Summary
Brain cells communicate with one another via local and long-range synaptic connections. Structural connectivity is the foundation for neural function. Brain-wide connectivity can be described at macroscopic, mesoscopic and microscopic levels. The mesoscale connectome represents connections between neuronal types across different brain regions. Building a mesoscale connectome requires a detailed understanding of the cell type composition of different brain regions and the patterns of inputs and outputs that each of these cell types receives and forms, respectively. In this review, I discuss historical and contemporary tracing techniques in both anterograde and retrograde directions to map the input and output connections at population and individual cell levels, as well as imaging and network analysis approaches to build mesoscale connectomes for mammalian brains.
The mammalian brain – including the human brain as ‘the most complex piece of organized matter in the known universe’ (http://www.alleninstitute.org/what-we-do/brain-science/), has millions to billions of neurons with trillions of synapses connecting them. One way the nervous system manages complexity is by grouping neurons into canonical ‘types’. Neurons can be grouped into an estimate of thousands of types, each located in specific brain regions. Brain regions and neuronal types are organized into four major functional systems: sensory, motor, cognitive and state [1]. Multiple subsystems exist within each major system, forming specific circuits and subserving specific functions. To understand how diverse forms of function emerge from the brain’s structural architecture, it is essential to identify the cell type components of brain circuits, determine the structural connections between those components, measure the physiological signals that pass between synaptically-coupled neurons, and precisely manipulate specific components to investigate their functional roles [2,3].
Brain connectivity can be described at at least three levels [3–5]. It is generally considered that the macroscale connectome describes inter-areal connections that can be inferred from fiber tracts using techniques such as diffusion tensor imaging (DTI). DTI is especially useful for generating macroscale connectomes from living human brains, where more invasive approaches cannot be applied [6]. The mesoscale connectome describes connections at the cellular level, between neuronal types across different brain regions. The microscale and/or nanoscale connectomes describe connections between individual neurons at the synapse level.
Microscale connectomes often rely on electron microscopy (EM) to provide the clearest evidence about the presence and location of synapses. Although whole-brain EM connectomics has been done in C. elegans [7–9], and is ongoing in other species such as fruit flies [10–12] or zebrafish larvae [13], the extremely high resolution as well as other experimental and computational requirements make it prohibitive to obtain an EM-based microscale connectome for a mammalian brain in the foreseeable future, and currently it is confined to the investigation of local circuits [14–18].
Mesoscale connectomes can be built using a variety of anatomical tracing approaches reviewed below at the brain-wide level. The cell type-based connections can be amenable to functional probing using the same cell type-targeting genetic strategies driving functional monitoring or manipulating tools. Mesoscale connectomes also bridge information collected at macroscale or microscale connectomic levels. Thus, mesoscale connectomics allows multiscale and structural-functional integration, and has been widely employed in neural circuit studies. In the text that follows I discuss techniques that enable important new insights at this level of abstraction. Many of the powerful genetically-based techniques have been developed in and most applicable to the mouse brain. It is our hope that at least some of these methods can be extended in the near future to other, especially primate brains.
Traditional tracers
Historically, inter-areal connections are mapped using a variety of tract tracers. These tract tracers can be made of chemical compounds, glycoproteins, radioactively tagged amino acids or fluorescently conjugated beads that can be transported along axon fibers in either directions (see [19,20] for a comprehensive overview). Commonly-used tracers that move principally in the anterograde direction, i.e., from the soma to the tips of the axons, include biotinylated dextran amine (BDA), Fluoro-ruby (FR) and Phaseolus vulgaris-leucoagglutinin (PHA-L). Commonly used retrograde tracers, i.e., tracers principally propagate from the axon terminals of a neuron back towards its soma and/or dendrites, include cholera toxin B fragment (CTB), Fluoro-gold (FG), and retrobeads. Wheat germ agglutinin (WGA) has been used as a trans-synaptic tracer that can be genetically targeted and can cross synapses in both retrograde (i.e., from postsynaptic to presynaptic side) and anterograde (i.e., from presynaptic to postsynaptic side) directions. Although not selective for neuronal types, these traditional tracers are still widely used in circuit studies to label axonal projections from specific regions (in the anterograde direction), or specific neuronal populations defined by projection targets (in the retrograde direction), often in conjunction with the use of newer generation of recombinant virus-based tracers that are more amenable to cell type specific targeting in genetic model organisms such as the mouse.
Virus-mediated anterograde and retrograde tracing
Harnessing the power of nature, a plethora of neurotropic viruses have been modified to support a wide range of neuroscience applications including neuroanatomical connectivity mapping. Readers should refer to an excellent recent review [19] that more systematically describes all the major types of recombinant viral vectors for neuroanatomical studies. Here I focus on the most recent technical advances in viral tools for mesoscale connectivity mapping.
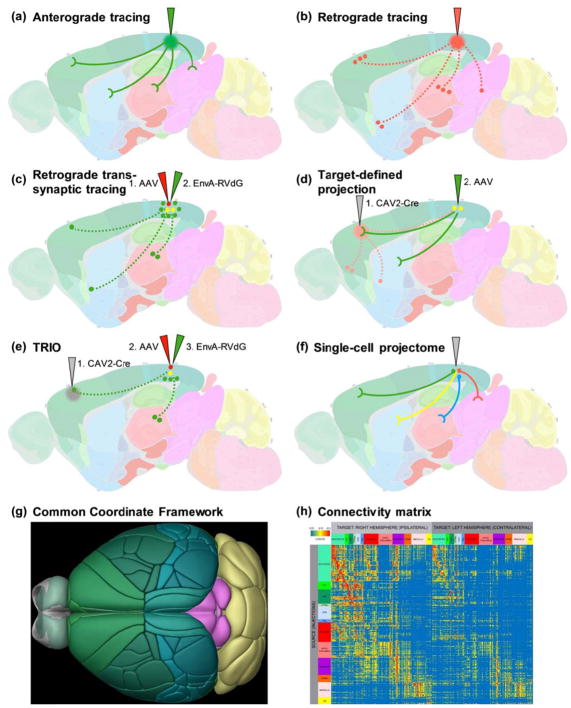
In anterograde tracing, cells at the viral injection site are directly infected by the virus and express genes that code for fluorescent proteins, which travel through the entire extent of axon fibers revealing the target areas to which the infected neurons project (Fig. 1a). In retrograde tracing, axons terminals at the viral injection site take-up the virus; the virus is transported back to the soma from the axon terminals and then leads to expression of fluorescent proteins in the infected cell (Fig. 1b). The first virus widely used for anterograde tracing was adeno-associated virus (AAV). It can express fluorescent proteins in a Cre and/or other recombinase (e.g. Flp) dependent manner using double-inverted recombination site cassettes (called DIO or FLEx) [21–23]. In addition, a suite of Brainbow AAVs have been generated that could enable simultaneous multi-color labeling and tracing [24].
Figure 1.
Representative approaches for mesoscale connectomics. (a) In anterograde tracing, traditional or viral anterograde tracer is injected in the source region of interest and tracer-labeled long-range axonal projections and terminals in other parts of the brain are mapped. (b) In retrograde tracing, traditional or viral retrograde tracer is injected in the target region of interest and tracer-labeled somata or nuclei of input neurons in other parts of the brain are mapped. Dotted lines indicate that axon fibers of input neurons may or may not be labeled by the tracer. (c) In the most common scenario of monosynaptic, retrograde trans-synaptic tracing, AAV helper virus(es) (red) expressing TVA and RV-G proteins under Cre control is first injected into the target region of interest of a Cre-driver-containing animal, and then EnvA-pseudotyped, G-deleted rabies virus (RVdG) (green) is injected into the same region. Only neurons infected with both AAV and rabies virus (called starter cells, which are turned from red to yellow) will produce rabies viral particles that will be transported across synapses and label the somata of presynaptic input neurons (green cells) both locally and in the long distance. (d) In target-defined projection mapping, CAV2-Cre (or another retrograde virus, gray) is first injected into a target region of interest, and it will be taken up retrogradely by neurons projecting to the target region (pale red paths and cells); then a Cre-dependent AAV is injected into one of the source regions of interest, and it will label only the neurons (yellow) projecting to the initial target region as they contain CAV2-derived Cre. The axonal projections of these target-defined neurons will then be visualized (green fibers). Pale red indicates that CAV2-Cre infected cells may or may not be visualized, depending on whether the animal contains a Cre-reporter or not. (e) In TRIO (tracing the relationship between input and output), two steps of retrograde tracing are involved. CAV2-Cre (or another retrograde virus, gray) is first injected into a target region of interest, and it will be taken up retrogradely by neurons projecting to the target region. Then a (set of) Cre-dependent AAV helper virus(es) is injected into one of the source regions of interest, to express TVA and RV-G proteins only in the neurons (red) projecting to the initial target region as they contain CAV2-derived Cre. Finally, EnvA-pseudotyped, G-deleted rabies virus (RVdG) (green) is injected into the same source region. Only neurons infected with CAV2-Cre, AAV and rabies virus (note these are target-defined starter cells, which are turned from red to yellow) will produce rabies viral particles that will be transported across synapses and label the somata of presynaptic input neurons (green cells) both locally and in the long distance. (f) In single-cell projectome, individual neurons (green, red and blue) in the source region of interest are sparsely and strongly labeled by transgenic or viral expression of a fluorescent protein, and their full extent of dendritic and axonal arbors are imaged and reconstructed to reveal each neuron’s unique set of projection targets (green, red and blue fibers). Sometimes two neurons with otherwise different targets may also share one or more common target(s) (as indicated by the yellow fiber). Alternatively, individual neurons can also be densely labeled by barcoded anterograde viruses, and their individual projection targets resolved through the barcodes. (g) The mouse brain Common Coordinate Framework (CCF) is a digital 3-D reference atlas for image data registration and anatomical delineation. (h) Segmented and quantified mesoscale connectomic datasets can be used to generate connectivity matrix for neural network analysis (adapted from Ref [53]).
AAVs have different serotypes (capsid proteins) with different degrees of neurotropism that can be utilized for cell-specific and direction-specific targeting. For example, AAVs have low rates of retrograde transport; through directed evolution, a new capsid variant rAAV2-retro was developed that enables efficient retrograde labeling [25*]. Similarly, using a cell type-specific capsid selection method, AAV variants were developed that can be delivered systemically and infect majority of neuronal types throughout the brain (AAV-PHP.B and AAV-PHP.eB, with the latter having enhanced efficiency), or infect preferentially peripheral neurons (AAV-PHP.S) [26*,27*].
Other viruses that have been used in circuit tracing studies include sindbis virus, canine adenovirus-2 (CAV-2), herpes simplex virus-1 (HSV-1), and rabies virus. Sindbis virus allows robust labeling of individual neurons and their anterograde axonal projections [28,29]. CAV-2 displays potent retrograde transport but modest transgene expression level, and CAV-2 vectors carrying Cre or Flp have been used in retrograde tracing studies [30–32*]. Replication-incompetent HSV-1 and rabies virus have also been used as retrograde tracers and can deliver strong transgene expression to infected neurons [19].
While AAV and CAV-2 facilitate relatively safe and stable long-term expression of proteins in host cells, sindbis virus, HSV-1 and rabies virus display cytotoxicity within days or weeks after infection. Efforts have been made to engineer new viral vectors with reduced toxicity, such as the self-inactivating rabies virus [33*] and the double-deletion mutant rabies virus RVΔGL [34*]. Both versions lead to diminished transgene expression, and thus their best use is to express Cre (or other) recombinase which doesn’t need to be abundant to work effectively in combination with Cre-dependent transgenic lines or viral vectors in retrograde tracing. It has also been observed that different retrograde viruses, e.g. rAAV2-retro, CAV-2 and RVΔGL, have differential neurotropism and do not label all input cell types equally [34*]. Thus, it will be important to compare tracing results from different retrograde viruses in order to obtain a more comprehensive picture.
Trans-synaptic tracing
The anterograde and retrograde tracing techniques described thus far enable one to map connections between cell populations and regions. To establish connections between input and recipient cell populations requires trans-synaptic (or trans-neuronal) tracing, in which the viral tracer is transported across synapses in either the anterograde or retrograde direction and label both pre- and post-synaptic neurons simultaneously. Monosynaptic trans-synaptic tracing, i.e., the viral tracer can only jump across one step of synaptic connections, is critical to reveal directly connected cell populations.
Both rabies and pseudorabies virus (PRV) have been used for polysynaptic retrograde trans-synaptic tracing. Monosynaptic, retrograde trans-synaptic tracing by EnvA-pseudotyped G-deleted rabies (RVdG) is a powerful tool that has been widely used to map interconnected neurons in a variety of circuits throughout the nervous system (see [35] for an overview) (Fig. 1c). For example, as a technical tour-de-force, rabies tracing from a single starter cell has revealed exquisite details of anatomical and functional correlations among interconnected neurons in microcircuits [36–38].
Recent efforts have focused on reducing cytotoxicity and improving trans-synaptic efficiency of the rabies virus. Replacing the glycoprotein of the original SAD-B19 strain with an optimized version oG [39*], or using the G-deleted CVS-N2c strain of rabies [40*], improves efficiency of trans-synaptic spread and, in the latter case, prolongs cell health. As mentioned above, self-inactivating [33*] or double-deletion mutant [34*] rabies virus can also reduce cytotoxicity; note, however, that these forms of rabies have yet to be modified to act in a trans-synaptic manner.
In the anterograde direction, the H129 strain of HSV-1 has been used to label neurons that are postsynaptic to the infected cells, i.e., as a polysynaptic anterograde trans-synaptic tracer. A Cre-dependent, polysynaptic, anterograde trans-synaptic H129 variant was developed by replacing the endogenous viral TK gene with a floxed TK expression cassette [41]. A monosynaptic, anterograde trans-synaptic version of H129 was recently developed by also deleting the endogenous TK gene and supplementing it from a helper virus [42*]. The vesicular stomatitis virus (VSV) has been engineered to express either the rabies virus glycoprotein (RV-G) or its own glycoprotein (VSV-G), which can then trans-synaptically label neuronal circuits in either retrograde or anterograde direction, respectively [43]. A drawback of these approaches is that the rapid onset of toxicity in both H129 and VSV viruses has limited their widespread use. One potential way to avoid, or overcome, this drawback is to use versions of Cre-expressing AAV1 or AAV9 that exhibit anterograde trans-synaptic spread properties [44].
Viral expression of cross-synaptic molecular tools provides another means to visualize synaptic connections between defined cell populations. The pre- and postsynaptic components of the mammalian version of GRASP (GFP-reconstitution across synaptic partners), mGRASP [45], can be expressed in two different neuronal populations and synapses between the two populations can be revealed through reconstituted GFP. Though currently only working in fruit flies, trans-Tango is a molecular anterograde trans-synaptic mapping system in which presynaptic ligand induced postsynaptic receptor activation is converted into reporter expression in postsynaptic cells through site-specific proteolysis [46*].
Combinatorial and functional circuit mapping
The array of anterograde, retrograde and trans-synaptic viral tools can be combined, and sometimes coupled with transgenic lines, to enable even more sophisticated circuit mapping approaches (for a comprehensive overview see [47]). For example, combining a retrograde tracer (e.g., CAV2-Cre) with an anterograde tracer (e.g., Cre-dependent AAV) can assess target-defined projection specificity [48] (Fig. 1d). The ‘tracing the relationship between input and output’ (TRIO) method combines CAV2-Cre with Cre-dependent rabies tracing, or CAV2-FLEX-Flp with Flp-dependent rabies (cTRIO), and enables trans-synaptic input tracing from specific subsets of neurons based on their projection and/or cell type [32*] (Fig. 1e). In a similar way, double retrograde and single-cell trans-synaptic tracing from visual cortex to dorsolateral geniculate nucleus to retina allowed examination of information encoding in directly connected circuit pathway across three distinct structures [49*].
Combining tracing with in vivo functional imaging (e.g., wide-field, one- or two-photon calcium imaging, fiber photometry, or fMRI) allows mapping of circuit dynamics in behaving animals. Circuit components activated during behavior can also be mapped through immediate early gene (IEG) activation [50]. Optogenetic or chemogenetic perturbation can further probe behavioral or functional consequences of activation/inactivation of the specific circuit [47]. Optogenetic activation of the axon terminals of presynaptic neurons while recording from potential postsynaptic neurons in the target area, in methods like subcellular channelrhodopsin-assisted circuit mapping (sCRACM) [51], allows establishment of pre- and postsynaptic connections and further investigation of the strengths and other functional properties of such connections.
Imaging, image registration and data analysis
To facilitate standardized data generation and comparison, and to enable the creation of brain-wide connectomic maps, it is important to establish standardized imaging platforms. Given that mesoscale connectomes are largely fluorescence-based datasets, high-throughput fluorescence imaging platforms have been deployed, including section-based epifluorescence slide scanners and whole-brain serial two-photon (STP) tomography [52–55]. To enable more efficient whole-brain imaging, many tissue clearing methods have been developed (see [56] for an overview), including CLARITY [57], expansion microscopy (ExM) [58], iDISCO [59], etc. Related imaging techniques are also being developed, such as long working-distance objectives with high refractive index matching the clearing agent. Light-sheet microscopy is well suited for imaging large-volume cleared tissues at a much faster rate, albeit with lower spatial resolution, potentially enabling processing of much larger number of samples [60*,61]. Some clearing methods, e.g., iDISCO [59], CLARITY [62], ExM [63], and ExFISH [64], are compatible with antibody or mRNA labeling, further enriching data content and potentially allowing determination of cell type identity of mapped neurons.
The mammalian brain can be parcellated into hundreds of distinct anatomical regions, each expected to contain a unique set of cell types. To generate a meaningful connectivity map, the anatomical location of labeled cells and their inputs/outputs need to be accurately determined. Tissues with anatomical context, e.g. having counterstaining or intrinsic background fluorescence, are more readily amenable for anatomical annotation, whereas cleared tissues present a much greater challenge. To facilitate efficient annotation of the large numbers of datasets needed for comprehensive coverage, and especially to facilitate comparison across these datasets, co-registration of all the individual datasets into a common reference space is highly desirable. Commonly used mouse or rat reference atlases include the Paxinos’ [65,66] and the Allen Reference Atlas [67]. By co-aligning 1,675 individual STP tomography whole-brain datasets from the Allen Mouse Brain Connectivity Atlas, a mesoscale connectome for the mouse brain [53], an average template brain was generated, from which a new 3-D reference atlas, a Common Coordinate Framework (CCF), was built (Fig. 1g). The current version, CCF v3, consists of 43 cortical areas with their associated layers, 330 subcortical gray matter areas, 82 fiber tracts, and 8 ventricle and associated structure volumes, all delineated natively in 3-D (http://atlas.brain-map.org/). Different labs have developed methods to register and map a variety of whole-brain multi-plane datasets into this CCF, including aMAP [68], ClearMap [60*], qBrain [69*], and WholeBrain [70*]. These approaches dramatically facilitate qualitative and quantitative comparison across circuit mapping datasets for the adult mouse.
Systematic analysis of the large-scale, mesoscale connectomics datasets requires signal segmentation in addition to registration with parcellated reference atlases described above. A raw connectivity matrix between source and target areas can then be generated from the quantitative signal strengths (Fig. 1h). Statistical or graph theory-based network analysis can be performed to identify network characteristics and organization, such as node degree, clustering, modularity and centrality [71]. Computational models can be built from structural connectivity to predict function and generate hypotheses [72,73].
Cell type characterization and development of cell type targeting tools
Mesoscale connectomics requires a solid foundation of cell types across all brain regions. However, our understanding of cell types in the brain remains far from complete. Large-scale efforts are underway to systematically characterize brain cell types in the mouse at transcriptomic, morphological and physiological levels [74]. Integrating these multi-dimensional properties of individual cells is essential for deriving a unified cell type taxonomy [75].
Single-cell transcriptomic profiling is powerful as it provides the genetic identity of cell types along with marker genes [76]. Targeting these marker genes to make driver lines provides genetic access to novel cell types, which can then be used for connectivity mapping and other functional studies. Single-cell epigenomic profiling [77,78] promises to identify cell type specific enhancer elements, or more broadly cis-regulatory modules (CRMs), useful for targeting specific cell types. If these CRMs can be utilized in viral vectors, as shown in the case of GABAergic neuron specific mDlx enhancer [79], cell type specific targeting will become much more efficient and can also be applied to other genetically less accessible species.
Transcriptomic profiles can also be used for cell type identification in mesoscale connectomic studies through spatial transcriptomic approaches [80], such as multiplexed fluorescence in situ hybridization (FISH). This is particularly relevant for retrograde and trans-synaptic tracing studies where relevant input or output cells are only revealed after tracing; post hoc multiplexed FISH would then be useful to obtain their cell type identities.
Single cell projectome and barcode connectomics
Connectivity itself is also likely to be an important criterion for definition of a cell type. At the single cell level, the full extent of the dendritic and axonal morphology of a neuron can be considered a partial surrogate for its connectivity, as it contains substantial, though not all, input/output connectional information. In fact, it was the neuronal morphologies that Cajal used to define a ‘neuron’ and the polarity of its information flow, and to discover neuronal diversity [81]. However, our understanding of single neuron morphologies, especially for projection neurons with their axons extending much beyond the local area their somata reside in, is still extremely limited, hindering our progress in understanding cell types. Thus, it is critical to incorporate into mesoscale connectomes the single cell morphologies or single cell axonal projections throughout the entire brain, which can be properly named as a single cell projectome (Fig. 1f).
Recent development of high-throughput fluorescence microscopy such as the MouseLight [82*] and the fMOST [83] systems allows imaging of whole mouse brains at sub-micron resolutions. The imaging technologies combined with genetic sparse labeling methods can generate high-resolution whole brain datasets that enable the reconstruction of full morphologies of single neurons, already revealing rich diversity. A major challenge now is to develop automatic or semi-automatic reconstruction algorithms that will substantially reduce the time-consuming morphology reconstruction efforts. Obtaining a large-scale single cell projectome dataset that spans all regions of the brain will transform our understanding of neuronal diversity and connectional specificity.
An alternative approach is barcode connectomics [84], in which barcoded DNA sequences are incorporated into viral vectors for projection/connectivity mapping. In the MAPseq method [85*], using a barcoded sindbis virus library, each virally infected neuron expresses a unique barcode and each neuron’s projection pattern can be distinguished from that of other neurons through sequencing of the barcodes in micro-dissected brain tissues. Since thousands of neurons can be uniquely barcoded simultaneously within the same brain, this approach can potentially map the axonal projection patterns from a large number of neurons simultaneously in a highly efficient manner.
Outlook
Recent technological advances in virus engineering, imaging and genomics are rapidly transforming the landscape of connectomics and enabling us to conduct comprehensive and multi-faceted structure-function studies of brain-wide neuronal networks. I expect that mesoscale connectomics will be dramatically accelerated by our increased understanding of cell types in the brain, and through the mutually synergistic interaction between cell type characterization and connectomics, uncovering underlying principles for both is within sight.
Acknowledgments
I am grateful to Gabe Murphy, Julie Harris, Christof Koch and Liqun Luo for valuable comments on the manuscript. This work was supported by the Allen Institute for Brain Science, and National Institutes of Health grants U01MH105982 and U19MH114830. I thank the Allen Institute founder, Paul G. Allen, for his vision, encouragement, and support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
- 1.Swanson LW. Brain architecture: understanding the basic plan. 2. New York, NY: Oxford University Press; 2012. [Google Scholar]
- 2.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanson LW, Lichtman JW. From Cajal to Connectome and Beyond. Annu Rev Neurosci. 2016;39:197–216. doi: 10.1146/annurev-neuro-071714-033954. [DOI] [PubMed] [Google Scholar]
- 4.Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Essen DC. Cartography and connectomes. Neuron. 2013;80:775–790. doi: 10.1016/j.neuron.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. The connectome of a decision-making neural network. Science. 2012;337:437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- 8.Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol. 2011;7:e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 10.Berck ME, Khandelwal A, Claus L, Hernandez-Nunez L, Si G, Tabone CJ, Li F, Truman JW, Fetter RD, Louis M, et al. The wiring diagram of a glomerular olfactory system. Elife. 2016:5. doi: 10.7554/eLife.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohyama T, Schneider-Mizell CM, Fetter RD, Aleman JV, Franconville R, Rivera-Alba M, Mensh BD, Branson KM, Simpson JH, Truman JW, et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature. 2015;520:633–639. doi: 10.1038/nature14297. [DOI] [PubMed] [Google Scholar]
- 12.Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, Wetzel AW, Scott Champion A, Graham BJ, Randlett O, et al. Whole-brain serial-section electron microscopy in larval zebrafish. Nature. 2017;545:345–349. doi: 10.1038/nature22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 15.Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- 16.Lee WC, Bonin V, Reed M, Graham BJ, Hood G, Glattfelder K, Reid RC. Anatomy and function of an excitatory network in the visual cortex. Nature. 2016;532:370–374. doi: 10.1038/nature17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan JL, Berger DR, Wetzel AW, Lichtman JW. The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus. Cell. 2016;165:192–206. doi: 10.1016/j.cell.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt H, Gour A, Straehle J, Boergens KM, Brecht M, Helmstaedter M. Axonal synapse sorting in medial entorhinal cortex. Nature. 2017;549:469–475. doi: 10.1038/nature24005. [DOI] [PubMed] [Google Scholar]
- 19.Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat. 2015;9:80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan WM. The emergence of modern neuroanatomy and developmental neurobiology. Neuron. 1998;20:413–426. doi: 10.1016/s0896-6273(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 21.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 25*.Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. Developed rAAV2-retro that allows efficient retrograde labeling of projection neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sanchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. Developed AAV-PHP.eB and AAV-PHP.S that are more efficient in infecting CNS or PNS neurons via systemic delivery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. Developed the first AAV variant, AAV-PHP.B, that can infect CNS neurons efficiently via systemic delivery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh S, Larson SD, Hefzi H, Marnoy Z, Cutforth T, Dokka K, Baldwin KK. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 2011;472:217–220. doi: 10.1038/nature09945. [DOI] [PubMed] [Google Scholar]
- 29.Kuramoto E, Ohno S, Furuta T, Unzai T, Tanaka YR, Hioki H, Kaneko T. Ventral medial nucleus neurons send thalamocortical afferents more widely and more preferentially to layer 1 than neurons of the ventral anterior-ventral lateral nuclear complex in the rat. Cereb Cortex. 2015;25:221–235. doi: 10.1093/cercor/bht216. [DOI] [PubMed] [Google Scholar]
- 30.Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PE, Kremer EJ, Palmiter RD. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci U S A. 2006;103:8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pivetta C, Esposito MS, Sigrist M, Arber S. Motor-circuit communication matrix from spinal cord to brainstem neurons revealed by developmental origin. Cell. 2014;156:537–548. doi: 10.1016/j.cell.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 32*.Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, et al. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. Developed the TRIO and cTRIO tracing methods that enable trans-synaptic input tracing from specific subsets of neurons based on their projection and cell type. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Ciabatti E, Gonzalez-Rueda A, Mariotti L, Morgese F, Tripodi M. Life-Long Genetic and Functional Access to Neural Circuits Using Self-Inactivating Rabies Virus. Cell. 2017;170:382–392. e314. doi: 10.1016/j.cell.2017.06.014. Developed a self-inactivating DeltaG-rabies virus that has substantially reduced cytotoxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Chatterjee S, Sullivan HA, MacLennan BJ, Xu R, Hou Y, Lavin TK, Lea NE, Michalski JE, Babcock KR, Dietrich S, et al. Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat Neurosci. 2018. doi: 10.1038/s41593-018-0091-7. Developed the double-deletion mutant rabies virus with substantially reduced cytotoxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callaway EM, Luo L. Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velez-Fort M, Rousseau CV, Niedworok CJ, Wickersham IR, Rancz EA, Brown AP, Strom M, Margrie TW. The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron. 2014;83:1431–1443. doi: 10.1016/j.neuron.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wertz A, Trenholm S, Yonehara K, Hillier D, Raics Z, Leinweber M, Szalay G, Ghanem A, Keller G, Rozsa B, et al. PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science. 2015;349:70–74. doi: 10.1126/science.aab1687. [DOI] [PubMed] [Google Scholar]
- 39*.Kim EJ, Jacobs MW, Ito-Cole T, Callaway EM. Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Rep. 2016;15:692–699. doi: 10.1016/j.celrep.2016.03.067. Developed an optimized glycoprotein, oG, that increases the tracing efficiency of rabies virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, Losonczy A. Rabies Virus CVS-N2c(DeltaG) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron. 2016;89:711–724. doi: 10.1016/j.neuron.2016.01.004. Developed the CVS-N2c(DeltaG) strain which exhibits reduced cytotoxicity and enhanced trans-synaptic neuronal transfer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Zeng WB, Jiang HF, Gang YD, Song YG, Shen ZZ, Yang H, Dong X, Tian YL, Ni RJ, Liu Y, et al. Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol Neurodegener. 2017;12:38. doi: 10.1186/s13024-017-0179-7. Developed a H129-DeltaTK-tdT strain that transmits to postsynaptic neurons aided by a TK-expressing helper virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mundell NA, Beier KT, Pan YA, Lapan SW, Goz Ayturk D, Berezovskii VK, Wark AR, Drokhlyansky E, Bielecki J, Born RT, et al. Vesicular stomatitis virus enables gene transfer and transsynaptic tracing in a wide range of organisms. J Comp Neurol. 2015;523:1639–1663. doi: 10.1002/cne.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, Zhang LI. AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron. 2017;93:33–47. doi: 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Myers E, Magee JC. mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat Methods. 2012;9:96–102. doi: 10.1038/nmeth.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Talay M, Richman EB, Snell NJ, Hartmann GG, Fisher JD, Sorkac A, Santoyo JF, Chou-Freed C, Nair N, Johnson M, et al. Transsynaptic Mapping of Second-Order Taste Neurons in Flies by trans-Tango. Neuron. 2017;96:783–795. e784. doi: 10.1016/j.neuron.2017.10.011. Developed the trans-Tango technique for anterograde transsynaptic circuit tracing and manipulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerner TN, Ye L, Deisseroth K. Communication in Neural Circuits: Tools, Opportunities, and Challenges. Cell. 2016;164:1136–1150. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gore BB, Soden ME, Zweifel LS. Manipulating gene expression in projection-specific neuronal populations using combinatorial viral approaches. Curr Protoc Neurosci. 2013;65:4 35 31–20. doi: 10.1002/0471142301.ns0435s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Rompani SB, Mullner FE, Wanner A, Zhang C, Roth CN, Yonehara K, Roska B. Different Modes of Visual Integration in the Lateral Geniculate Nucleus Revealed by Single-Cell-Initiated Transsynaptic Tracing. Neuron. 2017;93:767–776. e766. doi: 10.1016/j.neuron.2017.01.028. Conducted a targeted single-cell-initiated transsynaptic tracing study to label the retinal ganglion cells that provide input to individual principal cells in the mouse LGN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeNardo L, Luo L. Genetic strategies to access activated neurons. Curr Opin Neurobiol. 2017;45:121–129. doi: 10.1016/j.conb.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015;10:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen F, Tillberg PW, Boyden ES. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 60*.Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. Developed a pipeline for brain activity mapping via IEG expression using immunostaining, light-sheet fluorescence imaging, and an automated software program called ClearMap. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed Intact-Tissue Transcriptional Analysis at Cellular Resolution. Cell. 2016;164:792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Church GM, et al. Nanoscale imaging of RNA with expansion microscopy. Nat Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 4. New York, NY: Academic Press; 2012. [Google Scholar]
- 66.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 7. London, UK: Academic Press; 2014. [Google Scholar]
- 67.Dong HW. The Allen Reference Atlas: a Digital Color Brain Atlas of the C57BL/6J Male Mouse. Hoboken, New Jersey: John Wiley & Sons; 2008. [Google Scholar]
- 68.Niedworok CJ, Brown AP, Jorge Cardoso M, Osten P, Ourselin S, Modat M, Margrie TW. aMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nat Commun. 2016;7:11879. doi: 10.1038/ncomms11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69*.Kim Y, Yang GR, Pradhan K, Venkataraju KU, Bota M, Garcia Del Molino LC, Fitzgerald G, Ram K, He M, Levine JM, et al. Brain-wide Maps Reveal Stereotyped Cell-Type-Based Cortical Architecture and Subcortical Sexual Dimorphism. Cell. 2017;171:456–469. e422. doi: 10.1016/j.cell.2017.09.020. Developed the qBrain mapping platform and used it to document the distributions of inhibitory cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Furth D, Vaissiere T, Tzortzi O, Xuan Y, Martin A, Lazaridis I, Spigolon G, Fisone G, Tomer R, Deisseroth K, et al. An interactive framework for whole-brain maps at cellular resolution. Nat Neurosci. 2018;21:139–149. doi: 10.1038/s41593-017-0027-7. Developed the WholeBrain software program to quantify and spatially map multidimensional data, and the Openbrainmap program to support visualization and sharing of data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 72.Bassett DS, Sporns O. Network neuroscience. Nat Neurosci. 2017;20:353–364. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang XJ, Kennedy H. Brain structure and dynamics across scales: in search of rules. Curr Opin Neurobiol. 2016;37:92–98. doi: 10.1016/j.conb.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H. The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron. 2017;96:542–557. doi: 10.1016/j.neuron.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng H, Sanes JR. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci. 2017;18:530–546. doi: 10.1038/nrn.2017.85. [DOI] [PubMed] [Google Scholar]
- 76.Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, Awatramani R. Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci. 2016;19:1131–1141. doi: 10.1038/nn.4366. [DOI] [PubMed] [Google Scholar]
- 77.Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21:432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lein E, Borm LE, Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017;358:64–69. doi: 10.1126/science.aan6827. [DOI] [PubMed] [Google Scholar]
- 81.Ramón y Cajal S. Oxford University Press, translator. In: Histologie Du Système Nerveux de L’homme & Des Vertébrés. Swanson N, Swanson LW, translators. Paris: Maloine; 1909. 1995. [Google Scholar]
- 82*.Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, Chandrashekar J. A platform for brain-wide imaging and reconstruction of individual neurons. Elife. 2016;5:e10566. doi: 10.7554/eLife.10566. Developed the MouseLight platform for imaging of complete tissue volumes that enables the visualization and reconstruction of long-range axonal arbors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gong H, Xu D, Yuan J, Li X, Guo C, Peng J, Li Y, Schwarz LA, Li A, Hu B, et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat Commun. 2016;7:12142. doi: 10.1038/ncomms12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zador AM, Dubnau J, Oyibo HK, Zhan H, Cao G, Peikon ID. Sequencing the connectome. PLoS Biol. 2012;10:e1001411. doi: 10.1371/journal.pbio.1001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85*.Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, Zador AM. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron. 2016;91:975–987. doi: 10.1016/j.neuron.2016.07.036. Developed the MAPseq technique that can map the projections of many single neurons by labeling them with barcode mRNA which can be resolved by sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]