Abstract
Trisomy for human chromosome 21 (Hsa21) results in Down syndrome (DS), one of the most genetically complex conditions compatible with human survival. Assessment of the physiological consequences of dosage-driven overexpression of individual Hsa21 genes during early embryogenesis and the resulting contributions to DS pathology in mammals are not tractable in a systematic way. A recent study looked at loss-of-function of a subset of Caenorhabditis elegans orthologs of Hsa21 genes and identified ten candidates with behavioral phenotypes, but the equivalent over-expression experiment has not been done. We turned to zebrafish as a developmental model and, using a number of surrogate phenotypes, we screened Hsa21 genes for effects on early embyrogenesis. We prepared a library of 164 cDNAs of conserved protein coding genes, injected mRNA into early embryos and evaluated up to 5 days post-fertilization (dpf). Twenty-four genes produced a gross morphological phenotype, 11 of which could be reproduced reliably. Seven of these gave a phenotype consistent with down regulation of the sonic hedgehog (Shh) pathway; two showed defects indicative of defective neural crest migration; one resulted consistently in pericardial edema; and one was embryonic lethal. Combinatorial injections of multiple Hsa21 genes revealed both additive and compensatory effects, supporting the notion that complex genetic relationships underlie end phenotypes of trisomy that produce DS. Together, our data suggest that this system is useful in the genetic dissection of dosage-sensitive gene effects on early development and can inform the contribution of both individual loci and their combinatorial effects to phenotypes relevant to the etiopathology of DS.
Keywords: Hsa21, zebrafish, over expression
Down syndrome (DS) occurs in about one of 700 live births due to trisomy for human chromosome 21 (Hsa21) (Parker et al. 2010). The consequent 1.5 fold over expression of most genes on Hsa21 can result in more than 80 clinical phenotypes, many of which originate during prenatal development and vary in both severity and penetrance (Epstein et al. 1991; Kahlem et al. 2004; Deutsch et al. 2005). Among the most consistent features are cognitive impairment, characteristic craniofacial dysmorphism, smaller and hypocellular brain and Alzheimer histopathology [Roper and Reeves (2006); Aït Yahya-Graison et al. (2007)]. Individuals with DS also have a greatly increased risk of congenital heart disease, Hirschsprung disease and acute megakaryoblastic leukemia in children. However, the incomplete penetrance of many DS phenotypes indicates that trisomy 21 is not sufficient to cause most of these conditions, suggesting an important role for allelic variation of Hsa21 genes and additional modifier genes, as well as potential environmental and stochastic factors (Yang et al. 1999; Locke et al. 2010; Li et al. 2012). Estimates of the gene content on Hsa21 range from 300-600 genes/transcripts, of which 162 have been identified as well-conserved in other mammals (Sturgeon and Gardiner 2011). Understanding how trisomy for these genes affects the presentation of the phenotypes in DS is a major focus for research into this condition.
A major challenge in understanding mechanisms of gene action in DS is that trisomy 21 is present from conception and every cell is affected, causing effects throughout development. Trisomic genes may have a primary effect directly on cellular function or may secondarily affect expression and regulation of disomic genes. Trisomy-induced changes in one cell type could alter interactions with neighboring cells, thus initiating cascades of primary and secondary effects (Potier et al. 2006; Roper and Reeves 2006). A functional screen is further complicated by the large number of Hsa21 genes/transcripts. Use of mouse models trisomic for different segments of Hsa21-orthologous sequences supports to an extent the idea that different genetic segments correlate with some specific phenotypes, although independent replication of phenotypes has yielded conflicting results in some cases (Salehi et al. 2007; Gardiner et al. 2010; Herault et al. 2017), but even the smallest segmental trisomy still contains many genes. The effort and cost to systematically engineer individual transgenic mouse models of all conserved genes on Hsa21 would be prohibitive, to say nothing of the analysis of the possible combinations of genes. Further, events early in embryogenesis are difficult to access in mammals. However, previous studies have shown that the expression and/or suppression in zebrafish embryos of genes that map to disease-associated duplications and deletions in people can distinguish potent drivers of pathology (Golzio et al. 2012; Dauber et al. 2013; Golzio and Katsanis 2013; Carvalho et al. 2014; Lopez-Rivera et al. 2017). Motivated by such studies, we systematically over-expressed in zebrafish embryos each of 164 Hsa21 cDNAs representing 163 genes and assessed their effects on early development.
Recently, a screen to examine the effects of down-regulating orthologs of 47 Hsa21 genes was performed in Caenorhabditis elegans (Nordquist et al. 2018). Ten of these conserved genes exhibited neurobehavioral phenotypes: COL18A1, CBS, DONSON, EVA1C, N6AMT, NCAM2, POFUT2, PDXK, RUNX1 and SYNJ1 (Nordquist et al. 2018). Of these ten genes, five were shown to be essential for development based on the lethality phenotype seen in mouse knock-out models. The C. elegans screen identified three genes that were previously uncharacterized (DONSON, N6AMT and PDXK) as having a phenotype, providing new information about DS related genes and showing that these types of expression-based screens can provide a valuable resource to the DS research community. The knockdown screen in worms for all of the likely Hsa21 orthologs provided insights into gene function, but an over-expression screen in vertebrates that might more directly relate to over-expression of trisomic genes in DS has not been done. We systematically over-expressed in zebrafish embryos each of 164 Hsa21 cDNAs representing 163 genes and assessed their effects on early development.
Materials And Methods
Hsa21 Gene Expression Library preparation
cDNAs were selected from lists of conserved genes on Hsa21 (Gardiner et al. 2003; Kahlem et al. 2004). For 84 genes, plasmids containing the gene in the pENTR221 entry vector were obtained through the Invitrogen UltimateORF collection. Of the remaining genes, 49 were subcloned from a variety of vectors into one of the Invitrogen Gateway entry vectors (for complete list of original vectors and sources see Table S1) and 40 genes were commercially cloned into the pCS2DEST vector. Entry vector clones were selected using kanamycin and then sequenced to confirm correct insertion of the gene. All genes in the entry vector were subcloned into the pCS2DEST vector (Addgene) using LR clonase as previously described by (Katzen 2007). Genes in the pCSDEST vector were selected by ampicillin. The entire pCS2+ vector clone set, named the Hsa21 Gene Expression Set, is available through Addgene (https://www.addgene.org/kits/reeves-hsa21-set/); individual clones are available, as well (https://www.addgene.org/Roger_Reeves/). A limited set of HH pathway-related genes was used as a training set for the system including recognition of the U-shaped somite phenotype.
Bioinformatics
Comparisons of human, mouse, and zebrafish orthologs and ohnologs was performed using MouseMine (www.mousemine.org, accessed October 1, 2017 (Motenko et al. 2015). Briefly, Hsa21 gene symbols for the 163 genes in this screen were uploaded as a list in MouseMine and interrogated against the mouse and zebrafish using the HomoloGene data set from NCBI and the PANTHER data set from MGI (Mi et al. 2010). These lists were used to compile the ortholog and ohnolog lists in Table S1.
In Vitro Transcription of mRNA
Plasmids were transcribed in vitro using the mMessage mMachine SP6 kit (Ambion, Austin, TX). Plasmids were linearized and purified by precipitation. Transcribed sequence reactions were treated with DNAse1 and mRNA was purified with lithium chloride. mRNA quality and quantity were confirmed with a formaldehyde agarose gel and the Nanodrop8000, respectively.
Zebrafish maintenance and injections
All procedures were approved by the Johns Hopkins University Animal Care and Use Committee, protocol no. FI15M197. Zebrafish were raised in the FINZ center at the Institute for Genetic Medicine (Johns Hopkins University) as described previously (Westerfield 2000). Zebrafish were maintained at 28°. Male and female Tubingen zebrafish were placed in the same breeding tank in the morning and embryos were collected 30 min later. One hundred embryos were then injected at the 1-4 cell blastula stage using a Zeiss Stemi 2000 microscope and PV820 Pneumatic picopump injector. All genes were injected at 50pg or 100pg and most were injected a second time at a different dose (Table S2). All of those producing a phenotype were re-injected at 100pg. Embryos were raised to 5 days post fertilization and then phenotyped using a Nikon SMZ1500 microscope and imaged with NIS Elements Imaging Software. After imaging, embryos were fixed in 4% PFA overnight then transferred to 100% Methanol for storage at -20°. For low dosage experiments, SOD1 and RRP1 were injected at 2pg, 5pg and 10pg and examined at 5 dpf.
Morpholino Rescue
Translation-blocking antisense morpholinos (MO) were designed against the human sequence for the genes SOD1, RWDD2B, and CCT8, designed to bind to the ATG start codon of the mRNA using Gene Tools (Philomath, OR): Hs-SOD1 5′-GCACGCACACGGCCTTCGTCGCCAT-3′; Hs-RWDD2B 5′-GCTGCATGGACAGCTCAATTTTCAT-3′; and Hs-CCT8 5′-GAGCCTTGGGAACGTGAAGCGCCAT-3′. The MOs were checked using BLAST for sequence specificity to the human homolog and insure that they were unique in the either the human or zebrafish genomes. For each gene, 100 embryos were injected with 2ng MO, 100 embryos were injected with 100pg of mRNA and 100 embryos were injected with both 2ng MO and 100pg mRNA; 100 uninjected embryos were used as a control. Embryos were examined at 5 dpf for Hs-SOD1 and Hs-RWDD2B, 4 dpf for Hs-CCT8, and 24 hr. post-fertilization (hpf) for Dr-JAM2.
Combinatorial Injections
RNA from C21ORF84 was co-injected with RNA from the following genes: SOD1, RWDD2B, RRP1, PCBP3, POFUT2, and YBEY. Each gene was injected individually at 100pg into 100 embryos, and then co-injected at 100pg of each RNA (200pg RNA). Embryos were phenotyped at 24 hpf for the presence of U shaped somites and cyclopia. Each co-injection was performed twice. A core set of experiments was carried out independently at a different institution (NAZ and CCL) injecting 200 pg mRNA into 100 embryos of the Tubingen line using the pairs listed and phenotyping at 24 hpf.
The combinatorial strategy was repeated using SOD1 as the reference gene and co-injected with the same genes listed above. In this case, 50pg of each RNA was injected individually into 100 embryos each, and then 50pg each of both RNAs were co-injected into 100 embryos, with 100 control embryos. Embryos were examined at 24hpf for the presence of U-shaped somites.
Statistical Tests
For all injections, penetrance differences were examined using a Fisher’s Exact test with P < 0.05 required for significance.
Data Availability
The pCS2+ vector clone set, named the Hsa21 Gene Expression Set, is available through Addgene (https://www.addgene.org/kits/reeves-hsa21-set/). Supplemental files available at Figshare. Table S1 contains lists of all Hsa21 cDNAs included in this study along with cloning information, and zebrafish ohnologs/orthologs. Table S2 contains details of all injections for the whole clone set. Table S3 describes the 23 initial candidates and phenotypes. Table S4 contains information about LINCs and ORFs included in the clone-set. Figure S1 contains dosage curves for 10 of the candidate genes. S2 shows low dose RNA injections for SOD1 and RRP1. S3 shows pairwise injections for SOD1 and 6 other candidate genes. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6089324.
Results
Development of the clone set and initial screen
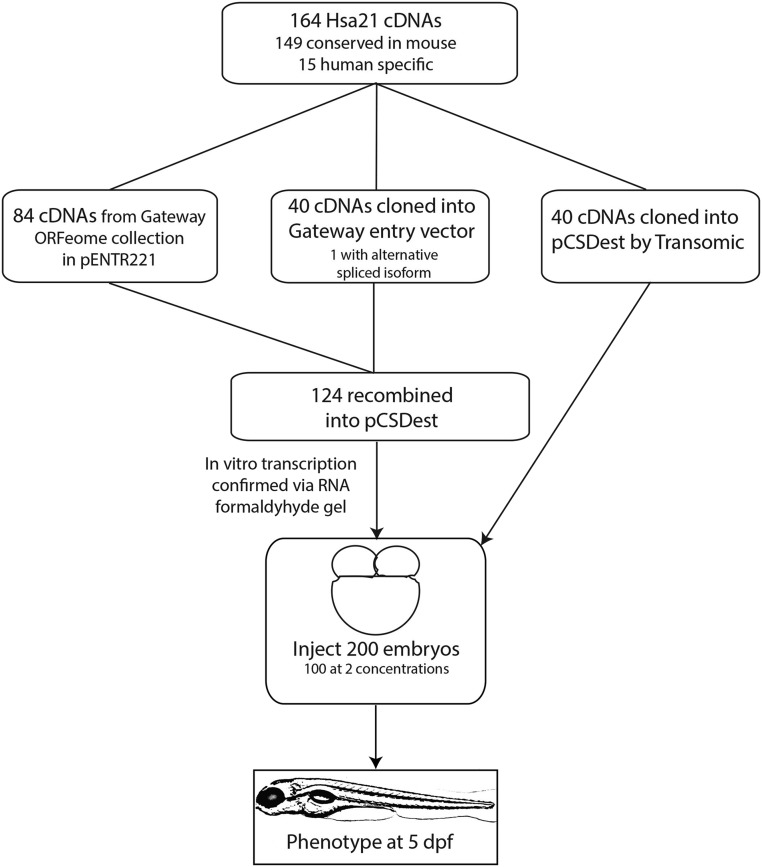
In a detailed annotation of transcripts from Hsa21, Sturgeon and Gardiner identified 162 genes that are highly conserved with mouse (Sturgeon and Gardiner 2011). We assembled a set of Hsa21 cDNA clones consisting of 148 of these conserved genes plus 15 human-specific genes (Gardiner et al. 2003; Sturgeon and Gardiner 2011). The 15 genes that are not conserved include so-called DSCR genes, long intergenic non-coding RNA, and Hsa21 open reading frames. One gene, SYNJ1, is represented by two splice isoforms, for a total of 164 cDNAs (Table S1). The majority of clones are from the InVitrogen UltimateORF collection library (now Thermo Fisher Scientific’s Ultimate ORF Clones), subcloned into the pCS2+ destination vector (Figure 1) (Rupp et al. 1994). Thus, we selected highly conserved genes from this carefully annotated set for which cDNA could be obtained and which could be cloned into the pCS2+ vector and transcribed. Additional sources and vectors are described in Table S1.
Figure 1.
Flowchart showing steps of making the Hsa21 Gene Expression Clone-set and the screen in zebrafish.
Next we used Mouse Mine to interrogate the human gene list and compare it to homology data sets, HomoloGene and Panther, to identify orthologs and ohnologs between zebrafish and mouse (Table S1) (Motenko et al. 2015; Mi et al. 2010). In zebrafish, 125 of the 163 Hsa21 genes (77%) are conserved; 35 of those (28%) are represented by more than one zebrafish ortholog (ohnolog). The number of Hsa21 genes conserved with zebrafish is similar to published reports for all human genes (71%), but the proportion of Hsa21 genes with ohnologs is less than the 47% rate for all human genes (Howe et al. 2013).
Zebrafish Screen
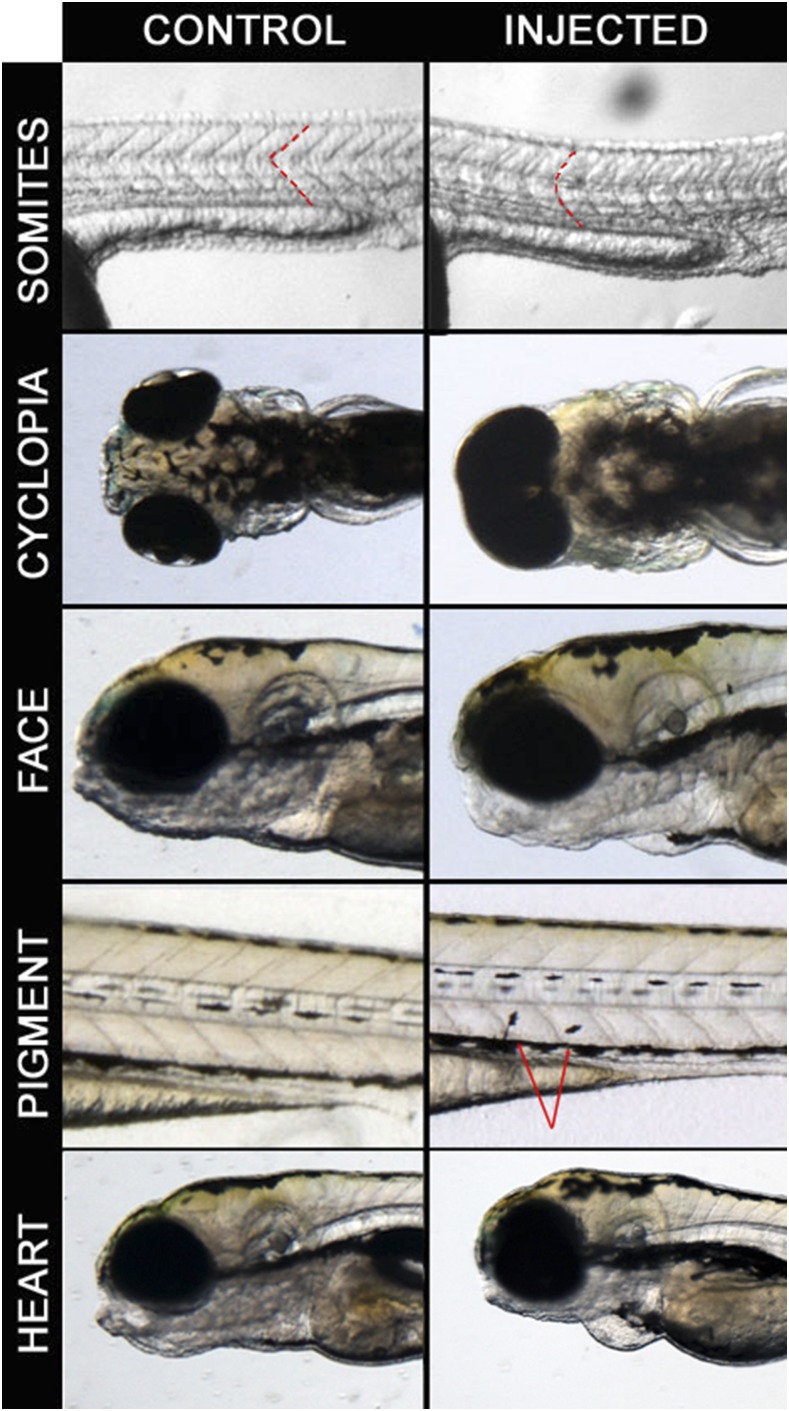
We synthesized mRNAs from the Hsa21 cDNA clones and injected each one into 100 zebrafish embryos at the 1-2 cell stage at 10, 50 and/or 100 pg (Table S2), ranges that have been used in similar studies previously (Golzio et al. 2012). More than 50,000 embryos were screened for the presence of gross morphological changes present at frequencies greater than controls, typically at five days post fertilization (dpf), but also earlier if mandated by the presence of clear pathologies. We focused primarily on three broad phenotypic classes: a) U-shaped somites and cyclopia, two phenotypes associated frequently but not exclusively with defects in the ciliome and Shh signaling pathway; b) craniofacial abnormalities and pigment differences, which may be related to aberrations affecting neural crest cells; and c) pericardial edema, which can have a number of causes including a structural heart defect (Figure 2, Table 1). We recorded the number of surviving embryos, the phenotype, and the percentage that were affected (Table 2).
Figure 2.
Examples of phenotypes observed in the screen. Control embryos are on the left panel and injected embryos are on the right panel. Somites: RWDD2B 100pg injected embryos at 24 hpf with dashed lines to highlight somitic boundaries. Cyclopia: C21ORF84 100pg injected embryos at 5 dpf. Pigment cell migration: CCT8 100pg injected embryos at 4 dpf, arrows indicating melanocytes. Heart: JAM2 100pg injected embryos at 48 hpf.
Table 1. Twenty-four candidates from the first pass of screen.
| Phenotype category | Description | Number Candidatesa |
|---|---|---|
| U-shaped Somites | Somites with characteristic U shape | 8 |
| Cyclopia | Single large eye | 6 |
| Craniofacial abnormalities | Small/missing mandible; skull abnormalities | 4 |
| Pigment abnormalities | Floating melanocytes; reduced pigment in eye | 3 |
| Heart | Pericardial edema | 1 |
| Other | Tail/fin abnormalities, embryo lethality | 2 |
See Supplemental Table 3.
Table 2. Final Candidate list of genes that produced a phenotype consistently.
| Gene Symbol | Phenotype | Penetrance | Expression in mouse (Reymond et al. 2002) |
|---|---|---|---|
| SOD1 | U-somites | 10–35% | Expressed ubiquitously at E10.5, strongly expressed in muscles at E14.5 |
| RWDD2B | U-somites | 15–31% | Expressed ubiquitously at E10.5 |
| RRP1 | U-somites | 15–40% | Weakly expressed in somites at E10.5 (Armit et al. 2012) |
| PCBP3 | U-somites | 8–13% | Expressed in brain and spinal cord at E14.5 |
| YBEY | U-somites | 12–19% | Expressed ubiquitously at E10.5 |
| C21ORF84 | U-somites | 9–23% | human specific (Gardiner et al. 2003) |
| C21ORF84 | Cyclopia | 0–7% | human specific (Gardiner et al. 2003) |
| POFUT2 | Cyclopia | 0–4% | Strong in face and pharyngeal arches at E9.5 (Armit et al. 2012) |
| CBR3 | Craniofacial | 7–10% | Expressed ubiquitously at E10.5, strong in cartilage at E14.5 |
| CCT8 | Pigment | 15–40% | Expressed ubiquitously at E10.5 |
| JAM2 | Pericardial Edema | 20–60% | High expression in human heart (Cunningham et al. 2000) |
| ERG | Embryonic Lethality | 64–93% | Expressed in the pharyngeal arches and limb buds at E10.5 (Carvalho et al. 2014) |
Of the 164 RNAs, 24 showed a phenotype after the initial screen (Supplemental Tables 2 and 3); the remaining 140 did not yield a significantly increased number of affected embryos compared to untreated. The lack of phenotype from 140 cDNAs is a strong control itself for general mutagenic or toxic effects of RNA or its preparation, thus, consistent with previous studies (Golzio et al. 2012), our approach is not generally toxic to zebrafish embryos. Expression of 14 of these 24 genes gave a phenotype consistent with perturbation of the ciliome/Shh pathway (eight with U-shaped somites and a partially overlapping set of six with cyclopia). An additional seven genes resulted in phenotypes that may be due to neural crest defects (four with craniofacial abnormalities and three with pigment differences). Finally, one gene induced pericardial edema, one resulted in dysmorphic fins, and one exhibited elevated lethality (Table 1 and Table S2). Of the 24 first-round candidates, 19 are conserved in both mouse and zebrafish; four are conserved between human and mouse but not zebrafish (BACH1, Clic6, MAP3K7CL, SPATC1L); and one gene (C21ORF84 also known as LINC00313) is human specific. Six of the 19 conserved Hsa21 genes have ohnologs in zebrafish (Table S1).
Fresh mRNA was prepared from these first-round candidates and injections were repeated. Eleven candidates recapitulated the original phenotypes robustly, seven from the Shh group, two with neural crest related phenotypes, and the single genes resulting in pericardial edema and embryonic lethality, respectively (Figure 2, Table 2, Supplemental Tables 2 and 3). Of the 11 sec-round candidates conserved in zebrafish, two have ohnologs in zebrafish (JAM2 and CBR3), i.e., they were involved in the partial genome duplication that characterizes the D. rerio genome.
Phenotypic rescue via Morpholinos
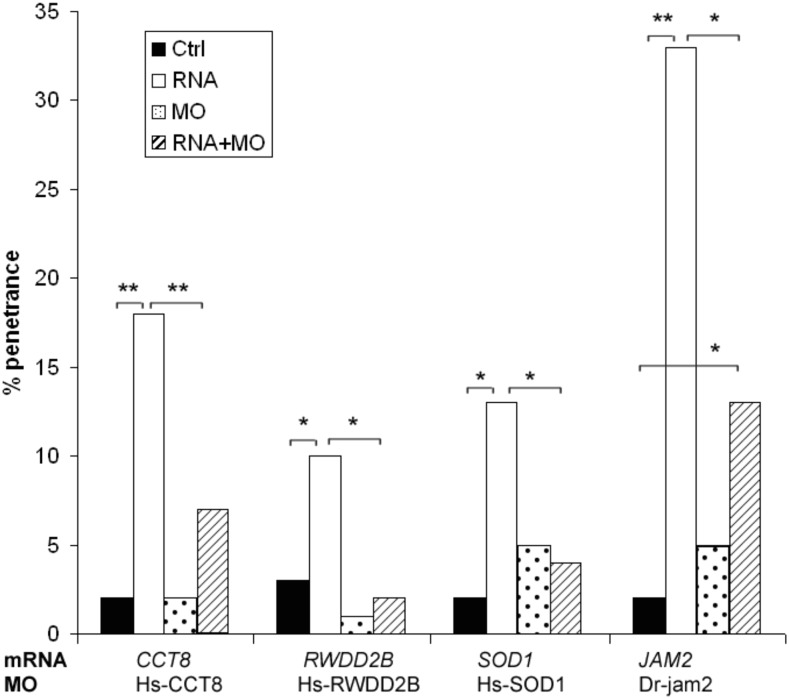
The first indication that the phenotypes observed in repeated injections are due to the expression of the specific RNA and not to a general RNA effect is that there are at least 140 clones that produced no phenotype in this screen. Next, we selected randomly four of the candidate genes and performed a rescue experiment using morpholino (MO)-based knockdown. We designed translation-blocking MOs against the human copies of SOD1, RWDD2B or CCT8 to target the ATG start site of the gene to suppress specifically the introduced human mRNA. We also designed MOs to knock down the endogenous zebrafish orthologs of JAM2, using a previously validated MO (Powell and Wright 2011). One hundred embryos were injected with 2ng MO and 100pg of RNA. For all four genes, injection of the RNA alone produced significantly higher penetrance than the uninjected controls, the MO alone, or the MO+RNA injected embryos (P < 0.05 Figure 3). For SOD1, RWDD2B, and CCT8 the MO+RNA was not significantly different from the controls. JAM2 MO+RNA showed significantly lower penetrance of heart edema relative to RNA-injection alone (Li et al. 2016). The JAM2 experiment suggests that this phenotype is a product of additive effects of the human cDNA with its fish orthologs/ohnologs.
Figure 3.
Candidate genes (SOD1, RWDD2B, CCT8, and JAM2) coinjected with translational blocking morpholino. Hs-RWDD2B, Hs-CCT8 and Hs-SOD1 MOs were targeted against the human mRNA, while jam2 MOs were targeted against the zebrafish ortholog, DR-jam2. 100pg RNA was injected alone, 2 ng MO alone, or both were coinjected. * P < 0.05, ** P < 0.01. JAM2 data adapted from (Li et al. 2016).
Dosage and Gene expression patterns
This screen was designed to identify Hsa21 genes with an effect on broad aspects of early embryonic development and not as a study of human dosage effects on fish (see Discussion). For those cDNAs producing phenotypes, penetrance was generally correlated with but not directly proportional to penetrance (Table S2). In the case of ERG, however, the frequency of embryonic lethality was proportional to RNA dose. Approximately 1/3 of embryos (24/67) survived after injection with 10 pg ERG RNA, while 14% survived injection with 50 pg and 6.5% (24/358) were alive two days after injection of 100 pg. Retrospective examination also showed somewhat elevated mortality following injection of either of the additional two ETS family transcription factors in the Hsa21 clone set, ETS2 and GABPA (Table S2). HMGN1 also showed a trend toward higher lethality.
Among the final candidates with repeatable phenotypes, only ERG showed increased lethality with increasing RNA concentrations. The 10 remaining candidates were injected at three or more concentrations ranging from 10pg to 200pg but no correlation between dosage and penetrance was observed (Figure S1). The mRNA concentrations that we used are consistent with those commonly reported (Takeda et al. 1994; Golzio et al. 2012). We also chose two genes, SOD1 and RRP1, to look for effects of low RNA doses. These genes were injected at 2pg and 5pg, and the penetrance of U-shaped somites was examined. Both genes showed low penetrance of the phenotype at these low doses, although each reproducibly produced a phenotype after injection of 50-100pg (Figure S2).
Nine of the eleven candidates have zebrafish homologs, the exceptions being C21ORF84 and CBR3. For eight of these genes, representative in situ hybridization data are available in ZFIN (Thisse and Thisse 2004). In most cases, the structure(s) affected by RNA injection is consistent with the in situ expression data. For example, sod1, rrp1 and ybey are expressed in somites, cct8 is expressed ubiquitously, jam2 shows expression in the region of the developing heart and erg is expressed in the vasculature (Table 2). pofut2 is expressed in the brain and eye (Ohata et al. 2009); knockdown of a C. elegans homolog of this gene produced a neuromuscular phenotype (Nordquist et al. 2018). pcbp3 is expressed in the retina and telencephalon beginning at the Prim-15 stage (Thisse and Thisse 2004). No information about rwdd2b expression was available in ZFIN. Ten of the eleven candidates have mouse orthologs whose expression has been examined at mid-gestation showing that the genes are expressed in the corresponding tissues during embryonic development in mouse (Table 2, (Reymond et al. 2002; Armit et al. 2012)). C21ORF84 is a human specific lncRNA (Gardiner et al. 2003).
Combinatorial injections
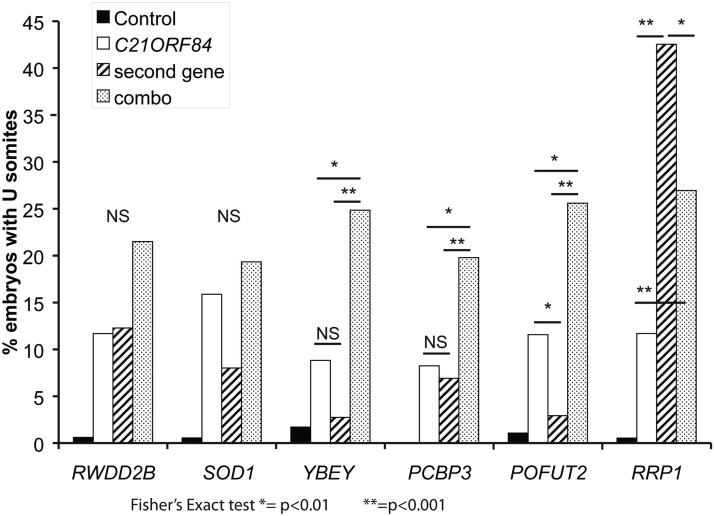
DS is a contiguous gene defect with effects on development that exceed those of individual genes (e.g., (Salehi et al. 2006; Sussan et al. 2008)). Examination of all pairwise interactions of 163 genes would require more than 13,000 pairs, posing a logistical challenge even in this system. We took an initial step toward understanding this process with respect to a small set of candidate genes. We first examined the subset of genes that produced possible Shh-related phenotypes in our screen. SOD1, RWDD2B, YBEY, PCBP3, and RRP1 all produced U-shaped somites, POFUT2 resulted in cyclopia and C21ORF84-injected embryos showed both phenotypes. We used C21ORF84 as the reference gene to pair with the other six genes. For each set of pairwise injections, C21ORF84 was injected alone at 100pg, the second gene was injected alone at 100pg, and both genes were injected together at 100pg each.
Injection of C21ORF84 plus YBEY, PCBP3 or POFUT2 showed a significant increase in the frequency of embryos with U-shaped somites (Figure 4, P < 0.05 for each combination). For two genes, SOD1 and RWDD2B, there was not a significant difference between the individual injections and the combinatorial injection. RRP1 alone had a penetrance of 42%, the highest of all genes tested. This frequency was reduced significantly in embryos injected with RRP1 and C21ORF84 together.
Figure 4.
Pairwise combinatorial injections of Shh candidate genes. C21ORF84 was coinjected with 6 other genes to look for synthetic effects. C21ORF84 was injected individually at 100pg RNA, the second gene was injected individually at 100pg RNA and then the two were injected together, 100pg each, for a total of 200pg RNA.
We repeated the pairwise injections using a different reference gene from this group, SOD1. As in the previous experiment, SOD1 was injected individually and in combination with one of the other six genes, and the embryos were examined for the presence of U-shaped somites. In contrast to C21ORF84, SOD1 did not show an interaction with any of the other six genes (Figure S3). Finally, combinatorial injections using C21ORF84 were repeated with freshly prepared RNAs at a separate institution (by NAZ and CCL) as a means of independent replication. We observed a similar increase in affected embryos upon injection of C21ORF84 with YBEY and PCBP3 compared to each gene alone.
We also selected candidate gene sets for combinatorial injection based on reported roles in other systems and assessed them for possible combinatorial effects on developmental phenotypes in zebrafish. Several genes associated previously with heart anomalies in Down syndrome, SH3BGR, DCSR6 and ADAMTS1, were co-injected (30pg each). This resulted in cyclopia in 3.6% and pericardial edema in a non-overlapping 3.6% of embryos, whereas no controls were observed to have either edema or cyclopia, a trend though not formally significant (Fisher’s exact test, P = 0.06 for either cyclopia or edema). Other Hsa21 gene combinations have been implicated in the high frequency of congenital heart disease in DS, as well (Ferencz et al. 1989). However, neither injection of all three collagens together (COL6A1, COL6A2, COL18A1) nor co-injection of DSCAM and SH3BGR produced a significant frequency of heart or other gross anatomical defects.
Discussion
We have developed a study set of Hsa21 gene expression clones (available from AddGene, see Methods) and used it to conduct the first large-scale study of the effects of Hsa21 gene expression on early vertebrate embryogenesis. Previous analyses of Hsa21 gene expression in early development used in situ hybridization (Gitton et al. 2002; Reymond et al. 2002) or microarrays to examine the localization, timing, and/or levels of Hsa21 gene up-regulation (Kahlem et al. 2004). Here, we used a functional assay in zebrafish embryos to find candidate genes with effects on early development in a systematic, unbiased approach. This stage of development is difficult to study in mammals. Of the 163 genes (164 cDNAs) assayed, only eleven genes consistently showed an effect. These genes are implicated in ciliome/Shh signaling, neural crest cell (NCC) generation and differentiation, heart development, and/or embryonic lethality. ERG has recently been shown by the Knock-Out Mouse Phenotyping Project (KOMP2) to be required during mammalian embryogenesis; embryos homozygous for a null allele of Erg are lethal prior to E15.5 (www.mousephenotype.org accessed March 27, 2017 (Brown and Moore 2012)). Several of the genes have little or no known function and none have been implicated previously in either Shh signaling or NCC generation and development.
Clearly, many additional phenotypes could be considered in a “next gen” zebrafish screen using the cDNA clone set described here. A handful of the 80 or so perturbations that occur more frequently in DS are highly penetrant (Epstein et al. 1991). These include characteristic craniofacial dysmorphology, the early onset of Alzheimer histopathology, cerebellar hypoplasia, resistance to many solid tumors, and intellectual disability (including detailed assessments in mouse models at the gene, phenotype, morphological and physiological levels). Screening in fish with appropriate genetic manipulations to mark and/or affect specific cells or tissues would allow deeper investigation of specific focused questions. Creating transgenic fish with candidates identified in this or other screens could support a quantitative approach to issues of gene dosage.
There are several caveats to interpretation of this large scale screen. A negative result in the screen does not rule out a contribution of that gene to DS. First, some human genes could fail to produce an effect due to inefficient translation related to the fact that these mammalian mRNAs may not be properly processed in fish. Some human proteins may simply be hypo- or non-functional in zebrafish. Except for SYNJ1, we selected a single isoform for each gene represented in the cDNA clone set. Next, DS is a contiguous gene syndrome, with many interactions among the over-expressed genes and with disomic genes at different stages of development (Potier et al. 2006; Roper and Reeves 2006). The exposure to human RNA is transient and although previous studies suggest that RNA is present through the first days of development, we would expect a high degree of variability in how and when the RNA arrives in a cell and is translated. While we injected a large number of embryos for every candidate and were thorough in replication, especially of the final interacting candidates, this is an inherently noisy biological system with multiple technical steps that could produce artifacts. It is important to note that the output of the injections is a value for penetrance – which is incomplete in every case – and introduces another opportunity for failure to replicate. Throughout the procedures (cloning and its validation, acceptance or not of a day’s injections based on batch results, interpretation of the severity of phenotypes vis-à-vis inclusion or exclusion of candidates, etc.) we have erred on the side of minimizing false positive findings.
Finally, this is not a study of the effects of dosage imbalance that occur in DS per se. Indeed, it would be difficult to define what dose of a human mRNA might result in human protein levels that, together with the orthologous fish proteins, replicate the functional stoichiometry of DS in a fish embryo. Genes with ohnologs that function in a complementary manner in fish represent an even more complicated situation when additional expression of proteins that have related but non-identical function is introduced via the human gene product. We did not observe a lower frequency of phenotypes among genes with ohnologs than in the Hsa21 gene set as a whole, but the numbers are very small. What we present is instead a screen for genes whose effects in this assay suggest candidates to pursue in far more labor-intensive studies of dosage-sensitive effects in early mammalian development.
We have successfully used information from this screen in one such application with Jam2 (Li et al. 2016). The consistent occurrence of heart edema in zebrafish after injection of JAM2 RNA in this screen led us to examine this candidate in conjunction with increased penetrance of heart defects in trisomic mouse models. Ts65Dn “Down syndrome” mice are trisomic for about 104 genes orthologous to Hsa21 (Das and Reeves 2011). Breeding a null allele of the disomic gene, Creld1, onto Ts65Dn significantly increased penetrance of septal defects in the heart from 4 to 33% (Li et al. 2012). However, putting the same null allele on a related trisomy (Ts1Cje) that was trisomic for 81 of the genes triplicated in Ts65Dn had no impact on penetrance. Jam2 was one of the 23 genes that are trisomic in Ts65Dn but not Ts1Cje, and one of 14 of these 23 that are expressed prenatally and/or in heart. Based on its effect on heart development in the zebrafish screen, we tested its role in mice by introducing a null allele of Jam2 to produce Ts65Dn;Creld1+/−;Jam2+/− mice (returning Jam2 to the normal two copies on an otherwise identical trisomic background). Instead of the expected increase in penetrance of heart defects observed in Ts65Dn;Creld1+/−, trisomy for all of the Ts65Dn genes except Jam2 resulted in penetrance in these mice that was the same as Ts65Dn alone (i.e., 4%) (Li et al. 2016). Thus Jam2 is required in this model as a trisomic potentiator of the disomic modifier of heart disease penetrance, Creld1. This Jam2-Creld1 interaction was the first demonstration of this type of genetic relationship in Down syndrome. It would not have been possible to pursue this relationship for all 14 candidate genes that are trisomic in Ts65Dn but not in Ts1Cje in mice.
In this screen, it was somewhat surprising that several Hsa21 genes that have been associated with robust phenotypes in mouse models of DS did not produce a phenotype. For example, DSCAM, a cell adhesion molecule that is involved in cell recognition (Garrett et al. 2012), has been implicated in both heart and neurogenesis defects based on work done in mice as well as in Drosophila (Grossman et al. 2011; Zhu et al. 2011) but did not produce a phenotype in our zebrafish screen. DYRK1A, a dual specificity kinase expressed during early neurogenesis that has been a target in pilot studies for treatment of cognitive deficits in DS (De la Torre et al. 2014; de la Torre et al. 2016), also produced no phenotype. Many types of refined screens with greater sensitivity and specificity are possible, taking advantage of transgenically marked zebrafish lines to ask specific questions about development of specific structures. Furthermore, our identification of phenotypes associated with pathways previously identified as central to multiple manifestations of trisomy, such as Shh signaling (Roper et al. 2009; Currier et al. 2012; Das et al. 2013), support the use of D. rerio for this type of large-scale systematic screen. This system represents a useful screening tool to identify individual candidate genes that may be significant drivers of DS phenotypes.
We observed that some Hsa21 genes that produce phenotypes on their own can interact in an additive manner, some have no apparent interaction and one pair had a compensatory interaction. Compensatory interaction implies that in some cases overexpression of one gene can balance the increased expression of another. To date, several genes of major effect have been associated with manifestations of Down syndrome in the mouse models. However, no single gene has been identified that is sufficient to produce completely a complex developmental phenotype of DS, consistent with the understanding of DS as a product of complex multi-gene interactions. Given the large number of possible gene-gene interactions on chr21 alone, the system described here provides a useful way to interrogate more complex interactions of non-contiguous genes from the earliest stages of development.
Acknowledgments
We thank Valerie DeLeon, Deborah Andrew, and Steven Leach for their advice and support. The work was supported by R01 HD038384 and the Lumind-RDS Foundation (to RHR). This work is submitted in partial fulfillment of the requirements for a Ph.D. (SE).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6089324.
Communicating editor: B. Andrews
Literature Cited
- Aït Yahya-Graison E., Aubert J., Dauphinot L., Rivals I., Prieur M., et al. , 2007. Classification of human chromosome 21 gene-expression variations in down syndrome: impact on disease phenotypes. Am. J. Hum. Genet. 81: 475–491. 10.1086/520000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armit C., Venkataraman S., Richardson L., Stevenson P., Moss J., et al. , 2012. emouseatlas, emage, and the spatial dimension of the transcriptome. Mamm. Genome 23: 514–524. 10.1007/s00335-012-9407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Moore M. W., 2012. The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23: 632–640. 10.1007/s00335-012-9427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho C. M., Vasanth S., Shinawi M., Russell C., Ramocki M. B., et al. , 2014. Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. Am. J. Hum. Genet. 95: 565–578. 10.1016/j.ajhg.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham S. A., Arrate M. P., Rodriguez J. M., Bjercke R. J., Vanderslice P., et al. , 2000. A novel protein with homology to the junctional adhesion molecule. characterization of leukocyte interactions. J. Biol. Chem. 275: 34750–34756. 10.1074/jbc.M002718200 [DOI] [PubMed] [Google Scholar]
- Currier D. G., Polk R. C., Reeves R. H., 2012. A sonic hedgehog (shh) response deficit in trisomic cells may be a common denominator for multiple features of down syndrome. Prog. Brain Res. 197: 223–236. 10.1016/B978-0-444-54299-1.00011-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I., Park J. M., Shin J. H., Jeon S. K., Lorenzi H., et al. , 2013. Hedgehog agonist therapy corrects structural and cognitive deficits in a down syndrome mouse model. Sci. Transl. Med. 5: 201ra120 10.1126/scitranslmed.3005983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I., Reeves R. H., 2011. The use of mouse models to understand and improve cognitive deficits in down syndrome. Dis. Model. Mech. 4: 596–606. 10.1242/dmm.007716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber A., Golzio C., Guenot C., Jodelka F. M., Kibaek M., et al. , 2013. Scrib and puf60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 93: 798–811. 10.1016/j.ajhg.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre R., de Sola S., Hernandez G., Farre M., Pujol J., et al. , 2016. Safety and efficacy of cognitive training plus epigallocatechin-3-gallate in young adults with down’s syndrome (tesdad): a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 15: 801–810. 10.1016/S1474-4422(16)30034-5 [DOI] [PubMed] [Google Scholar]
- De la Torre R., De Sola S., Pons M., Duchon A., de Lagran M. M., et al. , 2014. Epigallocatechin-3-gallate, a dyrk1a inhibitor, rescues cognitive deficits in down syndrome mouse models and in humans. Mol. Nutr. Food Res. 58: 278–288. 10.1002/mnfr.201300325 [DOI] [PubMed] [Google Scholar]
- Deutsch S., Lyle R., Dermitzakis E. T., Attar H., Subrahmanyan L., et al. , 2005. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum. Mol. Genet. 14: 3741–3749. 10.1093/hmg/ddi404 [DOI] [PubMed] [Google Scholar]
- Epstein C. J., Korenberg J. R., Anneren G., Antonarakis S. E., Ayme S., et al. , 1991. Protocols to establish genotype-phenotype correlations in down syndrome. Am. J. Hum. Genet. 49: 207–235. [PMC free article] [PubMed] [Google Scholar]
- Ferencz C., Neill C. A., Boughman J. A., Rubin J. D., Brenner J. I., Perry L. W., 1989. Congenital cardiovascular malformations associated with chromosome abnormalities: an epidemiologic study. The Journal of pediatrics. 114(1): 79–86. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Fortna A., Bechtel L., Davisson M. T., 2003. Mouse models of down syndrome: how useful can they be? comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene 318: 137–147. 10.1016/S0378-1119(03)00769-8 [DOI] [PubMed] [Google Scholar]
- Gardiner K., Herault Y., Lott I. T., Antonarakis S. E., Reeves R. H., et al. , 2010. Down syndrome: from understanding the neurobiology to therapy. J. Neurosci. 30: 14943–14945. 10.1523/JNEUROSCI.3728-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett A. M., Tadenev A. L., Burgess R. W., 2012. Dscams: restoring balance to developmental forces. Front. Mol. Neurosci. 5: 86 10.3389/fnmol.2012.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitton Y., Dahmane N., Baik S., Ruiz i Altaba A., Neidhardt L., et al. , 2002. A gene expression map of human chromosome 21 orthologues in the mouse. Nature 420: 586–590. 10.1038/nature01270 [DOI] [PubMed] [Google Scholar]
- Golzio C., Katsanis N., 2013. Genetic architecture of reciprocal cnvs. Curr. Opin. Genet. Dev. 23: 240–248. 10.1016/j.gde.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C., Willer J., Talkowski M. E., Oh E. C., Taniguchi Y., et al. , 2012. Kctd13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485: 363–367. 10.1038/nature11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman T. R., Gamliel A., Wessells R. J., Taghli-Lamallem O., Jepsen K., et al. , 2011. Over-expression of dscam and col6a2 cooperatively generates congenital heart defects. PLoS Genet. 7: e1002344 10.1371/journal.pgen.1002344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herault Y., Delabar J. M., Fisher E. M. C., Tybulewicz V. L. J., Yu E., et al. , 2017. Rodent models in down syndrome research: impact and future opportunities. Dis. Model. Mech. 10: 1165–1186. 10.1242/dmm.029728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., et al. , 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503 (erratum: Nature 505: 248) 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlem P., Sultan M., Herwig R., Steinfath M., Balzereit D., et al. , 2004. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 14: 1258–1267. 10.1101/gr.1951304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F., 2007. Gateway((r)) recombinational cloning: a biological operating system. Expert Opin. Drug Discov. 2: 571–589. 10.1517/17460441.2.4.571 [DOI] [PubMed] [Google Scholar]
- Li H., Cherry S., Klinedinst D., DeLeon V., Redig J., et al. , 2012. Genetic modifiers predisposing to congenital heart disease in the sensitized down syndrome population. Circ Cardiovasc Genet 5: 301–308. 10.1161/CIRCGENETICS.111.960872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Edie S., Klinedinst D., Jeong J. S., Blackshaw S., et al. , 2016. Penetrance of congenital heart disease in a mouse model of down syndrome depends on a trisomic potentiator of a disomic modifier. Genetics 203: 763–770. 10.1534/genetics.116.188045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke A. E., Dooley K. J., Tinker S. W., Cheong S. Y., Feingold E., et al. , 2010. Variation in folate pathway genes contributes to risk of congenital heart defects among individuals with down syndrome. Genet. Epidemiol. 34: 613–623. 10.1002/gepi.20518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rivera E., Liu Y. P., Verbitsky M., Anderson B. R., Capone V. P., et al. , 2017. Genetic drivers of kidney defects in the digeorge syndrome. N. Engl. J. Med. 376: 742–754. 10.1056/NEJMoa1609009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., et al. , 2010. Panther version 7: improved phylogenetic trees, orthologs and collaboration with the gene ontology consortium. Nucleic Acids Res. 38: D204–D210. 10.1093/nar/gkp1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motenko H., Neuhauser S. B., O’Keefe M., Richardson J. E., 2015. Mousemine: a new data warehouse for mgi. Mamm. Genome 26: 325–330. 10.1007/s00335-015-9573-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordquist S. K., Smith S. R., Pierce J. T., 2018. Systematic functional characterization of human 21st chromosome orthologs in Caenorhabditis elegans. G3 (Bethesda) 8: 967–979. 10.1534/g3.118.200019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata S., Kinoshita S., Aoki R., Tanaka H., Wada H., et al. , 2009. Neuroepithelial cells require fucosylated glycans to guide the migration of vagus motor neuron progenitors in the developing zebrafish hindbrain. Development 136: 1653–1663. 10.1242/dev.033290 [DOI] [PubMed] [Google Scholar]
- Parker S. E., Mai C. T., Canfield M. A., Rickard R., Wang Y., et al. , 2010. Updated national birth prevalence estimates for selected birth defects in the united states, 2004–2006. Birth Defects Res. A Clin. Mol. Teratol. 88: 1008–1016. 10.1002/bdra.20735 [DOI] [PubMed] [Google Scholar]
- Potier M. C., Rivals I., Mercier G., Ettwiller L., Moldrich R. X., et al. , 2006. Transcriptional disruptions in down syndrome: a case study in the ts1cje mouse cerebellum during post-natal development. J. Neurochem. 97: 104–109. 10.1111/j.1471-4159.2005.03624.x [DOI] [PubMed] [Google Scholar]
- Powell G. T., Wright G. J., 2011. Jamb and jamc are essential for vertebrate myocyte fusion. PLoS Biol. 9(12): e1001216 10.1371/journal.pbio.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., Marigo V., Yaylaoglu M. B., Leoni A., Ucla C., et al. , 2002. Human chromosome 21 gene expression atlas in the mouse. Nature 420: 582–586. 10.1038/nature01178 [DOI] [PubMed] [Google Scholar]
- Roper R., Reeves R., 2006. Understanding the basis for down syndrome phenotypes. PLoS Genet. 2: e50 10.1371/journal.pgen.0020050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R. J., VanHorn J. F., Cain C. C., Reeves R. H., 2009. A neural crest deficit in down syndrome mice is associated with deficient mitotic response to sonic hedgehog. Mech. Dev. 126: 212–219. 10.1016/j.mod.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp R. A., Snider L., Weintraub H., 1994. Xenopus embryos regulate the nuclear localization of xmyod. Genes Dev. 8: 1311–1323. 10.1101/gad.8.11.1311 [DOI] [PubMed] [Google Scholar]
- Salehi A., Delcroix J. D., Belichenko P. V., Zhan K., Wu C., et al. , 2006. Increased app expression in a mouse model of down’s syndrome disrupts ngf transport and causes cholinergic neuron degeneration. Neuron 51: 29–42. 10.1016/j.neuron.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Salehi A., Faizi M., Belichenko P. V., Mobley W. C., 2007. Using mouse models to explore genotype-phenotype relationship in down syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 13: 207–214. 10.1002/mrdd.20164 [DOI] [PubMed] [Google Scholar]
- Sturgeon X., Gardiner K. J., 2011. Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm. Genome 22: 261–271. 10.1007/s00335-011-9321-y [DOI] [PubMed] [Google Scholar]
- Sussan T., Yang A., Li F., Ostrowski M., Reeves R., 2008. Trisomy represses apc(min)-mediated tumours in mouse models of down’s syndrome. Nature 451: 73–75. 10.1038/nature06446 [DOI] [PubMed] [Google Scholar]
- Takeda H., Matsuzaki T., Oki T., Miyagawa T., Amanuma H., 1994. A novel pou domain gene, zebrafish pou2: expression and roles of two alternatively spliced twin products in early development. Genes Dev. 8: 45–59. 10.1101/gad.8.1.45 [DOI] [PubMed] [Google Scholar]
- Thisse, B., and C. Thisse, 2004 Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submission (http://zfin.org).
- Westerfield M., 2000. The Zebrafish Book: a guide for the laboratory use of zebrafish (Danio rerio), Ed. 5th University of Oregon Press, Eugene, OR. [Google Scholar]
- Yang Q., Sherman S. L., Hassold T. J., Allran K., Taft L., et al. , 1999. Risk factors for trisomy 21: maternal cigarette smoking and oral contraceptive use in a population-based case-control study. Genet. Med. 1: 80–88. 10.1097/00125817-199903000-00004 [DOI] [PubMed] [Google Scholar]
- Zhu K., Xu Y., Liu J., Xu Q., Ye H., 2011. Down syndrome cell adhesion molecule and its functions in neural development. Neurosci. Bull. 27: 45–52. 10.1007/s12264-011-1045-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The pCS2+ vector clone set, named the Hsa21 Gene Expression Set, is available through Addgene (https://www.addgene.org/kits/reeves-hsa21-set/). Supplemental files available at Figshare. Table S1 contains lists of all Hsa21 cDNAs included in this study along with cloning information, and zebrafish ohnologs/orthologs. Table S2 contains details of all injections for the whole clone set. Table S3 describes the 23 initial candidates and phenotypes. Table S4 contains information about LINCs and ORFs included in the clone-set. Figure S1 contains dosage curves for 10 of the candidate genes. S2 shows low dose RNA injections for SOD1 and RRP1. S3 shows pairwise injections for SOD1 and 6 other candidate genes. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6089324.