Abstract
Gene expression can be silenced by proximity to heterochromatin blocks containing centromeric α-satellite DNA. This has been shown experimentally through cis-acting chromosome rearrangements resulting in linear genomic proximity, or through trans-acting changes resulting in intranuclear spatial proximity. Although it has long been been established that centromeres are nonrandomly distributed during interphase, little is known of what determines the three-dimensional organization of these silencing domains in the nucleus. Here, we propose a model that predicts the intranuclear positioning of centromeric heterochromatin for each individual chromosome. With the use of fluorescence in situ hybridization and confocal microscopy, we show that the distribution of centromeric α-satellite DNA in human lymphoid cells synchronized at G0/G1 is unique for most individual chromosomes. Regression analysis reveals a tight correlation between nuclear distribution of centromeric α-satellite DNA and the presence of G-dark bands in the corresponding chromosome. Centromeres surrounded by G-dark bands are preferentially located at the nuclear periphery, whereas centromeres of chromosomes with a lower content of G-dark bands tend to be localized at the nucleolus. Consistent with the model, a t(11; 14) translocation that removes G-dark bands from chromosome 11 causes a repositioning of the centromere, which becomes less frequently localized at the nuclear periphery and more frequently associated with the nucleolus. The data suggest that “chromosomal environment” plays a key role in the intranuclear organization of centromeric heterochromatin. Our model further predicts that facultative heterochromatinization of distinct genomic regions may contribute to cell-type specific patterns of centromere localization.
INTRODUCTION
How are genomes organized in the nucleus, and what is the role of genome organization on cellular functions? These fundamental questions in cell biology are attracting increased attention as the genomes of higher eukaryotes are being sequenced. Diverse models, ranging from highly random to highly organized, have been proposed for the organization of interphase chromatin (for reviews see Manuelidis, 1990; Haaf and Schmid, 1991; Cremer et al., 1993). Recent evidence suggests that interphase chromatin is organized in large loops, several megabase pair in size (Sachs et al., 1995; Yokota et al., 1995; Ostashevsky, 1998). While within each loop chromatin is randomly folded, specific loop-attachment sites may impose a constrained backbone structure (Yokota et al., 1995; Marshall et al., 1997; Ostashevsky, 1998, 2000; Cremer et al., 2000).
At present, it is well established that both mitotic chromosomes and interphase chromatin are composed of distinct functional domains (for recent reviews, see Cockell and Gasser, 1999; Belmont et al., 1999; Cremer et al., 2000). Each domain occupies a specific spatial position and replicates at a precise time during S phase. In metaphase chromosomes, the domains are identified as alternate transverse bands along the chromosome length (reviewed by Sumner, 1990). Shortly after mitosis, the chromosomal domains decondense and are repositioned in the nucleus, where they are designated as either euchromatin or heterochromatin (for reviews see Manuelidis, 1990; Haaf and Schmid, 1991; Craig and Bickmore, 1993). Heterochromatin represents chromatin that remains condensed throughout the cell cycle except during its replication, which occurs late in S phase. Heterochromatin includes constitutive heterochromatin, which is almost entirely composed of noncoding, tandemly repeated, satellite DNA sequences, and facultative heterochromatin, which mainly consists of potentially transcribable genes. Constitutively heterochromatic regions on metaphase chromosomes are designated C bands and are mostly localized at or adjacent to centromeric regions, whereas facultative heterochromatin resides in so-called G-dark bands (Craig and Bickmore, 1993). The G-dark bands comprise tissue-specific genes that are transcribed only in selected cell types (Manuelidis, 1990). Housekeeping genes, which are early replicating and actively transcribed in almost all cells, reside on G-light bands (also called R bands). During interphase, the vast majority of late replicating bands from most (if not all) chromosomes are localized at the nuclear periphery, with a smaller fraction present around the nucleolus or scattered in the nucleoplasm. In contrast, early replicating G-light bands appear to spread throughout the nuclear interior (Ferreira et al., 1997; Sadoni et al., 1999). Both intranuclear repositioning and replication timing of chromosomal domains are established in early G1 phase of the cell cycle, suggesting that spatial distribution within the nucleus is tightly coupled to the establishment of a replication timing program (Dimitrova and Gilbert, 1999). Given that during cellular differentiation there are changes in replication timing, which are often coupled to changes in transcriptional activity (see Dimitrova and Gilbert, 1999 and references therein), a key issue is whether spatial position within the nucleus plays an epigenetic regulatory role in gene expression.
Since long, heterochromatin is known to inactivate genes. For example, when a normally euchromatic gene is juxtaposed to heterochromatin by chromosome rearrangement, it can become transcriptionally silenced in a fraction of the cells. The mosaic expression of the transposed gene is called heterochromatic position-effect variegation, PEV (reviewed by Wakimoto, 1998). Although the classical explanation for PEV invokes the spreading of the heterochromatin state along the length of the chromosome into neighboring genes, there are cases of PEV for which a “trans-inactivation” mechanism has been proposed. A particularly well characterized example occurs when the insertion of a large block of heterochromatin into the coding sequence of the eye-color gene brown in Drosophila causes variegated inactivation of a normal copy of the gene present on a homologous chromosome. At defined stages of development, this insertion is shown to physically associate with centromeric chromatin on the same chromosome in a stochastic manner (Dernburg et al., 1996). Thus, in this case the association with heterochromatin responsible for variegation results from long-distance looping rather than from linear proximity along the chromosome. Additional examples of silencing trans-interactions between tissue-specific genes and centromeric heterochromatin were recently described in mammalian lymphoid cells (Brown et al., 1997, 1999).
Despite current evidence indicating that the intranuclear positioning of genomic loci relative to centromeric heterochromatin affects their transcriptional activity, very little is known about the principles governing the spatial distribution within the nucleus of centromeres per se. During interphase, centromeric heterochromatin is predominantly located either at the nuclear periphery or around the nucleolus (reviewed in Haaf and Schmid, 1991; Pluta et al., 1995). Is this distribution stochastic, or are there defined positional constraints for individual centromeres? Clearly, the centromeres of chromosomes that contain genes coding for rRNA (i.e., the nucleolar organizing region or NOR) are expected to associate with the nucleolus. However, the question remains for the centromeres of chromosomes without NOR. Are these randomly distributed between the nuclear periphery and the nucleolus? To address this question, we used fluorescence in situ hybridization, to differentially tag the centromeric heterochromatin of 15 human chromosomes, and confocal microscopy to determine their three-dimensional distribution pattern within the nucleus of quiescent lymphoid cells. Our results reveal that the positioning of centromeric heterochromatin relative to the nuclear envelope and the nucleolus tends to be specific for each chromosome. Most important, centromeric positioning can be predicted, taking into account the abundance of G-dark bands in the same chromosome. We propose a model for positioning of centromeres during interphase based on intra- and interchromosomal interactions between constitutive and facultative heterochromatin domains in the nucleus.
MATERIALS AND METHODS
Cells, In Situ Hybridization, and Confocal Microscopy
This study was performed using JVM-2 cells. This cell line was derived from a patient with B-prolymphocytic leukemia and has the following karyotype: 46,XX,-8, +t(3;8) (p12–13;q12), t(11;14)(q13;q32) (Melo et al., 1986). Cell culture and cell cycle synchronization were performed as previously described (Parreira et al., 1997).
Digoxigenin labeled α-satellite DNA probes specific to human chromosomes 1, 2, 4, 6, 7, 9, 10, 12, 16, 17, 18, 20, and 22 were purchased from Oncor (Gaithersburg, MD) and Boehringer Mannheim (Indianapolis, IN). Chromosome 11 painting probe was from Cambio (Cambridge, United Kingdom). A clone from the centromeric region of chromosome 15 (pHSr) was kindly provided by Prof. M. Nordenskjold (Karolinska Hospital, Stockholm, Sweden), and a clone for 28S ribosomal DNA (pBS28S; Rothblum et al., 1982) was a gift from Dr. L. Rothblum (Baylor College of Medicine, Houston, TX). Rabbit polyclonal antibodies against lamin B were kindly provided by Dr. S. Georgatos (University of Crete, Greece), and human anticentromere proteins autoimmune serum K55 was a gift from Dr. W. vanVenrooij (University of Nijmegen, The Netherlands).
The specificity of α-satellite DNA probes was monitored by in situ hybridization to metaphase spreads, as described (Carvalho et al., 1995). Interphase cells were adhered to poly-l-lisine coated coverslips and fixed/permeabilized with 3.7% formaldehyde and 0.5% Triton X-100 in HPEM (Ferreira et al., 1994). For in situ hybridization, cells were treated with 0.7% Triton X-100, in 0.1 M HCl on ice for 10 min, and denatured in 50% deionized formamide, 2x SSC, 50 mM phosphate buffer, pH 7.0 at 80°C for 30 min (adapted from O'Keefe et al., 1992). Hybridization was carried out at 37°C overnight, using a mix of digoxigenin-labeled α-satellite DNA probe (16 ng), biotin-labeled 28S rRNA probe (16 ng), and sheared sperm DNA (8 μg), in 8 μl of hybridization buffer (50% deionized formamide, 2x SSC, 10% dextran sulfate, 50 mM phosphates buffer, pH7.0). Posthybridization washes were in 50–62% formamide, 2X SSC, at 45°C. Biotin signals were detected with FITC-conjugated DN avidin (Vector Laboratories). Digoxigenin signals were detected with mouse antidigoxigenin antibody (Boehringer-Mannheim) and CY5-conjugated anti-mouse Ig (Jackson ImmunoResearch, West Grove, PA). Antilamin B antibodies were detected with a TexasRed-conjugated secondary Ig (Jackson ImmunoResearch).
Confocal microscopy was performed with a Zeiss (Oberkochen, Germany) laser scanning microscope LSM 410, with the use of excitation wavelengths of 488 nm (for FITC), 543 nm (for TexasRed), and 633 nm (for Cy5). A series of 12 equidistant optical sections were taken comprising each entire nucleus. The image size of an optical section was 191 x 191 pixels, corresponding on average to a lateral resolution of 0.13 μm/pixel. The axial distance between sections ranged from 750–950 nm.
Comparison with a Model of Uniform Random Chromatin Distribution
To calculate the expected distribution of the centromeres under a model of random uniform distribution, a computer program was developed using MATLAB5 (MathWorks, Natick, MA) and the Image Processing Toolbox v2.0. The algorithm used to estimate lamina contours was based on a method developed by Dias and Leitão (1996).
The program proceeds in two steps. First, it estimates the probability of observing centromeres in each nuclear region (i.e., lamina, nucleolus, or nucleoplasm nonadjacent to either lamina or nucleolus). This is done by identifying the contour of the lamina and the nucleoli for each nucleus and for each optical section. Then the contours are expanded to include all points within the average radius of centromere signals. Under a model of uniform chromatin distribution, the probability of observing a α-satellite signal in a region is given by the volume of that region relative to the volume of the nucleus. Thus, volume ratios were calculated for each nuclear region and were averaged over all nuclei stained with each chromosome specific probe.
The second step corresponds to the automatic classification of the position of the center of mass of the centromere signals according to the expanded contours detected by the program. This automatic classification is then compared with the expected distributions calculated in step 1.
To simplify the computational algorithms, both the automatic classification of the centromere positions and the volume ratio estimates used only the six equatorial sections of each nucleus (∼69% of the nuclear volume). All additional studies performed throughout the paper were based on a manual classification of the centromere position in 12 optical sections of each nucleus, thus covering the entire nuclear volume.
Statistics
Linear regressions and confidence intervals were calculated in SYSTAT 5 (SYSTAT, Evanston, IL). All remaining statistics were performed in Mathematica 3.0 (Wolfram Research, Champaign, IL).
For the linear regressions, centromere frequencies in each nuclear compartment were linearized with the use of the logistic transformation,
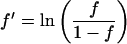 |
where f ′ stands for the linearized frequency and f for the original frequency.
RESULTS
Centromeric Heterochromatin Domains Occupy Defined Positions in the Nucleus
In this study we have used a mature B-cell line, which contains a diploid modal chromosomal number (JVM-2; Melo et al., 1986). All cells analyzed were synchronized at G0/G1, because it was previously shown that centromeric regions are repositioned in the nucleus during the cell cycle (Manuelidis, 1990; Ferguson and Ward, 1992; Shelby et al., 1996). For synchronization, the cultures were allowed to grow without medium change for 4–5 d, until more than 90% of the cells were in G0/G1, as monitored by incorporation of bromodeoxyuridine. Cellular viability at harvesting was systematically controlled and was always higher than 90%.
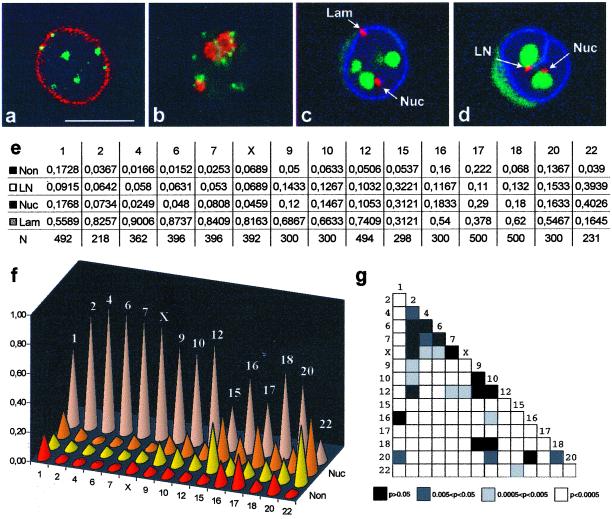
Centromeres in these quiescent human B-lymphoid cells are predominantly located near the nuclear periphery and around the nucleoli (Figure 1a, b). With the use of the confocal microscope and chromosome specific α-satellite DNA probes, the centromeres from 15 individual chromosomes were scored manually according to four categories (Lam, Nuc, LN, and Non). The α-satellite signals juxtaposed or superimposed to the lamina but not to nucleoli were classified in category Lam, whereas signals adjacent (i.e., juxtaposed or superimposed) to a nucleolus but not to the lamina were classified as Nuc (Figure 1c). The category LN includes signals simultaneously adjacent to both lamina and nucleolus (Figure 1d), and Non refers to signals that are nonadjacent to either lamina or nucleoli. Two to five independent hybridization experiments were performed for each chromosome probe and from each experiment 50 random nuclei were analyzed independently by two observers. No significant differences were detected between hybridization experiments (chi-square test, p > 0.05 for all chromosome probes).
Figure 1.
The intranuclear distribution of centromeric α-satellite DNA is chromosome specific. (a-d) JVM-2 cells were synchronized in G0/G1. Panels a and b depict single optical sections from cells double-labeled with a human autoimmune serum against centromeric proteins (green staining) and either a rabbit serum against lamin B (a, red staining) or a mouse mAb (72B9; Reimer et al., 1987) against the nucleolar protein fibrillarin (b, red staining). Panels c and d depict single optical sections from cells triple-labeled with a digoxigenin-coupled α-satellite probe for chromosome 1 (red staining), biotin-coupled probe for 28S rRNA (green staining), and rabbit antilamin B antibodies (blue staining). The α-satellite signals juxtaposed or superimposed on the lamina but not on nucleoli are classified in category Lam (c), whereas signals adjacent to a nucleolus but not to the lamina are classified as Nuc (c, d). The category LN (d) includes signals simultaneously adjacent to both lamina and nucleolus, and Non refers to signals that are nonadjacent to either lamina or nucleoli. Note that α-satellite DNA may be adjacent to the lamina even when it lies in the interior of the nucleus, due to invaginations of the nuclear envelope (d). Bar, 10 μm. Panel e shows the frequencies of α-satellite signals in each category; N is the number of signals analyzed. Panel f depicts graphically the distribution of α-satellite signals by the four categories. Panel g shows a chart measuring the similarity of α-satellite distribution for each pair of chromosomes. Each chart entry (small box) corresponds to the p-value of the chi-square test between the chromosome in that row and the chromosome in that column. Small p values, corresponding to statistically significant differences, are represented in light colors, and large p values, corresponding to similar centromere distributions, are represented in dark colors.
To test whether α-satellite sequences from homologous chromosomes tend to occupy the same compartment, the observed distributions were compared with what would be predicted if the homologous were distributed independently in the nucleus. With the exception of chromosomes 15 and 16, all chromosomes analyzed showed homologous independence (chi-square test, p > 0.05 for all chromosomes except 15 and 16). Chromosomes 20 and 22 were not included in the test because the hybridization signal was only detected in one chromosome from each pair (both in metaphase spreads and interphase nuclei). The inability to detect one of the two homologue centromeres from these chromosomes most likely reflects an array-length polymorphism, which is characteristic of human α-satellite DNA. In fact, human centromeres contain a highly variable number of α-satellite DNA monomeric units arranged in tandem arrays (Wevrick and Willard, 1989). This variability may explain why some centromeres are consistently seen with very bright signals, while others appear dimmer, and a few are not even visible.
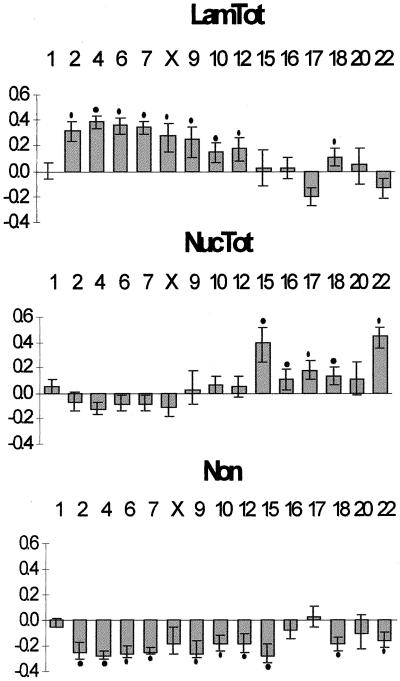
As depicted in Figure 1 (panels e, f), the distributions of the α-satellite sequences in the nucleus tend to be chromosome specific. To test if the observed differences were statistically significant, a chi-square test was applied pairwise between chromosomes (Figure 1g). Although a few chromosomes have similar centromere distributions, the vast majority shows significant differences (white and light gray boxes in Figure 1g). This argues against a model of random uniform chromatin distribution, which predicts a similar distribution for the α-satellite DNA from all chromosomes. According to such a model, the probability of any given portion of a chromosome to be located in a particular nuclear region depends only on the proportion of total nuclear volume occupied by that region. To understand how centromere positioning differs from a random model, we calculated the relative nuclear volume occupied by each compartment (see MATERIALS AND METHODS); the estimated ratios were then compared with the observed α-satellite DNA distribution (Figure 2). This comparison shows that centromeres from chromosomes 2, 4, 6, 7, X, 9, 10, 12, and 18 are nonrandomly localized at the nuclear periphery, whereas those from chromosomes 15, 16, 17, 18, and 22 associate nonrandomly with the nucleolus. Furthermore, a nucleoplasmatic localization of α-satellite DNA from most chromosomes is less frequent than expected from a random distribution.
Figure 2.
The distribution of centromeric α-satellite DNA in the nucleus is not random. Differences between the observed frequencies of α-satellite DNA distribution in the nucleus and the expected frequencies, according to a model of random uniform chromatin distribution. The categories Lam and LN were joined to produce LamTot, and categories Nuc and LN were merged into NucTot. In short, category LamTot corresponds to the totality of α-satellite signals in the lamina region, whereas NucTot represents the totality of α-satellite signals in the nucleolar region. The mean expected frequencies, according to a model of random uniform distribution, are LamTot, 52%; NucTot, 22%; and Non, 32% (see MATERIALS AND METHODS for details). Each bar in the graph represents the deviation of the observed from the expected frequencies. A positive bar means that the α-satellite signals are more frequently localized in that compartment than expected, whereas a negative bar means that the α-satellite signals are less frequent in that compartment than expected. The error bars represent the 95% confidence intervals (binomial distribution). A black dot indicates a significant difference between the observed and expected frequencies (one-sided binomial test, p < 0.05, with Bonferroni correction for multiple comparisons).
These results show that centromeric DNA is nonrandomly localized adjacent to either the lamina or the nucleolus, depending on constraints that are specific for each individual chromosome. This prompted us to search for chromosome-specific features that might correlate with the observed nonrandom distribution pattern.
Centromere Positioning within the Nucleus Correlates with the Presence of Dark G Bands in the Chromosome
A first inspection of Figure 1f suggests a relationship between chromosome size and α-satellite positioning. To investigate this apparent relationship, a linear regression analysis was performed, using chromosome physical length (Morton, 1991) as the predictor variable (variable Mbp in Figure 3b). Surprisingly, regression of the frequencies of α-satellite DNA association with the lamina or the nucleolus against chromosome length explains only a minor portion of the data (r2=33.6%, and r2=45.0%, respectively; Figure 3c). Furthermore, no apparent relationship exists between chromosome size and localization of α-satellite DNA in the nucleoplasm (r2=8.7%, p = 0.286; Figure 3c).
Figure 3.
 |
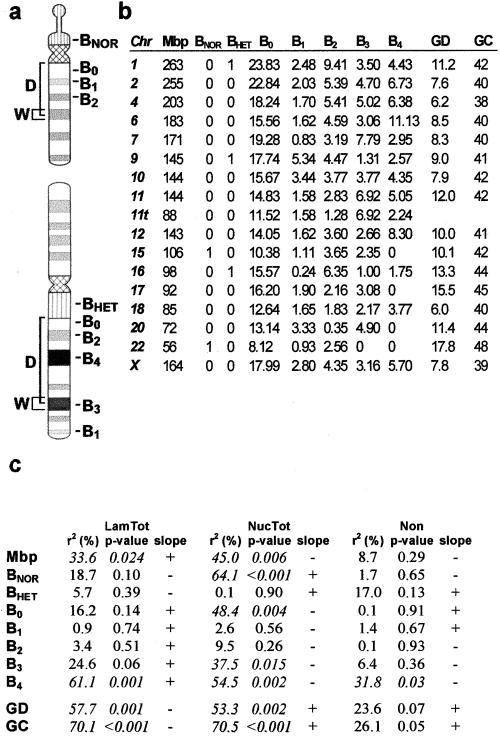
In face of these results, we next performed a systematic search of chromosomal-specific characteristics that might correlate better with the observed distributions of α-satellite sequences. The variables considered included presence/absence of a NOR (i.e., the Nucleolar Organizing Region, which contains the genes encoding for rRNA), presence/absence of pericentromeric constitutive heterochromatin (HET), and G-banding profile (Figure 3a, b).
To account for the G-banding pattern of each chromosome, we used the database reported by U. Francke (1994), which distinguishes five staining intensities at the 850 band resolution. These staining intensities were translated into a numerical scale from 0 (lightest bands) to 4 (darkest bands). According to the ISCN nomenclature, bands 0 correspond to G-light (or R) bands, and bands in the range 1–4 correspond to G-dark bands. We used five variables to describe the G-banding profile of each chromosome, one for each staining intensity. The value of each variable is the weighted sum of the widths of all bands of a given staining intensity (see Figure 3b and legend). The weights are the inverse of the square root of the genomic distance of the band center to the centromere. The rationale for this is that bands that are larger and/or closer to the centromere should exert more influence than smaller or more distant bands. We used the square root of the distance because of previous reports indicating a correlation between chromosomal genomic separation and mean-square interphase distance (Yokota et al., 1995: Ostashevsky, 1998).
The results of a linear regression analysis of α-satellite DNA distribution against each isolated variable are depicted in Figure 3c. The variable with higher explanatory power is B4. This corresponds to the darkest G bands. The second best overall explanatory variable is B3, representing less intensely stained G-dark bands. BNOR is highly significant for association of α-satellite DNA with the nucleolus. A stepwise regression confirmed the selection of variables B4, B3, and BNOR for a model of α-satellite DNA distribution. The regression coefficients for variables B3 and B4 are similar, suggesting that the two G-darkest band types have similar influences on the distribution of α-satellite DNA. Thus we use a single variable (B34=B3+B4) to account for the joint effect of these bands. Note that B34 is the weighted sum of the widths of all bands of staining intensities 3 or 4. A multiple regression with the use of simultaneously variables B34 and BNOR results in the after model,
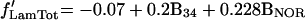 |
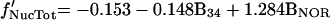 |
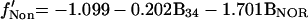 |
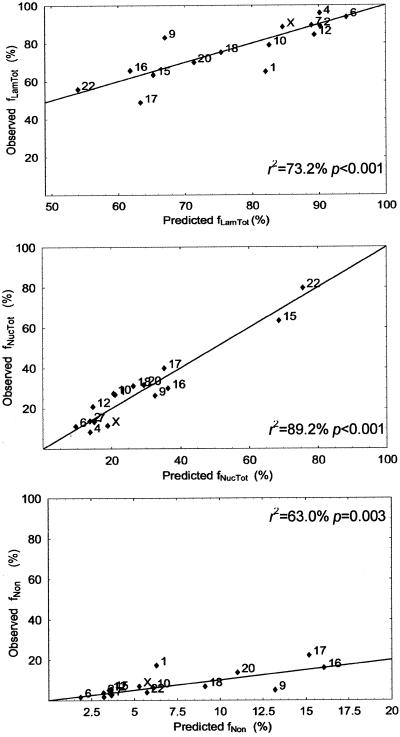
where f′ stands for the linearized frequency (see MATERIALS AND METHODS). This model shows an excellent fit to the data, being able to explain the majority of the variation of the centromere frequencies in the three nuclear compartments (r2LamTot = 73.2%, r2NucTot = 89.2%, r2Non = 63.0%).
After the recent publication of the human genome sequence (Venter et al., 2001), 2 additional parameters were introduced in our regression analysis: the average gene density and the base composition of each chromosome (Figure 3b). Because G-dark bands contain fewer genes and are less GC-rich than G-light bands (Craig and Bickmore, 1993), it is not surprising to observe a significant correlation between chromosome gene number, GC content, and centromere localization (Figure 3c). However, none of these variables alone adds any significant explanatory power to the above model (p > 0.025). The fit between observed frequencies and frequencies predicted with the use of this model is graphically depicted in Figure 4.
Figure 4.
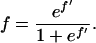 |
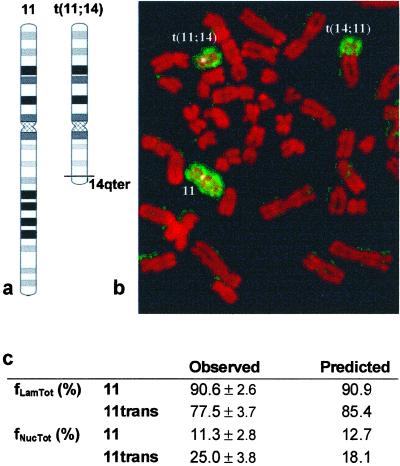
The picture that emerges from our analysis is that a centromere surrounded by G-dark bands will be preferentially located at the nuclear periphery (note the positive regression coefficient of B34 for f′LamTot), whereas a centromere that has no dark G bands in its vicinity will be mostly localized at the nucleolus (note the negative coefficient of B34 for f′NucTot). According to this model, if a chromosome looses G-dark bands (for example, as a consequence of a chromosomal translocation), its α-satellite DNA is expected to be less frequently localized at the nuclear periphery and more frequently associated with the nucleolus. To test this prediction, we compared the intranuclear distribution of α-satellite DNA from the normal chromosome 11 and the t(11; 14) (q13.3; q32.33) translocation present in JVM-2 cells. As depicted in Figure 5a, this translocation removes most of the bands of staining intensities 3 and 4 from the long arm of chromosome 11. The normal chromosome 11 and the two translocation products, t(11;14) and t(14; 11), are readily identified on metaphase chromosomes (Figure 5b). The α-satellite signals corresponding to the normal (11) and translocated t(11;14) chromosomes were scored for association with the lamina (fLamTot) and the nucleolus (fNucTot). Identification of the two signals during interphase was possible due to a fortuitous polymorphism that renders the centromere of the translocated chromosome much more intensely labeled than the centromere of the normal chromosome (see Wevrick and Willard, 1989). To investigate the possibility that length of α-satellite DNA arrays may influence centromere localization, we reanalyzed the distribution of chromosome 18 centromeres. In this chromosome pair, one centromere is consistently seen with a signal ∼twofold brighter than its homologue. Although the frequencies of association with the lamina and the nucleolus did not significantly differ between the two homologue centromeres, we found a 5% deviation of the brighter signal toward the nucleolus.
Figure 5.
A chromosomal translocation alters the positioning of centromeric α-satellite DNA. Scheme of the t(11;14)(q13;q32) translocation present in JVM-2 cells with resulting loss of dark G bands (a). Metaphase chromosomes (stained red) were hybridized with a painting probe for chromosome 11 (stained green). The normal chromosome 11 and the 2 translocation products, t(11;14) and t(14; 11), are indicated (b). The α-satellite signals corresponding to the normal (11) and translocated (t(11;14)) chromosomes were scored for association with the lamina (fLamTot), and the nucleolus (fNucTot) SD is indicated. Predicted distribution frequencies were calculated with the use of the model depicted in Figure 4.
The observed distribution of the centromeres of chromosomes 11 and t(11,14) agree with the predictions of the model (Figure 5c): centromeres of the truncated chromosome localize less frequently to the nuclear periphery and more frequently to the nucleolus than the normal counterpart. Again, we find a 7% deviation of the brighter signal toward the nucleolus, when compared with the expected value from the model.
Taken together, these data strongly suggest that the local banding environment of each chromosome plays an important role in the interphase organization of centromere heterochromatic domains. Clearly, additional, as yet unidentified factors contribute to this organization, as demonstrated by the incomplete fit between observed and predicted centromere distributions.
DISCUSSION
In this study we determined the three-dimensional distribution of centromeric heterochromatin from 15 human chromosomes in the nucleus of quiescent lymphoid cells. The results show that centromeric domains are not randomly localized in heterochromatic compartments of the nucleus. There is a defined probability for any particular centromeric domain being localized at either the nuclear periphery or surrounding the nucleolus, which differs significantly from chromosome to chromosome (Figure 1g). This implies that each individual chromosome constitutes a particular microenvironment in the interphase nucleus, which imposes specific positioning constraints on the centromere. In fact, there is a tight correlation between nuclear distribution of centromeric α-satellite DNA during interphase and presence of G-dark bands in the corresponding metaphase chromosome. Centromeres surrounded by G-dark bands are preferentially located at the nuclear periphery, whereas centromeres devoid of G-dark bands in their vicinity tend to be localized around the nucleolus. Moreover, a chromosomal translocation that removes G-dark bands from the chromosome causes a repositioning of α-satellite DNA, which becomes less frequently localized at the nuclear periphery and more frequently associated with the nucleolus.
What molecular mechanism(s) may be responsible for attracting some centromeres to the nuclear periphery and others to the nucleolus, depending on the banding context of each chromosome? At both the light and electron microscope, the nuclear periphery in most cell types is predominantly occupied by heterochromatin, which is closely associated with the nuclear lamina and the inner nuclear membrane (Paddy et al., 1990; Belmont et al., 1993; Marshall et al., 1996). Furthermore, late replicating bands from most (if not all) chromosomes localize to the nuclear periphery (Ferreira et al., 1997; Sadoni et al., 1999). Several lines of evidence indicate that chromatin anchorage to the nuclear envelope involves direct binding to both nuclear lamins and integral membrane proteins (see Pyrpasopoulou et al., 1996; Vlcek et al., 1999; Kourmouli et al., 2000). In particular, the lamin B receptor (LBR), a ubiquitous integral protein of the inner nuclear membrane, is thought to represent a major chromatin anchorage site at the nuclear envelope (Pyrpasopoulou et al., 1996). Interestingly, LBR decorates preferentially late replicating chromosomal bands but does not bind to centromeres (Pyrpasopoulou et al., 1996). This could imply that centromeres have no direct anchorage sites to the nuclear envelope.
From our study emerged a model that establishes chromatin context as a main constraint for centromere localization in the cell nucleus: we propose that facultative heterochromatin/late replicating chromatin regions corresponding to chromosomal G-dark bands act as nuclear envelope-attachment sites, and determine the spatial distribution of each chromosome's centromere in interphase. In our model we consider that the strength of attraction toward the periphery is directly proportional to the extent of heterochromatic domains (i.e., wider and darker G bands are stronger attractors) and inversely proportional to the distance from the centromere. Indeed, chromosomes with high attraction values estimated from G3 and G4 bands (see Figure 3b) have their centromeres consistently more peripheral than would be expected from a random distribution (see Figure 2). In contrast, the centromeres from chromosomes 15, 16, 20, and 22, which have low contributions from G3 and G4 bands, appear localized at the nuclear periphery with a frequency similar to what would be expected from a random distribution (see Figure 3b and 2).
In good agreement with our model, the whole territory of chromosomes rich in G-dark bands (thus poorer in genes) tend to adopt a more peripheral localization, whereas most gene-rich chromosomes appear concentrated at the center of the nucleus (Croft et al., 1999; Boyle et al., 2001). This strongly suggests that intrachromosomal patterns of chromatin configuration might influence the spatial positioning within the nucleus not only of the centromere but also of the entire chromosome.
As expected, we found the centromeres of chromosomes with NOR consistently associated with nucleoli. Intriguingly, also chromosomes 16, 17, and 18, which are devoid of NOR, have their centromeres more frequently associated with the nucleolus than expected for a random distribution (Figure 2). This implies a mechanism responsible for targeting particular centromeric α-satellite DNA sequences to the nucleolus. Although the molecular signals involved are entirely unknown, it is noteworthy that in the human genome all rDNA loci are embedded in constitutive heterochromatin. Most likely as a result of this linear proximity along the chromosome, nucleoli are always tightly associated with heterochromatin in the interphase nucleus. Heterochromatin has a strong tendency to participate in homologous associations, and this feature plays a key role in maintaining alignment between homologous chromosomes during mitosis and meiosis (Renauld and Gasser, 1997). The basis for this stickiness probably relies on the repeated nature of DNA sequence, which provides multiple binding sites for specific proteins capable of forming multimeric complexes. The ability of heterochromatin domains (including centromeres) to interact with other heterochromatin domains located either on the same or on a distinct chromosome has been documented. For example, insertion of satellite DNA at the eye-color gene brown in Drosophila causes this locus to associate with centromeric heterochromatin on the same chromosome (Dernburg et al., 1996). It is also well established that centromeres on distinct chromosomes may physically interact with each other in the nucleus, forming the so-called chromocenters (Hilliker and Appels, 1989; Manuelidis, 1990; Bartholdi, 1991; Alcobia et al., 2000). Thus, it is possible that centromeric heterochromatin regions on chromosomes devoid of NOR interact with centromeres and/or other heterochromatin domains on NOR-containing chromosomes and are therefore targeted to the nucleolus. This is also consistent with our observation that centromeres with longer α-satellite sequences tend to associate more frequently with the nucleolus.
In conclusion, our results suggest a model for positioning of centromeres during interphase based on competitive interactions between heterochromatin domains with other heterochromatin domains and with the nuclear envelope. The establishment of specific physical contacts between late replicating chromatin and the nuclear envelope could explain the dominant effect of G-dark bands on targeting adjacent centromeres to the nuclear periphery. On chromosomes where the mass attraction effect of dark G bands toward the nuclear envelope is weaker, the centromere would participate in homologous interactions with other heterochromatin domains in the nucleus. As these have a tendency to aggregate around nucleoli, centromeres that are not located at the nuclear periphery become most frequently associated with the nucleolus.
Finally, it is important to note that the centromeres of a few chromosomes (1, 9 and 17) are either more or less frequently associated with the nuclear periphery than would be expected taking into account the G-dark band content of those chromosomes (Figure 4). What may be contributing for this deviation? It is well established that in cells of different lineage, a G-dark domain can acquire G-light replication and transcriptional characteristics (see Manuelidis, 1990; Craig and Bickmore, 1993). Since early replicating loci do not associate with the nuclear envelope (Ferreira et al., 1997; Sadoni et al., 1999), the tissue-specific transcriptional activation of certain G-dark chromatin regions would result in weaker attraction of the corresponding centromere to the nuclear envelope (i.e., the centromere would appear less frequently located at the nuclear periphery than predicted). Furthermore, the distribution of constitutive heterochromatin in the nucleus is also known to be cell-type specific (Manuelidis, 1990; Haaf and Schmid, 1991). Given its potential to form interactions with centromeric heterochromatin, it is possible that constitutive heterochromatin domains (which are variably located in the nucleus) may contribute to centromere attraction and therefore may distort the predicted association of some centromeres with the nuclear periphery. Thus, a corollary from the proposed model is that different cell types should show specific deviations from the predicted distribution, depending on tissue-specific patterns of gene expression and chromatin organization. Clearly, further studies addressing the spatial distribution of centromeres in the nucleus of other human cell types are needed to validate the model.
ACKNOWLEDGMENTS
We thank Prof. David-Ferreira for encouragement. We further wish to acknowledge Prof. Bioucas Dias for help with image processing, Prof. Dinis Pestana for help with statistical analysis, and Mrs. Helena Pina, Maria do Céu Santos, Maria João Gracias, and Célia Mestre for technical support. We are also grateful to our colleagues José Cordeiro and Nuno Gracias. This study was supported by Fundação para a Ciência e Tecnologia/PRAXIS XXI.
REFERENCES
- Alcobia I, Dilão R, Parreira L. Spatial associations of centromeres in the nucleus of hematopoietic cells. evidence for cell-type–specific organizational patterns. Blood. 2000;95:1608–1615. [PubMed] [Google Scholar]
- Bartholdi MF. Nuclear distribution of centromeres during the cell cycle of human diploid fibroblasts. J Cell Sci. 1991;99:255–263. doi: 10.1242/jcs.99.2.255. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Dietzel S, Nye AV, Strukov YG, Tumber T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal, and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Carvalho C, Telhada M, Carmo-Fonseca M, Parreira L. In situ visualization of immunoglobulin genes in normal and malignant lymphoid cells. J Clin Pathol Mol Pathol. 1995;48:M158–M164. doi: 10.1136/mp.48.3.m158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Gasser SM. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- Craig JM, Bickmore WA. Chromosome bands –flavours to savour. BioEssays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathieu U, Jauch A, Emmerich P, Scherthan H, Ried T, Cremer C, Lichter P. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kreth G, Koester H, Fink RHA, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Chromosome territories, interchromatin domain compartment, and nuclear matrix. an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;12:179–212. [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Dias J, Leitão J. Wall position and thickness estimation from sequences of echocardiogram images. IEEE Trans Med Imaging. 1996;15:25–38. doi: 10.1109/42.481438. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Ferguson M, Ward DC. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Paolella G, Ramos C, Lamond A. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke U. Digitized and differentially shaded human chromosome ideograms for genomic applications. Cytogenet Cell Genet. 1994;65:206–219. doi: 10.1159/000133633. [DOI] [PubMed] [Google Scholar]
- Haaf T, Schmid M. Chromosome topology in mammalian interphase nuclei. Exp Cell Res. 1991;192:325–332. doi: 10.1016/0014-4827(91)90048-y. [DOI] [PubMed] [Google Scholar]
- Hilliker AJ, Appels R. The arrangement of interphase chromosomes: structural and functional aspects. Exp Cell Res. 1989;185:297. doi: 10.1016/0014-4827(89)90301-7. [DOI] [PubMed] [Google Scholar]
- Kourmouli N, Theodoropoulos PA, Dialynas G, Bakou A, Politou AS, Cowell IG, Singh PB, Georgatos SD. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J. 2000;19:6558–6568. doi: 10.1093/emboj/19.23.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:730–739. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Melo JV, Brito-Babapulle V, Foroni L, Robinson DSF, Luzzatto L, Catovsky D. Two new cell lines from B-prolymphocytic leukemia: characterization by morphology, immunological markers, karyotype and Ig gene rearrangement. Int J Cancer. 1986;38:531–538. doi: 10.1002/ijc.2910380413. [DOI] [PubMed] [Google Scholar]
- Morton NE. Parameters of the human genome. Proc Natl Acad Sci USA. 1991;88:7474–7476. doi: 10.1073/pnas.88.17.7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostashevsky J. A polymer model for the structural organization of chromatin loops and minibands in interphase chromosomes. Mol Biol Cell. 1998;9:3031–3040. doi: 10.1091/mbc.9.11.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostashevsky J. Higher-order structure of interphase chromosomes, and radiation-induced chromosomal exchange aberrations. Int J Radiat Biol. 2000;76:1179–1187. doi: 10.1080/09553000050134410. [DOI] [PubMed] [Google Scholar]
- Paddy MR, Belmont AS, Saumweber H, Agard DA, Sedat JW. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62:89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- Parreira L, Telhada M, Ramos C, Hernandez R, Neves H, Carmo-Fonseca M. The spatial distribution of human immunoglobulin genes within the nucleus: evidence for gene topography independent of cell type and transcriptional activity. Hum Genet. 1997;100:588–594. doi: 10.1007/s004390050558. [DOI] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos S. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe NA, Tan EM. Monoclonal antibody from (New Zealand Black x New Zealand White) F1 mouse and some human scleroderma sera target a Mr 34000 nucleolar protein of the U3-ribonucleoprotein particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Renauld H, Gasser S. Heterochromatin: a meiotic matchmaker? Trends Cell Biol. 1997;7:201–205. doi: 10.1016/S0962-8924(97)01034-9. [DOI] [PubMed] [Google Scholar]
- Rothblum LI, Parker DL, Cassidy B. Isolation and characterization of rat ribosomal DNA clones. Gene. 1982;17:75–77. doi: 10.1016/0378-1119(82)90102-0. [DOI] [PubMed] [Google Scholar]
- Sachs RK, Van Den Engh G, Trask B, Yokota H, Hearst JE. A random-walk/giant-loop model for interphase chromosomes. Proc Natl Acad Sci USA. 1995;92:2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, Zink D. Nuclear organization of mammalian genomes: polar chromosome territories build up functionally distinct higher order compartments. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of a-satellite DNA domains visualized in situ in living human cells. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner AT. Chromosome banding. London, UK: Unwin Hyman; 1990. pp. 1–434. [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vlcek S, Just H, Dechat T, Foisner R. Functional diversity of LAP2α and LAP2β in postmitotic chromosome association is caused by an α-specific nuclear targeting domain. EMBO J. 1999;18:6370–6384. doi: 10.1093/emboj/18.22.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakota H, Engh G, Hearst J, Sachs RK, Trask BJ. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J Cell Biol. 1995;130:1239–1249. doi: 10.1083/jcb.130.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota H, Singer MJ, van den Engh GJ, Trask BJ. Regional differences in the compaction of chromatin in human G0/G1 interphase nuclei. Chromosome Res. 1997;5:157–166. doi: 10.1023/a:1018438729203. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Wevrick R, Willard HF. Long-range organization of tandem arrays of a-satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci USA. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]