Abstract
The cytoskeleton is the basic machinery that drives many morphogenetic events. Elongation of the C. elegans embryo from a spheroid into a long, thin larva initially results from actomyosin contractility, mainly in the lateral epidermal seam cells, while the corresponding dorsal and ventral epidermal cells play a more passive role. This is followed by a later elongation phase involving muscle contraction. Early elongation is mediated by parallel genetic pathways involving LET-502/Rho kinase and MEL-11/MYPT myosin phosphatase in one pathway and FEM-2/PP2c phosphatase and PAK-1/p21 activated kinase in another. While the LET-502/MEL-11 pathway appears to act primarily in the lateral epidermis, here we show that FEM-2 can mediate early elongation when expressed in the dorsal and ventral epidermis. We also investigated the early elongation function of FHOD-1, a member of the formin family of actin nucleators and bundlers. Previous work showed that FHOD-1 acts in the LET-502/MEL-11 branch of the early elongation pathway as well as in muscle for sarcomere organization. Consistent with this, we found that lateral epidermal cell-specific expression of FHOD-1 is sufficient for elongation, and FHOD-1 effects on elongation appear to be independent of its role in muscle. Also, we found that fhod-1 encodes long and short isoforms that differ in the presence of a predicted coiled-coil domain. Based on tissue-specific expression constructions and an isoform-specific CRISPR allele, the two FHOD-1 isoforms show partially specialized epidermal or muscle function. Although fhod-1 shows only impenetrant elongation phenotypes, we were unable to detect redundancy with other C. elegans formin genes.
Keywords: C. elegans, morphogenesis, epidermis, embryo, formin
Epithelial morphogenesis is a key aspect of animal development, shaping tissues, organs and the organism as a whole (Guillot and Lecuit 2013; Heisenberg and Bellaiche 2013). C. elegans embryonic elongation is a simple system to study epithelial morphogenesis. Midway through C. elegans embryonic development, the worm embryo undergoes dramatic changes to elongate from a spheroid into a long, thin worm (Sulston et al. 1983; Priess and Hirsh 1986; Chisholm and Hardin 2005; Vuong-Brender et al. 2016). These morphogenetic events take place without major changes in cell number or extensive cell migration. The embryo reduces its circumference by a factor of three and increases in length by a factor of four during the elongation process. Embryonic elongation can be divided into two main phases, early and late elongation. Early elongation occurs between the 1.2 and 2-fold stages (“fold” refers to the length of the embryo relative to the long axis of the eggshell) while late elongation occurs after 2-fold. Early elongation is mediated by cell shape changes in the single cell layer that surrounds the embryo, the epidermis (also referred to as the hypodermis in nematodes). Epidermal actomyosin contractility provides the main force that mediates early elongation (Priess and Hirsh 1986; Chisholm and Hardin 2005; Vuong-Brender et al. 2016). Late elongation depends on muscle cell contraction (Williams and Waterston 1994; Zhang et al. 2011).
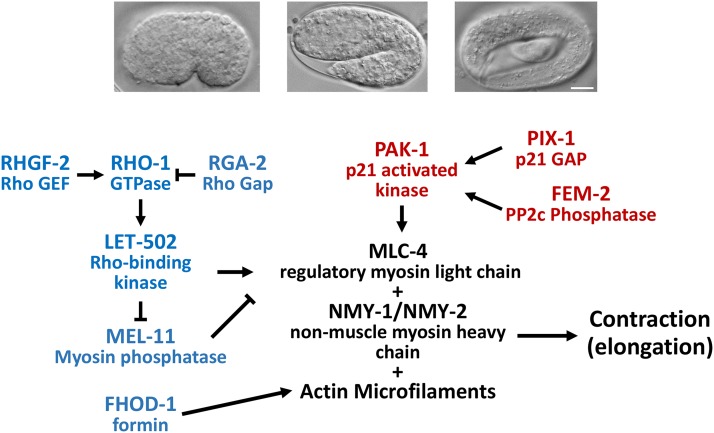
Two parallel pathways mediate early elongation (Wissmann et al. 1997; Piekny et al. 2000; Gally et al. 2009; Martin et al. 2014). One pathway involves Rho-binding kinase (ROCK, LET-502 in C. elegans), which activates non-muscle myosin II (NMY-1/NMY-2) by phosphorylating the regulatory myosin light chain (MLC-4, Figure 1). This activation is antagonized by a corresponding PP1c myosin phosphatase. However, activated ROCK/LET-502 also inactivates myosin phosphatase’s brake to contraction by phosphorylating the myosin phosphatase MYPT targeting subunit (MEL-11). Loss of let-502 thus results in hypocontraction and elongation arrest at the 1.2-fold stage. Conversely, mutations of mel-11, the inhibitor of contraction, result in hypercontraction and embryos often burst (Wissmann et al. 1997; Wissmann et al. 1999; Diogon et al. 2007). However, let-502; mel-11 double mutant embryos often elongate and grow to adulthood, albeit animals are sterile with abnormal body shapes (Wissmann et al. 1997). This indicates the presence of a parallel pathway, which includes p21 activated kinase/PAK-1 and its regulators PP2c phosphatase/FEM-2 and the CDC42/RAC-specific guanine-nucleotide exchange factor (GEF)/PIX-1 (Piekny et al. 2000; Diogon et al. 2007; Vanneste et al. 2013; Martin et al. 2014). PIX-1 and PAK-1 appear to be more active in the anterior of the embryo and the Rho and Rac pathways institute differing morphogenetic programs (Martin et al. 2016; Martin et al. 2017). Redundancy between the two parallel pathways is detected by strong enhancement of elongation defects seen when mutations of the different pathways are combined in double mutants.
Figure 1.
Model of early C. elegans embryonic elongation. Top, photomicrographs of 1.2 fold, 1.7-fold and 4-fold embryos, scale bar = 10 µm. Bottom, actomyosin contractility is triggered by phosphorylation of MLC-4/regulatory myosin light chain and inhibited by MEL-11/myosin phosphatase. Phosphorylation is mediated by kinases in two parallel pathways, PAK-1/p21 activated kinase (red; dorsal/ventral epidermal cells) and LET-502/Rho-binding kinase (blue; lateral epidermal cells). The actin nucleator FHOD-1/formin genetically acts in the LET-502 pathway.
Prior to the start of embryonic elongation, epidermal cells migrate to encase the entire embryo (Sulston et al. 1983; Williams-Masson et al. 1997). These cells are organized in three rows: (1) one row of epidermal cells straddling the dorsal midline along the anterior-posterior axis; (2) two rows (one on each side of the embryo) of lateral epidermal cells (also known as seam cells); and (3) two rows of ventral epidermal cells on each side of the embryo meeting at the ventral midline. The epidermal cell rows show different behaviors during early elongation as the lateral cells show stronger actinomyosin contractility while the dorsal/ventral cells play a more passive role (Priess and Hirsh 1986; Diogon et al. 2007; Gally et al. 2009; Vuong-Brender et al. 2017). This is reflected in the epidermal cytoskeleton. Actin microfilaments and microtubules become organized in circumferential patterns within the dorsal and ventral epidermal cells, but actin filaments and microtubules remain as a meshwork in the contractile lateral epidermal cells until the 2-fold stage (Priess and Hirsh 1986; Gally et al. 2009). After early elongation is completed, microfilaments in the lateral cells align along the circumferential axis. A number of components are implicated in this asymmetric behavior between the lateral or dorsal/ventral epidermis with contractility functions being enriched laterally (Figure 1). let-502 transcriptional reporters show higher expression in the lateral compared to dorsal/ventral cells with mel-11 showing the opposite pattern (Wissmann et al. 1999). The Rho activating guanine exchange factor (GEF/RHGF-2) and Rho inhibiting GTPase activating protein (GAP/RGA-2) are both expressed throughout the epidermis. However, lateral expression of RHGF-2 and dorsal/ventral expression of RGA-2 are each sufficient for embryonic elongation (Diogon et al. 2007; Chan et al. 2015). PIX-1 and PAK-1 act in the dorsal epidermis (Martin et al. 2014; Martin et al. 2016).
FHOD-1, which belongs to the formin family of proteins with nucleating and bundling functions, may also contribute to lateral vs. dorsal/ventral differences. During C. elegans elongation, fhod-1 mutants disrupt microfilament organization in the contractile lateral epidermis and result in early elongation arrest. However, these are low penetrance phenotypes, perhaps suggesting redundancy with other formins or actin organizing factors. fhod-1 genetically acts downstream of let-502/mel-11 and in parallel to pak-1/fem-2 (Figure 1) (Vanneste et al. 2013). Consistent with fhod-1 acting with let-502/mel-11 but not pix-1/pak-1, FHOD-1 shows strongest expression in the lateral epidermis, where let-502/mel-11 appears to act. However, FHOD-1 antibody staining is detected late in elongation, even though genetic effects are seen during early elongation.
Members of the formin protein family function during actin-mediated processes by influencing the assembly and elongation of unbranched actin filaments (Breitsprecher and Goode 2013; Bechtold et al. 2014; Kuhn and Geyer 2014; Pruyne 2016). Family members nucleate microfilaments and then bind to the growing (barbed) ends as a progressive cap that prevents filament termination by capping proteins. Formins have characteristic formin homology (FH) domains. FH1 recruits profilin-bound actin, FH2 is involved in formin dimerization, and the FH3/Diaphanous Inhibitory Domain/DID interacts with the C-terminal Diaphanous Autoregulatory Domain/DAD domain to cause autoinhibition. Formins can be activated by binding a Rac GTPase in a nucleotide-independent manner or by phosphorylation of the DAD domain by Rho-binding kinase (although the latter does not appear to be essential in C. elegans (Vanneste et al. 2013)). A coiled-coil domain is often found between the FH1 and FH3 domains. In addition to their role in actin modulation, formins have also been implicated in microtubule dynamics (Breitsprecher and Goode 2013; Kuhn and Geyer 2014).
fhod-1 is the sole C. elegans member of the FHOD subfamily of formins and is related to the human and mouse FHOD1 and FHOD3 (Mi-Mi et al. 2012; Pruyne 2016). FHODs are involved in stress fiber formation, cell spreading and adhesion, and myofibril maturation (Taniguchi et al. 2009; Iskratsch et al. 2010; Kan-O et al. 2012a; Breitsprecher and Goode 2013; Bechtold et al. 2014). In addition to actin assembly and stress fiber formation, FHOD bundles microfilaments in vitro (Alvarez and Agaisse 2013; Schonichen et al. 2013; Patel et al. 2018). FHOD-1 also functions in C. elegans muscle, where it is partially redundant with CYK-1 for formation of striated body wall sarcomeres (Mi-Mi et al. 2012; Mi-Mi and Pruyne 2015). FHOD-1 is localized near the Z lines (where the actin thin filaments are anchored into the plasma membrane) within the contractile lattice, and along the edges of growing body wall muscle cells. Actin thin filaments still form in fhod-1 mutants, but muscles are narrower and further apart, and Z lines attachments are defective. Finally, fhod-1 strongly enhances the spermathecal ovulation defects of the inverted formin exc-6/inft-1. The interaction may be indirect, with FHOD-1 acting in egg-laying muscles (Hegsted et al. 2016).
The C. elegans genome encodes six other formins that could potentially act in concert with fhod-1. In addition to the muscle, cyk-1 acts during embryonic cytokinesis and in the excretory cell (Severson et al. 2002; Mi-Mi et al. 2012; Shaye and Greenwald 2016). daam-1 and frl-1 participate in Wnt-mediated cell polarity of the B cell (Wu and Herman 2006). fozi-1 encodes a divergent formin required in cell fate specification of muscles and neurons (Johnston et al. 2006; Amin et al. 2007). In addition to the spermatheca, exc-6 functions in the excretory cell, as does the second inverted formin, inft-2 (Shaye and Greenwald 2015; Hegsted et al. 2016; Shaye and Greenwald 2016).
In this work, we address several aspects of elongation, including how fem-2 and fhod-1 contribute to the contractile differences between lateral and dorsal/ventral cells. We report that fem-2 expression in the dorsal/ventral epidermis is sufficient for elongation while fhod-1 acts laterally. We found that fhod-1 encodes two isoforms that differ in a predicted coiled-coil domain. These appear to have functions specialized for embryonic elongation or adult muscle differentiation, but there is a partial overlap between their functions. The impenetrant phenotype of fhod-1 mutants could indicate redundancy with other factors, but we were unable to detect genetic interactions with other C. elegans formins or actin nucleators. Finally, we examined whether muscles function in early elongation, in addition to their essential role later in the process. PAK-1 and PIX-1 have functions in the hemidesmosomes that link muscle to epidermis (Zhang et al. 2011), and FHOD-1 also has roles in embryonic muscle development (Mi-Mi et al. 2012; Mi-Mi and Pruyne 2015). Thus, it was theoretically possible that muscles may play a redundant role during early elongation, in parallel with the LET-502 pathway. We found this not to be the case.
Materials And Methods
Strains and alleles
C. elegans (var. Bristol) were maintained on NGM plates at 15° as described by Brenner (1974) unless otherwise specified. Temperature-sensitive (ts) strains were upshifted to 20°, 24° or 25° at the last larval (L4) stage. Complete broods from ≥ 4 hermaphrodites with a total of >100 offspring are reported. The following alleles were used. Descriptions can be found at Wormbase. Linkage Group I: let-502(sb118ts), fhod-1(tm2363 and sb123), wve-1(ok3308). Linkage Group II: mel-11(it26ts). Linkage Group III: fem-2(b245ts), frl-1(ok460), exc-6(gk386), fozi-1(ok1182), cyk-1(or36), pat-4(st551) (rescued by zpEx184[myo-3::pat-4(+)]). Linkage Group IV: wsp-1(gm324). Linkage Group V inft-2(ok1296), daam-1(tm2133). Alleles were often linked to visible morphological markers to facilitate strain construction and the following balancers were also used: hT2[bli-4(e937) let(q782) qIs48] (I;III), mnC1[dpy-10(e128) unc-52(e444)] II, qC1[dpy-19(e1259) glp-1(q339)] III, mls10[myo-2:gfp] V. Transgenic strains are listed in Table S1.
Scoring of elongation phenotypes
Elongation defects were examined using a dissecting stereomicroscope with twice the egg length being used to judge 2-fold arrest. Elongation arrest phenotypes were categorized into four groups, (i) unhatched embryos, (ii) early larval arrest, where embryos hatch but elongation arrests at or before the 2-fold stage, (iii) later larval arrest, in which embryos pass the 2-fold stage but usually arrest with a dumpy (Dpy) phenotype, and (iv) growth to L4/adulthood. Screening for redundancy between formins was carried out by RNA interference (RNAi) by feeding (Kamath et al. 2003). RNAi clones (www.lifesciences.sourcebioscience.com) were confirmed by sequencing. Three L3 or L4 larvae were transferred to the RNAi plate and subsequently transferred to a fresh RNAi plate every 12 hr. After removing the hermaphrodites, the brood on the third plate was scored, as RNAi should have taken effect. The plate was scored 24 and 48 hr after hermaphrodite removal to assess hatching and growth to L4/adult, respectively. All analyses were carried out at 25°, except as noted.
Transgenic rescue was assessed by measuring worm lengths. L4 hermaphrodites were shifted to the appropriate temperature overnight, and the now gravid animals were transferred to new plates to lay eggs for 2-3 hr. Progeny were photographed 45-48 hr later with a dissecting microscope and lengths were measured using Image J.
Isolation of fhod-1(sb123) deletion in exon 8
The long isoform of fhod-1 (C46H11.11; WormBase version WS262) includes the 474 bp exon 8, but not the short isoform. The WormBase gene model includes additional exons, but for simplicity, our numbering will only include those that we confirmed by sequencing. To create a fhod-1 allele defective for the long isoform, we used CRISPR to create an out of frame deletion in this exon. eft-3::Cas9, pJA58[dpy-10(cn64) gRNA] and a dpy-10(cn64) repair single-stranded oligonucleotide (University of Calgary Core DNA Services) were used as described for “Co-CRISPR” (Arribere et al. 2014). The fhod-1 exon 8 gRNA PCR product was created using PCR “stitching”. PCR’s on pJW1285 (pha-1 gRNA(F+E), a gift from Jordan Ward, Addgene) using oJW1787 + exon8rev and oJW1790 + exon8fwd were stitched together, substituting fhod-1 exon 8 sequences for those of pha-1 and gel-purified (oligonucleotides are listed in Table S2). Gravid wild-type hermaphrodites were injected with 50 ng/μL eft-3::Cas9, 25 ng/μL pJA58(dpy-10(cn64) gRNA), 500 nM dpy-10(cn64) oligonucleotide (Arribere et al. 2014) and 25 ng/μL fhod-1 exon 8 gRNA. Dpy and Rol F1 progeny were screened by PCR and polyacrylamide gel electrophoresis for deletions. A 7 bp deletion was confirmed by sequencing and out-crossed to remove the dpy-10 co-CRISPR allele.
Immunostaining and microscopy
For DIC microscopy, hermaphrodites were moved to fresh plate and allowed to lay eggs for a few hours. Embryos were mounted with a drop of M9 buffer on a 2% agar pad (Sulston et al. 1983). Embryos were examined with a Carl Zeiss Axio Imager 2 microscope and images were taken by AxioCam camera collected with Axiovision software (Zeiss).
For measurement of body-wall muscle (BWM) and body dimensions, adult worms were stained for F-actin with Alexa 568-phalloidin as described previously (Mi-Mi et al. 2012). For each strain, the lateral width of phalloidin-stained BWM, the gap width between BWM quadrants, the number of oblique striations per BWM cell, and the width of the body were analyzed in three replicates of 20 animals each, as described previously (Mi-Mi et al. 2012). L4 worms were stained for α-actinin and for FHOD-1 using monoclonal MH35 and polyclonal DPMSP2 antibodies, respectively, as previously described (Mi-Mi et al. 2012). All images were acquired at room temperature on an Eclipse 90i upright microscope (Nikon) through a CFI Plan Apochromat 40x/NA 1.0 oil immersion objective with a digital monochrome charge-coupled device camera (Cool-SNAP HQ2; Photometrics) driven by NIS-Elements AR software (version 3.1; Nikon). Measurements of dimensions were performed using NIS-Elements AR. Images processed linearly through Photoshop CS4 (Adobe) to enhance contrast.
Isolation of RNA and RT-PCR
Gravid hermaphrodites were treated with alkaline hypochlorite using standard protocols and embryos were collected using a sucrose suspension. Total RNA was isolated from both gravid adults and embryos using the TRIzol Plus RNA Purification System (Invitrogen). Reverse transcription PCR (RT-PCR) was carried out using the Pure High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Two sets of primers were designed at the N-terminus and C-terminus of fhod-1 cDNA (Nterm-F/R and Cterm-F/R, Table S2). To clone the full cDNA of fhod-1, we used PCR primer sets that covered the length of the predicted cDNA (FhdNR.3, Shrt.1F/R, Shrt.2F/R). Embryonic lysate was tested for expression of adult-specific vit-2 to assess contamination from adult RNA (Vit2-F/R) (Heine and Blumenthal 1986). PCR products of the appropriate size were extracted from gels and were sequenced to confirm their identity. Sequence alignments of FHOD proteins from different organisms were carried out using ClustalW2. Protein domain predictions were carried out using InterPro tool EMBL-EBI and the COILS program (Lupas et al. 1991).
fem-2 and fhod-1 tissue-specific rescue
Tissue-specific expression constructs used information from Wormbase WS244 data freeze. Primers for cloning of the fhod-1, fem-2, elt-3, ceh-16 and myo-3 promoters, all with the unc-54 3′UTR, are shown in Table S2. The fem-2 cDNA pDP#DH014 plasmid (Hansen and Pilgrim 1998) was provided by D. Pilgrim. The full-length cDNA was amplified by PCR using the two primers fem2-F and fem2-R, which introduce the cut sites of PstI and NotI, respectively. The PCR product was digested with PstI/NotI and then ligated into pBS II (SK+) previously digested with same enzymes. The 3′ UTR of unc-54 was cloned from wild-type genomic DNA using unc54-F and unc54-F primers, which introduce NotI and SacII restriction sites, respectively, and introduced to the 3′ end of the fem-2 cDNA in the pBS II (SK+) vector. Promoters of fem-2 (femPr-F/R, 344 bp), elt-3 (elt3-F/R, ∼1.9 kb), ceh-16 (cehl6-F/R, ∼2.9 kb) and myo-3 (myo3-F/R, ∼1.9 kb) were amplified by PCR from genomic DNA and digested by KpnI and PstI, which cut sites introduced by the two primer sets. These were ligated separately into the pBS II (SK+) vector that had the fem-2 cDNA and unc-54 3′ UTR. Each of the four constructs was injected into the fem-2(b245); fhod-1(tm2363) (HR1438) strain at 2 ng/μL, together with transformation markers of 20 ng/μL of the gut-specific reporter marker (elt-2::GFP::LacZ) (Fukushige et al. 1998), 50 ng/μL of the dominant Roller marker pRF4 (rol-6(su1006)) and 48 ng/μL pBS II (SK+). For every fem-2 cDNA plasmid, two independent transgenic lines were isolated, which are listed in Table S1.
fhod-1a(long) and fhod-1b(short) cDNAs (with and without exon 8, respectively) were isolated and cloned into pBS KS+ with the unc-54 3′UTR as described above. The native fhod-1 promoter (∼5.3 kb) was isolated from wild-type genomic DNA using oRS63 + oRS70. The ceh-16 and elt-3 promoters were isolated using restriction enzyme digestions from ceh-16::rhgf-2::unc-54 and elt-3::rhgf-2::unc-54 plasmids respectively (Chan et al. 2015). The myo-3 promoter was isolated from wild-type genomic DNA as described earlier for fem-2. Rescue plasmids were injected at 2 ng/μL with 50 ng/μL pRF4 (rol-6(su1006)) (Mello et al. 1991). Lines sbEx242 through sbEx249 also included 45 ng/μL pTG96 (sur-5::gfp) (Gu et al. 1998), which marks transgenic worms as GFP+ in all (or most) cells. Constructs were injected into fhod-1(tm2363) let-502(sb118) I (HR 1485). Transgenic strains are listed in Table S1.
Western blot analysis of FHOD-1
For each strain analyzed, mixed-age worm populations were washed from 6 un-starved 60-mm NGM plates using M9 buffer/0.1% triton X-100. The samples were enriched for adult worms by transferring them to 1.7-mL tubes and for four times, allowing the heavier adults to settle by gravity on ice for 2 min before removal of buffer. Adult worms were suspended with 2 volumes 2X SDS-sample buffer, boiled for 3.5 min, disrupted for 30 sec using a tissue grinder, and boiled 3 more minutes. Boiled extracts were drawn 8 times through an insulin syringe to shear genomic DNA before loading for SDS-PAGE. Extracts were normalized and probed for the FHOD-1 FH2 domain using the antibody DPMSP2 as described previously (Mi-Mi et al. 2012).
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6199286.
Results
fem-2 expression in dorsal and ventral cells can mediate elongation
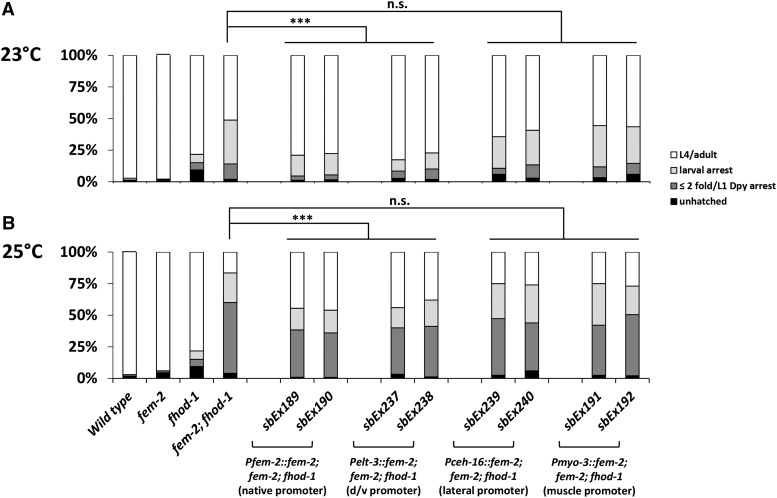
In addition to its well-established function in sex determination (Pilgrim et al. 1995; Chin-Sang and Spence 1996), fem-2/PP2c phosphatase acts during embryonic elongation, in parallel to fhod-1 and the mel-11/let-502 pathways (Figure 1) (Piekny et al. 2000; Vanneste et al. 2013). Other genes that genetically act in the fem-2 branch of the elongation pathway, including pak-1 and its activator pix-1, act in dorsal/anterior epidermis during elongation (Martin et al. 2014; Martin et al. 2016). To determine if fem-2 acts similarly, we expressed a fem-2 cDNA using drivers that are specifically active in lateral cells (ceh-16) or dorsal/ventral cells (elt-3). These promoters are expressed before elongation begins (Gilleard et al. 1999; Cassata et al. 2005; Diogon et al. 2007; Gally et al. 2009; Chan et al. 2015; Quintin et al. 2016). Transgenes of these constructs were compared to the same fem-2 cDNA driven by its native promoter, which is able to rescue the fem-2 sex determination phenotype (Pilgrim et al. 1995; Hansen and Pilgrim 1998). fem-2 mutant alleles have a low penetrance elongation phenotype (Figure 2) (Piekny et al. 2000), with a frequency of < 5%; however, the elongation phenotype for the ts allele fem-2(b245) can be sensitized in double mutants with fhod-1 (Vanneste et al. 2013). This fem-2; fhod-1 has elongation defects of 50% at 23° and 80% at 25° (Figure 2A,B). Since C. elegans transgenes are typically not expressed maternally (Kelly et al. 1997) and fem-2 elongation defects show both maternal and zygotic effects (Piekny et al. 2000), we confirmed that zygotic expression of fem-2(+) can rescue fem-2; fhod-1 elongation defects (Figure S1A).
Figure 2.
Tissue-specific rescue of fem-2. Extrachromosomal arrays contained constructs of the fem-2 cDNA driven by the fem-2 promoter, the elt-3 promoter (dorsal/ventral epidermal cells), the ceh-16 promoter (lateral epidermal cells) or the myo-3 promoter (body wall muscle). Two independent lines for each construct were cultured at 23° (A) or 25° (B) and scored for growth to the indicated stages as described in MATERIALS and METHODS. As fem-2 elongation phenotypes are low penetrance, fhod-1(tm2363) was included to sensitize the strains. At both temperatures, the native and dorsal/ventral promoters rescued elongation phenotypes to similar degrees (*** = P < 0.001 by One Way ANOVA of the animals that grew to L4/adult) while the lateral and muscle promoters did not (n.s. = non-significant, P > 0.05).
fem-2 expression appears to be sufficient in the dorsal/ventral epidermis for elongation. Animals were transferred daily until they ceased laying embryos, the hatching rate was scored a day after removal of hermaphrodites, and growth was assessed under the dissecting scope the following day. Both Pfem-2::fem-2 and Pelt-3::fem-2 partially rescued elongation defects at 23°. The proportion of worms growing to the L4/adult stage, which includes the rescued animals, increased from 51% in the fem-2; fhod-1 to 79% and 80% for Pfem-2::fem-2 and Pelt-3::fem-2, respectively (Figure 2A). Similar results were seen at 25°, where the proportion of L4/adults was elevated from 16% in controls to 45% and 43% in Pfem-2::fem-2 and Pelt-3::fem-2 transgenic strains, respectively (P ≤ 0.001). In contrast, introducing Pceh-16 (lateral) and Pmyo-3 (muscle) constructs driving the fem-2 cDNA had no significant effect in the elongation phenotype at either temperature (P > 0.05). We conclude that dorsal/ventral expression of fem-2 is at least partially sufficient for successful elongation, consistent with the dorsal/ventral functions observed for pak-2 and pix-1.
fhod-1 encodes multiple isoforms
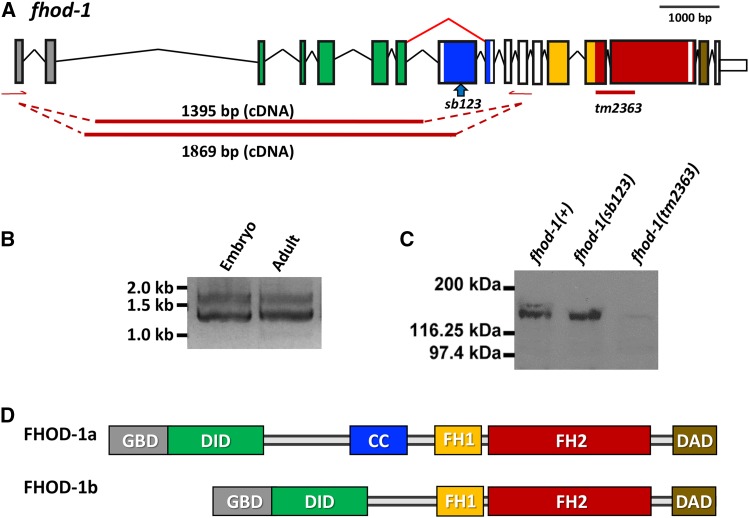
We next examined the role of fhod-1 during C. elegans elongation. We detected two major RT-PCR products for fhod-1 in both the adult and embryonic total RNA (Figure 3A,B). Sequencing showed that the shorter product lacked 474 bp encoded by exon 8 (WormBase WS262 indicates additional exons, but for simplicity, our numbering only includes those exons that we detected). The two predicted proteins shown in Figure 3 differ by 158 amino acids, corresponding to products of 1346 (FHOD-1a) and 1188 (FHOD-1b) amino acids, and the expected proteins are detected by Western blot (Figure 3C,D). Although conservation of the primary sequence outside nematodes is limited, the region unique to the C. elegans long form includes an 83 amino acid predicted coiled-coil region that is similarly predicted in FHOD-1 orthologs in human, Drosophila, and C. briggsae (Figure S2). Two other fhod-1 regions have shorter predicted coiled-coils that are common to both C. elegans isoforms, including 29 residues between positions 309 and 337 (exon 7) and 22 amino acids from 1085 and 1106 (exon 15). These are also found in the other species.
Figure 3.
fhod-1 gene structure and expression. (A) Diagram of the intron/exon structure of the fhod-1 gene. Exons are color coded to correspond to the protein domains shown in (D). Positions of the sb123 7 bp deletion and the 367 bp deletion tm2363 are indicated. The primers (red arrows) for RT-PCR (B) detected 1.4 kb and 1.8 kb bands in both gravid adults and embryos. Sequencing showed they differed by the inclusion of exon 8 in the longer form. (C) Western blots revealed two species of approximately 173 kD and 158 kD in fhod-1(+). The upper band was missing in fhod-1(sb123), which creates a frameshift mutation in exon 8. Both isoforms are absent from the out of frame deletion allele tm2363. (D) Domain structure of the long and short isoforms FHOD-1a and FHOD-1b, respectively, resulting from alternative splicing of exon 8. GBD, Guanine nucleotide binding domain, DID, diaphanous inhibitory domain, CC, predicted coiled-coil, FH1, formin homology domain 1, FH2, formin homology domain 2, DAD, diaphanous autoregulatory domain. The WormBase gene model includes additional exons, but for simplicity, our numbering will only include those that we confirmed by sequencing.
Differential functions of long and short fhod-1 isoforms during elongation
To investigate differing functions of fhod-1a and fhod-1b, we generated fhod-1(sb123), a 7 bp CRISPR deletion allele in the alternatively spliced exon 8. This results in an out of frame product, creating a stop codon after 26 amino acids. If translated, the predicted protein would lack the FH1, FH2 and DAD domains. Western blots demonstrated that the longer isoform is absent from mixed stage fhod-1(sb123) while the abundance of the short form was not altered (Figure 3C,D). Thus sb123 likely represents a null or strong loss of function specific for fhod-1a but would retain fhod-1b(+) function.
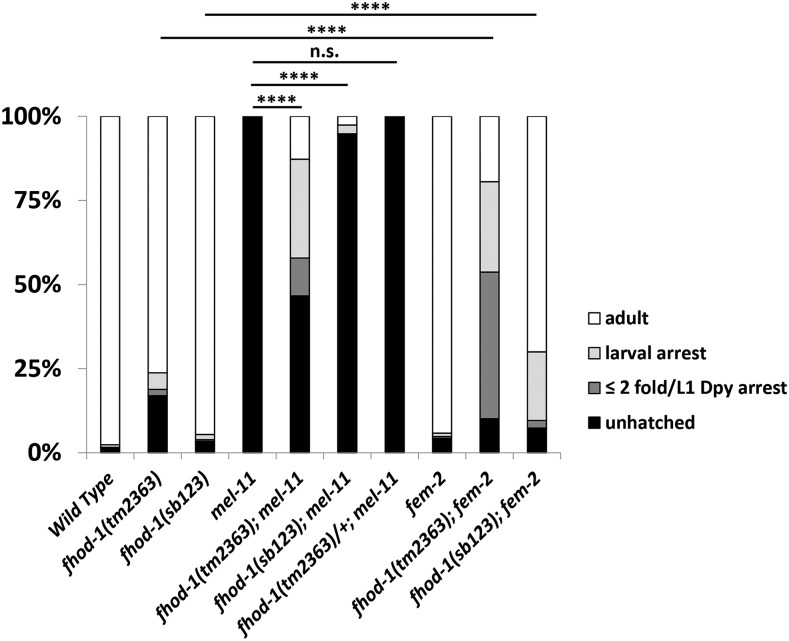
We compared animals lacking fhod-1a to loss of both fhod-1 isoforms in the tm2363 allele. fhod-1(tm2363) is an out of frame deletion of the FH2 domain that affects both isoforms and has little or no protein by immunofluorescence (Mi-Mi et al. 2012) and lacks both isoforms by western blot (Figure 3A,C). This allele has impenetrant elongation defects with 19% showing arrest by the 2-fold stage (Figure 4) (Vanneste et al. 2013). Only 4% of fhod-1(sb123) arrested at this stage, indicating that fhod-1(sb123) can mediate elongation. As a more sensitive test, we examined genetic interactions with elongation pathway mutants. All mel-11 embryos die at the restrictive temperature of 25° due to hypercontraction, but this can be suppressed by mutations such as fhod-1(tm2363) that lessen the contractile force (Vanneste et al. 2013). In particular, 53% of the embryos from fhod-1(tm2363); mel-11 hatched, with 13% surviving to L4/adult compared to 0% for mel-11 controls (Figure 4, P < 0.0001). In contrast, sb123 showed much weaker suppression of mel-11, with only 6% hatching and 3% growing to L4/adult. Nevertheless, this level of suppression is significant as hatched embryos represented 40/731 of the fhod-1(sb123); mel-11 progeny laid. In comparison, only 1/3026 of control mel-11 embryos hatched (P < 0.0001). Together, the data indicate that fhod-1(sb123) retains substantial wild-type elongation function, implying that fhod-1b(+) primarily mediates elongation, with fhod-1a(+) playing a minor role.
Figure 4.
fhod-1(sb123), which encodes only the short isoform, has little effect on elongation in mel-11 or fem-2 backgrounds. Indicated genotypes were scored for growth to the indicated stages as described in MATERIALS and METHODS. While the fhod-1(tm2363) null allele rescues mel-11 lethality at 20°C, sb123 does so weakly and tm2363/+ does not at all. **** = P < 0.0001 and P > 0.99 respectively, by One Way ANOVA of hatching rates. Likewise, at 25° tm2363 enhances fem-2 elongation defects, while effects of sb123 are milder. **** = P < 0.0001 by One Way ANOVA of growth to L4/adulthood.
Since total FHOD-1 would be decreased slightly in fhod-1(sb123) due to the loss of FHOD-1a, we asked if a simple 50% decrease in fhod-1 function in heterozygotes could rescue mel-11. However, none of the embryos of fhod-1(tm2363)/+; mel-11 hatched, indicating a lack of suppression (Figure 4, 1/564 hatched, P > 0.99).
As another assessment of fhod-1a function during elongation, we made double mutants with fem-2, which alone has impenetrant hypoelongation phenotypes. The null allele fhod-1(tm2363) strongly enhanced fem-2 (Figure 4) (Vanneste et al. 2013), with only 19% growing to L4/adult compared to the values for the single mutants of 76% for fhod-1 and 94% for fem-2. In contrast, 70% of fhod-1(sb123); fem-2 grew to L4/adult, again suggesting that fhod-1(sb123) retains substantial wild-type elongation function. Nevertheless, the genetic interactions of fhod-1(sb123) with fem-2, albeit much weaker than that for the null allele, was significant compared with the fhod-1(sb123) control (P < 0.0001). These data again indicate that the short isoform fhod-1b has the major role during elongation, with the long isoform fhod-1a playing an ancillary role.
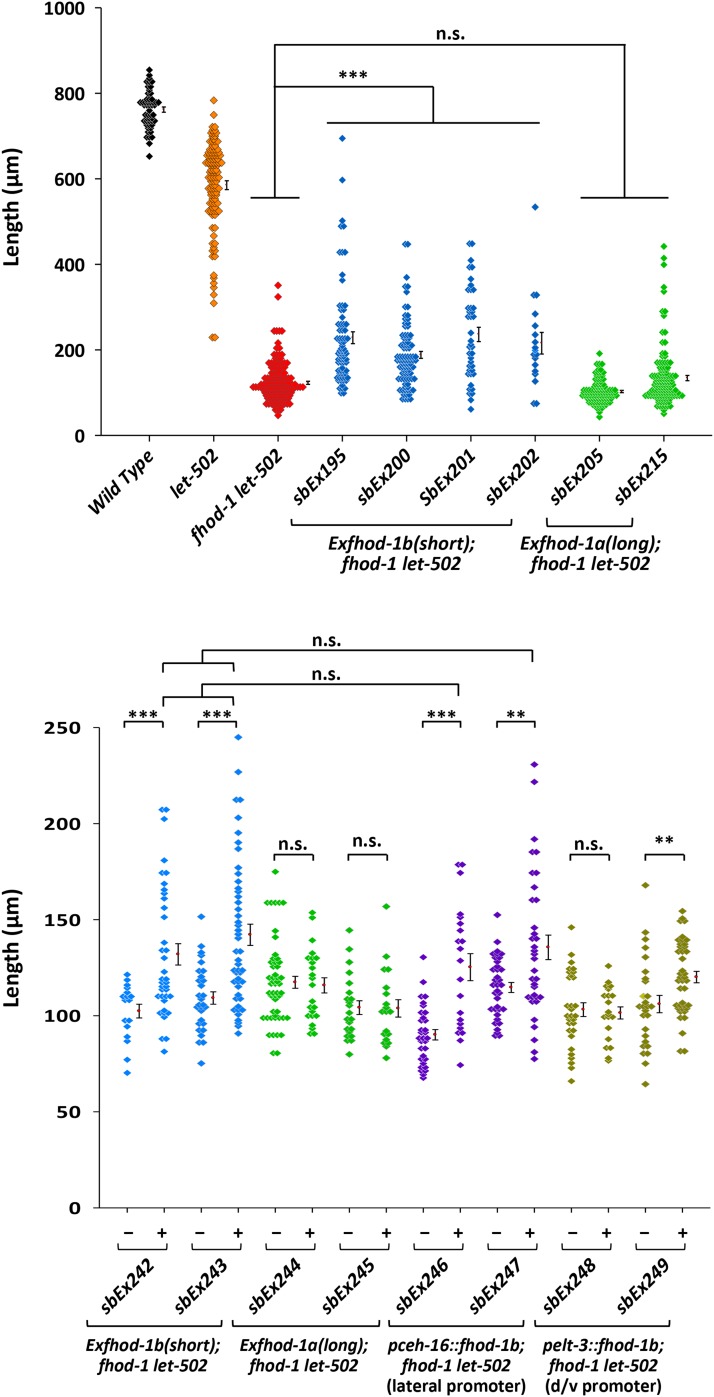
It is possible that the differing phenotypes for fhod-1a and fhod-1b stem not from the presence or absence of alternatively-spliced exon 8, but rather tissue-specific splicing to yield only fhod-1b in the epidermis. To test this, we created cDNA transgenics for the short and long form, driven by the endogenous fhod-1 promoter. Because of the low penetrance phenotype of even the more severe fhod-1(tm2363) allele, we injected constructs into the sensitized let-502(sb118) fhod-1(tm2363) background. let-502(sb118) is a hypomorphic, ts mutation that strongly enhances the null fhod-1(tm2363) allele (Vanneste et al. 2013). Because transgenes usually do not show maternal expression (Kelly et al. 1997), we first confirmed that zygotic fhod-1(+) effectively rescued the strain (Figure S1B). As a sensitive, quantitative metric to assess rescue by the long and short fhod-1 isoforms, gravid adults were brooded for 2-3 hr at the semi-permissive temperature of 24° and 1 or 2 days later body length was measured. We found that 2 days of growth amplified the differences between transgenic animals and the non-growing fhod-1let-502 controls (Figure S3). Figure S4 shows that the more subjective scoring scheme using 24 hr cohorts, as presented in Figures 2 and 4, yielded comparable results to the of scoring 2-3 hr cohorts, as used in Figure 5. Slow growing/arrested larvae invariably had lumpy and dumpy body shapes characteristic of elongation mutants. Four independent fhod-1b transgenic lines showed substantially better growth than controls (P < 0.001), while two fhod-1a transgenic lines did not (Figure 5A). Thus, fhod-1b, but not fhod-1a, appears to be at least partially sufficient in the epidermis for elongation, consistent with the fhod-1(sb123) results.
Figure 5.
Transgenic rescue by fhod-1 isoforms and tissue-specific rescue. Hermaphrodites were cultured at the semi-permissive temperature of 24° and progeny lengths of semi-synchronized broods were measured 2 days after egg laying (see MATERIALS and METHODS). The null fhod-1(tm2363) was sensitized with the ts allele let-502(sb118). There was little overlap in lengths of let-502 and fhod-1 let-502 controls. (A) Independent extrachromosomal transgenes containing the native fhod-1 promoter driving fhod-1b cDNA without exon 8 (sbEx195, sbEx200, sbEx201 and sbEx202) resulted in far more animals overlapping control let-502 lengths than did cDNAs encoding the longer isoform fhod-1a (sbEx205, sbEx215). (B) Transgenic lines marked with SUR-5::GFP enabled scoring of transgenic progeny in comparison to sibs who did not inherit the array. Rescue was seen among GFP+ progeny for the short (sbEx242, sbEx243) but not the long (sbEx244, sbEx245) fhod-1 isoform under the control of the native fhod-1 promoter. The short isoform was then driven using lateral-specific pceh-16 or dorsal/ventral-specific pelt-3 promoters. Rescue was seen for both lines using the lateral promoter (sbEx247, sbEx248) and was not significantly different from the control lines (sbEx242, sbEx243) that employed the native promoter. One dorsal/ventral line (sbEx249) did rescue, but none of the progeny fell outside the range of the control sibs. Unhatched embryos are ∼50 µm. Error bars = standard error of the mean. Because the lengths did not show normal distributions, the Kruskal-Wallis One Way Analysis of Variance on Ranks was used rather than ANOVA. *** = P < 0.001, ** = P < 0.01, n.s. = P > 0.05, not significant. Supplemental Figure 4 shows comparison of this method of assessing growth to scoring with the dissecting microscope used in Figures 2 and 4.
Because of the mosaic nature of extrachromosomal transgenes, the incomplete rescue seen in Figure 5A could be due to some progeny of transgenic animals not inheriting the transgenes. To test this, we made transgenic lines in the fhod-1let-502 background that included the ubiquitous marker sur-5::gfp (Gu et al. 1998) and compared GFP+ to GFP- progeny. Figure 5B again shows rescue by the short fhod-1b isoform but not by the long fhod-1a. For fhod-1b, the GFP+ progeny showed significantly more growth than their GFP- sibs (P < 0.001), but rescue was again incomplete. Unlike the rescued animals shown in Figure 5A, no GFP+ progeny in Figure 5B grew to >250 µm 2 days after laying, indicating that the sur-5::gfp marker may impede growth and/or transgene function.
We conclude from the loss-of-function experiments with the CRISPR generated fhod-1(sb123) mutant (Figure 4), and the transgenic rescue work (Figure 5A,B), that the short isoform fhod-1b is primarily responsible for elongation, with the long isoform fhod-1a playing a supplemental role.
fhod-1 acts in lateral epidermal cells during elongation
We next asked where fhod-1b expression is sufficient to mediate elongation. We previously found fhod-1 microfilaments defects only in lateral cells. Immunostaining with antibodies to FHOD-1 had highest expression in lateral cells, albeit expression was not detected until late in elongation, even though the impenetrant elongation arrest phenotype was visible by light microscopy during early elongation (Vanneste et al. 2013). To determine if fhod-1 expression in lateral cells is indeed required for elongation, we drove the fhod-1b cDNA using drivers that are specifically active in lateral cells (ceh-16) or dorsal/ventral cells (elt-3). As shown in Figure 5B, lateral expression of fhod-1b partially rescued fhod-1let-502 elongation (P < 0.001 for sbEx246 and < 0.01 for sbEx247). Notably, the two rescuing lines were not significantly different from either of the two control fhod-1b lines driven by the native promoter (P > 0.1). One dorsal/ventral line (sbEx248) was not rescued while a second line (sbEx249) showed a slight increase in mean body length (P < 0.01). However, unlike other rescued lines in Figure 5, there were no animals above the control range. We also created one transgenic line that did not contain sur-5::gfp, and this did not show rescue (Figure S4). Thus, lateral expression appears to be more important for elongation, although dorsal/ventral fhod-1 may play a secondary role.
fhod-1(sb123) indicates that the long isoform functions in muscle
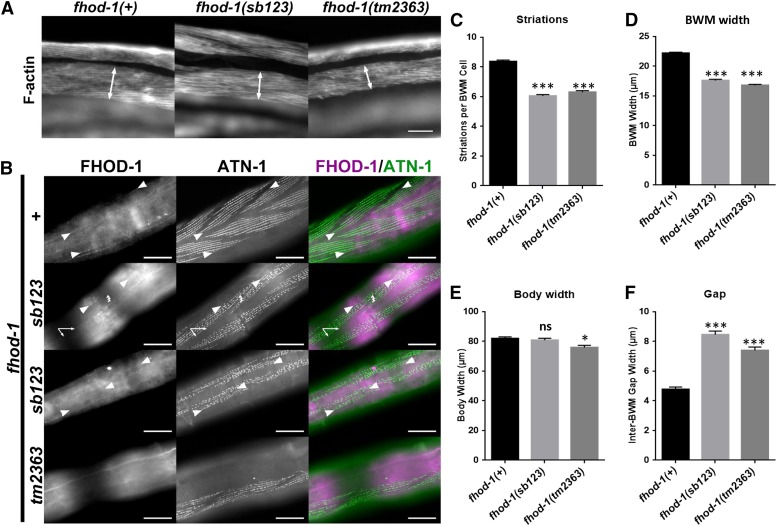
We asked if the two fhod-1 isoforms differ in their muscle function. Wild-type body-wall muscle (BWM) cells assemble 8-10 striations from embryogenesis through young adulthood. In null fhod-1(tm2363), addition of BWM striations through larval development is slower than wild type, resulting in only 6-7 striations per cell, and narrow BWM (Mi-Mi et al. 2012). Staining of F-actin in fhod-1(sb123) animals with fluorescent phalloidin showed that these mutants are nearly indistinguishable from fhod-1(tm2363) animals in terms of having fewer striations and narrower BWM than wild type (P < 0.001, Figure 6A,C,D). This was not a reflection of overall reduction in body size, as there were only modest differences in body width between wild-type and fhod-1(sb123) worms, although fhod-1(tm2363) animals were slightly narrower (Figure 6E).
Figure 6.
Muscle defects in fhod-1(sb123), which encodes only the short isoform, are as severe as the null allele fhod-1(tm2363). (A) Fluorescent phalloidin-stained F-actin in young adult stage animals. The arrow indicates the width of body wall muscles (BWM), which have fewer (C) and narrower (D) striations in both sb123 and tm2363. (B) L4 larva stage animals double-stained for FHOD-1 and Z-line marker ATN-1, showing that FHOD-1 is expressed in puncta along the edges of F-actin–rich body wall muscles cell contractile lattices (large arrowheads) and in faint striations across the lattices (small arrowheads). FHOD-1 levels are reduced in fhod-1(sb123) and absent in fhod-1(tm2363). The small arrows indicate non-specific stain by anti-FHOD-1 noted previously (Mi-Mi et al. 2012). Body width (E) was similar to wild type in both mutants while the gap between muscles (F) was similarly increased in both alleles. t-tests of mutants relative to wild type, * P ≤ 0.05, *** P ≤ 0.001, ns = P > 0.05, not significant. Scale bar, 100 µm.
Thin filaments in BWM cells are anchored primarily to dense bodies that are rich in α-actinin/ATN-1 (Moulder et al. 2010). In wild-type muscle, dense bodies are evenly distributed along striations, while in fhod-1(tm2363) muscle, dense body spacing is irregular (Mi-Mi et al. 2012). In fhod-1(sb123) muscle, dense bodies are also irregularly spaced, similar to fhod-1(tm2363) (Figure 6B). Thus, in terms of BWM cell morphology, the effects of fhod-1(sb123) closely resemble those of FH2-disrupting fhod-1(tm2363). This implies that the fhod-1a long isoform, which is deficient in fhod-1(sb123), is required in muscle. In wild type, FHOD-1 localizes in BWM from mid-larval development through young adulthood, concentrating along the lateral edges of muscle cells (Figure 6B, large arrowheads), with a much fainter, diffuse presence along striations (small arrowheads). FHOD-1 was also visible in these locations in fhod-1(sb123) BWM cells, but much more faintly, and less consistently. Thus, endogenous FHOD-1B has a limited capacity to assemble but apparently is not sufficient for normal morphology.
FHOD-1 is also required for the proper positioning of BWM. During embryogenesis, BWM cells migrate from lateral positions to the dorsal or ventral surfaces, where they form dorsal or ventral pairs of muscle. The gap between muscles in each pair are wider in fhod-1(tm2363) worms than in wild-type animals, suggesting mutant BWM cells do not migrate the full distance from their lateral positions during embryogenesis (Mi-Mi et al. 2012). In fhod-1(sb123) mutants, the gap not only is wider than wild-type, but surprisingly, is consistently wider than in fhod-1(tm2363) mutants (Figure 6B, F).
To test whether the defective BWM phenotype of fhod-1(sb123) worms reflects just a partial reduction in FHOD-1 expression levels, we compared BWM of wild-type animals to those heterozygous for fhod-1(tm2363). Among three replicated experiments, there were no consistent differences in BWM width, number of striations in individual BWM cells, or in width of the gaps between BWMs (data not shown).
The short isoform can function in muscle when expressed as a transgene
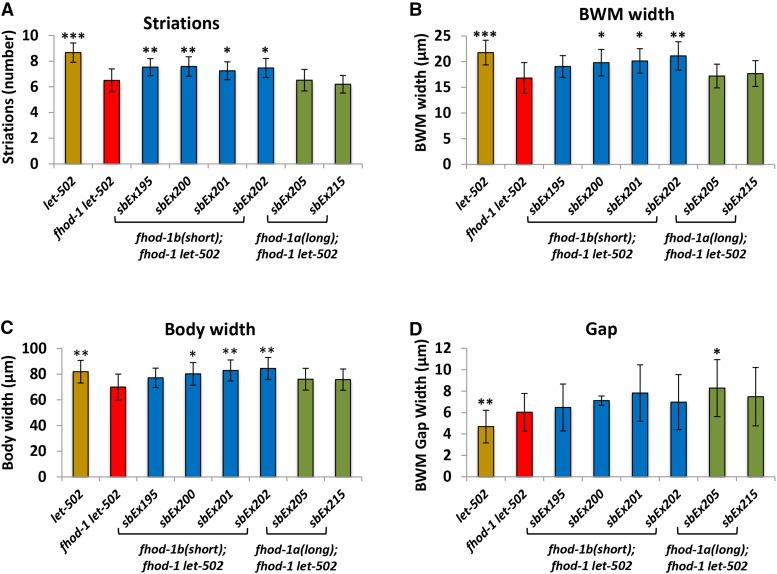
While the results from the fhod-1(sb123) allele showed that the long isoform is necessary for muscle function, transgenic expression of only the short fhod-1b isoform was sufficient to at least partially rescue some muscle phenotypes. Using the same transgenic strains used in Figure 5 to drive the long and short isoforms by the native fhod-1 promoter, muscle striations, BWM width and body width were often rescued by the short isoform fhod-1b (Figure 7A-C). Surprisingly, the long isoform was not sufficient to rescue these phenotypes. Additionally, transgenics for neither isoform were not able to rescue the muscle gap phenotype (Figure 7D). These data may indicate that expression levels in muscle, in addition to (or rather than) the coiled-coil region present in fhod-1a, are responsible for the fhod-1(sb123) muscle phenotypes (see Discussion).
Figure 7.
Transgenic rescue of the muscle defects by fhod-1a and fhod-1b. The same transgenic lines used in Figure 5A were scored for muscle phenotypes and are color coded similarly in the two figures. Extrachromosomal transgenes containing the native fhod-1 promoter driving fhod-1 cDNA without exon 8 (sbEx195, sbEx200, sbEx201 and sbEx202) rescued most fhod-1 let-502 muscle defects. cDNAs encoding the longer isoform fhod-1a (sbEx205, sbEx215) for the most part did not rescue. (A) striations, (B) body wall muscle width, (C) body width, (D) gap between muscle cells.
Redundancy of fhod-1 with other formins or actin nucleators
The incomplete penetrance of fhod-1 single mutant phenotypes suggests redundancy with other genes that regulate actin nucleation for elongation. Thus, we examined the other C. elegans formins as well as genes from the Arp2/3 actin nucleation pathway for elongation defects and genetic interaction with fhod-1. All analyses were carried out at 25° and putative formin strong to null alleles were used (Mi-Mi et al. 2012). Double mutants of fhod-1 and a gene acting in parallel would show synergistic genetic interactions, but as described below, we did not detect any.
C. elegans has six other formin family genes (see Introduction). exc-6 was the best candidate for acting in parallel with fhod-1 as exc-6 is the only formin mutant that exhibited an early elongation arrest similar to that of fhod-1, as shown by microscopic analysis of the 20% of embryos that did not hatch. Additionally, fhod-1; exc-6 adults often show a rolling phenotype (Mi-Mi et al. 2012), which is characteristic of weak let-502 alleles (Piekny et al. 2000). However, frequencies of unhatched embryos and elongation arrested larvae of fhod-1; exc-6 appeared additive rather than synergistic (Figure S6). Because cyk-1 acts in parallel to fhod-1 in muscle (Mi-Mi et al. 2012; Mi-Mi and Pruyne 2015), we examined the double mutant for elongation defects, but again the interactions appeared additive rather than synergistic (cyk-1 is homozygous maternal-effect lethal and so cyk-1/+; fhod-1 was examined). RNAi knockdown of let-502, which enhances fhod-1 (Vanneste et al. 2013), was used to further sensitize fhod-1; exc-6 and fhod-1; cyk-1/+ but showed only additive enhancement (Figure S6, combinations of let-502(RNAi) and cyk-1/+ did result in high percentages of unhatched embryos, but these may stem from the requirement of both genes for cytokinesis in the early embryo (Swan et al. 1998; Piekny and Mains 2002)). We extended the analysis to triple formin knockdowns. Treatment of the fhod-1; exc-6 with RNAi to fozi-1, daam-1 or cyk-1 had no synergistic effects (Figure S7). Similarly, let-502(RNAi) feeding of inft-2(ok1296), frl-1(ok460), fozi-1(ok1182) and exc-6(gk386) mutants had additive rather than synergistic interactions, suggesting that these formins do not function in the let-502 elongation pathway.
The ARP2/3 complex may potentially act in parallel to formins to nucleate microfilaments during elongation. We examined the elongation phenotype of the two Arp2/3 complex activators WVE-1/WAVE and WSP-1/WASP (Sawa et al. 2003). let-502(RNAi) had little effect on those two strains (Figure S7).
The role of muscle cells in early elongation
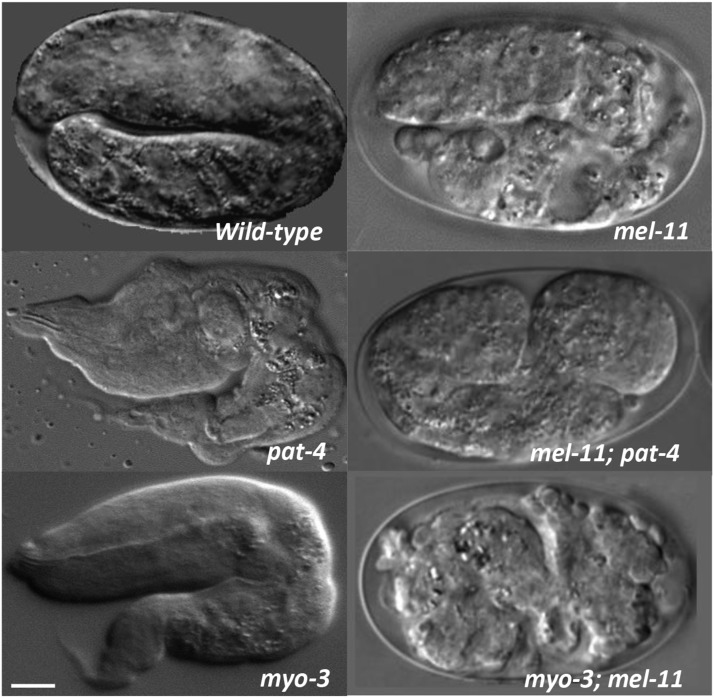
Early elongation is redundantly controlled by the let-502/mel-11 and fem-2/pak-1 pathways, while late elongation (after the 2-fold stage) depends on muscle contraction. Since pak-1 and its activator pix-1 function in muscle/epidermal attachments during late elongation and fhod-1 functions in muscle formation (Zhang et al. 2011; Mi-Mi et al. 2012), it is formally possible that muscle function also has a role in early elongation (i.e., in parallel to let-502/mel-11). To test this possibility, we took advantage of the fact that mutations in genes acting during early elongation (e.g., let-502, fem-2, fhod-1) suppress the hypercontraction phenotype of mel-11. We asked if compromising muscle function will act similarly to rescue mel-11. Mutations in muscle genes encoding two biochemically different products were tested for mel-11 suppression. pat-4/integrin-linked kinase (Mackinnon et al. 2002) encodes a focal adhesion protein necessary for muscle development and myo-3 encodes myosin heavy chain A (Miller et al. 1986). Despite their relatively late phenotypes (Pat, paralyzed arrest at two-fold stage), transcripts of both genes are detected earlier (Hashimshony et al. 2015). If mutations of either gene suppress mel-11, progeny of the double mutants should exhibit the Pat phenotype rather than arresting due to the hypercontraction of mel-11. mel-11; pat-4/+ embryos all exhibited the mel-11 hypercontraction phenotype (n = 63), rather than the expected 25% Pat phenotype (Figure 8). Similarly, the hypercontraction phenotype was epistatic to Pat among the progeny of mel-11(it26); myo-3/+ (n = 100). Additionally, myo-3 did not enhance early elongation defects of let-502(RNAi) (data not shown), again indicating that myo-3 does not mediate early elongation. As shown earlier, driving fem-2 cDNA in BWM cells did not rescue the fem-2 elongation phenotype (Figure 2). In conclusion, we found no evidence for a role of muscle cells during the early phase of elongation.
Figure 8.
Muscle mutants do not suppress the mel-11 hypercontraction defect. Top row, wild type is comparted to mel-11, which shows a disorganized embryo, with posterior cells not encased in the epidermis in this particular embryo. Middle row, pat-4 arrests at 2-fold while pat-4; mel-11 shows the mel-11 phenotype. Bottom row, myo-3 arrests at 2-fold while myo-3; mel-11 shows the mel-11 phenotype. Experiments were done at 25°C and anterior is to the left and dorsal to the top in all images. Scale bar = 10 µm.
Discussion
Morphogenesis within a population of cells is carried out with exquisite precision, with individual cells often implementing different subprograms. For example, some cells may remain rigid, while adjacent cells change their forms, or cells may assume complementary shapes. The actomyosin mediated elongation of the C. elegans embryo from a spheroid into a tube is a simple morphogenetic model that displays these properties. The C. elegans epidermis has two general cell types – lateral cells provide the primary contractile force while dorsal and ventral cells play a more passive role (Priess and Hirsh 1986; Diogon et al. 2007; Gally et al. 2009; Vuong-Brender et al. 2017). These differences may be in part mediated by components of the two parallel elongation pathways acting primarily in either lateral or dorsal/ventral cells. For example, previous transgenic experiments showed that RHGF-2/Rho GEF can rescue mutants when expressed laterally while RGA-2/Rho GAP acts dorsal/ventrally (Diogon et al. 2007; Chan et al. 2015). Based on phenotypic analysis, PIX-1/CDC42-RAC GEF and PAK-1/ p21 activated kinase have dorsal/ventral roles (Martin et al. 2014; Martin et al. 2016). It should be kept in mind that genes may not be entirely on or off in different cells types, but rather differ by high or low levels of activity, which could refine morphogenic changes. Here we have described additional genes with primarily lateral or dorsal/ventral functions: FEM-2/PP2c phosphatase can act dorsal/ventrally and the FHOD-1/Formin functions in the lateral cells.
FEM-2 primarily acts in the dorsal/ventral epidermis while FHOD-1 acts laterally
Based on transgenic rescue experiments, FEM-2 expression in the dorsal/ventral epidermal cells is at least partially sufficient to provide elongation function (Figure 2). Rescue by dorsal/ventral expression in the sensitized fem-2(-); fhod-1(-) background was not to the levels seen in fem-2(+); fhod-1(-), likely due to inefficient and/or mosaic transgene expression. However, rescue was comparable to the fem-2 transgene expressed by its native promotor. In dorsal/ventral epidermal cells, fem-2 likely interacts with pak-1 and pix-1 (Martin et al. 2014; Martin et al. 2016), which would be consistent with genetic interactions (Vanneste et al. 2013). In mammals, biochemical assays show that PP2C phosphatase, the FEM-2 homolog, is a regulator of PAK1 (Chan et al. 2008).
fhod-1 appears to act laterally, partially rescuing the sensitized fhod-1let-502 background (Figure 5). This is consistent with fhod-1 genetic interactions with the laterally active let-502 (Vanneste et al. 2013). Similar to what we observed with fem-2, fhod-1 rescue was incomplete, although lateral-only rescue was not statistically different from that of the native promoter. Incomplete rescue may reflect inherent limitations of transgenic gene expression. The ceh-16 and elt-3 promoters used here, as well in other studies of elongation gene function, typically result in incomplete rescue (Diogon et al. 2007; Gally et al. 2009; Lin et al. 2012; Chan et al. 2015), possibly because the expression levels and/or timing vary from that of the endogenous promoters. Although lateral fhod-1 expression is clearly most important during elongation, weak rescue was seen with one of three dorsal/ventral lines (sbEx249, Figure 5, Figure S5). Average length increased without increasing the length of any animals beyond the control range. This may indicate that while lateral fhod-1 function is most important, dorsal/ventral activity could provide supplemental function. Similarly, although expression of fem-2 in lateral epidermal cells or body wall muscle cells failed to rescue fem-2; fhod-1 (Figure 2), these tissues might supplement dorsal/ventral fem-2 mediated elongation to refine the process.
FHOD-1 isoforms may have specialized functions
FHOD-1 has two isoforms that may be partly specialized for epidermal or muscle function. Formin family proteins act by nucleation and bundling of linear actin filaments (Breitsprecher and Goode 2013; Bechtold et al. 2014; Kuhn and Geyer 2014; Pruyne 2016) and different functional FHOD isoforms (in different parts of the protein than we report here) are known in other species (Kan-O et al. 2012b; Iskratsch et al. 2013). Previously we showed that fhod-1 acts in the let-502 branch of the elongation pathway (Figure 1) as well as in striated muscle (Mi-Mi et al. 2012; Vanneste et al. 2013; Mi-Mi and Pruyne 2015). Here we found that fhod-1 encodes two isoforms that differ in the inclusion of the 474 bp exon 8, which includes a predicted coiled-coil in the longer fhod-1a allele. To explore functional differences between the predicted proteins, we created a CRISPR allele (sb123) that eliminates only the longer isoform. The results from fhod-1(sb123) showed that fhod-1b expression is primarily responsible for elongation: while the likely null allele fhod-1(tm2363) shows strong genetic interactions with mel-11 and fem-2 (Vanneste et al. 2013), these interactions were substantially weaker for sb123 (Figure 4). However, while weak, the genetic interactions were statistically significant, indicating the long isoform has a supplementary role during elongation. In muscle, the opposite pattern was seen, with fhod-1(sb123) and fhod-1(tm2363) showing similar phenotypes (Figure 6). Based on the mutant alleles, the short form is more important for elongation, while the long form is primarily required in muscle. Transgenic rescue, which demonstrates sufficiency rather than necessity, showed more complicated results. As expected from the fhod-1(sb123) data, transgenic expression of the short, but not the long form, partially rescued elongation (Figure 5). Unexpectedly, the short form at least partially rescued muscle phenotypes (Figure 7). While low levels of FHOD-1b were able to assemble into muscle in fhod-1(sb123) (Figure 6B), this was not sufficient to rescue. Perhaps overexpression, which commonly occurs for multicopy transgenes, leads to FHOD-1b muscle rescue. Why potential overexpression of FHOD-1a long form is not sufficient during either elongation or muscle formation is not clear. Because of the mosaic expression of transgenes, the difficulty in synchronizing large numbers of embryos, and the inability to distinguish between expression in different tissues, we did not measure protein levels by Western blots in transgenic strains. It is possible that the fhod-1(sb123) results from differences in isoform expression levels of the endogenous gene rather than differences in protein isoforms as fhod-1b is expressed at higher levels than fhod-1a, (Figure 3C). Arguing against this was the lack of elongation or muscle phenotypes when fhod-1 levels were decreased by 50% in heterozygotes (Figure 4 and data not shown).
fhod-1 elongation defects are incompletely penetrant ((Vanneste et al. 2013), Figure 4), suggesting redundancy with other actin nucleators. Testing mutants of the other six formin genes (daam-1, exc-6, inft-2, frl-1, fozi-1, and cyk-1) encoded by the C. elegans genome showed no dramatic genetic interactions with fhod-1 or let-502 (Supplemental Figures 6 and 7). Only two of these genes, exc-6 and cyk-1, had, at best, weak additive effects on fhod-1 elongation arrest rather than synergistic interactions predicted for genes acting in parallel. Sensitizing the background further with let-502 or testing triple formin mutants failed to detect strong interactions. These data indicate that fhod-1 may be involved in “fine tuning” rather than mediating an essential process, in concert with other formins. Alternatively, the FHOD family actin bundling activity (Schonichen et al. 2013; Bechtold et al. 2014; Schulze et al. 2014) may act in conjunction with unknown proteins. While our data found no evidence for redundancy, a caveat of this work is that RNAi and use of heterozygous strains for lethal mutations could have resulted in insufficient knockdown to detect genetic interactions.
Muscle genes are not involved in early elongation
pak-1 (Gally et al. 2009; Vanneste et al. 2013) and pix-1 mediate early elongation in parallel to let-502. Since pak-1 and pix-1 have roles in the attachments of muscle to the epidermis that are required during late elongation (Zhang et al. 2011), we asked if muscle has a previously unknown role during early elongation in addition to its later function after the 2-fold stage. However, epistasis experiments (Figure 8) indicated that muscle is likely not involved in early elongation, since compromising muscle does not block the early mel-11 hypercontraction phenotype.
Conclusion
Lateral and dorsal/ventral cells have different roles during elongation, and this is reflected by different genetic requirements. However, while wild-type expression in one cell type might not be strictly necessary for successful elongation, that expression may fine tune the final form or lend robustness to the process. It is likely that many levels of redundancy are involved in elongation.
Acknowledgments
We thank members of the MAINS, MCGHEE, HANSEN and PRUYNE labs for discussions during the course of this project. D. PILGRIM, I. CHIN-SANG and M. LABOUESSE provided constructs and strains. We thank H. KUROYANAGI for sharing information of fhod-1 splicing. This work was supported by funding from the Canadian Institute for Health Research to PEM, and from National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (Award R01AR06470) to DP. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6199286.
Communicating editor: D. Fay
Literature Cited
- Alvarez D. E., Agaisse H., 2013. The formin FHOD1 and the small GTPase Rac1 promote vaccinia virus actin-based motility. J. Cell Biol. 202: 1075–1090. 10.1083/jcb.201303055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N. M., Hu K., Pruyne D., Terzic D., Bretscher A., et al. , 2007. A Zn-finger/FH2-domain containing protein, FOZI-1, acts redundantly with CeMyoD to specify striated body wall muscle fates in the Caenorhabditis elegans postembryonic mesoderm. Development 134: 19–29. 10.1242/dev.02709 [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold M., Schultz J., Bogdan S., 2014. FHOD proteins in actin dynamics–a formin’ class of its own. Small GTPases 5: e973765 10.4161/21541248.2014.973765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D., Goode B. L., 2013. Formins at a glance. J. Cell Sci. 126: 1–7. 10.1242/jcs.107250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassata G., Shemer G., Morandi P., Donhauser R., Podbilewicz B., et al. , 2005. ceh-16/engrailed patterns the embryonic epidermis of Caenorhabditis elegans. Development 132: 739–749. 10.1242/dev.01638 [DOI] [PubMed] [Google Scholar]
- Chan B. G., Rocheleau S. K., Smit R. B., Mains P. E., 2015. The Rho guanine exchange factor RHGF-2 acts through the Rho-binding kinase LET-502 to mediate embryonic elongation in C. elegans. Dev. Biol. 405: 250–259. 10.1016/j.ydbio.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Chan P. M., Lim L., Manser E., 2008. PAK is regulated by PI3K, PIX, CDC42, and PP2Calpha and mediates focal adhesion turnover in the hyperosmotic stress-induced p38 pathway. J. Biol. Chem. 283: 24949–24961. 10.1074/jbc.M801728200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Sang I. D., Spence A. M., 1996. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes Dev. 10: 2314–2325. 10.1101/gad.10.18.2314 [DOI] [PubMed] [Google Scholar]
- Chisholm A. D., Hardin J., 2005. Epidermal morphogenesis. WormBook, ed. The C. elegans Research Community, WormBook 1–22. 10.1895/wormbook.1.35.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Diogon M., Wissler F., Quintin S., Nagamatsu Y., Sookhareea S., et al. , 2007. The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development 134: 2469–2479. 10.1242/dev.005074 [DOI] [PubMed] [Google Scholar]
- Fukushige T., Hawkins M. G., McGhee J. D., 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198: 286–302. [PubMed] [Google Scholar]
- Gally C., Wissler F., Zahreddine H., Quintin S., Landmann F., et al. , 2009. Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development 136: 3109–3119. 10.1242/dev.039412 [DOI] [PubMed] [Google Scholar]
- Gilleard J. S., Shafi Y., Barry J. D., McGhee J. D., 1999. ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Dev. Biol. 208: 265–280. 10.1006/dbio.1999.9202 [DOI] [PubMed] [Google Scholar]
- Gu T., Orita S., Han M., 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18: 4556–4564. 10.1128/MCB.18.8.4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot C., Lecuit T., 2013. Mechanics of epithelial tissue homeostasis and morphogenesis. Science 340: 1185–1189. 10.1126/science.1235249 [DOI] [PubMed] [Google Scholar]
- Hansen D., Pilgrim D., 1998. Molecular evolution of a sex determination protein. FEM-2 (pp2c) in Caenorhabditis. Genetics 149: 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T., Feder M., Levin M., Hall B. K., Yanai I., 2015. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature 519: 219–222. 10.1038/nature13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegsted A., Wright F. A., Votra S., Pruyne D., 2016. INF2- and FHOD-related formins promote ovulation in the somatic gonad of C. elegans Cytoskeleton (Hoboken) 73: 712–728. 10.1002/cm.21341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine U., Blumenthal T., 1986. Characterization of regions of the Caenorhabditis elegans X chromosome containing vitellogenin genes. J. Mol. Biol. 188: 301–312. 10.1016/0022-2836(86)90156-7 [DOI] [PubMed] [Google Scholar]
- Heisenberg C. P., Bellaiche Y., 2013. Forces in tissue morphogenesis and patterning. Cell 153: 948–962. 10.1016/j.cell.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Iskratsch T., Lange S., Dwyer J., Kho A. L., dos Remedios C., et al. , 2010. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 191: 1159–1172. 10.1083/jcb.201005060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Reijntjes S., Dwyer J., Toselli P., Degano I. R., et al. , 2013. Two distinct phosphorylation events govern the function of muscle FHOD3. Cell Mol Life Sci 70: 893–908. 10.1007/s00018-012-1154-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Jr., Copeland J. W., Fasnacht M., Etchberger J. F., Liu J., et al. , 2006. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development 133: 3317–3328. 10.1242/dev.02494 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kan-O M., Takeya R., Abe T., Kitajima N., Nishida M., et al. , 2012a Mammalian formin Fhod3 plays an essential role in cardiogenesis by organizing myofibrillogenesis. Biol. Open 1: 889–896. 10.1242/bio.20121370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan-o M., Takeya R., Taniguchi K., Tanoue Y., Tominaga R., et al. , 2012b Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One 7: e34765 10.1371/journal.pone.0034765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A., 1997. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics 146: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S., Geyer M., 2014. Formins as effector proteins of Rho GTPases. Small GTPases 5: e29513 10.4161/sgtp.29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Tran T., Hu S., Cramer T., Komuniecki R., et al. , 2012. RHGF-2 is an essential Rho-1 specific RhoGEF that binds to the multi-PDZ domain scaffold protein MPZ-1 in Caenorhabditis elegans. PLoS One 7: e31499 10.1371/journal.pone.0031499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J., 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Mackinnon A. C., Qadota H., Norman K. R., Moerman D. G., Williams B. D., 2002. C. elegans PAT-4/ILK Functions as an Adaptor Protein within Integrin Adhesion Complexes. Curr. Biol. 12: 787–797. 10.1016/S0960-9822(02)00810-2 [DOI] [PubMed] [Google Scholar]
- Martin E., Bonnamour G., Jenna S., 2017. AMPK and autophagy control embryonic elongation as part of a RhoA-like morphogenic program in nematode. Small GTPases 1–8. 10.1080/21541248.2017.1372868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Harel S., Nkengfac B., Hamiche K., Neault M., et al. , 2014. pix-1 Controls Early Elongation in Parallel with mel-11 and let-502 in Caenorhabditis elegans. PLoS One 9: e94684 10.1371/journal.pone.0094684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Ouellette M. H., Jenna S., 2016. Rac1/RhoA antagonism defines cell-to-cell heterogeneity during epidermal morphogenesis in nematodes. J. Cell Biol. 215: 483–498. 10.1083/jcb.201604015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Mi L., Pruyne D., 2015. Loss of Sarcomere-associated Formins Disrupts Z-line Organization, but does not Prevent Thin Filament Assembly in in Caenorhabditis elegans Muscle. J. Cytol. Histol. 6: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Mi L., Votra S., Kemphues K., Bretscher A., Pruyne D., 2012. Z-line formins promote contractile lattice growth and maintenance in striated muscles of C. elegans. J. Cell Biol. 198: 87–102. 10.1083/jcb.201202053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M., Stockdale F. E., Karn J., 1986. Immunological identification of the genes encoding the four myosin heavy chain isoforms of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 83: 2305–2309. 10.1073/pnas.83.8.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder G. L., Cremona G. H., Duerr J., Stirman J. N., Fields S. D., et al. , 2010. alpha-actinin is required for the proper assembly of Z-disk/focal-adhesion-like structures and for efficient locomotion in Caenorhabditis elegans. J. Mol. Biol. 403: 516–528. 10.1016/j.jmb.2010.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. A., Oztug Durer Z. A., van Loon A. P., Bremer K. V., Quinlan M. E., 2018. Drosophila and human FHOD family formin proteins nucleate actin filaments. J. Biol. Chem. 293: 532–540. 10.1074/jbc.M117.800888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A. J., Mains P. E., 2002. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J. Cell Sci. 115: 2271–2282. [DOI] [PubMed] [Google Scholar]
- Piekny A. J., Wissmann A., Mains P. E., 2000. Embryonic morphogenesis in Caenorhabditis elegans integrates the activity of LET-502 Rho-binding kinase, MEL-11 myosin phosphatase, DAF-2 insulin receptor and FEM-2 PP2c phosphatase. Genetics 156: 1671–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim D., McGregor A., Jackle P., Johnson T., Hansen D., 1995. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol. Biol. Cell 6: 1159–1171. 10.1091/mbc.6.9.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess J. R., Hirsh D. I., 1986. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117: 156–173. 10.1016/0012-1606(86)90358-1 [DOI] [PubMed] [Google Scholar]
- Pruyne D., 2016. Revisiting the Phylogeny of the Animal Formins: Two New Subtypes, Relationships with Multiple Wing Hairs Proteins, and a Lost Human Formin. PLoS One 11: e0164067 10.1371/journal.pone.0164067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin S., Wang S., Pontabry J., Bender A., Robin F., et al. , 2016. Non-centrosomal epidermal microtubules act in parallel to LET-502/ROCK to promote C. elegans elongation. Development 143: 160–173. 10.1242/dev.126615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Suetsugu S., Sugimoto A., Miki H., Yamamoto M., et al. , 2003. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116: 1505–1518. 10.1242/jcs.00362 [DOI] [PubMed] [Google Scholar]
- Schonichen A., Mannherz H. G., Behrmann E., Mazur A. J., Kuhn S., et al. , 2013. FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J. Cell Sci. 126: 1891–1901. 10.1242/jcs.126706 [DOI] [PubMed] [Google Scholar]
- Schulze N., Graessl M., Blancke Soares A., Geyer M., Dehmelt L., et al. , 2014. FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J. Cell Sci. 127: 1379–1393. 10.1242/jcs.134627 [DOI] [PubMed] [Google Scholar]
- Severson A. F., Baillie D. L., Bowerman B., 2002. A Formin Homology Protein and a Profilin Are Required for Cytokinesis and Arp2/3-Independent Assembly of Cortical Microfilaments in C. elegans. Curr. Biol. 12: 2066–2075. 10.1016/S0960-9822(02)01355-6 [DOI] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2015. The Disease-Associated Formin INF2/EXC-6 Organizes Lumen and Cell Outgrowth during Tubulogenesis by Regulating F-Actin and Microtubule Cytoskeletons. Dev. Cell 32: 743–755. 10.1016/j.devcel.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2016. A network of conserved formins, regulated by the guanine exchange factor EXC-5 and the GTPase CDC-42, modulates tubulogenesis in vivo. Development 143: 4173–4181. 10.1242/dev.141861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Swan K. A., Severson A. F., Carter J. C., Martin P. R., Schnabel H., et al. , 1998. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J. Cell Sci. 111: 2017–2027. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Takeya R., Suetsugu S., Kan O. M., Narusawa M., et al. , 2009. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 284: 29873–29881. 10.1074/jbc.M109.059303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste C. A., Pruyne D., Mains P. E., 2013. The role of the formin gene fhod-1 in C. elegans embryonic morphogenesis. Worm 2: e25040 10.4161/worm.25040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong-Brender T. T., Ben Amar M., Pontabry J., Labouesse M., 2017. The interplay of stiffness and force anisotropies drives embryo elongation. eLife 6: e23866 10.7554/eLife.23866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong-Brender T. T., Yang X., Labouesse M., 2016. C. elegans Embryonic Morphogenesis. Curr. Top. Dev. Biol. 116: 597–616. 10.1016/bs.ctdb.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Williams-Masson E. M., Malik A. N., Hardin J., 1997. An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development 124: 2889–2901. [DOI] [PubMed] [Google Scholar]
- Williams B. D., Waterston R. H., 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124: 475–490. 10.1083/jcb.124.4.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Ingles J., Mains P. E., 1999. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev. Biol. 209: 111–127. 10.1006/dbio.1999.9242 [DOI] [PubMed] [Google Scholar]
- Wissmann A., Ingles J., McGhee J. D., Mains P. E., 1997. Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev. 11: 409–422. 10.1101/gad.11.4.409 [DOI] [PubMed] [Google Scholar]
- Wu M., Herman M. A., 2006. A novel noncanonical Wnt pathway is involved in the regulation of the asymmetric B cell division in C. elegans. Dev. Biol. 293: 316–329. 10.1016/j.ydbio.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Zhang H., Landmann F., Zahreddine H., Rodriguez D., Koch M., et al. , 2011. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 471: 99–103. 10.1038/nature09765 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6199286.