Abstract
Oral health represents an essential component in the quality of life of people, being a determinant factor in general health since it may affect the risk of suffering other conditions, such as chronic diseases. Oral diseases have become one of the main public health problems, where dental caries is the condition that most affects oral health worldwide, occurring in about 90% of the global population. This condition has been considered a challenge because of its high prevalence, besides being a chronic but preventable disease which can be caused depending on the consumption of certain nutritional elements interacting simultaneously with different factors, such as socioeconomic factors. Based on this problem, an analysis of a set of 189 dietary and demographic determinants is performed in this work, in order to find the relationship between these factors and the oral situation of a set of subjects. The oral situation refers to the presence and absence/restorations of caries. The methodology is performed constructing a dense artificial neural network (ANN), as a computer-aided diagnosis tool, looking for a generalized model that allows for classifying subjects. As validation, the classification model was evaluated through a statistical analysis based on a cross validation, calculating the accuracy, loss function, receiving operating characteristic (ROC) curve and area under the curve (AUC) parameters. The results obtained were statistically significant, obtaining an accuracy ≃ 0.69 and AUC values of 0.69 and 0.75. Based on these results, it is possible to conclude that the classification model developed through the deep ANN is able to classify subjects with absence of caries from subjects with presence or restorations with high accuracy, according to their demographic and dietary factors.
Keywords: NHANES, oral health, dental caries, classification multivariate models, computer-aided diagnosis, artificial neural networks, deep learning, statistical analysis
1. Introduction
Chronic diseases are the main problems in public health worldwide. The pattern of disease has been transformed and oral diseases are considered one of the main public health problems due to its high incidence and prevalence in all regions of the world, and, as in all diseases, the greatest burden is on populations disadvantaged and socially marginalized; treatment of the conditions is extremely expensive and is not feasible in most low and middle income countries. This characteristic represents an important problem, since, according to the World Health Organization (WHO), oral diseases are the fourth most expensive cause to treat in the most industrialized countries [1].
Oral health is an essential component for quality of life of people due to its influence as a determinant factor in general health of individuals and communities becoming a relevant point in health care. Therefore, the serious repercussions in terms of pain and suffering, impairment of function and effect on quality of life should also be considered. An important aspect to consider is that oral diseases increase the risk of chronic diseases, such as: cardiovascular and cerebrovascular, diabetes mellitus and respiratory. On the other hand, epidemiological surveillance of oral diseases becomes important insofar as it provides useful elements for the planning, programming, organization, integration, control and direction of the oral health program, at the same time that it guides the attention to the population [2].
Caries is the most frequent condition and according to the WHO; it affects between 60% and 90% of children of school age between 5 to 17 years old. The determinants and conditions of oral health status are multicausal, multisectoral and interdisciplinary categories that encompass a series of situations related to the historical and political process that each country experiences. In addition, the report of the Organización Panamericana de la Salud (OPS) describes how the development of this condition depends on the frequency of carbohydrate consumption, the characteristics of the food, the time exposure, plaque removal and susceptibility of the guest, adding the few preventive measures in oral health and the difficulty of making use of specialized dental medical services. These factors interact simultaneously, and their variables correspond to different orders from biological processes, to complex historical-cultural structures and social relationships, socioeconomic level, educational level, among others, making health phenomena complex [3,4].
There are too many risk factors and important determinants related to the presence of caries, and it is clear that the greater the degree of risk exposure, the greater the probability of contracting or developing it. Due to the difficulty of representing the control of the incidence of caries due to its high prevalence and the large number of factors that can influence this state, at present, some studies have implemented algorithms and performed analyses based on computer-aided diagnosis (CAD) where prediction models are used when it is necessary to know in the future the behavior of highly related complex data; these have utilities in clinical and research dentistry [5].
An example of these algorithms are the artificial neural networks (ANNs), a method used for the prediction of diseases, such is the case of oral diseases, as well as being mathematic models based on a principle of learning that is based on the concepts of artificial intelligence and the biological response of the human brain. Likewise, they are involved in systems’ construction processes that allow classifying, modeling and predicting information. ANNs are semiparametric nonlinear models, which allow the integration of variables and easily handle large amounts of data compared to linear analyses. They have processing elements or neurons, which are the units of the system that can be adjusted or trained through a process of learning and generalization. The information of each of these neurons is grouped and processed [6].
According to the literature, dental caries incidence has been a problem that has been studied by a series of researchers, trying to identify caries risk determinants. Lam et al. [7] found that dental visits, brushing frequency, lower parental perceived importance of baby teeth, and weaning onto solids are determinants associated with plaque accumulation, validating these results specifically in children. In the work of Fernandes et al. [8], a cross-sectional study of 274 children and their mothers based on demographic/socio-economic status is presented; through a Poisson regression approach, an analysis was performed, obtaining that dental caries can be mainly found in mothers of children aged 1, demonstrating the relationship between demographic/socio-economic status and caries.
As mentioned in the text, there are different types of risk factors that are associated with genetic and nutritional data. Lips et al. [9] presented a work where the association between genetic polymorphisms and risk of dental caries is demonstrated for most of the salivary proteins. In addition, diabetic and hypertensive patients have dietary risk factors that can be caused by oral health problems. This affirmation was demonstrated in the work of Asif Ahmed et al. [10] through a statistical analysis using clinical data, in order to identify that patients with oro-dental problems were hemodynamically stressed.
In the present study, it is intended to analyze the determinants that affect this oral health situation based on dietary and demographic factors. Based on this general description of the oral health problem, specifically caries, the main contribution of this work is to determine that, given the socioeconomic and dietary features, using an ANN can determine if the patient presents an optimal state of health or with the presence of repairs or caries, obtaining a classification system that allows for knowing the differences in the features that affect the population, as well as looking for the future prediction of oral health.
This method is an important tool for human resources and dental services because it reflects information collected in the population groups studied, which could unveil that oral health problems are not simple conditions, but they are obtained from different processes that have their trigger, which can enhance or mitigate them, impacting positively or negatively on the state of oral and general health.
Related Work
The condition of dental caries has been described in the scientific literature under different terms, as a multifactorial disease that is characterized by localized and progressive demineralization of the inorganic portions of the tooth and the subsequent deterioration of its organic part, with a high degree of morbidity and high prevalence [11,12,13].
A related work that suggests CADx to improve oral health prediction is developed by Zhang et al. [14], where an autoregressive integrated moving average model is performed and a grey predictive model for the estimation of the national prevalence of early childhood caries from 2014 to 2018, finding that the highest annual prevalence would be of 55.8%, being attributed to the socioeconomic developments and the public health service. In the work of Asif et al. [15], a caries preventive tool based on the influence of genetic pattern using the frequency of occurrence of fingerprint patterns among children, using as a validation measure the p-value, finding that dermatoglyphics could be an appropriate method to explore the possibility of a noninvasive and early predictor for dental caries. Chapple et al. [16] performed a systematic appraisal to identify potential risk factors for caries and periodontal diseases using genetic, role of diet and nutrition risk factors, obtained as results that the genetic contribution to these conditions present an attributable risk up to 50%, while controlled diabetes and obesity are common acquired factors. Hayes et al. [17] had as an objective to determine the risk indicators associated with root caries conducting a prospective longitudinal study using a regression analysis with data related to hygiene habits, diet, smoking habits and education level and as the outcome the root caries experience, suggesting a correlation between root caries and the variables poor plaque control, xerostomia, coronal decay and exposed root surfaces, obtaining a prevention tool for the root caries condition. In the work of Sudhir et al. [18], evaluating Caries Management by Risk Assessment (CAMBRA) as a tool for caries risk prediction was proposed, using as validation parameter OR value and ROC curve, obtaining that CAMBRA was found to be 47.62% with a specificity of 80% and AUC was found to be 0.638, which means that it is valid and highly predictive in determining the caries risk. Finally, Twetman [19] proposed to summarise the findings of recent systematic reviews covering caries risk assessment, finding that caries risk assessment should be carried out at the subject’s first dental visit and reassessments should be done during childhood; multivariate models display a better accuracy than the use of single predictors; there is no clearly superior method to predict future caries and no evidence to support the use of one model, program, or technology before the other; and the risk category should be linked to appropriate preventive care with recall intervals based on the individual need.
This work is organized as follows: Section 2 briefly describes the materials and methods used for the development of the data analysis, including data preprocessing, data classification and data evaluation. Section 3 exposes the results obtained. Finally, results are discussed in Section 4 and concluded in Section 5.
2. Materials and Methods
In this section, the process that was carried on for the classification between subjects with presence of caries and subjects with absence was presented, based on the information obtained from a series of socioeconomic and nutritional features. The databases that were used in this work were obtained from the National Health and Nutrition Examination Survey (NHANES, 2013–2014). In addition, the description of the subjects and the methods used for their classification are presented.
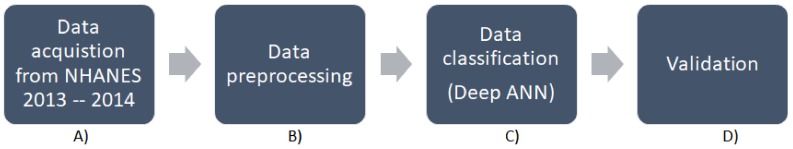
In Figure 1, the flowchart of the steps followed for the experimentation of this work is presented. Section (A) presents the data acquisition of the public databases “Demographic” and “Dietary” from NHANES 2013–2014. Section (B) represents the data preprocessing, where a feature reduction is performed due to the high amount of missing data and singular values that are presented, in addition to separating the data in two sets, one for training and one for testing. Section (C) refers to the data classification of subjects according to their oral health status. Finally, in Section (D), the evaluation of the ANN performance is presented, in order to know the accuracy with which the model classifies the subjects.
Figure 1.
Flowchart of the methodology followed.
2.1. Features Description
NHANES is a program of studies designed to evaluate the health and nutritional status of children and adults in United States, and it was founded by Centers of Disease Control and Prevention (CDC) and National Center for Health Statistics (NCHS). The surveys conducted by this program are unique, since it combines interviews and physical exams [20].
NHANES collects information from different types of data, and, in turn, this information is included in six main contexts; demographic, dietetic, examination, laboratory, questionnaire and limited access. These contexts are contained by the information described as follows:
Demographic: it provides individual, family and household level information in different topics (income of households and families, size of households and families, pregnancy status, among others).
Dietetic: it provides detailed information on dietary intake, in order to estimate the types and amount of food and beverages consumed, in addition to estimating the intake of energy, nutrients, and other food components.
Examination: it provides information of the health status, indicators of disease risk, and access to preventive and treatment services, from different aspects, including oral health.
Laboratory: it provides information of the results obtained from laboratory analysis of different components (components of urine, proteins, triglycerides, plasma, among others).
Questionnaire: it provides information of the data obtained from the interviews conducted through a system of computer-aided personal interview, of different topics (alcohol use, cardiovascular health, dermatology, among others).
Limited access: it provides similar information to that found in the questionnaire; however, it isn’t publicly available.
There were 189 features analyzed for this work, of which 188 belonged to demographic data, described in Appendix A, and dietary data, described in Appendix B. These features were used as input variables for the classification of subjects, while the remaining feature belonged to examination data, describing the oral health status of the subjects based on the presence/restoration or absence of caries, for which it was used as an output feature.
2.2. Subjects Description
The subjects of these databases were submitted to a series of different questionnaires, related to the different features. These subjects belong to different counties in the USA and they were randomly selected with a computer algorithm by NHANES.
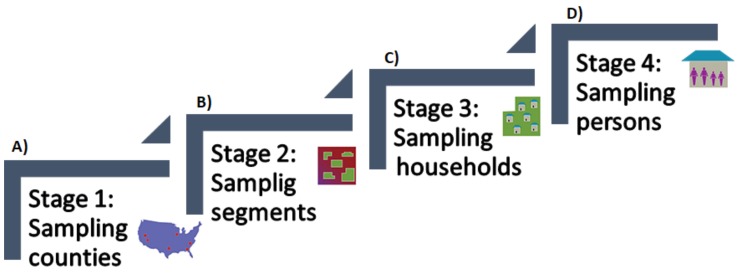
Figure 2 presents a flowchart of the randomly selection process followed by NHANES. (A) presents the first stage that corresponds to the sampling of counties. In this step, all the counties are divided into 15 groups according to their characteristics. Then, from each group, one county is selected, forming the 15 counties in the NHANES surveys for each year. (B) presents the second stage, corresponding to the sampling of segments, where from each county smaller groups are formed (with a large number of households in each group), selecting between 20 and 24 of these groups. (C) presented the third stage, which is referred to the sampling of households, where from all of the houses that correspond to the groups that were selected in the past stage, a sample of about 30 households are selected within each group. Finally, (D) presents the fourth stage, corresponding to the sampling of persons, where the NHANES interviewers go to the selected households and ask for the information of the surveys (age, race, and gender). The members of the households that responded to the surveys were randomly selected through a computer algorithm; they can be selected some, all, or none of the household members.
Figure 2.
Flowchart of the stages followed by NHANES 2013–2014 for the sampling of participants.
The main target population for NHANES is the non-institutionalized civilian resident population of the USA. The design of the population selection is based on the sampling of a larger number of specific subgroups that present particular public health characteristics, in order to increase the reliability and precision of health status indicators. NHANES started these design changes in 2011, including in its population the oversampled subgroups survey cycle:
Hispanic persons;
Non-Hispanic black persons;
Non-Hispanic Asian persons;
Non-Hispanic white and other* persons at or below 130% of the poverty level; and
Non-Hispanic white and other* persons aged 80 years and older.
This random selection of subjects avoid presenting any bias problem, favoring the important variety of demography characteristics of the subjects reducing the probability that the methodology followed could be influenced by the data used.
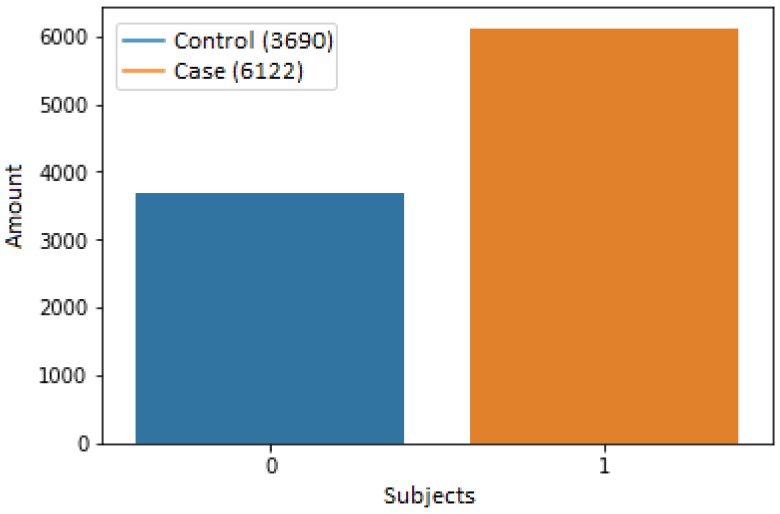
Figure 3 presents the total of subjects contained in the database used. From 9812 subjects, 3690 were control subjects and 6122 were case subjects. The subjects were in an age range of 0 to 80 years old; 4830 belonged to the masculine gender and 4982 to the feminine gender.
Figure 3.
Graph of the total number of subjects used, classified according to their oral health status. The subjects contained in the bar with ’0’ presented absence of caries, while the subjects contained in the bar with ’1’ presented presence/restoration of caries.
It is important to mention that this specific database was chosen in order to show the consistency of the data and the results obtained in the relationship to its background work. In this work [21] three different models obtained through a fast backward selection (FBS) of features are presented, each model corresponding to a different age group, then a posterior evaluation using a net reclassification improvement (NRI) technique is performed, besides the AUC parameter, obtaining a maximum true positive-true negative rate of 0.787. On the other hand, this work [22] presents a multivariate model obtained through a FBS method based on the p-value, in order to classify between three different classes, ”caries”, ”restorations” and ”control”; this model was evaluated using a statistical analysis, obtaining a maximum AUC value of 0.664. Finally, this work [23] presents a univariate analysis using a linear regression approach in order to classify between subjects with the presence of caries and with absence; then, from the most significant univariate models, a multivariate model contained by three features was developed, which obtained an AUC value of 0.572.
2.3. Data Analysis
The data analysis of this work was performed through a multivariate approach, subjecting the demographic and dietary features to a deep ANN for the classification of subjects according to their health and then a statistical validation was carried out in order to evaluate the results obtained from the ANN developed for these specific data.
2.3.1. Data Preprocessing
The data preprocessing consisted initially in the manual elimination of the features that presented a high percentage of missing values (>30%), represented as “Not a Number” (NaN). Then, of the remaining features, those that presented a low percentage of missing data (≤30%) were imputed with the ”rfimput” function, of the ”randomForest” package (version 4.6-12, 6 October 2015) [24], for R, which consists of replacing all NaN with the average of the values that present the column where the missing data is located.
The columns containing singular values were also removed because they didn’t provide significant information. Two values are singular if they have multiple values between them or if they present the same value in the whole column.
Finally, the database was separated into two sets, randomly and balanced selecting a set for the training stage, contained with 70% of the data and a set for the testing stage, contained with the remaining 30% of data.
2.3.2. Data Classification
The classification of subjects was performed according to the presence or absence of caries through a dense ANN that was designed based on the data of the subjects, using the package ”Keras”. ”Keras” is a high-level ANN application programming interface written in Python. It was developed with the approach of allowing fast experimentation, facilitating the creation of prototypes in a simple way [25].
ANNs seek a solution to a specific task, based on the correlation between features, through learning or training that resembles the behavior of biological neural networks. Using different layers composed of nodes or neurons, the ANNs look for a model of the relationship between the input features and the output feature. The number of nodes that exist for each layer is configurable, as is the number of layers and, depending on the data, there is usually a greater capacity of the network, the greater the number of layers and neurons; however, if there isn’t the right amount of these elements, problems of overfitting can be caused .
ANNs present three main elements: (1) a set of synapses or connections, characterized by a ”weight”, where the input signal is connected to a neuron through its product with the weight of that connection; (2) an adder, which adds the contributions of a signal weighted by all the weights; and (3) an activation function, which is equivalent to a transfer function, affecting the neurons, allowing for limiting the amplitude of the network output, providing a permissible range for the output signal in terms of finite values. Among the most common activation functions are the lineal, quadratic, geometric, logistic, and rectified linear unit function, among others.
On the other hand, an ANN is dense when each node of a layer is connected with every node of the next layer; while the recurrence term refers to the network presenting at least one feedback loop, which will provide a deeper impact on the learning capacity of the network, as well as on its performance, since it allows for optimizing its behavior through a parameter that gives knowledge of the behavior of the data and allows for guiding the adjustment of the configuration of the network to improve its accuracy.
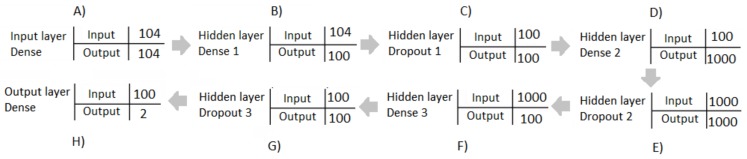
The ANN designed was composed by a series of dense and dropout hidden layers, as shown in the diagram of Figure 4. The input layer (A) was assigned 104 neurons, making reference to the 104 features of the dataset. The first dense hidden layer (B) was composed of 100 neurons, as was the first dropout hidden layer (C), which had a loss percentage of 50%. The second dense hidden layer (D) was assigned 1000 neurons, as was the second dropout hidden layer (E), which had a loss percentage of 25%. The third dense hidden layer (F) was composed of 100 neurons, as was the third dropout hidden layer (G), which had a loss of 50%. Finally, the fourth dense hidden layer was the output layer (H), which was characterized by two neurons, making reference to the two outputs or possible classifications.
Figure 4.
Diagram of the ANN designed.
The input layer and the dense hidden layers used as activation function, the Rectified Linear Unit (ReLU) function, which consists of assigning 0 to the values of the neurons that are <0, and to respect the value of the neurons when their values are ≥0, as shown in Equation (1) [26]:
| (1) |
The output layer used the Normalized Exponential function as an activation function, also known as ”Softmax”, which represents a general form of the logistic function and it is used to compress a vector of arbitrary values into a vector of real values in a range of [0, 1]. This function is shown in Equation (2), where represents the K-dimensional vector, z, of binary values [27]:
| (2) |
Finally, the dropout hidden layers were included in order to avoid overfitting problems, as mentioned before, and its performance is based on assigning the value of 0 to a percentage of the data and thus they are not taken into account for the classification of subjects in that layer. The data subset selected by these layers is changing with each iteration, causing that in each epoch the classification will be done omitting a different percentage of data. On the other hand, due to the recurrence of the ANN, it was possible to optimize its performance. The optimization algorithm, “Adam”, was selected, which bases its operation on stochastic gradient descent algorithms, making use of the average of the first and second moments of the gradients to adapt the rate of the learning parameter. Specifically, “Adam” calculates the exponential moving average of the gradient and the square gradient, and controls the decay rates of that moving average [28]. Some of the benefits that present “Adam” are that it is a method that is straightforward to implement, is computationally efficient, has little memory requirements, is invariant to diagonal rescaling of the gradients, and one of the most important points is that is suitable for problems that are large in terms of data or features. In addition, the parameters have intuitive interpretations and typically require little tuning [28].
It is important to mention that the number of iterations or epochs that the ANN will have for the optimization of its behavior, based on the recurrence, is also configurable and will depend on the type of data. To know what is the appropriate number of epochs, there are some parameters that allow for evaluating the behavior through each iteration, such as the loss function and the accuracy [29]. The number of the epochs that were selected through a series of tests with different numbers were 100 epochs, since they demonstrated having the best performance in terms of the accuracy of the ANN.
Based on the above, “keras” has the advantage that it allows for designing the architecture of the ANN according to the type of data, configuring the type of ANN, number of nodes, validation, loss function, among others [30,31].
2.3.3. Evaluation
Finally, in order to validate the results of classification obtained by the ANN, three parameters were evaluated; loss function, accuracy and ROC curve. The loss function and accuracy were calculated on each epoch, allowing to know if the performance of the ANN was improving, while the ROC curve was calculated based on the average of the general performance of the ANN.
ANNs are mostly trained using gradient methods through an iterative process of decreasing the loss function. A loss is designed to have the main property that the lower its value, the better the model that fits its data and, in addition, it will be differentiable, which will optimize the network directly, giving information of the capacity of the system. Therefore, the loss function is based on the search of the global minimum, which corresponds to the minimum error, based on a learning factor. Some of the most common techniques to calculate the loss function are the mean square error, mean absolute error, binary cross-entropy, and Poisson, among others [31].
The loss function that was used is preset in “keras” as “binary-crossentropy” and calculates the cross-entropy value for binary classification problems. This method uses the Kullback–Leibler distance, which is a measure between two density functions g and h, known as the cross-entropy between g and h, as shown in Equation (3). Its operation is based on iterations, generating a random set of values estimating the value that wants to be obtained and then actualizing the parameters in the next iteration to generate “better” values or more approximate, in terms of the Kullback–Leibler distance [32]:
| (3) |
On the other side, the accuracy allows for measuring the performance of the ANN through a non-differentiable function. This metric doesn’t allow to optimize the network, but it allows to select the model that shows the most suitable performance in the training of the network, and it is based on the calculation of the average of the differences that exist between the classification calculated by the ANN model and the true classification of the data, as shown in Equation (4), where the accuracy is reported as 1-. refers to the classification value that was calculated by the ANN, while refers to true classification value [33]:
| (4) |
In this work, the accuracy was calculated with the “binary-accuracy” function from “keras”, which calculates the average accuracy rate across all predictions for binary classification problems.
The ROC curve is a standard method that allows for evaluating the precision with which the model classifies, based on the relationship between sensitivity and specificity. Sensitivity is defined as the proportion of subjects with a condition that were classified as positive, which means the positive predictive values (); this value is calculated with Equation (5), where represents the number of true positives and represents the number of false positives [34,35]:
| (5) |
Specificity is defined as the proportion of subjects without a condition that was classified as negative; this means the negative predictive values (); this value is calculated with Equation (6), where represents the number of true negatives and represents the number of false negatives [34,35]:
| (6) |
The ROC curves were calculated to obtain the true positive and true negative rate for each class, for the macro-average precision and for the micro-average precision.
The macro-average precision is obtained through the sum of the true positives, false positives and false negatives of the system, for different sets, as shown in Equation (7), where refers to the true positives of the set one, refers to the true positives of the set two, refers to the false positives of the set one and refers to the false positives of the set two [36,37]:
| (7) |
On the other hand, the macro-average precision is a direct method; it takes the average of the accuracy of the system in different sets. It is calculated with Equation (8), where refers to the average of the set one and refers to the average of the set two [36,37]:
| (8) |
The analysis of this work was performed in Python (version 3.6), using the packages, “Keras” (version 2.1.5) [25], “Scipy” (version 1.0.0) [38], “Pandas” (version 0.22.0) [39] and “Sklearn” (version 0.19.1) [36].
3. Results
The two databases that were used in this work were contained by a total of 188 demographic and dietary features, besides a feature being used that was contained by the oral health status as an output feature for the classification of subjects.
After the data preprocessing, only 105 features were conserved, of which 25 belonged to demographic features and 79 to dietary features, the remaining feature refers to the output feature.
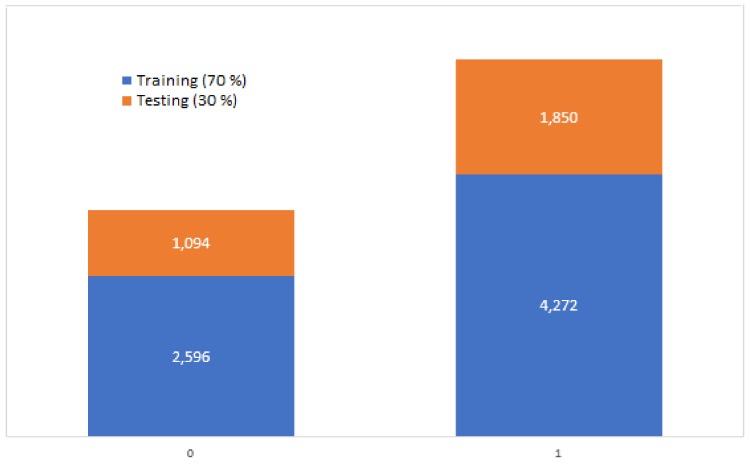
Of the total set of 9812 subjects, 3690 belonged to controls and 6122 belonged to cases. This dataset was divided into two subsets, one for training which was contained by 70% of the data (2596 controls/4272 cases), and one for testing which was contained with the remaining 30% of the data (1094 controls/1850 cases). These data are graphically shown in Figure 5.
Figure 5.
Graph of the number of subjects that belongs to each dataset, training and testing. The content in the bar with “0” presented a status of absence of caries, while the contest in the bar with “1” presented a status of presence/restoration of caries.
The dataset that was designed for the training stage was evaluated at each of the 100 epochs through the accuracy and loss function values. This number of epochs was selected through a comparison between the values obtained using different number of epochs; Table 1 presents the values obtained for ten different epochs.
Table 1.
Comparison of the accuracy and loss function values obtained using different number of epochs.
| Epochs | Accuracy | Loss Function | Processing Time (s) |
|---|---|---|---|
| 10 | 0.67 | 0.60 | 10.44 |
| 30 | 0.68 | 0.60 | 32.02 |
| 50 | 0.68 | 0.59 | 55.46 |
| 80 | 0.69 | 0.59 | 93.57 |
| 100 | 0.69 | 0.58 | 124.55 |
| 150 | 0.68 | 0.60 | 188.21 |
| 200 | 0.68 | 0.61 | 246.74 |
| 300 | 0.69 | 0.62 | 369.06 |
| 500 | 0.70 | 0.62 | 646.86 |
| 1000 | 0.70 | 0.66 | 4152.96 |
On the other hand, in Table 2, a comparison of results using different number of layers and neurons is shown. The number of epochs selected was 100, establishing this number according to the result obtained from the previous table. It is possible to observe that the most statically significant values of accuracy and loss function were obtained using an ANN designed with seven layers, four dense layers and three dropout layers. The dense layers were contained by 104, 1000, 100 and two neurons, in descendant order, while the dropout layers represented 0.50, 0.25 and 0.50 percentages, in descendant order too.
Table 2.
Comparison of the accuracy and loss function values obtained using different number of layers and neurons.
| Layers Dense/Dropout | Neurons | Accuracy | Loss Function | Processing Time (s) |
|---|---|---|---|---|
| 2/1 | 104>0.50>2 | 0.68 | 0.60 | 36.84 |
| 3/1 | 104>0.50>1000>2 | 0.68 | 0.59 | 71.10 |
| 3/2 | 104>0.25>1000>0.50>2 | 0.69 | 0.61 | 93.86 |
| 4/1 | 104>1000>0.50>100>2 | 0.68 | 0.73 | 117.63 |
| 4/2 | 104>0.50>1000>0.50>100>2 | 0.68 | 0.59 | 119.77 |
| 4/3 | 104>0.50>1000>0.25>100>0.50/2 | 0.69 | 0.58 | 124.55 |
| 5/1 | 104>100>1000>0.50>100>2 | 0.68 | 0.91 | 129.53 |
| 5/2 | 104>100>0.50>1000>0.25>100>2 | 0.68 | 0.65 | 131.66 |
| 5/3 | 104>100>0.50>1000>0.25>100>0.50>2 | 0.69 | 0.64 | 139.37 |
| 5/4 | 104>0.25>100>0.50>1000>0.25>100>0.50>2 | 0.68 | 0.59 | 138.38 |
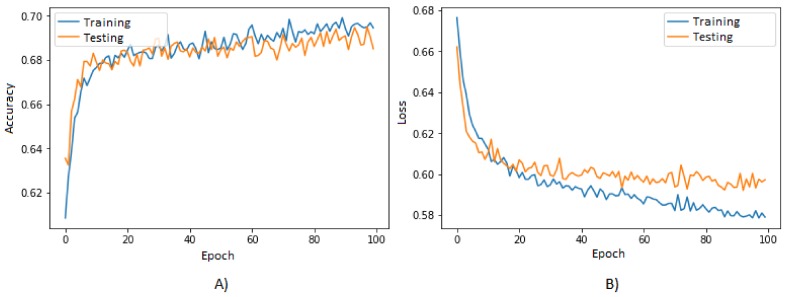
The graph of the performance of the accuracy is shown in Figure 6A, where the blue line represents the behavior of the training data, obtaining a final accuracy of 0.69, while the orange line represents the behavior of the testing data, obtaining a final accuracy of 0.68.
Figure 6.
Graphs of the performance of the accuracy (A) and the loss function (B) of the ANN in each epoch.
The graph of the performance of the loss function is shown in Figure 6B, where the blue line represents the behavior of the training data, obtaining a final value of 0.58, while the orange line represents the behavior of the testing data, obtaining a final value of 0.60.
Both accuracy and loss function values were obtained in order to know the performance of the classification of subjects on each iteration; nevertheless, the accuracy is a parameter that may show an optimistic response if the data presents any bias. Therefore, there were also measured the specificity and sensitivity parameters in order to ensure that the classification of subjects was statistically significant.
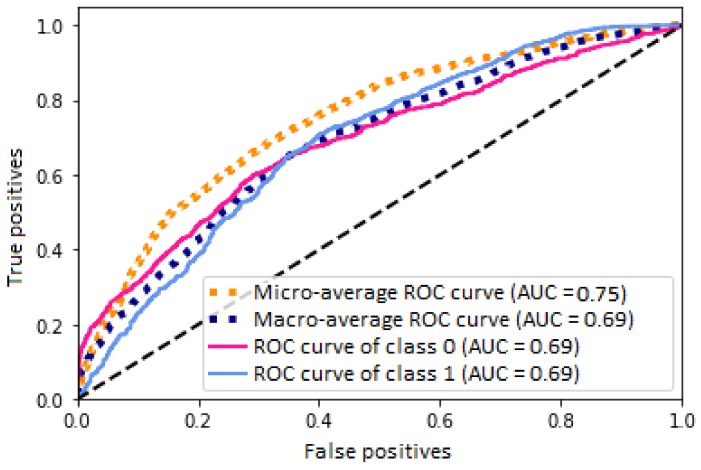
The graph of the Figure 7 shows the ROC curves of the mean performance of the ANN, where the pink line refers to the proportion of sensitivity and specificity for the class “0”, which are the subjects that presented absence of caries, obtaining an AUC value of 0.69. The light blue line refers to the proportion of sensitivity and specificity for the class “1”, which are the subjects that presented presence/restoration of caries, obtaining an AUC value of 0.69. The dotted orange line refers to the curve calculated with the micro-average of the proportion of sensitivity and specificity for the classification of subjects, obtaining an AUC value of 0.75; the dotted dark blue line refers to the curve calculated with the macro-average, obtaining an AUC value of 0.69.
Figure 7.
Graph of the ROC curves generated based on the average performance of the ANN in the classification of subjects.
4. Discussion
Of the two datasets that were obtained through the preprocessing step, it is possible to observe that the set of cases contains more data than the set of controls as it is observed in Figure 5; this was due to the fact that the cases included the subjects with presence of caries and with restorations. Nevertheless, the number of subjects that were contained on each dataset was enough to train the ANN, obtaining statistically significant results.
The data were separated in two sets based on an aleatory and balanced selection; one of the sets was contained with 70% of the data and the other dataset was contained with 30%, as shown in Figure 5. The dataset that was contained with the largest amount of data had the purpose of training the ANN. The dense ANN was trained during 100 epochs and was optimized through the optimization algorithm, “Adam”, which had the purpose of improving the performance of the ANN with feedback.
Finally, for the validation of the ANN modeling, the accuracy and the loss function in each of the epochs with the dataset that was contained with the least amount of data were obtained. The loss function was calculated using cross-entropy in order to know how the performance of the ANN was modified. The validation was carried out in both datasets, training and testing, in order to ensure that there was no adjustment in the classification of the subjects; that is, if the performance of these parameters is similar for both datasets, it means that the ANN is classifying based on a generalized model, since it manages to classify with an accuracy and a loss function similar in the training dataset and in the testing dataset.
In the graph of Figure 6A, it is observed that the performance of the accuracy is similar for both datasets, being slightly higher for the training data, which is normal since they are the data on which the ANN modeling is being based. As the performance is similar for both datasets, it is possible to know that the ANN model became generalized through the learning by being able to classify unknown data with a similar accuracy to that obtained with the training data. The final value obtained for the accuracy was 0.69, which is statistically significant since it means that at least 69% of the data was correctly classified.
On the other hand, in the graph of Figure 6B, it is observed that the performance of the loss function is also similar for both datasets. It is notable that, for the training dataset, this parameter is smaller compared to that obtained with the testing dataset; however, as in the case of the accuracy parameter, this performance is normal because the testing dataset was unknown for the model. In addition, having a similar performance allows for knowing that the model doesn’t have overfitting problems and that it manages to classify unknown data in a similar way as it classifies known data. The final value obtained for the accuracy had an average of 0.59. This value isn’t close to the ideal, which is zero; however, it is possible to observe that the data remains tending toward zero for both datasets, showing that the testing dataset decreases slower than the training dataset. This problem can be solved increasing the amount of data, removing features that are not significant or that are redundant for the model and increasing the size if the ANN or the number of epochs.
Although the graph of the loss function doesn’t present very favorable results, it allows for knowing that the modeling of the ANN is robust among the two datasets, in addition to continuing to decrease the loss function and reducing the error through the epochs, which is one of the main purposes of the training stage.
Finally, Figure 7 shows that all curves presented statistically significant values, obtaining AUC values ≥0.69. These curves are generated based on the proportion of true positives and true negatives, where for class “0”, class “1” and the macro-average, an AUC value of 0.69 was obtained, which implies that 69% of the subjects were classified appropriately for each of the classes and for their general average, while. for the curve generated for the micro-average, an AUC value of 0.75 was obtained, which implies that 75% of the subjects were classified appropriately when the measure was based in subsets of data. The ROC curves of class “0” and class “1” presented a very similar performance, besides obtaining the same AUC value, which means that the classification performance is equitable and the generalization of the model allows for classifying subjects of both classes.
On the other hand, the AUC value is greater for the micro-average than for the macro-average; this may be because the calculation of the macro-average is based in obtaining the true positives/true negatives proportion through the whole amount of data, while, for the micro-average, the calculation of the ROC curve is based in subsets of data, where some subsets can present better AUC values than obtained with the whole set, thus achieving a better average of classification.
Is it important to remark that this generalized model presents an impartial performance, since it was trained with data that cover a reasonable spectrum of the possible outcomes; in addition, the moment of time and geographical location are parameters that were taken into account in the initial data acquisition; regardless of whether the data was acquired in the USA, most of the subjects were from other regions, of which a percentage belongs to Mexican subjects, providing robustness in the results and avoiding any bias problem related to the structure, the moment of time and the geographical location of the data.
Finally, the results obtained allow for supporting the main motivation of this work, which was to develop a tool for human resources and dental services, since this model is able to give a statistically significant response of the possible development of caries based on information that doesn’t require any extra equipment for its acquisition, only the willingness of people to answer the demographic and diet surveys, providing support to the dental specialists in the work of reducing the high incidence of dental caries
Based on these discussions, it can be proposed as future work to add a stage in the methodology that consists of a step for the feature selection using a genetic algorithm approach. This stage will help to remove redundant information and correlated features, keeping only those features that provide the most significant information for the classification of subjects, besides reducing the computational cost. Additionally, for the validation of the feature selection, a forward selection and backward elimination may be used in order to test the accuracy of the set of features obtained through the selection process, ensuring the certainty of the model behavior.
5. Conclusions
According to the results obtained, it is possible to conclude that the amount of data that was used to model the classification of the subjects was adequate for the preliminary generalized learning of the ANN, basing this on the performance that the classification of cases had, being similar to the classification of controls. This performance was observed in the accuracy and loss function graphs, which allows for recognizing that the number of epochs that were assigned to the ANN was sufficient to maintain stable performance of the parameters in both datasets.
The accuracy achieved with this modeling was around 0.70, a value that is statistically significant since it implies that 70% of the time will be correctly classified to the subjects with presence or absence/restoration of caries.
The loss function decreased by approximately 10% since the beginning of the training, showing minor changes throughout the epochs, implying an approximation to the global minimum sought.
As a final evaluation, all the ROC curves obtained an AUC value statistically significant, implying that, around 70% of the time, subjects were correctly classified, according to the true positives and true negatives proportion, a value that corroborates the accuracy obtained. Nevertheless, a methodology was proposed based in convolutional neural networks in order to identify features in a spectral or spatial domain to obtain time-independent abstract features, looking for the improvement in the modeling for the classification of subjects.
Based on this, it is possible to classify subjects with the absence of caries from subjects with presence/restorations, through demographic and dietary data, with a statistically significant accuracy, demonstrating that the socioeconomic and nutritional status are important determinants in the development of caries.
Then, according to the results obtained, it was demonstrated that the demographic situation can significantly affect the prevalence of dental caries. Based on this, the analysis of oral health data from exclusively Mexican subjects is proposed, comparing those results with the ones obtained in this work, with the purpose of proving how demography can influence the oral health status.
On the other hand, even though the results do not show an ideal behavior, they give preliminary knowledge of the benefit that its implementation would have in a real environment, since the requirements for its use are minimal, no in-depth knowledge of the techniques used is required, results are presented quickly and the computational cost is very low, besides the accuracy obtained being statistically significant. In addition, an important point to take into account is that the performed experimental tests were based in a real production environment, since all the subjects that were part for the development of this work were real control and cases, and the demographic and dietary databases were obtained from real information.
The hardware tool that is required for the implementation of this work is a computer, while the free software tool that is required is Python, which is a programming language that allows for working quickly and integrating systems more effectively. As extra information, it is mentioned that the computer that was used for the development of this work was a laptop Acer Aspire F5-573-70LX 15.6” (Acer America Corporation, 333 West San Carlos Street, Suite 1500, San Jose, CA 95110), Intel Core i7-7500U 2.70 GHz (Plot 6, Bayan Lepas Technoplex Medan Bayan Lepas 11900 Bayan Lepas Penang, Malaysia, Georgetown, Pulau Pinang, Malasia), 16 GB, 1 TB + 128 GB Solid State Drive, Windows 10 Home (15010 NE 36th Street, Microsoft Campus, Building 92, Redmond, WA 98052), 64-bit; and the version of Python that was used is 3.6 [40].
Finally, based on the last point, it is evident that the implementation of the model proposed in this work could provide an easy, free and fast tool that helps the specialists in the preventive diagnosis of dental caries, besides offering an option that collaborates in the decrement of the incidence of this public health problem.
Appendix A
Table A1.
Description of demographic features.
| Feature | Description |
|---|---|
| AIALANGA | Language of the MEC ACASI Interview Instrument. |
| DMDBORN4 | In what country was Sample Person (SP) born? |
| DMDCITZN | Is SP a citizen of the United States? |
| DMDHHSIZ | Total number of people in the Household. |
| DMDHHSZE | Number of adults aged 60 years or older in the household. |
| DMDEDUC2 | Highest grade or level of school completed or the highest degree received. |
| DMDHHSZB | Number of children aged 6–17 years old in the household (HH). |
| DMDHSEDU | HH reference person’s spouse’s education level. |
| DMDMARTL | Marital status. |
| DMDFMSIZ | Total number of people in the Family. |
| DMDHHSZA | Number of children aged 5 years or younger in the household. |
| DMDHRGND | HH reference person’s gender. |
| DMDHREDU | HH reference person’s education level. |
| DMDEDUC3 | Highest grade or level of school completed or the highest degree received. |
| DMDHRAGE | HH reference person’s age in years. |
| DMDHRBR4 | HH reference person’s country of birth. |
| DMDHREDU | HH reference person’s education level. |
| DMDHRMAR | HH reference person’s marital status. |
| DMQADFC | Did SP ever serve in a foreign country during a time of armed conflict or on a humanitarian or peace-keeping mission? |
| DMDYRSUS | Length of time the participant has been in the US. |
| DMQMILIZ | Has SP ever served on active duty in the U.S. Armed Forces, military Reserves, or National Guard? |
| FIAINTRP | Was an interpreter used to conduct the Family interview? |
| FIALANG | Language of the Family Interview Instrument. |
| FIAPROXY | Was a Proxy respondent used in conducting the Family Interview? |
| INDFMPIR | A ratio of family income to poverty guidelines. |
| INDHHIN2 | Total household income (reported as a range value in dollars). |
| INDFMIN2 | Total family income (reported as a range value in dollars). |
| LATVPSU | Variance unit: PSU variable for variance estimation. |
| LATVSTRA | Variance unit: stratum variable for variance estimation. |
| MIAINTRP | Was an interpreter used to conduct the MEC CAPI interview? |
| MIALANG | Language of the MEC CAPI Interview Instrument. |
| MIAPROXY | Was a Proxy respondent used in conducting the MEC CAPI Interview? |
| RIAGENDR | Gender of the participant. |
| RIDAGEEX | Age in years of the participant at the time of examination. Individuals aged 959 months and older are topcoded at 959 months. |
| RIDAGEMN | ge in months of the participant at the time of screening. Reported for persons aged 24 months or younger at the time of exam. |
| RIDRETH3 | Recode of reported race and Hispanic origin information, with Non-Hispanic Asian Category. |
| RIDEXMON | Six month time period when the examination was performed. |
| RIDEXPRG | Pregnancy status for females between 20 and 44 years of age at the time of MEC exam. |
| RIDAGEYR | Age in years of the participant at the time of screening. |
| RIDRETH1 | Recode of reported race and Hispanic origin information. |
| RIDEXAGM | Age in months of the participant at the time of examination. |
| RIDSTATR | Interview and examination status of the participant. |
| SDMVPSU | Masked variance unit pseudo-PSU variable for variance estimation. |
| SDDSRVYR | Data release cycle. |
| SDMVSTRA | Masked variance unit pseudo-stratum variable for variance estimation. |
| WTLAF8YR | Subsample 8-year fasting weight for participants aged 12 years and older who were examined in the morning sessions in Los Angeles, CA, USA. |
| WTLAI8YR | Full sample 8-year interview weight for participants in Los Angeles, CA, USA. |
| WTLAM8YR | Full sample 8-year MEC exam weight for participants in Los Angeles, CA, USA. |
Appendix B
Table A2.
Description of dietary features.
| Feature | Description |
|---|---|
| WTDRD1 | Dietary day one sample weight. |
| DBQ095Z | Type of salt usually add to food at the table. |
| DR1TKCAL | Energy (kcal). |
| DR1TSUGR | Total sugars (g). |
| DR1TFIBE | Dietary fiber (g). |
| DR1TSFAT | Total saturated fatty acids (g). |
| DR1TMFAT | Total monounsaturated fatty acids (g). |
| DR1TPFAT | Total polyunsaturated fatty acids (g). |
| DR1TLYCO | Lycopene (g). |
| DR1TFA | Folic acid (g). |
| DR1TB12A | Added vitamin B12 (g). |
| DR1_300 | Was the amount of food that you ate yesterday much more than usual, usual, or much less than usual? |
| DR1_320Z | Total plain water drank yesterday—including plain tap water, water from a drinking fountain, water from a water cooler, bottled water, and spring water. |
| DR1_330Z | Total tap water drank yesterday—including filtered tap water and water from a drinking fountain. |
| DR1BWATZ | Total bottled water drank yesterday (g). |
| DR1DAY | Intake day of the week. |
| DR1DBIH | Number of days between intake day and the day of family questionnaire administered in the household. |
| DR1SKY | What type of salt was it? (Was it ordinary or seasoned salt, lite salt, or a salt substitute?). |
| DR1STY | Did SP add any salt to her/his food at the table yesterday? |
| DR1TACAR | Alpha-carotene (g). |
| DR1TBCAR | Beta-carotene (g). |
| DR1TCAFF | Caffeine (mg). |
| DR1TCALC | Calcium (mg). |
| DR1TCOPP | Copper (mg). |
| DR1TCRYP | Beta-cryptoxanthin (g). |
| DR1TFDFE | Folate as dietary folate equivalents (g). |
| DR1TFF | Food folate (g). |
| DR1TFOLA | Total folate (g). |
| DR1TLZ | Lutein + zeaxanthin (g). |
| DR1TM161 | MFA 16:1 (Hexadecenoic) (g). |
| DR1TM181 | MFA 18:1 (Octadecenoic) (g). |
| DR1TM201 | MFA 20:1 (Eicosenoic) (g). |
| DR1TM221 | MFA 22:1 (Docosenoic) (g). |
| DR1TMAGN | Magnesium (mg). |
| DR1TMOIS | Moisture (g). |
| DR1TNIAC | Niacin (mg). |
| DR1TNUMF | Total number of foods/beverages reported in the individual foods file. |
| DR1TP182 | PFA 18:2 (Octadecadienoic) (g). |
| DR1TP183 | PFA 18:3 (Octadecatrienoic) (g). |
| DR1TP184 | PFA 18:4 (Octadecatetraenoic) (g). |
| DR1TP204 | PFA 20:4 (Eicosatetraenoic) (g). |
| DR1TP205 | PFA 20:5 (Eicosapentaenoic) (g). |
| DR1TP225 | PFA 22:5 (Docosapentaenoic) (g). |
| DR1TP226 | PFA 22:6 (Docosahexaenoic) (g). |
| DR1TPOTA | Potassium (mg). |
| DR1TRET | Retinol (g). |
| DR1TS040 | SFA 4:0 (Butanoic) (g). |
| DR1TS060 | SFA 6:0 (Hexanoic) (g). |
| DR1TS100 | SFA 10:0 (Decanoic) (g). |
| DR1TS120 | SFA 12:0 (Dodecanoic) (g). |
| DR1TS140 | SFA 14:0 (Tetradecanoic) (g). |
| DR1TS180 | SFA 18:0 (Octadecanoic) (g). |
| DR1TSELE | Selenium (g). |
| DR1TSODI | Sodium (mg). |
| DR1TTHEO | Theobromine (mg). |
| DR1TVARA | Vitamin A as retinol activity equivalents (g). |
| DR1TVB1 | Thiamin (Vitamin B1) (mg). |
| DR1TVB12 | Vitamin B12 (g). |
| DR1TVB2 | Riboflavin (Vitamin B2) (mg). |
| DR1TVB6 | Vitamin B6 (mg). |
| DR1TVC | Vitamin C (mg). |
| DR1TVK | Vitamin K (g). |
Table A3.
Description of dietary features.
| Feature | Description |
|---|---|
| DR1TWS | When you drink tap water, what is the main source of the tap water? Is the city water supply; a well or rain cistern; a spring; or something else? |
| DR1TZINC | Zinc (mg). |
| DRABF | Indicates whether the sample person was an infant who was breast-fed on either of the two recall days. |
| DRD340 | During the past 30 days did you eat any types of shellfish listed on this card? |
| DRD350A | Clams eaten during the past 30 days. |
| DRD350AQ | Number of times clams were eaten in the past 30 days. |
| DRD350B | Crabs eaten during the past 30 days. |
| DRD350BQ | Number of times crab was eaten in the past 30 days. |
| DRD350C | Crayfish eaten during the past 30 days. |
| DRD350CQ | Number of times crayfish was eaten in the past 30 days. |
| DRD350D | Lobsters eaten during the past 30 days. |
| DRD350DQ | Number of times lobster was eaten in the past 30 days. |
| DRD350E | Mussels eaten during the past 30 days. |
| DRD350EQ | Number of times mussels were eaten in the past 30 days. |
| DRD350F | Oysters eaten during the past 30 days. |
| DRD350FQ | Number of times oysters were eaten in the past 30 days. |
| DRD350G | Scallops eaten during the past 30 days. |
| DRD350GQ | Number of times scallops were eaten in the past 30 days. |
| DRD350H | Shrimp eaten during the past 30 days. |
| DRD350HQ | Number of times shrimp was eaten in the last 30 days. |
| DRD350I | Other shellfish ( ex. octopus, squid) eaten during the past 30 days. |
| DRD350IQ | Number of times other shellfish (ex. octopus, squid) was eaten in the past 30 days. |
| DRD350J | Other unknown shellfish eaten during the past 30 days. |
| DRD350JQ | Number of times other unknown shellfish was eaten in the past 30 days. |
| DRD350K | Refused to give detailed information on shellfish eaten during the past 30 days. |
| DRD360 | During the past 30 days did you eat any types of fish listed on this card? |
| DRD370A | Breaded fish products eaten during the past 30 days. |
| DRD370AQ | Number of times breaded fish products were eaten in the past 30 days. |
| DRD370B | Tuna eaten during the past 30 days. |
| DRD370BQ | Number of times tuna was eaten in the past 30 days. |
| DRD370C | Bass eaten during the past 30 days. |
| DRD370CQ | Number of times bass was eaten in the past 30 days. |
| DRD370D | Catfish eaten during the past 30 days. |
| DRD370DQ | Number of times catfish was eaten in the past 30 days. |
| DRD370E | Cod eaten during the past 30 days. |
| DRD370EQ | Number of times cod was eaten in the past 30 days. |
| DRD370F | Flatfish eaten during the past 30 days. |
| DRD370FQ | Number of times flatfish was eaten in the past 30 days. |
| DRD370G | Haddock eaten during the past 30 days. |
| DRD370GQ | Number of times haddock was eaten in the past 30 days. |
| DRD370H | Mackerel eaten during the past 30 days. |
| DRD370HQ | Number of times mackerel was eaten in the past 30 days. |
| DRD370I | Perch eaten during the past 30 days. |
| DRD370IQ | Number of times perch was eaten in the past 30 days. |
| DRD370J | Pike eaten during the past 30 days. |
| DRD370JQ | Number of times pike was eaten in the past 30 days. |
| DRD370K | Pollock eaten during the past 30 days. |
| DRD370KQ | Number of times pollock was eaten in the past 30 days. |
| DRD370TQ | Number of times other type of fish was eaten in the past 30 days. |
| DRD370V | Refused to give detailed information on fish eaten during the past 30 days. |
| DRQSDIET | Are you currently on any kind of diet, either to lose weight or for some other health-related reason? |
| DRQSDT1 | What kind of diet are you on? (Is it a weight loss or a low calorie diet: low fat or cholesterol diet; low salt or sodium diet; sugar free or low sugar diet; low fiber diet; high fiber diet; diabetic diet; or another type of diet?). |
| DRD370L | Porgy eaten during the past 30 days. |
| DRD370LQ | Number of times porgy was eaten in the past 30 days. |
| DRD370M | Salmon eaten during the past 30 days. |
| DRD370MQ | Number of times salmon was eaten in the past 30 days. |
| DRD370N | Sardines eaten during the past 30 days. |
| DRD370NQ | Number of times sardines were eaten in the past 30 days. |
| DRD370O | Sea bass eaten during the past 30 days. |
| DRD370OQ | Number of times sea bass was eaten in the past 30 days. |
| DRD370U | Other unknown type eaten during the past 30 days. |
Table A4.
Description of dietary features.
| Feature | Description |
|---|---|
| WTDR2D | Dietary two-day sample weight. |
| DR1DRSTZ | Dietary recall status. |
| DR1TPROT | Protein (g). |
| DR1TCARB | Carbohydrate (g). |
| DR1TTFAT | Total fat (g). |
| DR1TCHOL | Cholesterol (mg). |
| DR1TFA | Folic acid (g). |
| DR1TCHL | Total choline (mg). |
| DR1TIRON | Iron (mg). |
| DBD100 | Frequency with which ordinary salt is added to the food on the table. |
| DRQSPREP | Frequency with which ordinary salt or seasoned salt is added in cooking or preparing foods in the household. |
| DR1TALCO | Alcohol (g). |
| DR1TS080 | SFA 8:0 (Octanoic) (g). |
| DR1TATOC | Vitamin E as alpha-tocopherol (mg). |
| DR1TATOA | Added alpha-tocopherol (Vitamin E) (mg). |
| DR1TVD | Vitamin D (D2 + D3) (g). |
| DR1TPHOS | Phosphorus (mg). |
| DR1TS160 | SFA 16:0 (Hexadecanoic) (g). |
| DRD370UQ | Number of times other unknown type of fish was eaten in the past 30 days. |
| DRD370V | Refused to give detailed information on fish eaten during past 30 days. |
| DRDINT | Indicates whether the sample person has intake data for one or two days. |
| DRQSDIET | Are you currently on any kind of diet, either to lose weight or for some other health-related reason? |
Author Contributions
C.E.G.-T. and L.A.Z.-C., performed the study. C.E.G.-T., L.A.Z.-C., and J.I.G.-T. performed the study design and data analysis. N.M.C.-L. and J.R.-G. contributed to materials and methods (selection of patients from database) used in this study. C.E.G.-T., J.I.G.-T., and J.M.C.-P. performed statistical analysis and statistical validation with critical feedback to authors. J.M.C.-P., R.M.-Q. and L.A.Z.-C. contributed with the neural network implementation used in this study. L.A.Z.-C. and H.G.-R. provide feedback from results. All authors interpreted findings from the analysis and drafted the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Oral Health. [(accessed on 5 June 2017)]; Available online: http://www.who.int/oral_health/disease_burden/global/en/
- 2.Ridao Marín D. Desarrollo de un Sistema de Ayuda a la Decisión para Tratamientos Odontológicos con Imágenes Digitales. Universidad de Málaga; Málaga, Spain: 2017. pp. 10–12. [Google Scholar]
- 3.Espinoza Solano M., León-Manco R.A. Prevalencia y experiencia de caries dental en estudiantes según facultades de una universidad particular peruana. Rev. Estomatol. Hered. 2015;25:187–193. doi: 10.20453/reh.v25i3.2621. [DOI] [Google Scholar]
- 4.Acuña Aguilar L.D., Porras Cerón D., Ríos Rueda L.D. Prevalencia de Lesiones Cariosas y Factores Asociados Presentes en Pacientes con SíNdrome de Down en las Fundaciones Fundown y san Luis Guanella de Bucaramanga. Universidad Santo Tomás; Bucaramanga, Colombia: 2017. pp. 12–16. [Google Scholar]
- 5.Gispert Abreu E.D.L.Á., Castell-Florit Serrate P., Herrera Nordet M. Salud bucal poblacional y su producción intersectorial. Rev. Cubana Estomatol. 2015;52:62–67. [Google Scholar]
- 6.Niño T.C., Guevara S.V., González F.A., Jaque R.A., Infante C. Uso de redes neuronales articiales en predicción de morfología mandibular a través de variables craneomaxilares en una vista posteroanterior. Univ. Odontol. 2016;35:1–28. [Google Scholar]
- 7.Lam C.U., Khin L., Kalhan A., Yee R., Lee Y., Chong M.F., Kwek K., Saw S., Godfrey K., Chong Y., et al. Identification of Caries Risk Determinants in Toddlers: Results of the GUSTO Birth Cohort Study. Caries Res. 2017;51:271–282. doi: 10.1159/000471811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes I., Sá-Pinto A., Marques L.S., Ramos-Jorge J., Ramos-Jorge M. Maternal identification of dental caries lesions in their children aged 1–3 years. Eur. Arch. Paediatr. Dent. 2017;18:197–202. doi: 10.1007/s40368-017-0286-8. [DOI] [PubMed] [Google Scholar]
- 9.Lips A., Antunes L.S., Antunes L.A., Pintor A.V.B., Santos D.A.B.D., Bachinski R., Küchler E.C., Alves G.G. Salivary protein polymorphisms and risk of dental caries: A systematic review. Braz. Oral Res. 2017;31 doi: 10.1590/1807-3107bor-2017.vol31.0041. [DOI] [PubMed] [Google Scholar]
- 10.Ahmed A., Ikram K., Masood H., Urooj M. Identification of relationship between oral disorders & hemodynamic parameters. Pak. Oral Dent. J. 2017;37:202–204. [Google Scholar]
- 11.Sarmiento R.V., Barrionuevo F.P., Huamán Y.S., Loyola M.C. Prevalencia de caries de infancia temprana en niños menores de 6 años de edad, residentes en poblados urbano marginales de Lima Norte. Rev. Estomatol. Herediana. 2011;21:79–86. doi: 10.20453/reh.v21i2.237. [DOI] [Google Scholar]
- 12.Oropeza-Oropeza C.D., Molina-Frechero N., Castañeda-Castaneira E., Zaragoza-Rosado C.D., Cruz Leyva C.D. Caries dental en primeros molares permanentes de escolares de la delegación Tláhuac. Rev. ADM. 2012;69:63–68. [Google Scholar]
- 13.Cardozo B.J., Gonzalez M.M., Pérez S.R., Vaculik P.A., Sanz E.G. Epidemiología de la caries dental en niños del Jardín de Infantes “Pinocho” de la ciudad de Corrientes. Rev. Fac. Odontol. 2017;9:35–41. [Google Scholar]
- 14.Zhang X., Zhang L., Zhang Y., Liao Z., Song J. Predicting trend of early childhood caries in mainland China: A combined meta-analytic and mathematical modelling approach based on epidemiological survey. Sci. Rep. 2017;7:6507. doi: 10.1038/s41598-017-06626-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asif S.M., Babu D.B., Naheeda S. Utility of Dermatoglyphic Pattern in Prediction of Caries in Children of Telangana Region, India. J. Contemp. Dent. Pract. 2017;18:490–496. doi: 10.5005/jp-journals-10024-2071. [DOI] [PubMed] [Google Scholar]
- 16.Chapple I.L., Bouchard P., Cagetti M.G., Campus G., Carra M.C., Cocco F., Nibali L., Hujoel P., Laine M.L., Lingstrom P., et al. Interaction of lifestyle, behaviour or systemic diseases with dental caries and periodontal diseases: Consensus report of group 2 of the joint EFP/ORCA workshop on the boundaries between caries and periodontal diseases. J. Clin. Periodontol. 2017;44:s39–s51. doi: 10.1111/jcpe.12685. [DOI] [PubMed] [Google Scholar]
- 17.Hayes M., Da Mata C., Cole M., McKenna G., Burke F., Allen P.F. Risk indicators associated with root caries in independently living older adults. J. Dent. 2016;51:8–14. doi: 10.1016/j.jdent.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Sudhir K.M., Kanupuru K.K., Fareed N., Mahesh P., Vandana K., Chaitra N.T. CAMBRA as a Tool for Caries Risk Prediction Among 12-to 13-year-old Institutionalised Children-A Longitudinal Follow-up Study. Oral Health Prev. Dent. 2016;14:355–362. doi: 10.3290/j.ohpd.a35621. [DOI] [PubMed] [Google Scholar]
- 19.Twetman S. Caries risk assessment in children: How accurate are we? Eur. Arch. Paediatr. Dent. 2016;17:27–32. doi: 10.1007/s40368-015-0195-7. [DOI] [PubMed] [Google Scholar]
- 20.National Health and Nutrition Examination Survey Data. 2013–2014. [(accessed on 5 November 2017)]; Available online: http://www.cdc.gov/nchs/nhanes.htm.
- 21.Zanella-Calzada L.A., Galván-Tejada C.E., Chávez-Lamas N.M., Gracia-Cortés M.D.C., Moreno-Báez A., Arceo-Olague J.G., Celaya-Padilla J.M., Galván-Tejada J.I., Gamboa-Rosales H. A Case–Control Study of Socio-Economic and Nutritional Characteristics as Determinants of Dental Caries in Different Age Groups, Considered as Public Health Problem: Data from NHANES 2013–2014. Int. J. Environ. Res. Public Health. 2018;15:957. doi: 10.3390/ijerph15050957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanella-Calzada L.A., Galván-Tejada C.E., Chávez-Lamas N.M., Galván-Tejada J.I., Celaya-Padilla J.M. Multivariate features selection from demographic and dietary descriptors as caries risk determinants in oral health diagnosis: Data from NHANES 2013–2014; Proceedings of the International Conference on Electronics, Communications and Computers (CONIELECOMP); Cholula, Mexico. 21–23 Feburary 2018; pp. 217–224. [Google Scholar]
- 23.Chávez-Lamas N.M., Zanella-Calzada L.A., Galván-Tejada C.E. An Analysis of Dietary and Demographic Data in Oral Health, Data from the National Health and Nutrition Examination Survey: A Preliminary Study. Adv. Comput. Netw. Appl. 2017;142:79–88. [Google Scholar]
- 24.Liaw A., Wiener M. Classication and regression by randomforest. R News. 2002;2:18–22. [Google Scholar]
- 25.Chollet F. Keras: Deep Learning Library for Theano and Tensorflow. [(accessed on 5 May 2018)];:145–151. Available online: https://keras.io/k.
- 26.Lomuscio A., Maganti L. An approach to reachability analysis for feed-forward relu neural networks. arXiv. 2017. 1706.07351
- 27.Carlini N., Wagner D. Towards evaluating the robustness of neural networks; Proceedings of the IEEE Symposium on Security and Privacy (SP); San Jose, CA, USA. 22–26 May 2017; pp. 39–57. [Google Scholar]
- 28.Kingma D.P., Ba J. Adam: A method for stochastic optimization. arXiv. 2014. 1412.6980
- 29.Haykin S.S. Neural Networks and Learning Machines. Volume 3. Pearson; Upper Saddle River, NJ, USA: 2009. pp. 1–46. [Google Scholar]
- 30.Chollet F. Deep Learning with Python. Volume 1. Manning Publications Co.; Greenwich, CT, USA: 2017. pp. 1–93. [Google Scholar]
- 31.Antona Cortés C. Bachelor’s Thesis. UAM, Departamento de Ingeniería Informática; Madrid, Spain: 2017. Herramientas Modernas en Redes Neuronales: La LibreríA Keras; pp. 21–38. [Google Scholar]
- 32.Helene Bischel S. Master’s Thesis. Universidad de Almería; Almería, Spain: 2015. El método de la EntropíA Cruzada. Algunas Aplicaciones; pp. 4–14. [Google Scholar]
- 33.Nye M., Saxe A. Are Efficient Deep Representations Learnable? UAM, Departamento de Ingeniería Informática; Madrid, Spain: 2017. pp. 1–4. [Google Scholar]
- 34.Lobo J.M., Jiménez-Valverde A., Real R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008;17:145–151. doi: 10.1111/j.1466-8238.2007.00358.x. [DOI] [Google Scholar]
- 35.Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 36.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 37.Rosa K.D., Shah R., Lin B., Gershman A., Frederking R. Topical clustering of tweets; Proceedings of the ACM SIGIR: SWSM; Beijing, China. 28 July 2011. [Google Scholar]
- 38.Jones E., Oliphant T., Peterson P. SciPy: Open Source Scientific Tools for Python. [(accessed on 5 May 2018)]; Available online: https://docs.scipy.org/doc/scipy.
- 39.McKinney W. Data Structures for Statistical Computing in Python; Proceedings of the 9th Python in Science Conference; Austin, TX, USA. 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- 40.Python Software Foundation Python Language Reference. Version 3.6 [(accessed on 5 June 2018)]; Available online: http://www.python.org.