Abstract
The yeast cyclin-dependent kinase Cdc28p regulates bud morphogenesis and cell cycle progression via the antagonistic activities of Cln and Clb cyclins. Cln G1 cyclins direct polarized growth and bud emergence, whereas Clb G2 cyclins promote isotropic growth of the bud and chromosome segregation. Using colony morphology as a screen to dissect regulation of polarity by Cdc28p, we identified nine point mutations that block the apical-isotropic switch while maintaining other functions. Like a clb2Δ mutation, each confers tubular bud shape, apically polarized actin distribution, unipolar budding, and delayed anaphase. The mutations are all suppressed by CLB2 overexpression and are synthetically lethal with a CLB2 deletion. However, defects in multiple independent pathways may underlie their common phenotype, because the mutations are scattered throughout the CDC28 sequence, complement each other, and confer diverse biochemical properties. Glu12Gly, a mutation that alters a residue involved in Swe1p inhibition of Cdc28p, was unique in being suppressed by deficiency of SWE1 or CLN1. With wild-type CDC28, filament formation induced by CLN1 overexpression was markedly decreased in a SWE1 deletion. These results suggest that Swe1p, via inhibition of Clb2p/Cdc28p, may mediate much of the effect of Cln1p on filamentous morphogenesis.
INTRODUCTION
From G1 exit to the completion of mitosis, the growth of a yeast cell is limited to the bud (Lew et al., 1997). Delivery of cell wall components to the bud surface is under close regulation during cell cycle progression (Lew and Reed, 1995). From bud emergence to the onset of mitosis, new cell wall is deposited primarily at the bud tip, causing tubular growth. In mitosis, a switch from apical to isotropic growth allows delivery of cell wall material over the entire bud surface, and the bud fattens to adopt a rugby-ball shape. The cyclin-dependent kinase Cdc28p not only maintains the order of DNA replication and chromosome segregation but also regulates actin localization (Lew and Reed, 1995; Mendenhall and Hodge, 1998; Pruyne and Bretscher, 2000a). Cell cycle regulation of cortical actin assembly and the resulting control of localized secretion underlie bud morphogenesis (Madden and Snyder, 1998; Chant, 1999; Pruyne and Bretscher, 2000a,b). The G1 cyclins Cln1p and Cln2p activate Cdc28p to promote S phase onset and restrict cortical actin to an apical distribution, leading to polarized secretion and tubular growth. In mitosis, the cyclins Clb1p and Clb2p activate Cdc28p to direct chromosome segregation and to redistribute actin throughout the bud, promoting isotropic growth until mitotic exit.
When activation of Cdc28p by Clb2p is slowed, both mitotic onset and the apical-isotropic switch are delayed (Lew and Reed, 1993). CLB2-deletion cells display a pattern of mitotic delay, tubular bud shape, unipolar budding, and delayed cell separation (Surana et al., 1991; Lew and Reed, 1993; Sheu et al., 2000). A similar phenotype is conferred by overexpression of Cln1p or Cln2p or by ectopic activation of the protein kinase Swe1p, which provides inhibitory phosphorylation on Cdc28p Tyr19. This response recapitulates the morphogenetic pattern of filamentous growth (Kron and Gow, 1995), a developmental option in budding yeast that is normally stimulated by nitrogen starvation (Gimeno et al., 1992). Indeed, mutations that delay the apical-isotropic switch uniformly confer an enhanced filamentous growth phenotype. For example, clb2 mutants exhibit dramatically enhanced filamentous growth (Ahn et al., 1999; Edgington et al., 1999). In addition, overexpression of CLN1 (Oehlen and Cross, 1998; Loeb et al., 1999), activation of Swe1p (Ahn et al., 1999; Edgington et al., 1999), inactivation of the Mih1p phosphatase that antagonizes Swe1p (Ahn et al., 1999), or loss of cell cycle-dependent expression of CLB2 (Hollenhorst et al., 2000; Zhu et al., 2000) all enhance filamentous differentiation. Directly linking CDC28 to filamentous growth and the apical-isotropic switch, the temperature-sensitive allele cdc28-1N (Pro250Leu) confers persistently polarized cell shape, slow mitotic progression, and constitutively enhanced filamentous growth at permissive temperature (Surana et al., 1991; Ahn et al., 1999). A second mutant, cdc28-127 (Cys127Tyr), is cold-sensitive, constitutively elongated (Blacketer et al., 1995; Edgington et al., 1999), and has enhanced filamentous growth. The defects in these mutants may derive from inadequate mitotic activity of Cdc28p, because overexpression of CLB2 suppresses the enhanced polarity and blocks filamentous growth in both the cdc28-1N and cdc28-127 mutants (Ahn et al., 1999; Edgington et al., 1999).
Like cell cycle mutations, defects in actin cytoskeletal components, bud site selection regulators, and other determinants of cell polarity confer dramatic filamentous growth phenotypes (Gimeno et al., 1992; Mösch and Fink, 1997; Cali et al., 1998). The sensitivity of filamentous growth to determinants of cell polarity and Cdc28p activity suggested that it would be a useful tool for exploring links between cell cycle regulation and morphogenesis. We hypothesized that screening a collection of CDC28 mutants for constitutive filamentous growth might reveal alleles with specific defects in the apical-isotropic switch. We have isolated new CDC28 alleles that enhance cell polarization without affecting essential functions. The mutants share a constellation of cell growth and cell polarity phenotypes. However, genetic analysis suggests that these mutants do not identify a single pathway but instead affect diverse functions of Cdc28p required for the apical-isotropic switch and mitotic progression.
MATERIALS AND METHODS
Yeast Strains, Media, and Molecular and Genetic Procedures
Yeast strains (Table 1) were derived in the Σ1287b background (Grenson et al., 1966; Gimeno et al., 1992). Genotypes were confirmed by marker segregation, Southern blot, and/or polymerase chain reaction (PCR) assay. Media and reagents were obtained from United States Biological (Swampscott, MA), Fisher (Pittsburgh, PA), and Sigma (St. Louis, MO) and were prepared as described by Ahn et al. (1999). YPG contains 2% galactose and 2% raffinose.
Table 1.
Yeast strains
| Straina | Genotype | Source |
|---|---|---|
| L5683 | MATα ura3-52 | G. Fink |
| L5977 | MATa ura3-52 his3 trp1 | G. Fink |
| SKY723 | MATa/MATα ura3-52/ura3-52 his3/his3 trp1/trp1 | |
| SKY750 | MATa/MATα cdc28Δ∷TRP1/cdc28Δ∷TRP1ura3-52/ura3-52 his3/his3 trp1/trp1 [pCT3 CDC28] | |
| SKY754 | MATa ura3-52 | Ahn et al., 1999 |
| SKY757 | MATa/MATα ura3-52/ura3-52 | Ahn et al., 1999 |
| SKY772 | MATa ura3-52 leu2 clb2Δ∷LEU2 | Ahn et al., 1999 |
| SKY778 | MATa/MATα ura3-52/ura3-52 leu2/leu2 clb2Δ∷LEU2/clb2Δ∷LEU2 | Ahn et al., 1999 |
| SKY786 | SKY862 clb2Δ∷LEU2/clb2Δ∷LEU2 | |
| SKY862 | SKY750 leu2/leu2 | |
| SKY940 | SKY862 mih1Δ∷LEU2/mih1Δ∷LEU2 | |
| SKY942 | SKY862 swe1Δ∷LEU2/swe1Δ∷LEU2 | |
| SKY962 | SKY862 leu2∷GAL1-CLB2∷LEU2/leu2 | Stueland et al., 1993 |
| SKY979 | MATα ura3-52 his3 | |
| SKY980 | MATa ura3-52 trp1 | |
| SKY1057 | MATa/MATα ura3-52/ura3-52 mih1Δ∷LEU2/mih1Δ∷LEU2 | |
| SKY1058 | MATa/MATα ura3-52/ura3-52 swe1Δ∷LEU2/swe1Δ∷LEU2 | |
| SKY2030 | SKY862 cln1Δ∷LEU2/cln1Δ∷LEU2 | |
| SKY2407 | MATα ura3-52 cdc28-1N | Ahn et al., 1999 |
| SKY2415 | MATa ura3-52 cdc28-1N | Ahn et al., 1999 |
| SKY2646 | SKY750 GAL-CLN1∷kanMX6/GAL-CLN1∷kanMX6 | |
| SKY2665 | MATa/MATα GAL-CLN1∷kanMX6b/GAL-CLN1∷kanMX6 | |
| SKY2666 | MATa/MATα GAL-CLN2∷kanMX6/GAL-CLN2∷kanMX6 | |
| SKY2667 | MATa/MATα swe1Δ∷LEU2/swe1Δ∷LEU2 GAL-CLN1∷kanMX6/CLN1 | |
| SKY2668 | MATa/MATα swe1Δ∷LEU2/swe1Δ∷LEU2 GAL-CLN2∷kanMX6/CLN2 | |
| SKY2669 | MATa/MATα SWE1-MYCc/SWE1-MYC | |
| SKY2670 | MATa/MATα SWE1-MYC/SWE1-MYC [pEG202 CDC28] | |
| SKY2671 | MATa/MATα SWE1-MYC/SWE1-MYC [pEG202 cdc28-E12G] | |
| SKY2672 | MATa/MATα SWE1-MYC/SWE1-MYC [pEG202 cdc28-E12K] | |
| SKY2673 | MATa/MATα swe1Δ∷LEU2/swe1Δ∷LEU2 |
All strains are Σ1287b derivatives and were generated in this study unless otherwise indicated. The his3, trp1, and leu2 alleles are hisG disruptions, as described by Liu et al. (1993).
Constructed by PCR mutagenesis using pFA6a-kanMX6 plasmid (Wach et al., 1994).
Constructed by PCR mutagenesis using pFA61-13Myc-kanMX6 plasmid (Longtine et al., 1998).
Shuffle Mutagenesis of CDC28 and Screen for Enhanced Cell Polarity Mutants
A CDC28 deletion was constructed from pRD47 (kind gift of R. Deshaies) by removing the coding sequences between an NdeI site engineered at the initiation codon and an HindIII site, leaving 51 carboxyl-terminal residues and flanking regions. BglII linkers were added and a BglII/BamHI TRP1 fragment was cloned into the plasmid. The knockout construct was then excised with PvuII and transformed into SKY723. The XhoI/SpeI fragment of pRD47 cloned into the SalI and XbaI sites of pCT3, a CEN URA3 plasmid (kind gift of C. Thompson), was transformed into the heterozygous cdc28::TRP1 diploid. Trp+, 5-fluoro-orotic acid (5-FOA)-sensitive meiotic segregants were mated to form SKY750, a diploid shuffle mutagenesis strain. These segregants were crossed (e.g., to SKY772 for the clb2Δ::LEU2 allele) or transformed and mated to form other diploid shuffle strains. Wild-type CDC28 and cdc28-1N were amplified with PCR primers, CDC28L, GGAATAGAATTATCGTTCTCG, and CDC28R, GAGCAATGAATTTGCGGAGG, the XhoI and MunI genomic sites were restricted, and the fragment was ligated into SalI/EcoRI-digested pRS413 (Sikorski and Hieter, 1989). Inserts were confirmed by sequencing. SKY750 transformed with pRS413-CDC28 or pRS413-cdc28-1N was transferred to 5-FOA medium at 22°C to evict pCT3-CDC28.
Error-prone PCR was used to construct a library of mutant CDC28 genes. Thirty-five cycles of PCR were performed with 1 min of annealing at 55°C and 2 min of extension at 72°C using a genomic DNA template, primers CDC28L and CDC28R, 50 μM MnCl2, and 5% 2-mercaptoethanol, in four reactions limiting dATP, dTTP, dCTP, or dGTP to 25 μM. The PCR products were pooled, digested with XhoI/MunI, ligated into SalI/EcoRI-digested pRS413, and electroporated into Escherichia coli DH5α to recover ∼106 clones, from which plasmid DNA was prepared. SKY750 transformed with the mutant library was plated onto 5-FOA medium. When colonies were pooled, plated onto low-nitrogen synthetic media, and incubated for 36 h at 30°C, nearly 0.5% of the colonies displayed enhanced filamentous growth. When the pRS413-cdc28 plasmids were recovered from 48 clones, retransformed into SKY750, and plated on 5-FOA, 45 yielded colonies with enhanced filamentous growth, of which 10 displayed a growth defect at 37°C. The DNA sequences of the CDC28 open reading frames (ORFs) were determined from the 35 non-ts CDC28 mutant candidates. Nine point mutants were selected for further analysis; their sequences were confirmed, and they were designated the cdc28ECP alleles. For further analysis, a URA3-marked integrating plasmid carrying each cdc28ECP allele was first constructed by subcloning the insert from pRS413 into pRS306 (Sikorski and Hieter, 1989). Haploid strains carrying the cdc28ECP alleles at the CDC28 genomic locus were then constructed by transforming SKY2407 and SKY2415 with the pRS306-cdc28ECP plasmids after digestion with HindIII, to direct integration and create a tandem duplication of cdc28ECP and cdc28-1N flanking URA3. After selection of 5-FOA–resistant segregants at 37°C, the CDC28 ORF was PCR amplified and sequenced to confirm the presence of the cdc28ECP allele and absence of the cdc28-1N allele. The haploid cdc28ECP integrants were backcrossed to CDC28 wild-type strain SKY979 or SKY980, and the cdc28ECP mutation was reconfirmed by sequencing in each segregant used for further analysis.
Structural Modeling
Atomic models of Cdc28p were based on the crystallographic structures of CDK2 in its inactive, dephosphorylated, and monomeric (De Bondt et al., 1993), active, phosphorylated and cyclin-bound (Russo et al., 1996b), and active, phosphorylated, and cyclin- and substrate-bound (Brown et al., 1999) forms. Monomeric and cyclin-bound models of Cdc28p were constructed manually with Biosym Insight II Homology Modeling software and also by subjecting the Cdc28p sequence to the SwissProt Modeling server after initial threading with the SwissPDBViewer (http://www.expasy.ch/spdbv/). When the Cdc28p models were compared, Cα traces of the Swiss and Biosym Cdc28p models based on 1FIN were superimposable with an RMS deviation of 0.78 Å and varied from the reference CDK2 structure by root mean square (RMS) values of 15.54 and 15.56 Å, respectively. These differences derive largely from surface loops that include “inserted” residues in Cdc28p.
Phenotypic Analysis
Imaging of yeast colonies growing on agar in plastic Petri dishes was performed with an Axiovert 25 (Zeiss, Oberkochen, Germany) with bright field illumination and a 32× LD Achroplan objective. Images were captured with a Pixera (Los Gatos, CA) Professional CCD camera at 1280 by 1024 resolution with Pixera VCS image acquisition software. Images were converted to gray scale, cropped, and assembled in Photoshop (Adobe Systems, Mountain View, CA). Analysis of cell elongation was performed at 22°C as described by Mösch and Fink (1997), with scores ranging from +/− (round) to ++++ (spindle shaped). Staining of fixed cells with DAPI, Calcofluor, and rhodamine or fluorescein phalloidin (Molecular Probes, Eugene, OR) and imaging were as described by Ahn et al. (1999). Polarization of actin in budded cells was scored based on the distribution of cortical actin patches as apical, isotropic, septal, or intermediate/indeterminate. For flow cytometry, yeast cells at 22°C were stained with propidium iodide and analyzed as described previously (Ahn et al., 1999). The G2/G1 ratio was determined from the ratio of the percentages of cells in two fluorescence intensity (FL-2) gates that included the majority of cells with G1 or G2/M DNA contents, respectively. To test sensitivity to mating pheromone, log-phase cultures were spread onto YPD medium onto which were then placed 1-cm-diameter glass fiber filters saturated with 10 μl of 3 mM α-factor in dimethyl sulfoxide. The diameters of cleared halos were measured after 2 d at 22°C.
Molecular and Biochemical Assays
For Northern analyses, 20-μg samples of total cellular RNA were separated on 1% agarose-formaldehyde gels and transferred to Nytran membranes (Schleicher & Schuell, Keene, NH). Probes for ACT1. CDC28, CLB2, CLN2, and CLN1 were labeled by PCR performed with [32P] dATP using the ORFs as templates. Whole-cell extracts were prepared and subjected to Western analysis as described before (Ahn et al., 1999). Yeast extracts were assayed for p13suc1-associated and Clb2p-associated Cdc28p histone H1 kinase activity by a modification of published methods (Surana et al., 1991; Amon et al., 1994). 32P labeling of histone H1 was detected by phosphorimager analysis (Molecular Dynamics, Sunnyvale, CA) of SDS-PAGE gels. Quantitation of band intensity with Imagequant (Molecular Dynamics) and IPLab (Scanalytics, Billerica, MA) showed that both kinase activities were specific and linear with respect to extract and incubation. H1 kinase activity that immunoprecipitated from wild-type cell lysate by anti-Clb2p antibody was completely absent from the clb2Δ::LEU2 mutant, confirming the specificity of this assay (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results). For the two-hybrid analysis, the CDC28 ORF was amplified from genomic DNA with primers CCTGAATTCATGAGCGGTGAATTAGC and GTGGTCGACTAATG CTTATGATTCTTGG, and the in-frame EcoRI and SalI (indicated in bold) restriction sites were used to clone CDC28 into the two-hybrid vectors pEG202 and pJG4–5 (Golemis et al., 1999). pEG202-CDC28 complements a cdc28 shuffle strain, and pJG4–5-CDC28 complements cdc28-4 and cdc28-1N. Plasmids were transformed into yeast strain EGY48 (James et al., 1996), and the transformants were examined for growth on selective media. Primers CGCGGATCCATGAGA TCTAGCGTGAATTAGCAAATTACAAAAGA and GTAATACGACTCACTATAGGGC were used to amplify the CDC28 ORF from a genomic clone. BamHI and PstI were then used to clone CDC28 into the GBDU-C1 and GAD-C1 two-hybrid vectors (James et al., 1996). Both complement cdc28-4 and cdc28-1N. GBDU-C1-CDC28 and GAD-C1-CDC28 were transformed into yeast strain pJ69–4α (James et al., 1996). Transformants were examined on selective medium to detect possible interactions. For coimmunoprecipitation analysis, extracts from strain L5977 carrying pEG202-CDC28 and pJG4–5-CDC28 were analyzed by immunoprecipitation with 12-CA5 anti-hemagglutinin (HA) antibody (Roche, Gipf-Oberfrick, Switzerland), SDS-PAGE, and transfer to a polyvinylidene difluoride membrane. With the use of standard procedures, the blots were blocked, incubated with 16 B12 anti-HA (Berkeley Antibody, Berkeley, CA) or anti-LexA (Golemis et al., 1999) antibody, probed with sheep anti-mouse-immunoglobulin (Ig)-horseradish peroxidase (HRP) conjugate (Amersham, Arlington Heights, IL) or donkey anti-rabbit- Ig-HRP conjugate (Amersham), respectively, and developed with enhanced chemiluminescence as described by the manufacturer (Pierce, Rockford, IL). In a separate experiment, the CDC28, cdc28-E12K (kind gift of D. Lew), and cdc28-E12G ORFs were cloned into the pEG202 vector. Each plasmid was transformed into a strain homozygous for Myc-tagged SWE1 (SKY2671). Incubation of extracts prepared from these strains with protein A-Sepharose beads was followed by immunoprecipitation with 9E10 anti-HA (Berkeley Antibody, Berkeley, CA), SDS-PAGE, and transfer to a nitrocellulose membrane. The blots were blocked, incubated with 1:1000 A14 anti-MYC antibody (Santa Cruz) or 1:5000 anti-LexA (gift of E. Golemis) antibody, probed with donkey anti-rabbit-Ig-HRP conjugate (Amersham), and developed with the use of enhanced chemiluminescence.
RESULTS
New CDC28 Point Mutations That Promote Polar Bud Growth
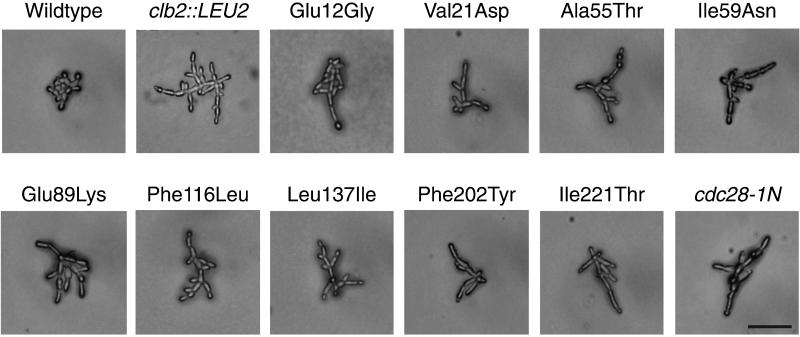
To investigate the role of the cyclin-dependent kinase in polarized morphogenesis, we mutagenized CDC28 and screened for altered filamentous growth in a Σ1278b background (see MATERIALS AND METHODS). We isolated nine point mutations in CDC28 that confer enhanced filamentation in otherwise wild-type cells (Figure 1). Like cdc28-1N or clb2Δ::LEU2 (Ahn et al., 1999), the cdc28ECP mutant diploids display a high proportion of highly elongated buds and other markers of enhanced cell polarity. Whereas budded wild-type cells score +/− (round) in a visual cell elongation assay and display ∼40% apically polarized actin distribution, cdc28-1N and clb2Δ::LEU2 each score ++++ (spindle shaped) and display 83 and 82% polar actin, respectively. In the same assays, each cdc28ECP mutant scored ++++ and displayed 71–81% apical polar actin distribution. By contrast to wild-type, very few of the remaining cells displayed actin patches distributed over the bud cortex. In addition, like cdc28-1N and clb2Δ::LEU2 (Ahn et al., 1999), the mutants perform unipolar budding, with the majority of mother cells displaying all bud scars localized to the same pole as the bud.
Figure 1.
Mutations in CDC28 that confer enhanced polar morphogenesis. Photomicrographs are of homozygous mutant cells grown to exponential phase in YPD liquid at 30°C. Bar, 50 μm.
Like cdc28-1N, the mutants remain filamentous even on rich medium and do not require an intact Ras-signaling pathway (Gimeno et al., 1992), because all retain their phenotypes in a RAS2-deficient shuffle strain (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results). However, by comparison to cdc28-1N, the defects are relatively specific to morphogenesis. The cdc28ECP mutants grow well at 14, 22, 30, and 37°C and they are similar to wild type in sensitivity to α-mating pheromone, as measured by halo assay; growth on plates containing 200 mM hydroxyurea, 1 M NaCl, or 12 μg/ml benomyl; and recovery from 25 mJ/cm2 254-nm UV irradiation.
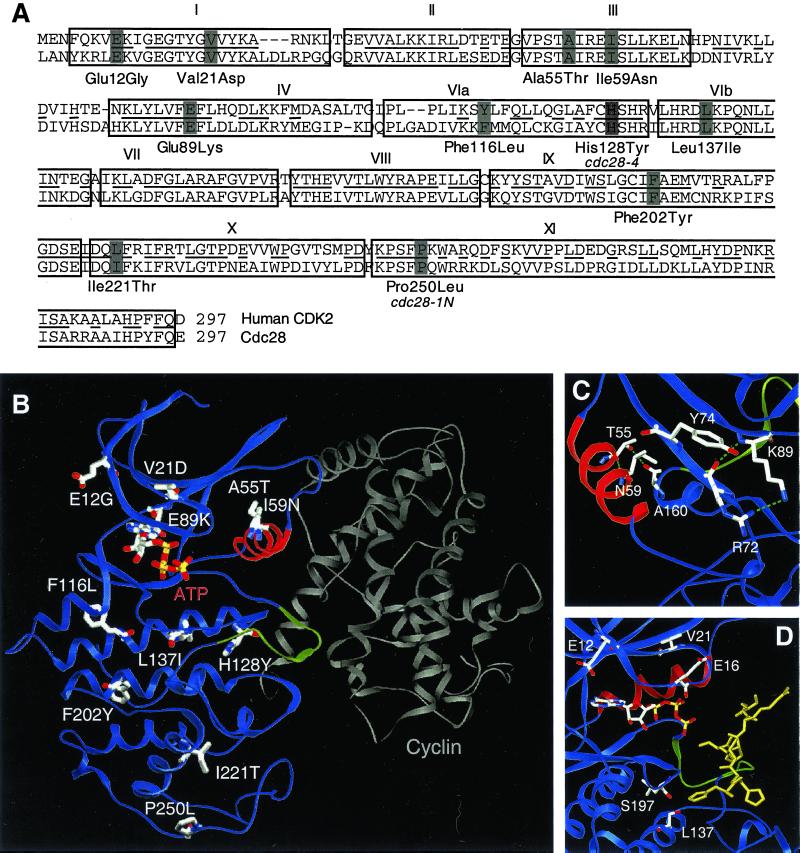
The Enhanced Cell Polarity Mutations Alter Structurally Distinct Residues
The cdc28ECP mutations are scattered throughout the primary sequence. Mutated residues include kinase consensus sites considered important for catalysis and CDK-specific residues shown to participate in regulation (Figure 2A). To gain further insight into the likely structural consequences of the mutations, we derived atomic models of Cdc28p from crystal structures of human CDK2 (Figure 2B). The models share a small amino-terminal domain composed primarily of β-sheet that includes Hanks domains I to III and a large α-helical carboxyl-terminal domain formed from subdomains VI through XI. The catalytic core, Hanks subdomains IV and V, forms a hinge between the two domains. Placing the cdc28ECP mutations onto the models revealed that seven of the nine residues are exposed on the solvent-accessible surface.
Figure 2.
Structural analysis of the cdc28ECP mutants. (A) Cdc28p and human CDK2 sequences aligned by Clustal (Higgins et al., 1996). Boxes delineate consensus kinase subdomains (Hanks et al., 1988) and bars decorate residues altered in cdc28-4, cdc28-1N, and the cdc28ECP mutants. (B) The residues altered in cdc28-4, cdc28-1N, and the cdc28ECP alleles are depicted on a ribbon diagram of Cdc28p modeled after CDK2 in active complex with cyclin A. The PSTAIRE helix is highlighted in red, the inhibitory T-loop in green, and cyclin A in gray. (C) Glu89Lys, Ala55Thr, and Ile59Asn, the mutations in or near PSTAIRE, are shown modeled into the inactive, monomeric form of the enzyme. The view shows the PSTAIRE helix (red) from the perspective of an “approaching” cyclin A. (D) The residues mutated in Leu137Ile, Glu12Gly, and Val21Asp are decorated in a model of Cdc28p with peptide substrate bound. The view shows the ATP as a stick diagram, PSTAIRE in red, the T loop in green, and the substrate peptide HHASPRK (Brown et al., 1999) in gold.
Based on this modeling, the Glu89, Ala55, and Ile59 side chains should directly contact cyclin. Rotation of the PSTAIRE helix upon cyclin binding moves the catalytic residue Glu51 into the active site and exposes hydrophobic residues including Ala55 and Ile59 at the CDK/cyclin interface (Figure 2C). The Ala55Thr or Ile59Asn substitution may stabilize the monomer and/or disrupt Van der Waals interactions with cyclin partners. Similarly, Glu89Lys may restrict mobility of the amino-terminal domain during cyclin binding and/or alter specificity toward cyclin partners. Based on Cdc28p modeled with bound ATP and peptide substrate (Figure 2D), Glu12, Val21, and Leu137 may influence kinase activity and/or interactions with substrates or binding partners. Glu12 and Val21 lie above the active site in the amino-terminal domain. In CDK2 and CDK6, the corresponding residues contact stoichiometric inhibitors (Russo et al., 1996a, 1998). Leu137 sits below the T loop. The additional methyl group in Leu137Ile may affect substrate contacts directly or by displacing neighboring residues such as Ser197. Phe116, Ile221, and Phe202 are all hydrophobic residues in the carboxyl-terminal domain (Figure 2B). In both inactive and cyclin-bound conformations, these side chains are predicted to be partially or completely buried at sites distant from known CDK protein-protein interaction surfaces.
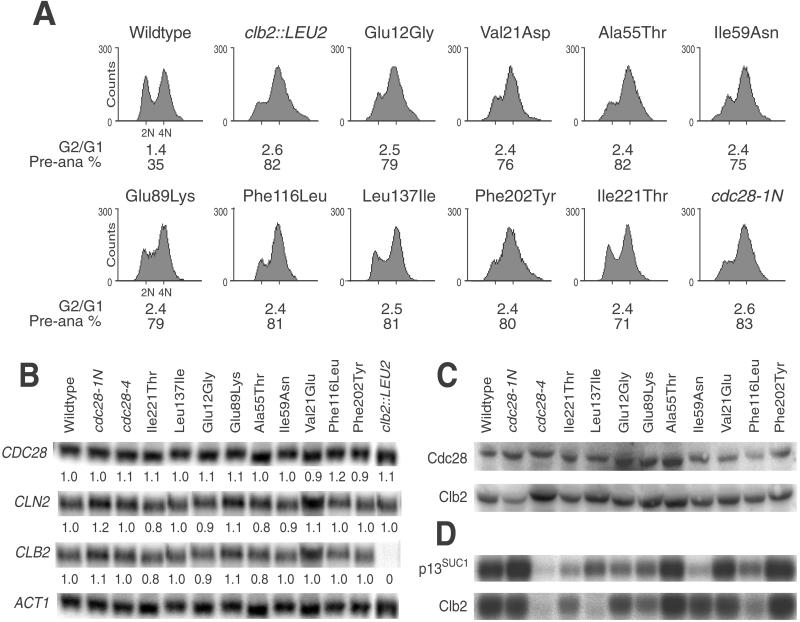
Cell Cycle and Biochemical Defects in cdc28ECP Mutants
Flow cytometry of cdc28ECP mutant diploids revealed a shift toward 4N DNA content equivalent to that of a clb2Δ::LEU2/clb2Δ::LEU2 mutant (Figure 3A). Moreover, disproportionately many large-budded cdc28ECP cells contain a single round or oblong nucleus at or near the bud neck (>75% for mutant, 35% for wild-type), consistent with a preanaphase delay. To test for altered expression or stability of the cdc28ECP mutant proteins or cyclins, we performed Northern and Western analyses. Expression analysis of CDC28, CLB2, and CLN2 in comparison to ACT1 in log-phase cultures revealed little or no change in the abundance of CDC28 or cyclin message (Figure 3B). In turn, Western analysis revealed amounts of Cdc28p and Clb2p comparable to those in wild type in each cdc28ECP mutant (Figure 3C). To test whether the cdc28ECP mutant proteins are specifically defective in activation by the Clb2p cyclin, we compared their Clb2p-associated H1 kinase activity to total p13Suc1-associated H1 kinase activity. Whole-cell lysates from cdc28-1N, cdc28-4, and the cdc28efg alleles were analyzed similarly (Figure 3D). As previously reported for W303-related strains (Surana et al., 1991), wild-type, cdc28-1N, and cdc28-4 strains yielded high Clb2p-/high p13Suc1-, high Clb2p-/high p13Suc1-, and low Clb2p-/low p13Suc1-associated kinase activities, respectively, in the Σ1278b background. In contrast, the cdc28ECP alleles yielded several different patterns of activity. None appeared to be profoundly catalytically defective, like cdc28-4. Val21Glu expressed activities indistinguishable from those in wild type, and two mutants, Ala55Thr and Phe202Tyr, appeared to have enhanced in vitro activities. Leu137Ile and Phe116Leu yielded low Clb2p-associated kinase activities but retained proportionately higher p13Suc1-associated kinase activities. Only one allele, Ile59Asn, yielded a markedly decreased p13Suc1-associated kinase activity, but this mutant expressed a near–wild-type level of Clb2p-associated activity.
Figure 3.
Similar cell cycle kinetics but distinct enzymatic properties of cdc28ECP mutants. Wild-type strain SKY757 and diploids homozygous for the indicated mutations were grown to exponential phase and analyzed as described in MATERIALS AND METHODS. (A) The cdc28ECP mutations confer a G2/M cell cycle shift. Flow cytometry of asynchronous cultures revealed a shift to higher DNA content, and microscopic examination revealed a high percentage of preanaphase (Pre-ana) cells. (B) The cdc28ECP mutations do not alter the expression of key cell cycle regulators. Total RNA was subjected to Northern analysis with the indicated probes. Expression levels were quantitated by phosphorimager analysis and are indicated as the relative ratios of expression of each transcript normalized to the level of ACT1. CLN1 and CLN2 yielded similar results. (C) Western analysis shows that the cdc28ECP alleles do not decrease Cdc28p or Clb2p abundance. (D) The cdc28ECP mutant proteins exhibit diverse enzymatic properties. Whole-cell extracts of the indicated strains were analyzed for p13suc1- and Clb2p-associated histone H1 kinase activity.
The cdc28ECP Mutants Define Multiple Complementation Groups Modulating Cell Morphogenesis
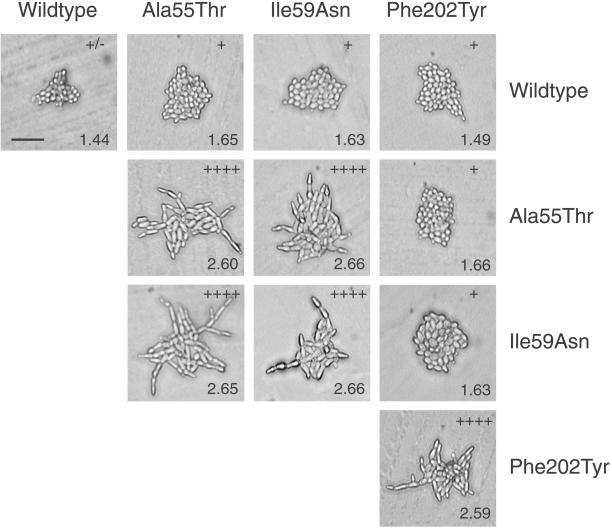
When haploids carrying the cdc28ECP alleles were mated to wild type, the cdc28ECP phenotypes were almost completely recessive (Figure 4, top row; Table 2). In turn, pairs of haploids carrying the same cdc28ECP allele yielded homozygous diploids indistinguishable from the original shuffle strain carrying the plasmid-borne allele (Figure 4). We then tested for possible interallelic complementation. Although heteroallelic combinations of Ala55Thr and Ile59Asn yielded highly polarized cells (Figure 4), indicating noncomplementation, other pairs of alleles displayed complementation for the enhanced cell polarity and G2/M-shift phenotypes (Figure 4; Table 2). This pattern of complementation suggested that Cdc28p might function as a dimer as had been observed for Cdc7p (Shellman et al., 1998). To test this possibility, the CDC28 ORF was cloned into bait-and-prey yeast two-hybrid expression vectors in each of two experimental systems. Both the bait-and-prey constructs complemented the essential functions of CDC28 in Σ1278b strains. However, when coexpressed in the appropriate strains, unlike Cdc7p, no interaction was observed as assayed by growth on selective media (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results). Moreover, no coimmunoprecipitation of an amino-terminal LexA DNA-binding domain Cdc28p fusion with a carboxyl-terminal HA epitope-tagged Cdc28p was detected when they were coexpressed in strain Σ1278b (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results).
Figure 4.
Complementation behavior of cdc28ECP mutations. Representative photomicrographs demonstrate cell morphology of homozygous and heteroallelic diploids after 24 h of incubation at 22°C on YPD agar. Polarization indices (+/− to ++++) and G2/G1 ratios (numbers) were determined as described in MATERIALS AND METHODS. Bar, 50 μm.
Table 2.
Phenotypes of homozygous and heteroallelic diploids
| Wild type | Glu12Gly | Val21Glu | Ala55Thr | Ile59Asn | Glu89Lys | Phe116Leu | Leu137Ile | Phe202Tyr | Ile221Thr | cdc28-1N | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | +/− | + | + | + | + | + | + | + | + | + | + |
| 1.44 | 1.56 | 1.61 | 1.65 | 1.63 | 1.61 | 1.55 | 1.39 | 1.49 | 1.53 | 1.6 | |
| Glu12Gly | + | ++++ | + | ++ | ++ | ++ | + | ++ | + | ++ | + |
| 1.62 | 2.62 | 1.62 | 1.83 | 1.81 | 1.71 | 1.63 | 1.76 | 1.64 | 1.61 | 1.71 | |
| Val21Glu | + | ++ | ++++ | ++ | ++ | ++ | ++ | + | ++ | ++ | + |
| 1.48 | 1.89 | 2.64 | 1.75 | 1.75 | 1.73 | 1.74 | 1.56 | 1.80 | 1.91 | 1.76 | |
| Ala55Thr | + | +++ | ++ | ++++ | ++++ | ++ | + | ++ | ++ | + | ++ |
| 1.64 | 1.99 | 1.78 | 2.65 | 2.66 | 1.90 | 1.66 | 1.86 | 1.80 | 1.71 | 1.91 | |
| Ile59Asn | + | + | ++ | ++++ | ++++ | + | + | ++ | ++ | ++ | ++ |
| 1.61 | 1.73 | 1.99 | 2.60 | 2.66 | 1.68 | 1.63 | 1.81 | 1.81 | 1.89 | 1.83 | |
| Glu89Lys | + | + | + | ++ | + | ++++ | ++ | + | ++ | ++ | + |
| 1.63 | 1.69 | 1.59 | 1.83 | 1.66 | 2.64 | 1.83 | 1.69 | 1.86 | 1.73 | 1.73 | |
| Phe116Leu | + | + | ++ | + | + | ++ | ++++ | + | + | ++ | + |
| 1.51 | 1.71 | 1.86 | 1.71 | 1.69 | 1.84 | 2.69 | 1.69 | 1.61 | 1.81 | 1.62 | |
| Leu137Ile | + | ++ | ++ | ++ | ++ | ++ | + | ++++ | + | ++ | + |
| 1.44 | 1.72 | 1.76 | 1.79 | 1.83 | 1.83 | 1.63 | 2.71 | 1.56 | 1.81 | 1.56 | |
| Phe202Tyr | + | ++ | + | + | + | + | + | ++ | ++++ | ++ | ++ |
| 1.56 | 1.88 | 1.65 | 1.68 | 1.69 | 1.69 | 1.61 | 1.87 | 2.59 | 1.88 | 1.83 | |
| Ile221Thr | + | + | ++ | ++ | ++ | ++ | + | ++ | ++ | ++++ | + |
| 1.56 | 1.61 | 1.72 | 1.83 | 1.81 | 1.79 | 1.67 | 1.87 | 1.87 | 2.66 | 1.65 | |
| cdc28-1N | + | ++ | + | ++ | ++ | ++ | ++ | + | ++ | + | ++++ |
| 1.58 | 1.78 | 1.60 | 1.91 | 1.85 | 1.83 | 1.87 | 1.59 | 1.81 | 1.62 | 2.65 |
Diploids were derived from pairs of the indicated MATa (top row) and MATα (left column) strains. Polarized growth on YPD agar and G2/G1 ratio in YPD liquid were assayed as described in MATERIALS AND METHODS. Non-complementing diploids (cell elongation = ++++, G2/G1 > 2.5) are shown in bold.
Genetic Interactions Of Cdc28ECP Alleles with Mitotic Cyclins
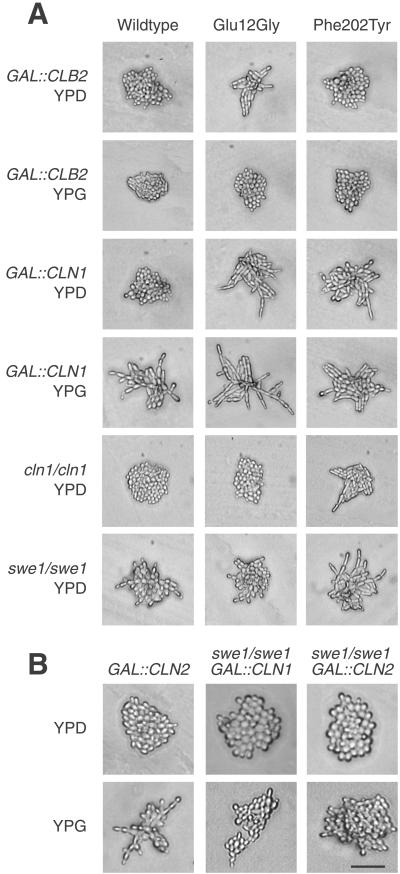
The morphogenetic and cell cycle phenotypes are consistent with the cdc28ECP alleles being deficient in functions of Cdc28p directed by Clb mitotic cyclins. To explore this possibility further, we constructed diploid shuffle strains carrying a GAL1 promoter fused to the CLB1, 2, 3, 4, or 5 gene (Stueland et al., 1993) or carrying a homozygous deletion of CLB1, 2, 3, 4, 5, or 6. Inducing GAL-CLB1 or GAL-CLB2 completely suppressed the cell elongation phenotype of the cdc28ECP alleles (Figure 5A, top 2 rows; Table 3; Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results), but little or no suppression was observed in strains carrying GAL-CLB3, GAL-CLB4, or GAL-CLB5. In addition, expression in a CLB1-deficient shuffle strain or in strains deficient for CLB5 and CLB6 or CLB3 and CLB4 did not markedly affect the cdc28ECP phenotypes (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results). However, a marked synthetic enhancement or synthetic lethality was observed in a CLB2 mutant shuffle strain (Table 3). Segregants presumed to carry a cdc28ECP allele in a clb2Δ::LEU2/clb2Δ::LEU2 shuffle strain isolated from 5-FOA and transferred to rich medium at 22°C grew only as abortive microcolonies of highly elongated cells. We used the integrated cdc28ECP alleles to confirm this result in a standard cross to a clb2Δ::LEU2 mutant. Viable meiotic segregants carrying both clb2Δ::LEU2 and cdc28-1N, Val21Asp, Ala55Thr, Ile59Asn, Phe116Leu, Leu137Ile, Phe202Tyr, or Ile221Thr were not recovered. Glu12Gly clb2Δ::LEU2 double mutants were slow growing, whereas Glu89Lys clb2Δ::LEU2 haploid cells demonstrated little growth defect. However, both Glu12Gly/Glu12Gly clb2Δ::LEU2/clb2Δ::LEU2 and Glu89Lys/Glu89Lys clb2Δ::LEU2/clb2Δ::LEU2 diploids could form only abortive microcolonies of highly elongated cells.
Figure 5.
Modulation of polarized growth by CLB2, CLN1, SWE1. (A) Homozygous diploid shuffle strains of the indicated genotypes expressing the indicated CDC28 alleles were incubated on YPD or YPG agar for 24 h at 22°C. The Phe202Tyr mutant shown is representative of the cdc28ECP alleles other than Glu12Gly. (B) Wild-type or swe1Δ mutant diploid cells carrying integrated GAL-CLN1 or GAL-CLN2 constructs were examined for polarized growth on synthetic medium containing galactose. Bar, 50 μm.
Table 3.
Phenotypes of cdc28 mutants with altered cyclin dosage
| Mutant | Polar morphogenesis when expressed in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type |
GAL-CLB2
|
clb2Δ
|
cln3Δ
|
GAL-CLN1
|
cln1Δ
|
swe1Δ
|
|||
| YPD | YPG | clb2Δ | cln3Δ | YPD | YPG | cln1Δ | swe1Δ | ||
| Wild type | +/− | + | + | ++++ | +/− | + | +++ | +/− | ++ |
| Glu12Gly | ++++ | ++ | + | L | ++ | +++ | +++++ | +/− | + |
| Val21Asp | +++ | ++ | + | L | + | ++ | +++ | +++ | ++++ |
| Ala55Thr | ++++ | ++ | + | L | ++ | ++ | +++ | ++++ | ++++ |
| Ile59Asn | ++++ | ++ | + | L | +++ | ++ | +++ | +++ | +++ |
| Glu89Lys | ++++ | ++ | + | L | ++ | ++ | +++ | +++ | ++++ |
| Phe116Leu | ++++ | ++ | + | L | ++++ | + | +++ | +++ | ++++ |
| Leu137Ile | +++ | ++ | + | L | ++ | + | +++ | +++ | ++++ |
| Phe202Tyr | ++++ | + | + | L | +++ | ++ | +++ | +++ | ++++ |
| Ile221Thr | +++ | ++ | + | L | ++ | ++ | +++ | +++ | +++ |
Shuffle diploid strains of the indicated genotypes were transformed with plasmid-borne cdc28ECP alleles. Polarized growth on YPD or YPG agar was assayed as described in MATERIALS AND METHODS. L, synthetic lethal.
Interactions of cdc28ECP Alleles with Mitotic Antagonists
Using additional diploid shuffle strains, we also tested the effects of modulating negative regulators of Clb2p/Cdc28p on polarized morphogenesis in cdc28ECP mutants. Results ranged from a SIC1-deficient strain that had only subtle effects on morphogenesis to CLN3 deficiency, which conferred a complex pattern of suppression (Table 3; Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished observations). Strikingly, genetic interactions with CLN1 and SWE1 were limited to a single mutant, Glu12Gly (Figure 5A, bottom four rows; Table 3): Induction of GAL-CLN1 markedly enhanced the polarity of this mutant, whereas CLN1 or SWE1 deficiency suppressed its elongation. Glu12Gly might sensitize Clb2p/Cdc28p to the antagonistic effects of Cln1p and Swe1p independently or via a single pathway. A related mutant, Glu12Lys, has been shown to resist the inhibitory effects of Swe1p, perhaps by altered Cdc28p-Swe1p interaction (McMillan et al., 1999). Thus, we examined whether Cdc28p-Glu12Gly, Cdc28p-Glu12Lys, or wild-type Cdc28 may interact differently with Swe1p through a coimmunoprecipitation test. Similar interaction of Swe1p-13Myc with LexA-Cdc28p-Glu12Gly, LexA-Cdc28p-Glu12Lys, and LexA-Cdc28p was detected (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results). Therefore, the importance of Swe1p in mediating the hyperfilamentous growth induced by Cdc28p-Glu12Gly may involve a direct or indirect mechanism. Next, we investigated the link to Cln1p. To establish the order of function, we examined the effects of GAL-CLN1 and GAL-CLN2 on cell polarity in SWE1- and MIH1-deficient strains carrying wild-type CDC28 (Figure 5B; Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished observations). Whereas GAL-CLN1 and GAL-CLN2 strongly enhanced polarized morphogenesis in the wild-type strain and in the mih1 mutant diploid, this effect was attenuated in the swe1 mutant, suggesting that CLN1 is upstream of SWE1 in a pathway to CDC28.
DISCUSSION
Multiple Functions of Cdc28p affecting Mitotic Progression and Polarized Morphogenesis
We used filamentous growth as a sensitive reporter of cell polarity to investigate the regulation of bud morphogenesis by the cyclin-dependent kinase Cdc28p. We isolated mutations in CDC28 that confer constitutively enhanced cell polarity (cdc28ECP) without critically affecting essential functions of the kinase. All of the mutants are characterized by apically polarized actin distribution, unipolar bud-site selection, and tube-like bud growth. These phenotypes are associated with a shift in cell cycle kinetics toward G2/M. This pattern is a close phenocopy of a deletion of the CLB2 mitotic cyclin gene. Indeed, the cdc28ECP phenotypes are suppressed by overexpression of CLB2. However, each cdc28ECP mutation is also synthetically lethal with a deletion of CLB2, indicating that the defects extend beyond a deficit in Clb2p-dependent Cdc28p activity per se. This pattern of morphogenetic phenotypes and genetic interactions is also like that of the well described mutant cdc28-1N, a ts allele with critical defects in mitotic progression. Based on the growth phenotypes of the mutants and their interactions with CLB2, a conservative hypothesis would be that the cdc28ECP mutations are each functionally equivalent to cdc28-1N, although they are weaker alleles that do not confer temperature sensitivity. However, our other genetic data suggest that the cdc28ECP alleles are fundamentally distinct from cdc28-1N and each other.
First, rather than identifying a single domain of Cdc28p, such as the Cks1p-binding site disturbed by the Pro250Leu mutation in cdc28-1N, the cdc28ECP mutations are scattered along the amino acid sequence and predicted to alter spatially distinct surfaces of the Cdc28p molecule. Certainly, residues distributed over the molecular surface might collaborate in a single process. Cyclins, via their binding to Cdc28p and the resulting conformational change, reorganize the catalytic site and recruit Cdc28p to particular substrates. By this reasoning, the structurally diverse cdc28ECP mutations might all disrupt functions of cyclins or other regulators. A genetic test of this model is to examine cells carrying combinations of mutations for their cell polarity and cell cycle phenotypes. Noncomplementation would strongly suggest a single pathway involving the two sites on the molecular surface. However, to our surprise, cells carrying nearly all the combinations of mutations exhibited a return to nearly normal cell shape and cell cycle parameters. These results favor a second model in which multiple independent activities of Cdc28p modulate cell polarity and mitotic progression. A single case of noncomplementation was observed, between Ala55Thr and Ile59Asn. The side chains of these two highly conserved residues lie adjacent to each other on the solvent-exposed surface of the PSTAIRE helix. Both residues are likely to participate in a hydrophobic interaction with cyclins. In the CDK2/cyclin A structure (Russo et al., 1996b), the Ala and Ile side chains form walls of a pocket occupied by the benzene ring of a Phe on the surface of cyclin A. Modeling suggests that the structurally equivalent residue is also a Phe in Clb2p but a Tyr in Cln1p (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results).
Alternatively, the observed pattern of interallelic complementation might derive from a mechanism other than the disruption of multiple independent functions. In particular, one explanation may be that Cdc28p functions as a dimer. In this case, the ability of two different alleles to form dimers may be the determinant of complementation. Interallelic complementation based on dimerization has been observed with Cdc7p (Shellman et al., 1998). However, our two-hybrid and coimmunoprecipitation tests were unable to detect interaction between Cdc28p monomers. Results consistent with monomeric Cdc28p being the physiologically active form have also been obtained by Sclafani and colleagues (Shellman, 1997). Thus, Cdc28p dimerization is not likely to underlie the pattern of interallelic complementation among the cdc28ECP mutants.
Thus, a picture of Cdc28p emerges as a polypeptide that via its multiple individual and combinatorial interactions with regulators and substrates performs a wide range of functions each independently required for mitotic progression. A similarly complex view of a conserved, globular protein with significant conformational flexibility has emerged from alanine-scanning mutagenesis of Act1 (Amberg et al., 1995; Cali et al., 1998). Based on this paradigm, the cdc28ECP complementation groups may reflect defects in binding various specific Cdc28p partners. Clearly, defining these partners will be of great interest.
Linking Cln1p and Swe1p-dependent Modulation of Polarity
Significantly, our studies suggest that the activities of Cln1p and Swe1p as morphogenetic regulators may be mediated through a single pathway. The enhanced cell polarity of one mutant, Glu12Gly, was markedly suppressed by deletion of SWE1. Via a Glu12Lys mutation that confers Swe1p resistance, the Glu12 residue was shown to be involved in Swe1p inhibition of Cdc28p activity (McMillan et al., 1999). Much like the nonphosphorylatable CDC28 allele Thr18Ala, Tyr19Phe (Ahn et al., 1999), Glu12Lys does not abrogate low-nitrogen or STE11-4-induced filamentous growth but confers resistance to GAL-SWE1 (Ahn, Tobe, Fitz Gerald, Anderson, Acurio, and Kron, unpublished results). Combined with the data of McMillan et al. (1999), our results suggest that much or all of the hyperpolarized phenotype of the Glu12Gly strain might derive from altered binding and/or phosphorylation of Cdc28p by Swe1p. Nonetheless, an indirect role for Swe1p in the Glu12Gly phenotype cannot be ruled out. With the use of a simple coimmunoprecipitation assay, we were unable to detect a difference in association between Swe1p and wild-type Cdc28p or Glu12Gly-mutant Cdc28p; nor did we observe lack of association of Swe1p with Glu12Lys-mutant Cdc28p.
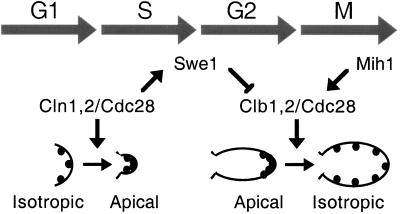
In further analyzing the Glu12Gly mutation, we made the surprising observation that hyperpolarization in Glu12Gly cells is abrogated in a CLN1-deficient mutant and markedly enhanced by CLN1 overexpression. Similar effects were not observed with any other cdc28ECP allele. Rather than invoking direct regulation of cell polarization or filamentous growth by CLN1, we infer that the effects of Cln1p may be mediated by Swe1p-dependent down-regulation of Clb2p/Cdc28p. We tested this model directly by introducing GAL-CLN1 and GAL-CLN2 into a swe1Δ background. Unlike the markedly filamentous and hyperpolarized phenotype on galactose media of a wild-type strain carrying GAL-CLN1 or GAL-CLN2, these cells remain similarly nonpolarized on glucose or galactose media. These results implicate negative regulation of Cdc28p by Swe1p as the relevant determinant of response to Cln1p dose and suggest a model for cell cycle control of the apical-isotropic switch (Figure 6). Here, Cln1p/Cdc28p complexes promote apical growth from G1 exit to mitotic onset via Swe1p-dependent inhibition of Clb2p/Cdc28p complexes. During mitosis, a combination of destruction of Cln1p and Swe1p, accumulation of Clb2p, and activation of Mih1p may release sufficient active Clb2p/Cdc28p to phosphorylate cytoskeletal targets and flip the switch to isotropic growth. This model offers a simple and attractive alternative to competitive regulation of cytoskeletal targets by Cln and Clb cyclins during S phase and G2. Further work to establish a biochemical pathway from Cln1p/Cdc28p to Clb2p/Cdc28p via Swe1p may provide insights regarding Swe1p regulation in normal growth and Swe1p activation in the morphogenesis checkpoint. In turn, placing Cln1p, Swe1p, and Clb2p in a single linear pathway reconciles previously incompatible models that propose each regulator as an independent determinant of filamentous growth.
Figure 6.
Regulation of yeast cell polarity by Cdc28p. Previous work has established antagonistic functions of Cln and Clb cyclins in morphogenesis. The polarization of cortical actin patches (black dots) and cell wall deposition to the nascent bud site at Start is Cln dependent, whereas the apical-isotropic switch leading to redistribution of actin and swelling growth of the bud in mitosis is Clb dependent. The results of this study suggest that the Cln cyclins may maintain polar growth and prevent the apical-isotropic switch by activating Swe1p, a tyrosine kinase that inhibits Clb-Cdc28p complexes.
ACKNOWLEDGMENTS
The authors wish to thank A. Amon, R. Deschaies, G. Fink, E. Golemis, P. James, D. Lew, H. Liu, A. Myers, S. Reed, P. Sorger, R. Sclafani, and the present and former members of the Kron and Fink laboratories for generously sharing reagents, unpublished results, enthusiasm, and advice. S.J.K. acknowledges his postdoctoral mentor, Dr. Gerald R. Fink, for generous support at the initiation of this project. B.T.T. is a trainee of the National Institutes of Health Medical Scientist Training Program. These studies were supported by the Arnold and Mabel Beckman Foundation, the Cancer Research Foundation, a Howard Hughes Medical Institute Research Resources for Medical Schools award, and National Science Foundation CAREER grant MCB9875976 to S.J.K.
REFERENCES
- Ahn SH, Acurio A, Kron SJ. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol Biol Cell. 1999;10:3301–3316. doi: 10.1091/mbc.10.10.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Blacketer MJ, Madaule P, Myers AM. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- Cali BM, Doyle TC, Botstein D, Fink GR. Multiple functions for actin during filamentous growth of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1873–1889. doi: 10.1091/mbc.9.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- Edgington NP, Blacketer MJ, Bierwagen TA, Myers AM. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol Cell Biol. 1999;19:1369–1380. doi: 10.1128/mcb.19.2.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Jr, Kolonin MG, Gyuris J, Brent R. Interaction trap/two-hybrid systems to identify interacting proteins. In: Ausubel F, Brent R, Kingston R, Moore D, Moore J, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 20. New York: John Wiley and Sons.; 1999. pp. 1.1–20.1.40. [Google Scholar]
- Grenson M, Mousset M, Wiame JM, Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Bose ME, Mielke MR, Muller U, Fox CA. Forkhead genes in transcriptional silencing, cell morphology, and the cell cycle: overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics. 2000;154:1533–1548. doi: 10.1093/genetics/154.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain design for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Gow NA. Budding yeast morphogenesis: signaling, cytoskeleton, and cell cycle. Curr Opin Cell Biol. 1995;7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR. Cell cycle control in Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. III. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- McMillan JN, Sia RA, Bardes ES, Lew DJ. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol Cell Biol. 1999;19:5981–5990. doi: 10.1128/mcb.19.9.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H-U, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000a;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J Cell Sci. 2000b;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996a;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996b;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumor suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- Shellman YG. Studies of Cdc28p and Cdc7p protein kinases. Ph.D. Dissertation. University of Colorado Health Sciences Center, Denver, CO; 1997. [Google Scholar]
- Shellman YG, Schauer IE, Oshiro G, Dohrmann P, Sclafani RA. Oligomers of the Cdc7/Dbf4 protein kinase exist in the yeast cell. Mol Gen Genet. 1998;259:429–436. doi: 10.1007/s004380050833. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Barral Y, Snyder M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5235–5247. doi: 10.1128/mcb.20.14.5235-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueland CS, Lew DJ, Cismowski MJ, Reed SI. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]