Abstract
We have previously identified a novel family of proteins called the GGAs (Golgi-localized, γ-ear-containing, ADP-ribosylation factor-binding proteins). These proteins consist of an NH2-terminal VHS domain, followed by a GAT domain, a variable domain, and a γ-adaptin ear homology domain. Studies from our own laboratory and others, making use of both yeast and mammals cells, indicate that the GGAs facilitate trafficking from the trans-Golgi network to endosomes. Here we have further investigated the function of the GGAs. We find that GGA-deficient yeast are not only defective in vacuolar protein sorting but they are also impaired in their ability to process α-factor. Using deletion mutants and chimeras, we show that the VHS domain is required for GGA function and that the VHS domain from Vps27p will not substitute for the GGA VHS domain. In contrast, the γ-adaptin ear homology domain contributes to GGA function but is not absolutely required, and full function can be restored by replacing the GGA ear domain with the γ-adaptin ear domain. Deleting the γ-adaptin gene together with the two GGA genes exacerbates the phenotype in yeast, suggesting that they function on parallel pathways. In mammalian cells, the association of GGAs with the membrane is extremely unstable, which may account for their absence from purified clathrin-coated vesicles. Double- and triple-labeling immunofluorescence experiments indicate that the GGAs and AP-1 are associated with distinct populations of clathrin-coated vesicles budding from the trans-Golgi network. Together with results from other studies, our findings suggest that the GGAs act as monomeric adaptors, with the four domains involved in cargo selection, membrane localization, clathrin binding, and accessory protein recruitment.
INTRODUCTION
GGAs (Golgi-localized, γ-ear–containing, ADP-ribosylation factor[ARFs]-binding proteins) are a recently identified family of ∼60- to 75-kDa proteins that contain a COOH-terminal domain with homology to the γ-adaptin ear and that bind to activated ARF (Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000; Poussu et al., 2000; Takatsu et al., 2000). The GGAs are found in the cytosol and associated with membranes, and gel filtration and ultracentrifugation studies indicate that at least in the cytosol the GGAs are monomeric (Dell'Angelica et al., 2000; Hirst et al., 2000). The GGAs have been localized to the trans-Golgi network (TGN) in mammalian cells, and gene disruption studies in yeast indicate that they facilitate trafficking from the late Golgi to the vacuole, via an endosomal intermediate (Black and Pelham, 2000; Dell'Angelica et al., 2000; Hirst et al., 2000; Takatsu et al., 2000; Zhdankina et al., 2001).
In mammalian cells, TGN to endosome traffic is also carried out by vesicles coated with clathrin and the AP-1 adaptor complex. AP-1 adaptors, which consist of γ-adaptin, β1-adaptin, μ1, and ς1 subunits, bind to cargo receptors, in particular mannose 6-phosphate receptors for lysosomal enzymes, at the TGN (Klumperman et al., 1993). Clathrin is then recruited onto the membrane by binding to AP-1, and the coated vesicles bud from the TGN and deliver their cargo to endosomes (reviewed by Lemmon and Traub, 2000). In yeast, clathrin has been implicated in TGN to endosome trafficking, but the role of the AP-1 complex is not clear. Cells lacking AP-1 subunits show no apparent phenotype, although synthetic effects have been reported with clathrin (Phan et al., 1994; Stepp et al., 1995; Yeung et al., 1999). The finding that the GGAs appear to facilitate a similar pathway to AP-1 and clathrin suggests that there may be an overlap in function and/or interactions.
So far, three mammalian GGAs and two yeast GGAs have been identified and characterized. All of the GGAs have a similar domain organization, with an NH2-terminal VHS domain, followed by a GATdomain, a variable hinge-like domain, and the γ-adaptin ear homology domain at the COOH-terminal end. The functions of these domains are beginning to be understood. VHS domains (for Vps27p, Hrs, and STAM, the first three VHS domain-containing proteins to be characterized) have been found in a number of proteins, and the crystal structures of two VHS domains have been solved (Mao et al., 2000; Misra et al., 2000), but until recently the function of the VHS domain was unknown. However, studies by Nielsen at al. (2001), Puertollano et al. (2001a), and Zhu et al. (2001) demonstrated that the VHS domains of mammalian GGAs bind to acidic-cluster-dileucine motifs found in the cytoplasmic tails of three proteins that cycle between the TGN and an endosomal compartment: sortilin, the cation-independent mannose 6-phosphate receptor and the cation-dependent mannose 6-phosphate receptor. These observations suggest that the GGAs select cargo for transport from the TGN to endosomes, via their VHS domains. It is not known whether other VHS domains are also involved in cargo recognition, although the VHS domains of Hrs, STAM, and Tom1 were unable to bind either of the mannose 6-phosphate receptor tails in the yeast two-hybrid system (Puertollano et al., 2001a).
The GAT domain (so called because it is found both in the GGAs and in the VHS domain-containing protein TOM1, i.e., GGA and TOM1) has been shown to mediate both the interaction of GGAs with activated ARF (Boman et al., 2000; Dell'Angelica et al., 2000) and the targeting of the GGAs to Golgi membranes (Dell'Angelica et al., 2000). The variable domain is the least conserved among the GGAs, in both sequence and length, but its amino acid content is similar to that of the adaptin hinge domains, suggesting that it too may function as a flexible linker connecting the NH2 and COOH domains. In addition, the variable domains of both yeast and mammalian GGAs contain potential clathrin-binding sites, similar to those found in adaptin hinges; however, GGAs are not detectable in purified clathrin-coated vesicles prepared from rat liver (Hirst et al., 2000). The COOH-terminal γ-adaptin ear-like domain, or GGA ear, may recruit accessory proteins onto the membrane, in the same way that the ear domain of the α-adaptin subunit of the AP-2 complex recruits accessory proteins onto the plasma membrane (Slepnev and De Camilli, 2000).
In this paper, we have further investigated the function of GGAs in yeast. We show that the GGAs are involved in α-factor processing as well as in carboxypeptidase (CPY) sorting, and we examine the roles of the various domains by constructing deletion mutants and chimeras. We also compare the phenotypes of cells that are deficient in GGAs, γ-adaptin, and clathrin. In addition, we use mammalian cells to address the question of whether the GGAs are associated with clathrin.
MATERIALS AND METHODS
Yeast Strains
All yeast strains were grown in yeast extract, peptone, dextrose medium (YPD) or yeast nitrogen base medium (YNB). Yeast transformations were performed using the lithium acetate method. Yeast strains used in this study are listed below.
Plasmid Construction
Standard molecular biology techniques and protocols for DNA manipulation were used throughout this study (Sambrook et al., 1989). A number of constructs were made, including deletion mutants and chimeras, and these are summarized in Figure 2a. All constructs were expressed from their own promoters by the inclusion of ∼250 bp of 5′- and 3′-untranslated sequence and then cloned into either the CEN vectors, pRS414 and pRS415, for expression at endogenous levels or into the 2μ vectors, pRS424 and pRS425, for overexpression.
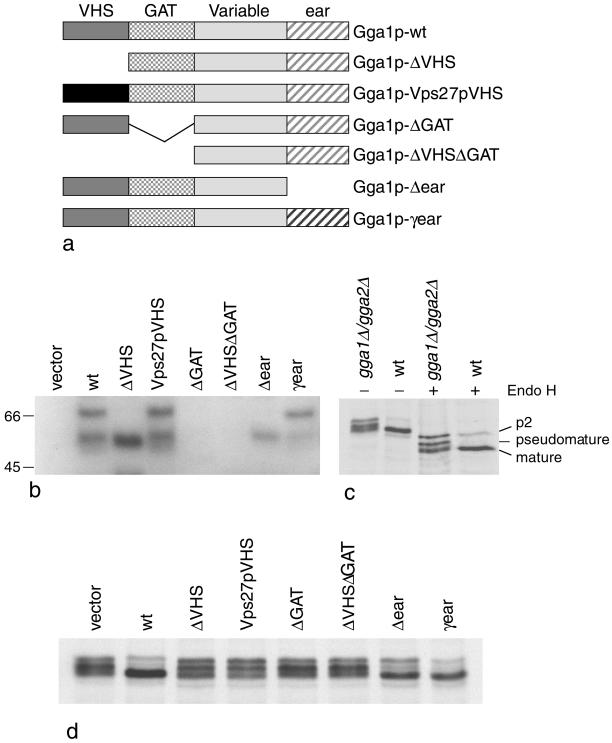
Figure 2.
(a) Diagrams of the Gga1p deletion mutants and chimeras. These constructs were all transformed into gga1Δ/gga2Δ cells. (b) Stability of the various constructs in yeast. Total extracts of cells expressing each of the constructs shown in a, as well as cells transformed with empty vector, were subjected to SDS-PAGE, loading equal amounts of protein in each lane, and Western blots were probed with anti-Gga1p. Labeled bands with the expected mobility are seen in most of the cells, with the exception of the cells transformed with the ΔGAT and ΔVHSΔGAT constructs, indicating that these constructs are unstable. The lower molecular weight band seen in some of the lanes presumably corresponds to a breakdown product. (c) CPY is aberrantly processed in gga1Δ/gga2Δ cells. Cells were labeled with 35S for 10 min and chased for 30 min, and then cell extracts were immunoprecipitated with anti-CPY. In the absence of endo H digestion, CPY in the gga1Δ/gga2Δ cells runs as a triplet. In the wild-type cells, most of the CPY comigrates with the lowest molecular weight band of the triplet, indicating that it has been processed to the mature form, although there is still a small amount of higher molecular weight (p2) CPY that has not yet been processed. In the presence of endo H, which removes N-linked oligosaccharides, the CPY in the gga1Δ/gga2Δ cells still runs as a triplet, indicating that the middle (pseudomature) band is the result of partial proteolysis rather than incomplete glycosylation. (d) CPY in cells expressing the constructs shown in a. The CPY in the cells transformed with empty vector runs as a triplet, whereas in the cells rescued with wild-type Gga1p most of the CPY runs at the mature position. The ΔVHS, ΔGAT, and ΔVHSΔGAT deletion mutants all fail to rescue the phenotype, although the mobility of the CPY is somewhat different in the cells expressing the ΔVHS construct. Unlike the other deletion mutants, the Δear mutant partially rescues the phenotype. The Vps27pVHS domain chimera does not rescue the phenotype; however, the γ-ear domain chimera appears to give full rescue.
Full-length GGA1 was cloned by polymerase chain reaction (PCR) from genomic DNA and transferred into pRS415 for expression at close to endogenous levels (Hirst et al., 2000). Domain deletion constructs were created by PCR using either genomic DNA (prepared from YPH500) or pRS415-GGA1 as a template, with the addition of restriction enzyme sites and start methionines where necessary.
For the Gga1pΔVHS construct, −250–0 and 484-1925 bp were amplified from pRS415-GGA1 with the addition of BamHI/SalI and SalI/PstI restriction sites, respectively, allowing rapid cloning by three-way ligation into pRS414 and pRS415. A similar protocol was used to construct all other chimeras. For the Gga1pΔGAT construct, −250–483 and 994-1925 bp were amplified; for the Gga1pΔVHSΔGAT construct, −250–0 and 994-1925 bp were amplified; and for the Gga1pΔear construct, −250–1323 bp were amplified, followed by a stop codon. For the Vps27pVHS domain chimera, −250–459 bp were amplified from VPS27, and for the γ-ear chimera, 2170–2765 bp were amplified from APL4. The hemagglutinin (HA)-tagged construct was made by PCR, adding a triple HA tag at the 3′-end.
To test whether the mutant proteins were stable, pellets of cells expressing the above constructs were ruptured with glass beads, extracted with sample buffer, and subjected to SDS-PAGE. Western blots of the gels were probed as previously described (Robinson and Pearse, 1986), using an antibody against full-length Gga1p generously provided by Annette Boman (University of Minnesota, Minneapolis, MN; Zhdankina et al., 2001).
Gene Disruptions
The APL4-deficient triple deletion mutant strain was constructed in the JHY3 strain (Hirst et al., 2000), which has a YPH500 strain background and is already deficient in GGA1 and GGA2. Primers were designed to flank the open reading frame of APL4 with 50 complementary bp, in addition to 25 bp of sequence complementary to the selectable marker TRP1 from Kluyveromyces lactis. The resulting PCR product was transformed into the haploid strain JHY3 and transformants selected for growth on −trp plates. A similar protocol was used to construct a CHC1-deficient strain.
Sorting Assays
Strains were tested for their ability to sort and process CPY, α-factor, and Kex2p by immunoprecipitation of radiolabeled cells. For most experiments, cells were grown overnight in selective media, cut back to 0.3 OD600 U the next morning, and then harvested in log phase at 0.6 OD600. For each experiment or time point, 5 OD units of cells were suspended in 1 ml of selective medium for radiolabeling.
The CPY-sorting assay has been described by Seaman et al. (1997). Briefly, cells were radiolabeled for 10 min and then chased for 30 min. Protein was recovered by trichloroacetic acid (TCA) precipitation, the cells were ruptured, and the total homogenate was immunoprecipitated for CPY. For some experiments the intracellular (I) and extracellular (E) fractions were separated before TCA precipitation. For this procedure cells were spheroplasted at 30°C, and then the resulting supernatant and cell pellet were subjected to TCA precipitation followed by immunoprecipitation. For α-factor secretion, cells were radiolabeled for 30 min, and the medium was TCA precipitated and immunoprecipitated for α-factor using an antibody that preferentially recognizes the precursor form.
Immunoprecipitations
Immunoprecipitations were carried out as described by Seaman et al. (1997). Samples were incubated overnight with antibodies against CPY (Seaman et al., 1997) or α-factor precursor (Wuestehube et al., 1996). The next morning, 50 μl of 50% protein A-Sepharose were added and the samples incubated for a further 2 h. Immunoprecipitates were washed sequentially with buffers containing Tris-buffered saline (TBS)/Triton/Tween-20, urea/TBS/Triton/Tween-20, TBS/Triton/Tween-20, 1% SDS, and phosphate-buffered saline, before the addition of sample buffer. Immunoprecipitations of HA-tagged proteins were performed using the mAb 12CA5 (anti-HA; Boehringer Mannheim, Indianapolis, IN) and recovered using protein G-Sepharose.
Halo Assay
This assay relies on a mating type a strain (α-tester strain) whose growth is arrested in the presence of mature α-factor. α-Tester strain (0.3 OD units) was suspended in 100 μl of water, mixed with 7 ml of molten YPD top agar, and then spread onto YPD plates. The cells to be tested were grown to log phase and 1 OD was suspended in 100 μl of water. Once the top agar had set, 5 μl of each strain to be tested were spotted onto the top agar. The plates were incubated overnight at 30°C, by which time the appearance of halos around the strains could be monitored. There was some variability in the sharpness of the halos from one experiment to another, so strains to be compared were always tested at the same time.
Immunofluorescence and Electron Microscopy
Immunofluorescence on yeast cells was performed as previously described (Hirst et al., 2000). For immunogold localization of GGAs in yeast, vps4 cells (SEY 4–1) were transformed with Gga1p-HA and Vps10p-myc (Cereghino et al., 1995) and then fixed for 1 h with 4% paraformaldehyde in 0.1 M sodium cacodylate, pH 7.2, at room temperature, pelleted, and embedded in gelatin. The cells were then prepared for ultrastructural immunocytochemistry essentially as described by Griffiths (1993). Briefly, the embedded cell pellets were infused with 1.7 M sucrose and 15% polyvinylpyrrolidone in phosphate-buffered saline overnight at 4°C and then frozen on aluminum stubs in liquid nitrogen. Frozen ultrathin sections were cut using a Reichert Ultracut S Ultramicrotome equipped with an FCS cryochamber attachment (Leica, Milton Keynes, United Kingdom). Sections were collected and labeled with rat monoclonal anti-HA (3F10; Boehringer Mannheim) and rabbit anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA), followed by goat anti-rat IgG and/or goat anti-rabbit IgG coupled to colloidal gold (Slot and Geuze, 1983). The sections were then contrasted by embedding them in freshly prepared 1.8% methyl cellulose and 0.3% uranyl acetate (Tokuyasu, 1978), allowed to air dry, and observed in a transmission electron microscope (CM100; Philips Electronic Instruments, Mahwah, NJ).
Mammalian Cells
To investigate the stability of membrane-associated GGAs, COS cells were frozen on dry ice, thawed, and incubated in cytosol buffer (25 mM HEPES-KOH, pH 7.0, 125 mM potassium acetate, 25 μM magnesium acetate, 100 μM EGTA, 1 mM dithiothreitol) for up to 5 min before fixing in methanol/acetone. Immunofluorescence double labeling was carried out as previously described, using affinity-purified rabbit polyclonal anti-GGA1 (Hirst et al., 2000) and mouse mAb 100/3 anti-γ-adaptin (Sigma, St. Louis, MO). Membrane stability was also investigated by Western blotting, using HeLa cells. The cells were scraped from the dish with a rubber policeman and drawn through a 21-gauge needle to break them up, centrifuged for 10 min at 3000 rpm in a benchtop microcentrifuge to remove unbroken cells and large particles, and then centrifuged for 45 min at 100,000 × g in an Optima-TLX ultracentrifuge (Beckman Coulter, Fullerton, CA) to pellet all remaining cell membranes. Blots of supernatant and pellets were probed with affinity-purified rabbit antibodies against γ-adaptin (Seaman et al., 1996), GGA1, or GGA2 (Hirst et al., 2000).
To compare the distribution of GGAs and clathrin, HeLa cells were fixed with 3% paraformaldehyde, permeabilized with 0.1% Triton X-100, and double labeled with rabbit anti-GGA1 (Hirst et al., 2000) and the mouse monoclonal anti-clathrin heavy chain antibody X22 (Brodsky, 1985). The cells were photographed using a MRC 1024 confocal microscope (Bio-Rad, Hercules, CA). Triple labeling was also performed. For these experiments, COS cells were fixed with methanol/acetone and then incubated with rabbit anti-GGA1 (Hirst et al., 2000) followed by Alexa488 protein A (Molecular Probes, Eugene, OR) diluted 1:100 for 1 h. The cells were then incubated with 10 μg/ml rabbit immunoglobulin (Ig) G (Sigma) for 30 min to block any free IgG-binding sites. Next the cells were incubated with affinity-purified rabbit anti-clathrin (Simpson et al., 1996), followed by Alexa594 protein A (Molecular Probes) diluted 1:300 for 1 h. This was followed by mAb 100/3 and then by Alexa350 goat anti-mouse IgG (Molecular Probes), which had been preadsorbed with rabbit IgG to block any cross-reacting antibodies. Controls included omitting the anti-clathrin (i.e., the second antibody) to make sure that the Alexa594 protein A was not binding to the first antibody. The cells were viewed using an Axioplan fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a CCD camera (Princeton Instruments, Trenton, NJ).
RESULTS
GGA-deficient Yeast Secrete α-Factor Precursor
We and others have previously shown that GGA-deficient yeast (gga1Δ/gga2Δ) have a defect in their ability to sort and process the vacuolar hydrolase CPY: >50% of their CPY is secreted into the extracellular medium instead of delivered to the vacuole, and the secreted CPY has an electrophoretic mobility different from mature CPY, indicating that it has been processed aberrantly (Dell'Angelica et al., 2000; Hirst et al., 2000; Zhdankina et al., 2001). Black and Pelham (2000) have shown that GGA-deficient yeast also missort the prevacuolar t-SNARE Pep12p. Clathrin-deficient yeast also missort Pep12p (Black and Pelham, 2000), and temperature-sensitive clathrin mutants secrete CPY when they are first increased to the nonpermissive temperature (Seeger and Payne, 1992). The similarity of the two phenotypes suggests that the GGAs and clathrin function either on the same pathway or on parallel pathways, transporting the same types of cargo from the TGN to an endosomal compartment.
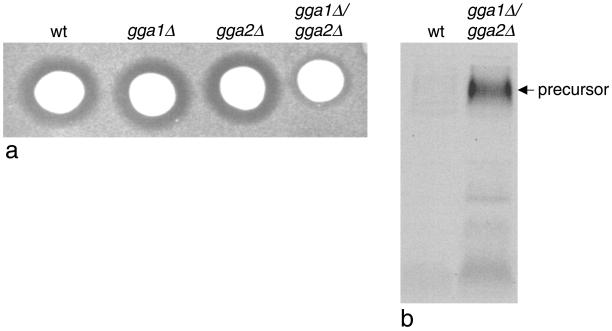
Clathrin-deficient yeast are also unable to process the pheromone α-factor, and this has been correlated with mislocalization of the α-factor-processing enzyme, Kex2p, which normally cycles back and forth between the late Golgi and an endosomal compartment (Payne and Schekman, 1989). Because of the similarities between clathrin-deficient yeast and GGA-deficient yeast, we investigated the processing of α-factor in the gga1Δ/gga2Δ strain using a halo assay. Colonies of either wild-type cells or cells lacking one or both of the GGA genes were spotted onto a lawn of mating type a cells. Active α-factor arrests the growth of the a cells, resulting in a halo around the colony. Figure 1a shows that wild-type cells and cells lacking just one of the two GGA genes (gga1Δ or gga2Δ) all have similar sized halos. However, in the gga1Δ/gga2Δ strain the halo size is significantly reduced, indicating that less active α-factor is secreted.
Figure 1.
GGA-deficient yeast have a defect in α-factor processing. (a) Colonies of wild-type (wt) cells, single-deletion mutants (gga1Δ and gga2Δ), or double-deletion mutants (gga1Δ/gga2Δ) were spotted onto a lawn of mating type a cells to test for the secretion of active α-factor. The reduced halo size surrounding the gga1Δ/gga2Δ colony when compared with the other three colonies indicates that less active α-factor is secreted in the double-deletion mutant. (b) Secreted α-factor was immunoprecipitated from the medium of metabolically labeled cells using an antibody that preferentially recognizes the precursor form. The gga1Δ/gga2Δ cells secrete mainly α-factor precursor.
To investigate this phenotype further, we performed an α-factor secretion assay. Cells were continuously radiolabeled for 30 min, and then the medium was collected and α-factor was immunoprecipitated using an antibody that preferentially recognizes the precursor form. Figure 1b shows that there is a significant increase in the amount of α-factor precursor secreted by gga1Δ/gga2Δ cells compared with the wild-type cells. This result indicates that in GGA-deficient yeast the Kex2p is unable to process α-factor to its mature form, most likely because it is mislocalized.
Domain Deletions and Substitutions
Having developed two assays for GGA function, we next constructed a number of domain deletion or substitution mutants to investigate the roles of the various domains in CPY sorting and α-factor processing (Figure 2a). The plasmids were all transformed into gga1Δ/gga2Δ cells, together with an empty vector control. To investigate the stability of the constructs, total cell extracts were subjected to SDS-PAGE, and Western blots were probed with an antibody increased against full-length Gga1p (Zhdankina et al., 2001; Figure 2b). There was no signal in the lane containing cells transformed with empty vector, whereas in cells transformed with the wild-type construct, a band of ∼70 kDa could be detected. This is somewhat higher than the predicted molecular weight of Gga1p, but similar findings have been reported using wild-type cells expressing endogenous protein (Zhdankina et al., 2001). In addition to the ∼70-kDa band, a ∼55-kDa band could also be detected, which was of variable intensity, depending on the sample. This band is not present in the empty vector lane and presumably corresponds to a breakdown product, most likely cut at the variable hinge region (the hinge domains of all the adaptins are very protease sensitive; Kirchhausen et al., 1989). The construct in which the VHS domain had been removed (Gga1p-ΔVHS) and the one in which the Gga1p VHS domain had been replaced by the Vps27p VHS domain (Gga1p-Vps27pVHS) both gave strong signals by Western blotting and ran with the expected mobilities. However, little or no signal could be detected in the lanes containing cells transformed with either of the two constructs missing the GAT domain (Gga1p-ΔGAT and Gga1p-ΔVHSΔGAT). Although it is possible that the antibody may primarily recognize the GAT domain and therefore may not react with these two proteins, the most likely explanation for the lack of signal is that the proteins are unstable. However, good signals were obtained both with the ear deletion construct (Gga1p-Δ-ear) and with the chimeric construct in which the Gga1p ear domain had been replaced by the yeast γ-adaptin ear domain (Gga1p-γ-ear).
To investigate whether the constructs were functional, we first analyzed the ability of the various strains to process CPY. We and others have previously shown that CPY is processed aberrantly in gga1Δ/gga2Δ cells, and this is illustrated in Figure 2c (left two lanes). In this experiment, cells were radiolabeled for 10 min and chased for 30 min, and then total cell extracts were immunoprecipitated with an antibody against CPY. In the wild-type cells, most of the CPY runs at the mature position, indicating that it has been proteolytically processed in the vacuole, whereas in the gga1Δ/gga2Δ cells there is a characteristic triplet of bands. Kinetic studies indicate that the middle band is the result of partial proteolytic processing rather than incomplete glycosylation (Hirst et al., 2000); however, this band has a similar mobility to the p1 form of CPY, i.e., protein that has been made in the ER but that has not yet received additional oligosaccharides in the Golgi complex. To confirm that the middle band is distinct from the p1 form, samples were treated with endoglycosidase H to remove all N-linked oligosaccharides. Under these conditions, the p1 form and the p2 (Golgi) form have identical mobilities by SDS-PAGE, whereas the mature form runs faster. Two bands can be seen in the endo H-treated wild-type lane, a strong band corresponding to the mature form and a weaker band corresponding to the p2 form (Figure 2c, fourth lane). In the gga1Δ/gga2Δ lane, the p2 band is stronger, the mature band is somewhat fuzzier, and most strikingly, there is a strong middle band that is well resolved away from the other two (Figure 2c, third lane). The mobility of this band after endo H treatment clearly demonstrates that it corresponds to pseudomature (i.e., incompletely or aberrantly processed) CPY rather than p1 CPY.
Figure 2d shows CPY immunoprecipitated from cells transformed with each of the constructs. In the cells transformed with empty vector, the characteristic triplet can be seen, whereas in cells expressing wild-type Gga1p, most of the CPY runs at the mature position. The Gga1p-ΔVHS construct did not restore the wild-type phenotype; however, there was a small but reproducible difference in the appearance of the triplet, with the three bands more equal in intensity and the lowest band migrating somewhat faster. Cells transformed with a chimeric Vps27pVHS domain construct showed the same phenotype as the cells expressing the ΔVHS domain construct, indicating that the two VHS domains are not functionally interchangeable. Cells transformed with the ΔGAT or ΔVHSΔGAT constructs showed the same phenotype as cells transformed with empty vector. However, because these two constructs appear to be unstable, we were unable to determine whether the proteins are truly nonfunctional or whether they are expressed at such low levels that they are unable to rescue. The Δ-ear construct produced a phenotype intermediate between the empty vector and the wild-type phenotype, indicating that this protein is partially functional. The γ-ear construct completely restored the wild-type phenotype, indicating that the γ-adaptin ear can functionally substitute for the GGA ear.
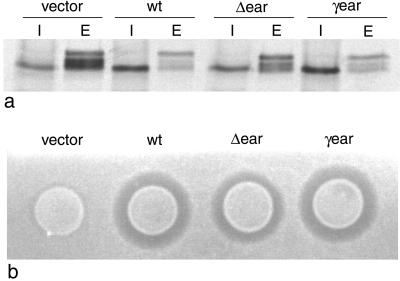
To investigate the phenotypes of the Δ-ear and γ-ear constructs further, we repeated the pulse-chase procedure and this time separated the cells into intracellular and extracellular fractions before immunoprecipitating with anti-CPY. Figure 3a confirms that the γ-ear chimeric construct completely restores CPY sorting, whereas the Δ-ear deletion construct improves the sorting efficiency when compared with cells transformed with empty vector: relatively more of the CPY is retained intracellularly and runs at the mature position, although sorting is not restored to wild-type levels. We also examined the ability of these two constructs to restore α -factor processing using the halo assay. Figure 3b shows that cells expressing wild-type Gga1p and the γ-ear chimera have similar sized halos, whereas the cells expressing the Δ-ear deletion construct have a larger halo than the cells transformed with empty vector but smaller than the cells expressing either the wild-type protein or the γ-ear chimera. Together, these results indicate that the GGA ear contributes to protein function but is not absolutely essential, and that the γ -adaptin ear can completely replace the GGA ear.
Figure 3.
a. Extracts from intracellular (I) and extracellular (E) fractions of pulse-chased gga1Δ/gga2Δ cells transformed with either empty vector, wild-type Gga1p (wt), the Δ-ear deletion mutant, or the γ-ear chimera were immunoprecipitated with anti-CPY. The γ-ear chimera gives full rescue of CPY sorting, whereas the Δ-ear deletion mutant gives partial rescue. (b) Secretion of active α-factor was investigated using the halo assay. Again, the γ-ear chimera gives full rescue of the wild-type phenotype, and the Δ-ear deletion mutant gives partial rescue
Deletion of γ-Adaptin and Clathrin
Why is the Gga1p construct lacking its ear domain partially functional, whereas constructs lacking the VHS and/or GAT domains are nonfunctional? If the role of the ear is to recruit accessory factors onto the late Golgi membrane, then one possibility is that this task can be performed by the γ-adaptin subunit of the AP-1 complex, Apl4p. Thus, the prediction would be that deleting the APL4 gene together with the two GGA genes should exacerbate the missorting phenotype and that in such cells the Δ-ear deletion construct should no longer be able to rescue the phenotype, even partially.
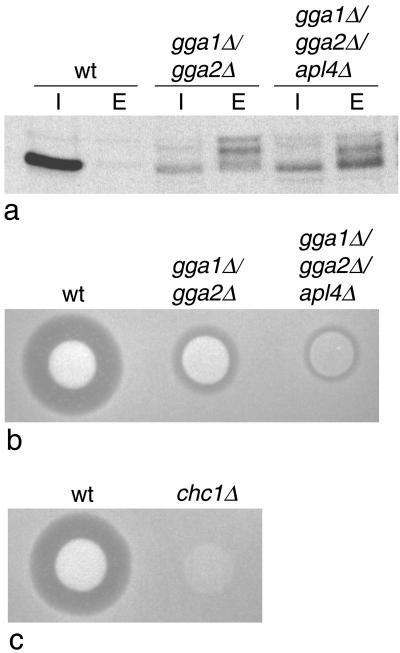
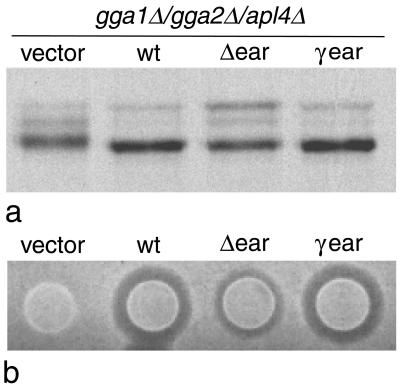
It has been previously shown that, if APL4 or any of the other yeast AP-1 subunit genes are deleted on their own, there is no discernible phenotype: the cells grow normally and both CPY sorting and α-factor processing are unimpaired (Phan et al., 1994; Stepp et al., 1995; Yeung et al., 1999). To determine the phenotype of a triple mutant, we deleted APL4 in the gga1Δ/gga2Δ strain. The cells were found to be completely viable, with no apparent change in growth rate (data not shown). We next tested the ability of the gga1Δ/gga2Δ/apl4Δ strain to sort and process CPY. Figure 4a shows that the amount of CPY secreted in the gga1Δ/gga2Δ/apl4Δ cells, relative to the amount retained intracellularly, is similar to that in the gga1Δ/gga2Δ cells. However, there is a change in the relative amounts of the different forms of CPY. In the triple mutant (gga1Δ/gga2Δ/apl4Δ), the predominant form of secreted CPY is low molecular weight. This band has a similar mobility to the mature form, but it has a somewhat fuzzier appearance and on some gels it could be seen to run more slowly (see Figure 5a), so we suspect that it may be another pseudomature form. There was less of the middle pseudomature form and considerably less of the p2 (uncleaved) form in the gga1Δ/gga2Δ/apl4Δ cells than in the gga1Δ/gga2Δ cells. These observations suggest that the missorted CPY is more prone to proteolytic degradation in cells deleted for APL4 as well as GGA1 and GGA2. We also tested the cells for α-factor processing, using the halo assay. Figure 4b shows that the halo size in the gga1Δ/gga2Δ/apl4Δ cells is significantly reduced when compared with the gga1Δ/gga2Δ cells, pointing to a role for γ-adaptin as well as the GGAs in Kex2p sorting and α-factor processing.
Figure 4.
Effect of deleting other coat components. (a) Wild-type (wt) cells, cells lacking both GGA genes (gga1Δ/gga2Δ), and cells lacking both GGA genes as well as γ-adaptin (gga1Δ/gga2Δ/apl4Δ) were pulse-chased, and CPY was immunoprecipitated from both intracellular (I) and extracellular (E) fractions. Although the ratios of extracellular to intracellular CPY are similar in the gga1Δ/gga2Δ and gga1Δ/gga2Δ/apl4Δ cells, the electrophoretic mobility of the secreted CPY is different in the triple mutant, suggesting that more proteolytic degradation has occurred. (b) Colonies of wild-type, gga1Δ/gga2Δ, and gga1Δ/gga2Δ/apl4Δ cells were spotted onto a lawn of mating type a cells and secretion of active α-factor was monitored using the halo assay. Deleting the APL4 gene together with the two GGA genes exacerbates the α-factor misprocessing phenotype. (c) The clathrin heavy chain (CHC1) gene was deleted in the same strain as the cells shown in b, and the halo assay was performed at the same time under identical conditions. The chc1Δ cells have a more severe phenotype than either the gga1Δ/gga2Δ cells or the gga1Δ/gga2Δ/apl4Δ cells.
Figure 5.
The gga1Δ/gga2Δ/apl4Δ cells were transformed with either empty vector, wild-type Gga1p (wt), the Δ-ear deletion mutant, or the γ-ear chimera and then assayed for both CPY processing and active α-factor secretion. (a) Total CPY was immunoprecipitated from pulse-chased cells. The γ-ear chimera produces the same phenotype as the wild-type construct, whereas the Δ-ear deletion mutant produces an intermediate phenotype. (b) Secretion of active α-factor was monitored using the halo assay. Again, the γ-ear chimera gives full rescue and the Δ-ear deletion mutant gives partial rescue. Thus, even in the absence of γ-adaptin, the Δ-ear deletion mutant is partially functional.
How do the triple mutants compare with clathrin mutants, which also show a defect in α-factor processing? To address this question, we deleted the clathrin heavy chain gene (CHC1) in the same strain background (YPH500) as our other mutants. It has been previously shown that depending on the strain background, deleting CHC1 is either lethal or causes the cells to grow more slowly. Similarly, we found that our chcΔ cells grew more slowly than any of our other mutants (data not shown). In addition, when we performed a halo assay, we found that no halo was visible (Figure 4c, which was performed under identical conditions to those shown in Figure 4b). We also attempted to knock out CHC1 in our gga1Δ/gga2Δ cells, but no colonies were obtained, perhaps because this particular combination may be lethal.
We then asked whether the phenotype of the gga1Δ/gga2Δ/apl4Δ cells could be rescued with the Δ-ear deletion construct. Figure 5 shows these cells transformed with either empty vector, the wild-type Gga1p construct, the Δ-ear deletion construct, or the γ-ear chimeric construct. As with the gga1Δ/gga2Δ cells, in the gga1Δ/gga2Δ/apl4Δ cells the wild-type construct and the γear chimeric constructs gave equally good rescue of both the CPY phenotype (a) and the α-factor phenotype (b). In addition, the Δ-ear deletion construct gave partial rescue of both phenotypes. However, in the halo assay, the Δ-ear deletion construct appeared to be somewhat less efficient at rescuing the phenotype in the gga1Δ/gga2Δ/apl4Δ cells than in the gga1Δ/gga2Δ cells (compare Figure 5b with Figure 3b). Nevertheless, the fact that the Δ-ear deletion construct gave even partial rescue in cells with no other obvious γ-ear-like domain suggests that, at least in yeast, this domain is not absolutely essential for either GGA or AP-1 function.
Localization of Yeast Gga1p
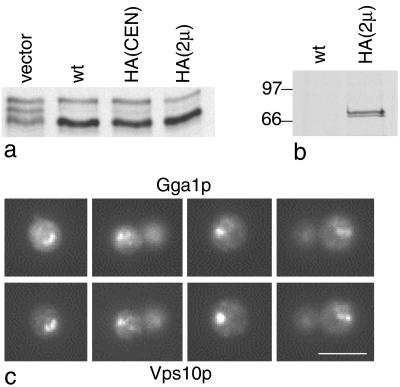
We and others have previously shown that in mammalian cells GGAs are localized on trans-Golgi cisternae and on the TGN (Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000). However, so far no GGA localization data in yeast have been reported. To determine whether GGAs in yeast are also localized on late Golgi membranes, we engineered an HA tag onto the COOH-terminal end of Gga1p. This construct was then transformed into gga1Δ/gga2Δ cells to see whether it was able to rescue the missorting phenotype. Figure 6a shows a CPY-sorting and -processing assay performed on cells transformed with empty vector, wild-type Gga1p, tagged Gga1p expressed at endogenous levels using a CEN vector, or tagged Gga1p expressed at higher than normal levels using a 2μ plasmid. All three constructs can be seen to rescue the phenotype equally well. To confirm that the tag is able to be recognized by the antibody, extracts of metabolically labeled cells expressing either wild-type Gga1p or tagged Gga1p were immunoprecipitated with an anti-HA antibody. Figure 6b shows that a doublet of the appropriate molecular weight is specifically brought down in the cells expressing the tagged construct.
Figure 6.
Epitope tagging of Gga1p. (a) An HA tag was engineered onto the COOH terminus of Gga1p and expressed at either endogenous levels (HA[CEN]) or higher than normal levels (HA[2μ]). Cells were pulse-chased and total CPY was immunoprecipitated. The tagged construct gives full rescue at both endogenous and high levels. (b) Cells expressing HA-tagged Gga1p on a 2μ plasmid were metabolically labeled and then immunoprecipitated with anti-HA, together with cells expressing wild-type Gga1p (wt) as a control. The autoradiograph shows that the tagged construct is recognized by anti-HA. (c) Localization of the tagged Gga1p in a vps4 mutant. Cells were double labeled for HA-tagged Gga1p (top) and myc-tagged Vps10p (bottom). Vps10p is known to go to the class E compartment in a vps4 mutant; therefore, the colocalization of Gga1p with Vps10p indicates that it also goes to the class E compartment. The reason for the use of a class E mutant is that late Golgi, endosomal, and vacuolar proteins all accumulate in an exaggerated prevacuolar compartment, the class E compartment, making the membrane localization of tagged Gga1p much easier to see. Scale bar, 5 μm.
When we attempted to localize the tagged Gga1p by immunofluorescence, we found that it was difficult to discern a pattern, possibly because in yeast the Golgi is highly dispersed and any labeling would be hard to see against the background of cytosolic Gga1p. Therefore, we made use of a class E vacuolar protein-sorting mutant, vps4. In class E mutants, late Golgi proteins such as Vps10p, together with endocytosed proteins and proteins destined for the vacuole, all accumulate in an exaggerated prevacuolar compartment called the class E compartment (Raymond et al., 1992; Rieder et al., 1996). Figure 6c shows vps4 cells coexpressing HA-tagged Gga1p and myc-tagged Vps10p and double labeled for HA (top) and myc (bottom). In these cells, much of the Gga1p colocalizes with Vps10p, indicating that both are associated with the class E compartment.
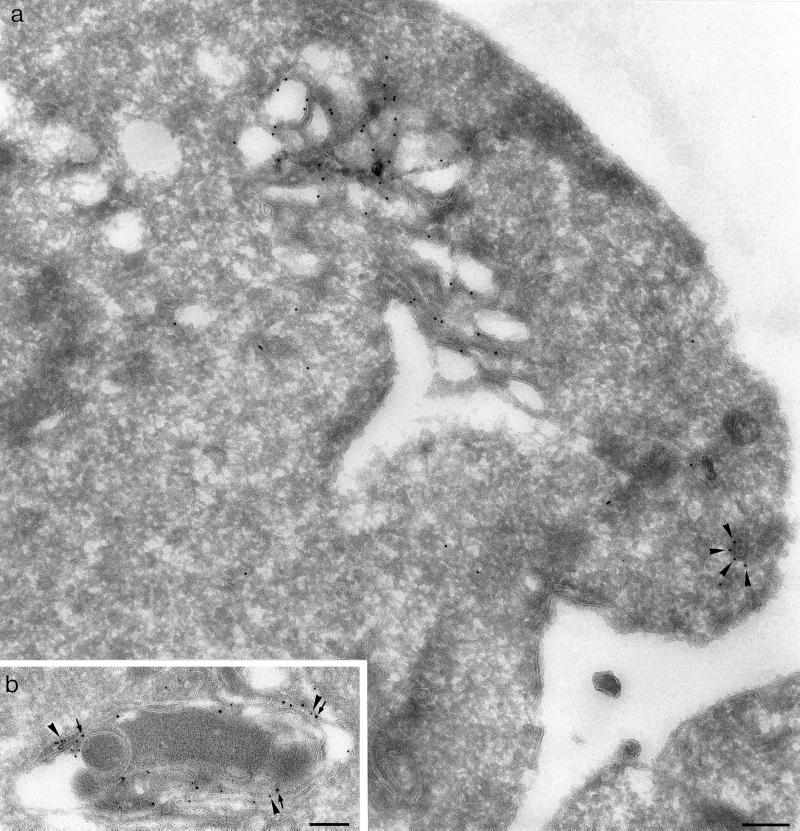
To confirm the localization of Gga1p at the ultrastructural level, immunogold electron microscopy labeling was performed on the vps4 cells expressing tagged Gga1p. Figure 7a shows an electron micrograph of such a cell, labeled with 10 nm gold. The cell contains stacks of multilamellar membranes, characteristic of the class E compartment, which are heavily labeled with 10 nm gold, indicating that Gga1p has been recruited onto this compartment. To confirm that this is indeed the class E compartment, cells coexpressing HA-tagged Gga1p and myc-tagged Vps10p were double labeled with 10 nm gold (HA) and 15 nm gold (myc). Figure 7b shows that both labels colocalize on the same membranes. This result indicates that in class E mutants the docking site for Gga1p, which is presumably normally in the late Golgi, moves to the class E compartment, together with other late Golgi proteins like Vps10p and Kex2p.
Figure 7.
Localization of Gga1p at the electron microscope level. (a) vps4 cells expressing HA-tagged Gga1p on a 2μ plasmid were labeled with anti-HA followed by protein A coupled to 10 nm gold. Most of the label is associated with a multilamellar compartment with the characteristic appearance of the class E compartment. A coated vesicular profile is also labeled (arrowheads). (b) vps4 cells coexpressing HA-tagged Gga1p and myc-tagged Vps10p were double labeled for HA (10 nm gold, arrowheads) and myc (15 nm gold, arrows). The colocalization of the two proteins on the same multilamellar compartment confirms that in a vps4 mutant GGAs are targeted to the class E compartment. Scale bars, 200 nm.
Mammalian GGAs and Clathrin
In our previous study, we suggested that GGAs were not associated with clathrin-coated vesicles, primarily because they could not be detected in purified clathrin-coated vesicle preparations from rat liver (Hirst et al., 2000). However, the present study, as well as studies by Black et al. (2000) and Costaguta et al. (2001), demonstrate that clathrin-deficient and GGA-deficient yeast have similar phenotypes. Moreover, a recent study by Puertollano et al. (2001b) of mammalian cells showed that GGAs could recruit clathrin onto TGN membranes. These findings caused us to consider alternative explanations for the absence of GGAs from purified clathrin-coated vesicles.
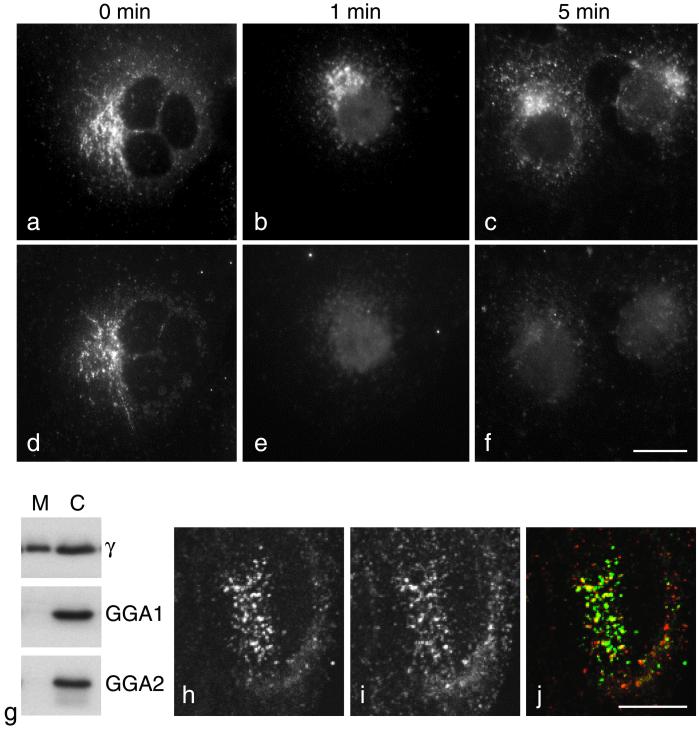
One possibility might be that the GGAs are not stably associated with membranes. To investigate the stability of membrane-associated GGAs, we permeabilized COS cells by freezing and thawing and then incubated them at 37°C in a physiological buffer. We have previously shown that this procedure causes cells to become leaky so that proteins and other molecules escape, although the overall architecture of the cell is preserved. Figure 8, a–f, shows such cells double labeled for the γ-adaptin subunit of AP-1 (a–c) and GGA1 (d–f). Immediately after freezing and thawing, both proteins have a punctate perinuclear pattern, although the two patterns are distinct (Figure 8, a and d). One minute after thawing the cells, the perinuclear GGA1 labeling has essentially disappeared (Figure 8e), although there is still AP-1 associated with the TGN at 1 min (Figure 8b) and at later time points as well (Figure 8c). We also investigated the relative stability of the membrane association of GGAs and AP-1 by homogenizing HeLa cells, spinning at low speed to remove unbroken cells, and then spinning at 100,000 × g to pellet cell membranes. Western blots of the membrane-containing pellets (M) and cytosol-containing supernatants (C) were then probed with antibodies against the γ-adaptin subunit of AP-1, GGA1, and GGA2 (Figure 8g). As expected, γ-adaptin was about equally distributed between membranes and cytosol. However, both GGA1 and GGA2 were almost exclusively (>95%) cytosolic.
Figure 8.
GGAs in mammalian cells. (a–f) COS cells were double labeled for the γ-adaptin subunit of AP-1 (a–c) and GGA1 (d–f). In cells that were fixed immediately after freezing and thawing (a and d), both proteins have a punctate perinuclear distribution, although the two patterns are distinct. In cells that had been incubated in a physiological buffer for either 1 min (b and e) or 5 min (c and f) after freezing and thawing, much of the γ-adaptin is still associated with the membrane (b and c), but membrane-associated GGA1 is no longer detectable (e and f). Scale bar, 20 μm. (g) HeLa cells were homogenized, unbroken cells and nuclei were pelleted, and the remaining postnuclear supernatant was centrifuged at 100,000 × g for 45 min. Blots of the membrane-containing pellets (M) and cytosol-containing supernatants (C) were then probed with antibodies against γ-adaptin, GGA1, and GGA2. Nearly half of the γ-adaptin remains in the membrane-containing pellet; however, at least 95% of the GGA1 and GGA2 are in the cytosol. H–j. HeLa cells were double labeled with antibodies against GGA1 (h) and clathrin (i) and then photographed with a confocal microscope. There is significant colocalization between the two proteins, as shown in the merged image (j; GGA1 is labeled green, clathrin is labeled red). Scale bar, 10 μm.
The extreme lability of membrane-associated GGAs when compared with AP-1 may explain why GGAs are not detectable in purified clathrin-coated vesicles while AP-1 is enriched, because the procedure we use for preparing clathrin-coated vesicles takes several hours (Pilch et al., 1983). However, membrane-associated GGAs can be seen in fixed cells by immunofluorescence. Thus, we double labeled fixed HeLa cells with antibodies against GGA1 and clathrin and then viewed them in a confocal microscope. Figure 8, h–j, shows that there is considerable overlap between the two patterns. They are not identical, because much of the clathrin (Figure 8i) is associated with other compartments such as the plasma membrane, and in addition not all of the GGA labeling (Figure 8h) coincides with clathrin (unlike AP-1 and AP-2, in which virtually all of the AP-positive structures are also positive for clathrin). However, there does appear to be more colocalization between GGAs and clathrin than between GGAs and AP-1.
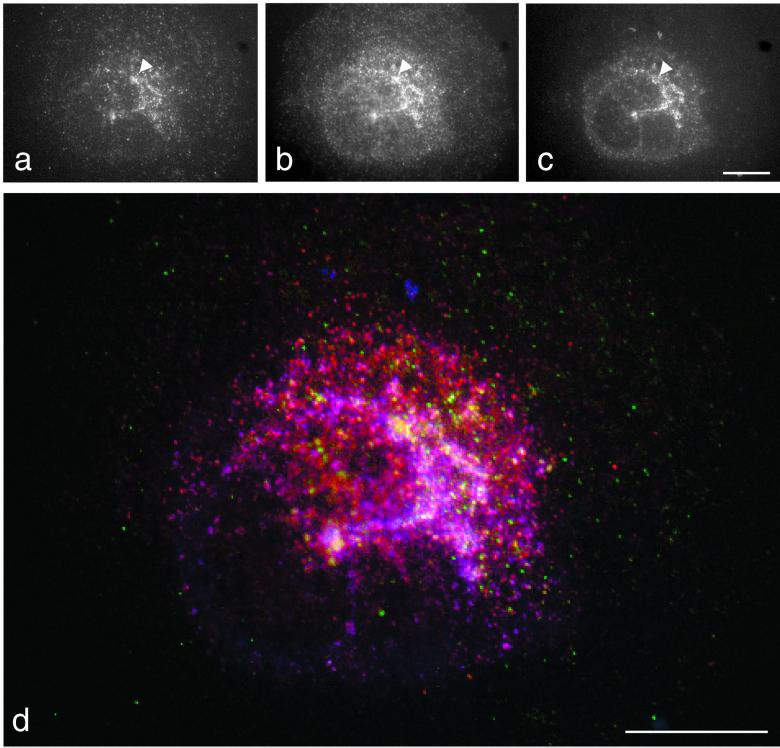
To compare the distribution of GGAs, clathrin, and AP-1 in the same cell, we used a novel triple-labeling procedure (Figure 9). This involved incubating fixed and permeabilized COS cells first with rabbit anti-GGA1, followed by protein A coupled to a green fluorescent dye (Figure 9a), then with rabbit anti-clathrin, followed by protein A coupled to a red fluorescent dye (Figure 9b), and finally with mouse monoclonal anti-γ-adaptin, followed by goat anti-mouse IgG coupled to a blue fluorescent dye (Figure 9c). The cells were then viewed using a conventional fluorescence microscope and CCD camera. Suitable controls were carried out to ensure that there was no protein A labeling of the “wrong” antibody. Figure 9 shows that all three patterns are similar; however, there are a number of structures that are positive for GGA1 and clathrin but not for γ-adaptin (Figure 9, arrowhead). Merging the three images (Figure 9d) shows structures that are positive for both clathrin and γ-adaptin (purple-pink) and for both clathrin and GGA1 (yellow). These results suggest that two populations of clathrin-coated vesicles bud from the TGN: one with clathrin and AP-1 and one with clathrin and GGA1 (and presumably other GGAs as well).
Figure 9.
Triple labeling of COS cells. COS cells were fixed in methanol/acetate and then labeled sequentially with rabbit anti-GGA1 followed by Alexa488 protein A (a), rabbit anti-clathrin followed by Alexa594 protein A (b), and mouse anti-γ-adaptin followed by Alexa350 goat anti-mouse. (d) Merging of the three images, with GGA1 labeled in green, clathrin labeled in red, and γ-adaptin labeled in blue. Overlap between clathrin and GGA1 appears yellow, and overlap between clathrin and γ-adaptin appears purple-pink. The presence of discrete structures labeled either for clathrin and GGA1 or for clathrin and γ-adaptin indicates that GGA1 and AP-1 are segregated from each other but both are associated with clathrin. Scale bars, 20 μm.
DISCUSSION
The GGAs are the most recent addition to the collection of vesicle coat proteins. Although their discovery was only reported last year, work conducted independently in a number of different laboratories, making use of both yeast genetics and mammalian cell biology, has enabled rapid progress to be made in learning about their function. The GGAs have been shown to be localized mainly to the TGN in mammalian cells, to bind activated ARF, and to facilitate trafficking to the yeast vacuole via an endosomal intermediate (Black and Pelham, 2000; Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000). In the present study, we have examined the yeast mutant phenotype in greater detail and have found that GGA-deficient yeast also secrete α-factor precursor, probably because of an inability to sort the α-factor–processing enzyme Kex2p. We have also shown that the VHS domain is essential for protein function and that the Vps27p VHS domain will not substitute. In contrast, the ear domain contributes to protein function but is not absolutely required, and the GGA ear domain can be replaced by the γ-ear domain. We have found that deleting the γ-adaptin gene as well as the two GGA genes exacerbates the missorting phenotype, that GGAs in yeast most likely localize to late Golgi membranes, and that in mammalian cells GGAs show significant colocalization with clathrin but less colocalization with AP-1.
While this paper was under review, several very important studies of the GGAs in both yeast and mammalian cells were published. Costaguta et al. (2001) demonstrated that yeast GGAs and clathrin interact both genetically and biochemically. They also found, as we did, that deleting AP-1 subunits together with GGAs produces a more severe phenotype than deleting GGAs alone. Puertollano et al. (2001b) showed that GGAs can recruit clathrin onto Golgi membranes in mammalian cells, and they found that the two colocalize at the electron microscope level as well as at the light microscope level. Another study by Puertollano et al. (2001a), as well as studies by Nielsen et al. (2001) and Zhu et al. (2001), established a role for the GGA VHS domain. All three studies showed that the VHS domains of mammalian GGAs bind to acidic-cluster-dileucine–sorting signals in the cytoplasmic tails of proteins that traffic from the TGN to endosomes. In addition, Puertollano et al. (2001a, b) showed that expressing a truncated construct consisting of the GGA1 VHS and GAT domains caused acidic-cluster-dileucine–containing proteins to become trapped in the TGN. Thus, the results of the present study can now be interpreted in light of the new findings.
Of particular interest is the elucidation of the role of the GGA VHS domain. The prediction from these findings is that a construct lacking this domain should still be able to form vesicles in a GGA-dependent manner, but cargo selection would be impaired. Our results show that CPY is still mainly secreted in cells expressing the ΔVHS domain mutant, but the CPY appears to be processed slightly differently in these cells when compared with gga null cells (Figure 2b). It is possible that the CPY in the ΔVHS domain-expressing cells encounters different proteases, or the same proteases but under different conditions, from those that it encounters either in wild-type cells or in gga null mutants. In mammalian cells, only the GGA VHS domains bound to acidic-cluster-dileucine sequences in the yeast two-hybrid system; other VHS domains did not, including the VHS domain from Hrs, the mammalian orthologue of Vps27p. Similarly, we find that yeast cells expressing the Vps27pVHS chimera have a phenotype identical to cells expressing the ΔVHS deletion mutant, indicating that the two VHS domains are not functionally interchangeable. So far only the binding specificity of mammalian VHS domains has been determined; it is not yet known precisely what yeast GGA VHS domains recognize, although presumably they also also have a role in cargo recognition. It is also not known what other VHS domains might interact with. However, there is no evidence that Vps27p acts as a coat protein or plays any role in cargo selection; instead it has been implicated in multivesicular body formation (Piper et al., 1995). Thus, it is possible that cells expressing the VHS domain chimera might package the wrong set of proteins, i.e., those that bind to the Vps27p VHS domain rather than the GGA VHS domain, when they form GGA-dependent transport vesicles at the TGN. Clearly, more studies will be needed to establish the role of the VHS domain in different proteins.
It is harder to draw conclusions about the constructs missing the GAT domain because of their apparent instability. However, the GAT domain has been shown to be both necessary and sufficient for recruiting GGAs onto Golgi membranes (Dell'Angelica et al., 2000; Puertollano et al., 2001b), so presumably both the ΔGAT and the ΔVHSΔGAT constructs would be unable to be recruited onto the membrane and therefore nonfunctional. The GAT domain has also been shown to interact with ARF (Boman et al., 2000; Dell'Angelica et al., 2000), and Puertollano et al. (2001b) have shown by mutagenesis that ARF binding and membrane recruitment are coupled. However, the same ARF, ARF1, can facilitate the recruitment of a number of different coat proteins onto membranes, including coatomer, AP-1, and AP-3 as well as the GGAs (Ooi et al., 1998). Because these proteins all have different distributions in the cell, this suggests that, even though ARF contributes to membrane binding, there must be other factors as well. In our previous studies of AP complexes, we have attempted to identify targeting signals on the complexes and targeting machinery in the cell (Robinson, 1993; Seaman et al., 1993; Page and Robinson, 1995; West et al., 1997). These studies have proved difficult, in part because the AP complexes are heterotetramers and our data indicate that targeting involves more than one subunit. The GGAs may be much more amenable to this type of study, because they are monomeric with a targeting domain already defined. Although the association of GGAs with the membrane is extremely labile, we have found that we can drive exogenous GGAs onto Golgi membranes by the addition of ATP, an ATP-regenerating system, and GTPγS (Hirst et al., 2000); thus, we now have the basis for an in vitro system that could be used to study targeting machinery. In addition, we have been able to localize GGAs in yeast, something that has not yet been done with any of the AP complexes, so this also raises the possibility of the use of yeast genetics to determine how the GGAs are recruited onto membranes.
Unlike the VHS and GAT domains, we find that the GGA ear domain is not absolutely required for protein function, although it improves sorting efficiency. We have proposed that the function of this domain is to recruit accessory proteins onto the membrane (Hirst et al., 2000). In the case of the AP-2 complex, in which multiple α-ear–binding partners have been identified, these accessory proteins appear to be just as important as the AP complex itself for vesicle formation (Slepnev and De Camilli, 2000), so it is surprising that the GGA ear seems to be dispensible. However, several of the α-ear–binding partners can interact with more than one component of the coat (e.g., AP180 has been shown to interact not only with the α-ear but also with the β2-ear and with clathrin; Owen et al., 2000). Similarly, any partners for the GGA ear may have other ways of getting onto the membrane. Our studies of mammalian cells indicate that the GGA ears bind to a subset of those proteins that bind to the γ-ear (Hirst et al., 2000), and consistent with this finding, a construct consisting of Gga1p with the γ-ear gave full functional rescue. Although we cannot rule out the possibility that the γ-ear simply helped the construct to fold properly, we believe that a more likely explanation for this observation is that the γ-ear was able to recruit accessory proteins normally recruited by the GGA ear. So far, the only protein that has been shown to bind to the γ-ear in vivo is γ-synergin (Page et al., 1999). γ-Synergin can also interact with GGA ears in vitro; however, it does not coimmunoprecipitate or colocalize with either GGA1 or GGA2, suggesting that this interaction may not be physiologically relevant (Hirst et al., 2000; but see also Takatsu et al., 2000). In addition, there is no obvious homologue of γ-synergin in yeast. There are a number of other proteins that bind to the γ-ear in GST pulldowns, at least three of which also appear to bind to the GGA ear (Hirst et al., 2000), so it will be important to identify these proteins and establish their function.
When we deleted the γ-adaptin gene, APL4, together with the two GGA genes, we found that the phenotype was exacerbated, even though deleting APL4 on its own has no obvious phenotype. CPY delivery to the vacuole did not seem to be impaired in the triple mutant: in the gga1Δ/gga2Δ cells and the gga1Δ/gga2Δ/apl4Δ cells the ratio of intracellular to extracellular CPY was the same. However, the extracellular CPY in the two strains had different electrophoretic mobilities: there was relatively less of the p2 band in the gga1Δ/gga2Δ/apl4Δ cells and relatively more of a lower molecular weight band that could either be mature CPY or another pseudomature form. The aberrant proteolysis that occurs in GGA-deficient yeast clearly provides clues into GGA function, although at present it is difficult to interpret them. Most other vps mutants secrete CPY only in the p2 form, and the secretion of pseudomature form(s) in the GGA-deficient cells suggests that their endosomal/vacuolar system is dysfunctional, with degradative enzymes in compartments destined for exocytosis.
The results of the halo assay in the gga1Δ/gga2Δ/apl4Δ cells were more clear-cut. The halos were significantly smaller in these cells than in gga1Δ/gga2Δ cells, indicating that less mature α-factor had been secreted. Costaguta et al. (2001) have also found that, if AP-1 subunit genes are deleted as well as GGA genes, more α-factor precursor is secreted, and in addition they demonstrated that transport of CPS to the vacuole is more strongly impaired in such cells. Together, these observations indicate that, if the GGAs are missing, the AP-1 complex can to some extent compensate and vice versa. Initially we performed the triple knockout experiment because we suspected that the γ-ear might help to recruit accessory proteins onto the membrane in the absence of the GGA ear; however, even in the gga1Δ/gga2Δ/apl4Δ cells the Δ-ear deletion construct was still partially able to rescue the missorting phenotype. This result indicates that the ability of the AP-1 complex to compensate for the GGAs is not just a function of the γ-ear but rather that the GGAs and AP-1 may act on parallel pathways.
Do the GGAs, like AP-1, act together with clathrin? In our previous study, we suggested that they did not, primarily because of their absence from purified clathrin-coated vesicles. Here we show that the association of GGAs with the membrane is much less stable than the association of AP-1 with the membrane, and this could explain why the GGAs cannot be detected in purified clathrin-coated vesicle preparations. Indeed, AP-1, AP-2, and clathrin are unusual in that they remain membrane associated for hours or even days after cells are disrupted, which makes it possible to purify clathrin-coated vesicles from tissue homogenates; in contrast, purification of COPI- or COPII-coated vesicles is much less straightforward and relies on treatment with nonhydrolyzable GTP analogues to drive the coats onto the membrane (Malhotra et al., 1989; Barlowe et al., 1994). In the present study we find significant colocalization between GGA1 and clathrin by immunofluorescence. Two other recent studies (Costaguta et al., 2001; Puertollano et al., 2001b) also indicate that the GGAs interact with clathrin in vivo. This interaction helps to explain why deleting subunits of all of the putative AP complexes in yeast has a very mild phenotype, whereas deleting clathrin has a very severe one (Huang et al., 1999; Yeung et al., 1999), because the GGAs could act as alternative clathrin adaptors on the late Golgi to endosome pathway. Similarly, in mammalian cells, a dominant negative clathrin mutant, consisting of the “hub” domain only, is extremely toxic (Liu et al., 1998), whereas AP-1–deficient cells grow normally (Meyer et al., 2000), so again the GGAs may be acting as alternative adaptors, enabling TGN to endosome traffic to occur even in the absence of AP-1. However, although the phenotype of the gga1Δ/gga2Δ/apl4Δ cells in the halo assay was more severe than that of the gga1Δ/gga2Δ cells, it was still not as severe as the phenotype of the chc1Δ cells. This suggests either that clathrin can function to some extent on its own or alternatively that there may be additional clathrin adaptors in yeast that have not yet been identified.
What is the precise function of the GGAs? Our study, together with others that have recently been published, indicates that they act as monomeric adaptors, with each of their four domains performing a different function (Figure 10). The VHS domain would be involved in cargo selection, like the μ-subunit (Ohno et al., 1995; Owen and Evans, 1998) and β-subunit (Rapoport et al., 1998) of an AP complex. The GAT domain would bind ARF, a property that has been attributed to both the γ- and the β-subunits of AP-1 (Austin et al., 2000). The GAT domain would also target the protein to the appropriate membrane, which in the AP-1 and AP-2 complexes has been shown to involve the γ- or α-subunit (both NH2-terminal and ear domains; Robinson, 1993; Page and Robinson, 1995; Gaidarov and Keen, 1999), the μ-subunit (Meyer et al., 2000), and possibly the ς-subunit as well (Page and Robinson, 1995). The variable hinge-like domain would bind clathrin, like the hinge domains of the β-subunit (Shih et al., 1995; Dell'Angelica et al., 1998; ter Haar et al., 2000) and γ/α-subunit (Goodman and Keen, 1995; Morgan et al., 2000; Doray and Kornfeld, 2001). The ear domain would recruit accessory proteins, like the α- and β-ear domains of the AP-2 complex (Owen et al., 2000; Slepnev and De Camilli, 2000). Thus, a GGA molecule would be able to perform all of the functions of an AP complex but with one polypeptide instead of four.
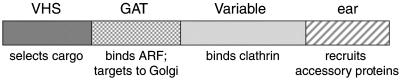
Figure 10.
Schematic diagram showing the four distinct GGA domains and their predicted functions. The VHS domain has been shown to bind to the cytoplasmic tails of proteins that traffic from the TGN to endosomes (Nielsen et al., 2001; Puertollano et al., 2001a; Zhu et al., 2001), indicating that it is involved in cargo selection. The GAT domain both binds ARF and targets the protein to the Golgi apparatus (Boman et al., 2000; Dell'Angelica et al., 2000; Puertollano et al., 2001b). The variable domain binds clathrin (Costaguta et al., 2001; Puertollano et al., 2001b). The ear domain is likely to recruit accessory proteins, by analogy with the α-adaptin and γ-adaptin ear domains (Page et al., 1999; Slepnev and De Camilli, 2000).
In this respect, the GGAs may be similar to the nonvisual arrestins at the plasma membrane. Arrestin-2 and arrestin-3 can bind both cargo (G protein-coupled receptors) and clathrin and are thought to act as alternative adaptors (Goodman et al., 1996). However, there are also important differences between the nonvisual arrestins and the GGAs. The GGAs appear to have the ability not only to bind cargo and clathrin but also to associate with the appropriate membrane and to recruit accessory proteins, two properties that have not been reported for the arrestins. Moreover, the nonvisual arrestins colocalize with AP-2 and there is evidence that they bind to AP-2 as well as clathrin (Laporte et al., 1999), whereas the GGAs appear to function essentially independently of AP-1.
Why does the cell need both GGAs and AP-1? One possibility is that they select different types of cargo at the TGN, in the same way that the nonvisual arrestins and AP-2 select different types of cargo at the plasma membrane. However, both GGAs and AP-1 have been shown to interact with mannose 6-phosphate receptors in vitro, even though they recognize different motifs (Glickman et al., 1989; Höning et al., 1997; Puertollano et al., 2001a), and both have been shown to colocalize with mannose 6-phosphate receptors at the TGN (Klumperman et al., 1993; Puertollano et al., 2001a). Alternatively, the GGAs and AP-1 may be recruited onto different subdomains of the TGN (Ladinsky et al., 1994), facilitating traffic from essentially distinct compartments. This would be consistent with our observation that the GGAs and AP-1 have distinct distributions by immunofluorescence. There is also evidence that the GGAs and AP-1 may facilitate traffic in different directions, because mannose 6-phosphate receptors in AP-1–deficient cells are impaired in their ability to recycle back to the TGN (Meyer et al., 2000), whereas expression of a dominant negative GGA mutant in mammalian cells (Puertollano et al., 2001 a, b), or deletion of GGAs in yeast (Costaguta et al., 2001), impairs the export of such proteins from the TGN. Further studies of the GGAs, AP-1, and clathrin, using both yeast and mammalian systems, should help to clarify the functional relationship between these three coat components.
ACKNOWLEDGMENTS
We thank Matthew Seaman for the vps4 strain and anti-Vps10p and for invaluable input into the project, Annette Boman for the antibody against Gga1p, Claus Munck Petersen for sharing unpublished results, Rainer Duden for the original YPH500 strain and α-factor antibody, and Matthew Seaman, Rainer Duden, Paul Luzio, John Kilmartin, and members of the Robinson laboratory for reading the manuscript and for helpful discussions. This work was supported by grants from the Wellcome Trust and the Medical Research Council.
REFERENCES
- Austin C, Hinners I, Tooze SA. Direct and GTP-dependent interaction of ADP-ribosylation factor 1 with clathrin adaptor protein AP-1 on immature secretory granules. J Biol Chem. 2000;275:21862–21869. doi: 10.1074/jbc.M908875199. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Black MW, Pelham HR. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Zhang CJ, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM. Clathrin structure characterized with monoclonal antibodies. I. Analysis of multiple antigenic sites. J Cell Biol. 1985;101:2047–2054. doi: 10.1083/jcb.101.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino J-L, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCostaguta C, Stefan CJ, Bensen ES, Emr DD, Payne GS. Yeast Gga proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Kornfeld S. γ subunit of the AP-1 adaptor binds clathrin: implications for cooperative binding in coated vesicle assembly. Mol Biol Cell. 2001;12:1925–1935. doi: 10.1091/mbc.12.7.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I, Keen JH. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J Cell Biol. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman JN, Conibear E, Pearse BMF. Specificity of binding of clathrin adaptors to signals on the mannose-6-phosphate/insulin-like growth factor II receptor. EMBO J. 1989;8:1041–1047. doi: 10.1002/j.1460-2075.1989.tb03471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman OB, Keen JH. The alpha chain of the AP-2 adaptor is a clathrin binding subunit. J Biol Chem. 1995;270:23768–23773. doi: 10.1074/jbc.270.40.23768. [DOI] [PubMed] [Google Scholar]
- Goodman OB, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Fine Structure Immunocytochemistry. Berlin: Springer-Verlag; 1993. [Google Scholar]
- Hirst J, Lui WWY, Bright NA, Totty N, Seaman MNJ, Robinson MS. A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the TGN and the vacuole/lysosome. J Cell Biol. 2000;149:67–79. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höning S, Sosa M, Hille-Rehfeld A, von Figura K. The 46-kDa mannose 6-phosphate receptor contains multiple binding sites for clathrin adaptors. J Biol Chem. 1997;272:19884–19890. doi: 10.1074/jbc.272.32.19884. [DOI] [PubMed] [Google Scholar]
- Huang KM, D'Hondt K, Riezman H, Lemmon SK. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 1999;18:3897–3908. doi: 10.1093/emboj/18.14.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Nathanson KL, Matsui W, Vaisberg A, Chow EP, Burne C, Keen JH, Davis AE. Structural and functional division into two domains of the large (100- to 115-kDa) chains of the clathrin-associated protein complex AP-2. Proc Natl Acad Sci USA. 1989;86:2612–2616. doi: 10.1073/pnas.86.8.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Hille A, Veenendaal T, Oorschot V, Stoorvogel W, von Figura K, Geuze HJ. Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J Cell Biol. 1993;121:997–1010. doi: 10.1083/jcb.121.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh RJ, Howell KE. HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J Cell Biol. 1994;127:29–38. doi: 10.1083/jcb.127.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson SS, Caron MG, Barak LS. The beta2-adrenergic receptor/beta arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Natl Acad Sci USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- Liu SH, Marks MS, Brodsky FM. A dominant-negative clathrin mutant differentially affects trafficking of molecules with distinct sorting motifs in the class II major histocompatibility complex (MHC) pathway. J Cell Biol. 1998;140:1023–1037. doi: 10.1083/jcb.140.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Serafini T, Orci L, Shepherd JC, Rothman JE. Purification of a novel class of coated vesicles mediating biosynthetic protein transport through the Golgi stack. Cell. 1989;58:329–336. doi: 10.1016/0092-8674(89)90847-7. [DOI] [PubMed] [Google Scholar]
- Mao Y, Nickitenko A, Duan X, Lloyd TE, Wu MN, Bellen H, Quiocho FA. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–456. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phsophate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Beach BM, Hurley JH. Structure of the VHS domain of human Tom1 (target of myb 1): insights into interactions with proteins and membranes. Biochemistry. 2000;39:11282–11290. doi: 10.1021/bi0013546. [DOI] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Hao W, Augustine GJ, Lafer EM. A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J Neurosci. 2000;20:8667–8676. doi: 10.1523/JNEUROSCI.20-23-08667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Madsen P, Christensen EI, Nykjaer A, Gliemann J, Kaspar-Biermann D, Pohlmann R, Petersen CM. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LJ, Robinson MS. Targeting signals and subunit interactions in coated vesicle adaptor complexes. J Cell Biol. 1995;131:619–630. doi: 10.1083/jcb.131.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LJ, Sowerby PJ, Lui WWY, Robinson MS. γ-Synergin: an EH domain-containing protein that interacts with γ-adaptin. J Cell Biol. 1999;146:993–1004. doi: 10.1083/jcb.146.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne GS, Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Phan HL, Finlay JA, Chu DS, Tan P, Kirchhausen T, Payne GS. The S. cerevisiae APS1 gene encodes a homologue of the small subunit of the mammalian clathrin AP-1 complex: evidence for functional interaction with clathrin at the Golgi complex. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch PF, Shia MA, Benson RJ, Fine RE. Coated vesicles participate in the receptor-mediated endocytosis of insulin. J Cell Biol. 1983;96:133–138. doi: 10.1083/jcb.96.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poussu A, Lohi O, Lehto VP. Vear, a novel Golgi-associated protein with VHS and gamma-adaptin “ear” domains. J Biol Chem. 2000;275:7176–7183. doi: 10.1074/jbc.275.10.7176. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001a;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Randazzo PA, Presley JF, Hartnell LM, Bonifacino JS. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001b;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- Rapoport I, Chen YC, Cupers P, Shoelson SE, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Assembly and targeting of adaptin chimeras in transfected cells. J Cell Biol. 1993;123:67–77. doi: 10.1083/jcb.123.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Pearse BMF. Immunofluorescent localization of 100K coated vesicle proteins. J Cell Biol. 1986;102:48–54. doi: 10.1083/jcb.102.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seaman MNJ, Ball CL, Robinson MS. Targeting and mistargeting of plasma membrane adaptors in vitro. J Cell Biol. 1993;123:1093–1105. doi: 10.1083/jcb.123.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Marcusson EG, Cereghino J-L, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Sowerby PJ, Robinson MS. Cytosolic and membrane-associated proteins involved in the recruitment of AP-1 adaptors onto the trans-Golgi network. J Biol Chem. 1996;271:25446–25451. doi: 10.1074/jbc.271.41.25446. [DOI] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih W, Gallusser A, Kirchhausen T. A clathrin binding site in the hinge of the β2 chain of mammalian AP-2 complexes. J Biol Chem. 1995;270:31083–31090. doi: 10.1074/jbc.270.52.31083. [DOI] [PubMed] [Google Scholar]
- Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev VI, De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ. The Use of Protein A-Colloidal Gold (PAG) Complexes as Immunolabels in Ultrathin Frozen Sections, C. A.C. Chichester, UK: John Wiley & Sons; 1983. [Google Scholar]
- Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon S. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Yoshino K, Nakayama K. Adaptor gamma ear homology domain conserved in gamma-adaptin and GGA proteins that interact with gamma-synergin. Biochem Biophys Res Commun. 2000;271:719–725. doi: 10.1006/bbrc.2000.2700. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Harrison S C, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT. A study of positive staining of ultrathin frozen sections. J Ultrastruct Res. 1978;63:287–307. doi: 10.1016/s0022-5320(78)80053-7. [DOI] [PubMed] [Google Scholar]
- West MA, Bright NA, Robinson MS. The role of ADP-ribosylation factor and phospholipase D in adaptor recruitment. J Cell Biol. 1997;138:1239–1254. doi: 10.1083/jcb.138.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestehube LJ, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung BG, Phan HL, Payne GS. Adaptor complex-independent clathrin function in yeast. Mol Biol Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdankina O, Strand NL, Redmond JM, Boman AL. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Doray B, Poussu A, Lehto V-P, Kornfeld S. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science. 2001;292:1716–1718. doi: 10.1126/science.1060896. [DOI] [PubMed] [Google Scholar]