as cell surface proteins of C. ulcerans 809 and BR-AD22.
Author Contributions
M.B. and S.G. prepared the extracellular and cell surface protein fractions, which were analyzed by mass spectrometry by B.A. and J.H. Zymography was carried out by M.B., who also prepared the figures of the manuscript. A.B. designed the experiments. The manuscript was written by M.B. and A.B.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tauch A., Sandbote J. The family Corynebacteriaceae. In: Rosenberg E., DeLong E., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes. Springer; Berlin/Heidelberg, Germany: 2014. pp. 239–277. [Google Scholar]
- 2.Gilbert R., Stewart F.C. Corynebacterium ulcerans: A pathogenic microorganism resembling Corynebacterium diphtheriae. J. Lab. Clin. Med. 1927;12:756–761. [Google Scholar]
- 3.Sangal V., Hoskisson P.A. Corynephages: Infections of the infectors. In: Burkovski A., editor. Diphtheria and Its Etiological Agents. Springer; Dordrecht, The Netherlands: 2014. pp. 67–82. [Google Scholar]
- 4.Wagner K.S., White J.M., Crowcroft N.S., De Martin S., Mann G., Efstratiou A. Diphtheria in the United Kingdom, 1986-2008: The increasing role of Corynebacterium ulcerans. Epidemiol. Infect. 2010;138:1519–1530. doi: 10.1017/S0950268810001895. [DOI] [PubMed] [Google Scholar]
- 5.Dias A.A., Santos L.S., Sabbadini P.S., Santos C.S., Silva F.C., Jr., Napoleão F., Nagao P.E., Villas-Bôas M.H., Hirata R., Jr., Guaraldi A.L. Corynebacterium ulcerans diphtheria: An emerging zoonosis in Brazil and worldwide. Rev. Saude Publica. 2011;45:1176–1191. doi: 10.1590/S0034-89102011000600021. [DOI] [PubMed] [Google Scholar]
- 6.Burkovski A. Pathogenesis of Corynebacterium diphtheriae and Corynebacterium ulcerans. In: Singh S.K., editor. Human Emerging and Re-Emerging Infections. Volume 2. John Wiley & Sons/Wiley Blackwell Press; Hoboken, NJ, USA: 2016. pp. 697–708. [Google Scholar]
- 7.Hacker E., Azevedo Antunes C., Mattos-Guaraldi A.L., Burkovski A., Tauch A. Corynebacterium ulcerans—An emerging human pathogen. Future Microbiol. 2016;11:1191–1208. doi: 10.2217/fmb-2016-0085. [DOI] [PubMed] [Google Scholar]
- 8.Berger A., Teutsch B., Heinzinger S., Sing A. Corynebacterium ulcerans—Ein Emerging Pathogen? Daten des Konsiliarlabors für Diphtherie 2011–2016. Epid. Bull. 2018;8:83–86. [Google Scholar]
- 9.Zakikhany K., Efstratiou A. Diphtheria in Europe: Current problems and new challenges. Future Microbiol. 2012;7:595–607. doi: 10.2217/fmb.12.24. [DOI] [PubMed] [Google Scholar]
- 10.Meinel D.M., Margos G., Konrad R., Krebs S., Blum H., Sing A. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin-encoding pathogenicity island. Genome Med. 2014;6:113. doi: 10.1186/s13073-014-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meinel D.M., Konrad R., Berger A., König C., Schmidt-Wieland T., Hogardt M., Bischoff H., Ackermann N., Hörmansdorfer S., Krebs S., et al. Zoonotic transmission of toxigenic Corynebacterium ulcerans strain, Germany, 2012. Emerg. Infect. Dis. 2015;21:356–358. doi: 10.3201/eid2102.141160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trost E., Al-Dilaimi A., Papavasiliou P., Schneider J., Viehoever P., Burkovski A., de Castro Soares S., Silva Almeida S., Alves Dorella F., Miyoshi A., et al. Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genom. 2011;12:383. doi: 10.1186/1471-2164-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias A.A., Silva F.C., Pereira G.A., Souza M.C., Camello T.C., Damasceno J.A., Pacheco L.G., Miyoshi A., Azevedo V.A., Hirata R., Jr., et al. Corynebacterium ulcerans isolated from an asymptomatic dog kept in an animal shelter in the metropolitan area of Rio de Janeiro, Brazil. Vector Borne Zoonotic Dis. 2010;10:743–748. doi: 10.1089/vbz.2009.0132. [DOI] [PubMed] [Google Scholar]
- 14.Mattos-Guaraldi A.L., Sampaio J.L., Santos C.S., Pimenta F.P., Pereira G.A., Pacheco L.G., Miyoshi A., Azevedo V., Moreira L.O., Gutierrez F.L., et al. First detection of Corynebacterium ulcerans producing a diphtheria-like toxin in a case of human with pulmonary infection in the Rio de Janeiro metropolitan area, Brazil. Mem. Inst. Oswaldo Cruz. 2008;103:396–400. doi: 10.1590/S0074-02762008000400014. [DOI] [PubMed] [Google Scholar]
- 15.Dias A.A., Silva F.C., Jr., Santos L.S., Ribeiro-Carvalho M.M., Sabbadini P.S., Santos C.S., Filardy A.A., Miyoshi A., Azevedo V.A., Hirata R., Jr., et al. Strain-dependent arthritogenic potential of the zoonotic pathogen Corynebacterium ulcerans. Vet. Microbiol. 2011;153:323–331. doi: 10.1016/j.vetmic.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Hacker E., Ott L., Hasselt K., Mattos-Guaraldi A.L., Tauch A., Burkovski A. Colonization of human epithelial cell lines by Corynebacterium ulcerans from human and animal sources. Microbiology. 2015;161:1582–1591. doi: 10.1099/mic.0.000121. [DOI] [PubMed] [Google Scholar]
- 17.Hacker E., Ott L., Schulze-Luehrmann J., Lührmann A., Wiesmann V., Wittenberg T., Burkovski A. The killing of macrophages by Corynebacterium ulcerans. Virulence. 2016;7:45–55. doi: 10.1080/21505594.2015.1125068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson-Louredo L., Ramos J.N., Peixoto R.S., Santos L.S., Antunes C.A., Ladeira E.M., Santos C.S., Vieira V.V., Boas M.H., Hirata R., Jr., et al. Corynebacterium ulcerans isolates from humans and dogs: Fibrinogen, fibronectin and collagen-binding, antimicrobial and PFGE profiles. Anton. Leeuwenhoek. 2014;105:343–352. doi: 10.1007/s10482-013-0080-5. [DOI] [PubMed] [Google Scholar]
- 19.Hansmeier N., Chao T.C., Kalinowski J., Pühler A., Tauch A. Mapping and comprehensive analysis of the extracellular and cell surface proteome of the human pathogen Corynebacterium diphtheriae. Proteomics. 2006;6:2465–2476. doi: 10.1002/pmic.200500360. [DOI] [PubMed] [Google Scholar]
- 20.Hansmeier N., Chao T.C., Daschkey S., Müsken M., Kalinowski J., Pühler A., Tauch A. A comprehensive proteome map of the lipid-requiring nosocomial pathogen Corynebacterium jeikeium K411. Proteomics. 2007;7:1076–1096. doi: 10.1002/pmic.200600833. [DOI] [PubMed] [Google Scholar]
- 21.Pacheco L.G., Slade S.E., Seyffert N., Santos A.R., Castro T.L., Silva W.M., Santos A.V., Santos S.G., Farias L.M., Carvalho M.A., et al. A combined approach for comparative exoproteome analysis of Corynebacterium pseudotuberculosis. BMC Microbiol. 2011;11:12. doi: 10.1186/1471-2180-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poetsch A., Haußmann U., Burkovski A. Proteomics of corynebacteria: From biotechnology workhorses to pathogens. Proteomics. 2011;11:3244–3255. doi: 10.1002/pmic.201000786. [DOI] [PubMed] [Google Scholar]
- 23.Rees M.A., Kleifeld O., Crellin P.K., Ho B., Stinear T.P., Smith A.I., Coppel R.L. Proteomic characterization of a natural host-pathogen interaction: Repertoire of in vivo expressed bacterial and host surface-associated proteins. J. Proteome Res. 2015;14:120–132. doi: 10.1021/pr5010086. [DOI] [PubMed] [Google Scholar]
- 24.Silva W.M., Seyffert N., Ciprandi A., Santos A.V., Castro T.L., Pacheco L.G., Barh D., Le Loir Y., Pimenta A.M., Miyoshi A., et al. Differential exoproteome analysis of two Corynebacterium pseudotuberculosis biovar ovis strains isolated from goat (1002) and sheep (C231) Curr. Microbiol. 2013;67:460–465. doi: 10.1007/s00284-013-0388-4. [DOI] [PubMed] [Google Scholar]
- 25.Silva W.M., Seyffert N., Santos A.V., Castro T.L., Pacheco L.G., Santos A.R., Ciprandi A., Dorella F.A., Andrade H.M., Barh D., et al. Identification of 11 new exoproteins in Corynebacterium pseudotuberculosis by comparative analysis of the exoproteome. Microb. Pathog. 2013;61–62:37–42. doi: 10.1016/j.micpath.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Silva W.M., Carvalho R.D., Soares S.C., Bastos I.F., Folador E.L., Souza G.H., Le Loir Y., Miyoshi A., Silva A., Azevedo V. Label-free proteomic analysis to confirm the predicted proteome of Corynebacterium pseudotuberculosis under nitrosative stress mediated by nitric oxide. BMC Genom. 2014;15:1065. doi: 10.1186/1471-2164-15-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiery E., Poetsch A., Moosbauer T., Amin B., Hofmann J., Burkovski A. A proteomic study of Clavibacter michiganensis subsp. michiganensis culture supernatants. Proteomes. 2015;3:411–423. doi: 10.3390/proteomes3040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 29.Tjalsma H., Lambooy L., Hermans P.W., Swinkels D.W. Shedding & shaving: Disclosure of proteomic expressions on a bacterial face. Proteomics. 2008;8:1415–1428. doi: 10.1002/pmic.200700550. [DOI] [PubMed] [Google Scholar]
- 30.Löwer M., Weydig C., Metzler D., Reuter A., Starzinski-Powitz A., Wessler S., Schneider G. Prediction of extracellular proteases of the human pathogen Helicobacter pylori reveals proteolytic activity of the Hp1018/19 protein HtrA. PLoS ONE. 2008;3:e3510. doi: 10.1371/journal.pone.0003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suvajdzic L., Potkonjak A., Milanov D., Lako B., Kocic B., Milic N., Cabarkapa I. A proposal of a diagnostic protocol for isolation of Corynebacterium ulcerans from cow’s milk. Acta Sci. Vet. 2012;40:1039. [Google Scholar]
- 32.Burkovski A. Cell envelope of corynebacteria: Structure and influence on pathogenicity. ISRN Microbiol. 2013:935736. doi: 10.1155/2013/935736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tauch A., Burkovski A. Molecular armory or niche factors: Virulence determinants of Corynebacterium species. FEMS Microbiol. Lett. 2015;362:1–6. doi: 10.1093/femsle/fnv185. [DOI] [PubMed] [Google Scholar]
- 34.Burkovski A. The role of corynomycolic acids in Corynebacterium-host interaction. Anton. Leeuwenhoek. 2018 doi: 10.1007/s10482-018-1036-6. [DOI] [PubMed] [Google Scholar]
- 35.Fels U., Gevaert K., Van Damme P. Proteogenomics in aid of host-pathogen interaction: A bacterial perspective. Proteomes. 2017;5:26. doi: 10.3390/proteomes5040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.