Abstract
Bayesian models for genomic prediction and association mapping are being increasingly used in genetics analysis of quantitative traits. Given a point estimate of variance components, the popular methods SNP-BLUP and GBLUP result in joint estimates of the effect of all markers on the analyzed trait; single and multiple marker frequentist tests (EMMAX) can be constructed from these estimates. Indeed, BLUP methods can be seen simultaneously as Bayesian or frequentist methods. So far there is no formal method to produce Bayesian statistics from GBLUP. Here we show that the Bayes Factor, a commonly admitted statistical procedure, can be computed as the ratio of two normal densities: the first, of the estimate of the marker effect over its posterior standard deviation; the second of the null hypothesis (a value of 0 over the prior standard deviation). We extend the BF to pool evidence from several markers and of several traits. A real data set that we analyze, with ours and existing methods, analyzes 630 horses genotyped for 41711 polymorphic SNPs for the trait “outcome of the qualification test” (which addresses gait, or ambling, of horses) for which a known major gene exists. In the horse data, single marker EMMAX shows a significant effect at the right place at Bonferroni level. The BF points to the same location although with low numerical values. The strength of evidence combining information from several consecutive markers increases using the BF and decreases using EMMAX, which comes from a fundamental difference in the Bayesian and frequentist schools of hypothesis testing. We conclude that our BF method complements frequentist EMMAX analyses because it provides a better pooling of evidence across markers, although its use for primary detection is unclear due to the lack of defined rejection thresholds.
Keywords: association analysis, single marker regression, QTL, GWAS, Bayesian regression, GenPred, Shared Data Resources, Genomic Selection
Bayesian models including simultaneously all marker effects are becoming very popular for GWAS analysis (Habier et al. 2011; Wang et al. 2012, 2016; Moser et al. 2015). The most frequently used prior for marker effects is the normal distribution, known as RRBLUP or SNP-BLUP (Habier et al. 2007; VanRaden 2008), which is equivalent to GBLUP (VanRaden 2008), also known in the human literature as GCTA analysis (Yang et al. 2010). GBLUP is simple and can be generalized to marker data missing in a large fraction of individuals in the so-called Single Step methods (Aguilar et al. 2010; Christensen 2012), and also to multiple traits, or complex models (random regression, genotype by environment, etc.). Because of the equivalence of GBLUP and SNP-BLUP, it is straightforward to obtain from Single Step methods estimates of marker effects for complex traits like, e.g., multiple trait maternal effects (Lourenco et al. 2015) or genotype-environment models (Jarquín et al. 2014).
Therefore, GWAS can be done exploiting results of GBLUP (Wang et al. 2012; Dikmen et al. 2013; Casiró et al. 2017). Most of these works (e.g., (Wang et al. 2012; Dikmen et al. 2013) do not report classical statistics neither p-values, whereas standard GWAS by fixed regression “one marker at a time” (e.g., EMMAX (Kennedy et al. 1992; Kang et al. 2010; Teyssèdre et al. 2012)) yields a normal test, i.e., dividing the estimate of the marker effect by its standard error of the estimate, with associated p-values. Remarkably, Gualdrón-Duarte et al. (2014) and Bernal-Rubio et al. (2016) proved that in (SS)GBLUP or SNP-BLUP, dividing the estimate of the marker effect by its standard error is mathematically equivalent to fixed regression EMMAX, even if markers are estimated as random effects in GBLUP and as fixed effects in EMMAX. In addition, Chen et al. (2017) generalized the single marker EMMAX test to a multiple marker test that considers simultaneous sets of markers. In this test, signals from neighboring markers are pooled to create a single p-value measuring strength of association.
This paper has two objectives. The first one is to show that, in addition to previous frequentist tests (single marker and multiple marker EMMAX) it is possible to obtain from GBLUP analysis single marker and multiple marker Bayes Factors (BF) as strength of evidence for the presence or absence of a QTL. In short, the BF is the ratio of probabilities of the data given two competing models (Kass and Raftery 1995) and has been often used in QTL mapping (Heath 1997; Varona et al. 2001; Wakefield 2009, 2012; Varona 2010; Habier et al. 2011; Legarra et al. 2015). The BF empirically seems to provide a consistent procedure across traits and species (Legarra et al. 2015). In current Markov Chain MonteCarlo implementations, computation of the BF require indicator variables for “null” or “non null” effects of markers (Habier et al. 2011; Legarra et al. 2015) and does not include the extensively used GBLUP and SNP-BLUP. In this work, we show how the BF can be easily computed from results of SNP-BLUP or GBLUP, for evidence of a single loci or a set of loci (possibly contiguous). The resulting BF considers correctly both the estimated effect and its incertitude, at one or several loci.
The second objective is to illustrate properties of these two procedures (single and multiple marker EMMAX and BF), plus a Bayesian multiple marker regression (BayesCPi), by analysis of a challenging small horse real data set with presence of a known, yet barely significant, major gene (DMRT3) for gait.
Material and Methods
Distributions of marker effect estimates
The methods use the prior (before observing the data) and posterior (estimates and associated errors) distributions of marker effects assuming a priori multivariate normality (i.e., SNP-BLUP or GBLUP). Most theory can be found in (Gualdrón Duarte et al. 2014; Bernal Rubio et al. 2016; Chen et al. 2017) and we include it in the Appendix for completion. We will assume throughout that variance components are known; this is a frequent assumption that allows obtaining of closed-form estimators. In particular, variance components can be estimated beforehand (e.g., by REML), or (making strong assumptions) they can be borrowed from pedigree analysis. In either case, a point estimate is used “as if” it was exact, which results in optimistic results. The main notation that we need is a vector of marker effects a normally distributed with a priori mean 0 and variance , and their prediction error variance .
EMMAX tests of association from GBLUP
This section is a reminder of (Gualdrón Duarte et al. 2014; Bernal Rubio et al. 2016; Chen et al. 2017) and we include it here for completeness.
Single marker:
The single marker EMMAX procedure is a normal test obtained, in our notation, with the statistic
where is the marker estimate of the locus under consideration, obtained from a single SNP-BLUP evaluation (or an equivalent model), and where is the frequentist distribution (over conceptual repeated sampling of ) of the SNP-BLUP estimator of the effect . Somewhat surprisingly, the numerical value of t is the same as if a was fit as a “fixed regression” GWAS and therefore the distribution of t under the null is (Bernal Rubio et al. 2016). For instance, assume that is the a priori variance of marker effects. Output of the SNP-BLUP gives an estimate of the marker effect with a standard deviation of the posterior distribution . With these numbers, the frequentist . Thus, which yields a p-value of 0.006.
Multiple marker:
Consider a subset of n markers (possibly consecutive), starting at marker i. The statistic is a quadratic form , where is the frequentist covariance of these marker effects. Chen et al. (2017) proved that under multivariate normality the quadratic form x follows a chi-square distribution of n degrees of freedom, which yields p-values for the multiple marker EMMAX. Alternatively, derivation of the Hotelling-t squared test, that tests whether a set of correlated sample means are simultaneously different from zero, yields the same result. The previous normal test for the single marker EMMAX is also equivalent to the chi-squared test. Matrix Σ takes into account uncertainty and collinearity of marker estimates.
For instance, consider two markers with effects (similar effects) with (estimates of effects are negatively correlated because of linkage disequilibrium) and . The quadratic form has value with p-value 0.27. The evidence given by the p-value lowers because the two effects are correlated.
Bayes Factors from GBLUP
In this section we include our original derivations.
Single marker:
There are two competing models in the BF: that the marker i with effect “has some effect” (1H: ) or «has 0 effect» (H0: ), and the BF can be written as
The BF measures whether the data is more probable under either of the hypothesis. This can be written, alternatively, as
| (1) |
where is the effect of the marker. Typically, this involves a complex MCMC integration. In the particular case of multivariate normality with known variances, Varona et al. (2001), Varona (2010) showed that the expression (1) is equal to
| (2) |
where is the density of a priori evaluated at , and is the density of a posteriori evaluated at . Computation of BF using (2) is straightforward because is a normal density. In particular, is the density of knowing that there is an estimate with a certain a posteriori variance (e.g., different for each data set). In algebraic form this is
| (3) |
where is the density of in the normal distribution with mean and variance . Consider the same example as before: , , . The BF is thus, in R code:
dnorm(0,0,sqrt(0.2))/dnorm(0,0.5,0.05)
which is 20.76 in the log10 scale. According to Kass & Raftery (1995) this is « Very Strong » evidence.
Multiple marker:
Evidence from several consecutive markers in a segment can be pooled together using the BF. Expression (3) is generalized to several SNP markers (markers from i to n) as:
| (4) |
where MVN is the density of a multivariate normal distribution and is the posterior (co)variance matrix between the marker estimates. Posterior covariance matrix , which is a submatrix of , takes into account colinearity between markers caused by LD. In this case, the BF tests whether a set of markers are all simultaneously 0, against the alternative that some of them (if not all) are different from zero.
Consider the same example as before: two markers with effects , , . The BF can be computed in R as
dmvnorm(c(0,0),mean = c(0,0),sigma = diag(0.2,2))/dmvnorm(ahat,mean = c(0,0),sigma = Caa) yielding a BF of 1.65 in the log10 scale, lower than the single marker analysis. In a way, this reflects that there is a confusion of marker effects.
Multiple trait:
Above methods can be easily extended to the multiple trait case. Multiple trait genomic predictions can be done from Bayesian regressions, SNP-BLUP or (Single Step) GBLUP (Tsuruta et al. 2011; Jia and Jannink 2012; Maier et al. 2015). Then, the EMMAX tests or the BF for several traits (and possibly markers) simultaneously is very similar to the “Several markers” case considering joint estimates of marker effects for the n traits, the a priori covariance among marker effects for the n traits , and the a posteriori covariance matrix of marker effect estimates . Vector a can include either one or several markers. Typically is a function of , the genetic covariance among traits.
Data:
We used a horse real data set to explore and illustrate the properties of the procedures. We also did a limited number of simulations but we chose not to present them as this was extensively done in (Chen et al. 2017).
A single base polymorphism at the gene DMRT3 in chromosome 23 has a strong effect on horse ambling gaits (Andersson et al. 2012). In French trotters, a SNP marker (marker BIEC2-620109 on chromosome 23 at position 22967656 bp) in strong disequilibrium with this polymorphism has a strong effect in qualification at the race (Ricard 2015; Brard and Ricard 2015). In this work we reanalyzed the same data set, which contains 630 horses and 41711 polymorphic SNP markers. The trait was “outcome of the qualification test”, with a heritability of 0.56. The major gene was not discovered in this data set, and therefore there is no bias due to discovery. We tried the following methods for GWAS:
Bayes factors with the mixture model BayesCPi: (Habier et al. 2011) fixing a priori that only 0.1% of the markers have an effect (see (Legarra et al. 2015) for a full description). This method provides BFs, although our implementation only considers single markers.
Single marker and multiple marker EMMAX tests: as presented in this work, computed via MCMC, up to segments of 100 consecutive markers.
Bayes factors from SNP-BLUP: as presented in this work, computed via MCMC, up to segments of 100 consecutive markers.
EMMAX was fitted using blupf90 (Misztal et al. 2002) and homemade scripts, whereas the other used our software GS3 (available at https://github.com/alegarra/gs3), using “OPTION Bayes Factor”. After completion of the analysis, we produced Manhattan plots based on BF and the other statistics; for EMMAX we used Bonferroni corrections to claim genowide significance; for Bayesian procedures, we did not address thresholds for declaring detection; this point will be addressed in the discussion.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6241928.
Results
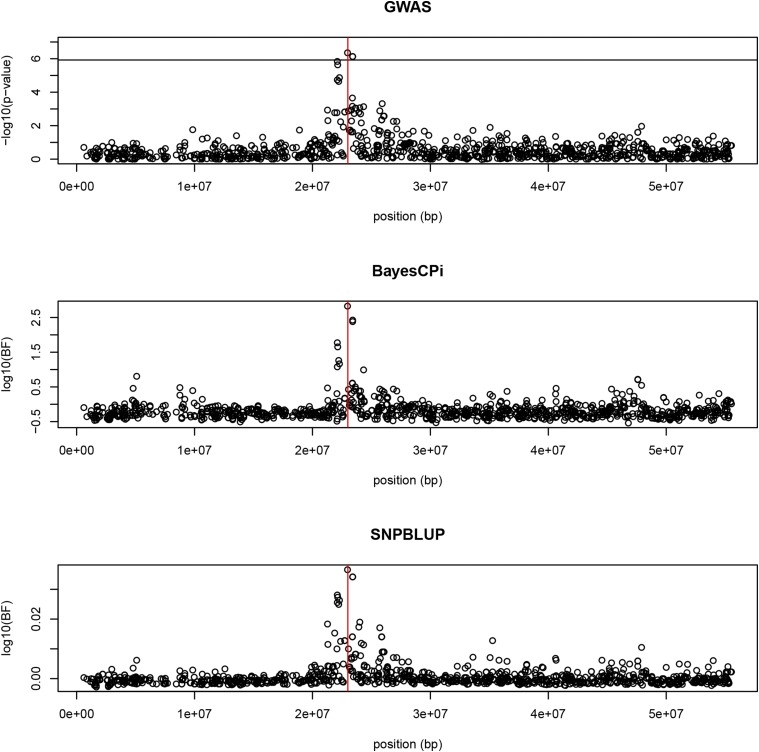
Figure 1 shows results from the single marker association test (EMMAX), the Bayesian multi-marker mixture model GWAS (BayesCPi) and the single marker BF. All three methods point to the SNP (BIEC2-620109 at position 22967656 bp) closest and most associated to the causal gene, and the four significant markers in the EMMAX single marker regression are in LD with each other. This reproduces the results in Ricard (2015). The EMMAX yields significant p-values at the Bonferroni level. Concerning BF, a threshold of 150 (2.17 in the log10 scale) has been suggested (Legarra et al. 2015), and the BayesCPi analysis in Figure 2 does reach this threshold, but this is not the case in the single marker BF using GBLUP.
Figure 1.
Results (from top to bottom) of single marker regression EMMAX, Bayes Factor for BayesCPi, and Bayes Factor for SNP-BLUP. Bonferroni rejection threshold in EMMAX is 5.9.
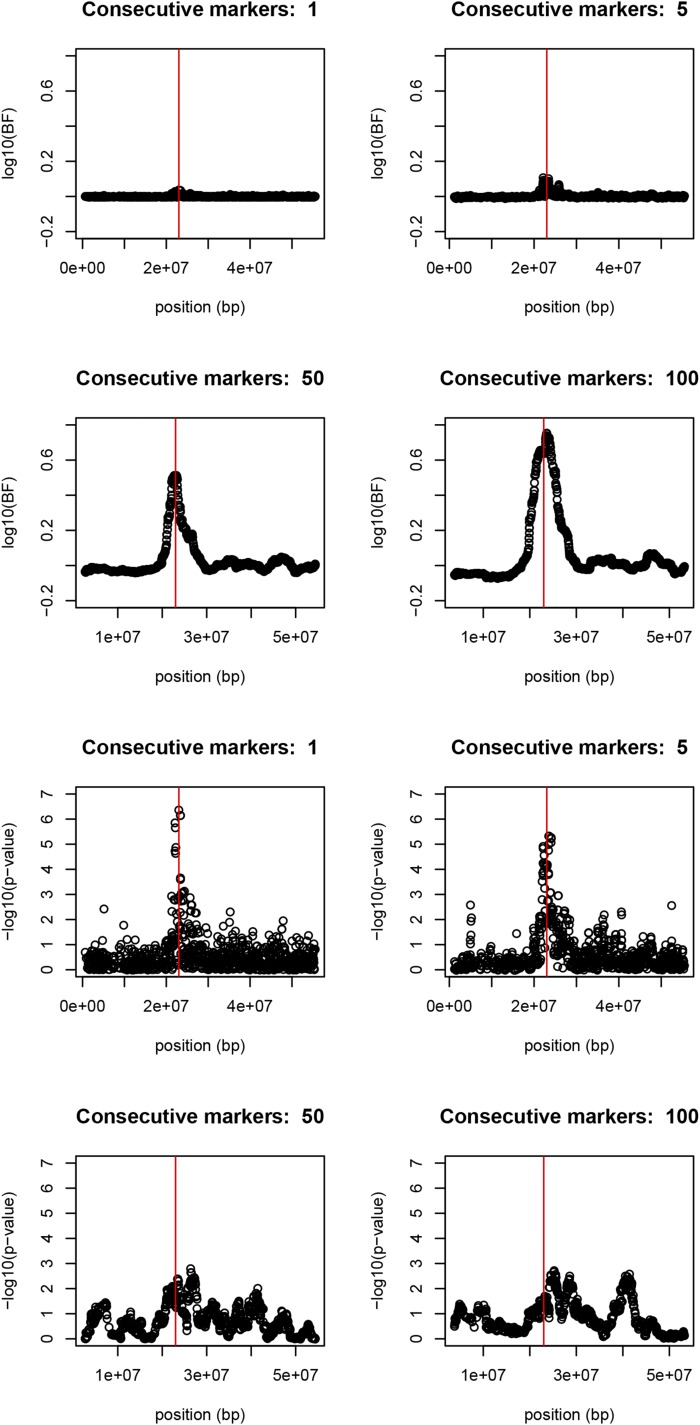
Figure 2.
Bayes factor profiles (top) and p-values (bottom) for qualification test in French trotters, chromosome 23. The location of the causal mutation marked with a red vertical bar. Bonferroni rejection threshold is 5.9.
In both analyses (BayesCPi and single marker BF), a large number of markers fall below the threshold of 0 in log10(BF), in other words, BF < 1. This means that for those regions the hypothesis that these markers have an effect is less likely that the hypothesis that they do not have an effect.
Figure 2 shows that evidence of the causal gene increases when using BF across consecutive markers. Systematically, the same location (BIEC2-620109) is spotted. It can be seen that the strength of evidence increase dramatically with increasing consecutive numbers, reaching the suggested “suggestive” threshold of BF > 3 (Kass and Raftery 1995), but not the much higher threshold of 150 suggested (Legarra et al. 2015). On the other hand, evidence from EMMAX does actually decrease, becomes non-significant, and, moreover, the highest peak deviates from the true location. This is at first sight a rather surprising result that will be discussed later.
Discussion
The standard test for GWAS by association analysis is the single marker association analysis (e.g., Kruglyak 1999). Association analysis can account for genetic relationships (Kennedy et al. 1992), population structure (Kang et al. 2010) and also to a part of individuals not genotyped (Legarra and Vitezica 2015). An alternative is to fit multiple marker simultaneously in the form of Bayesian regression (e.g., Fernando and Garrick 2013). Legarra et al. (2015) did not see qualitative differences of Bayesian regressions and association analysis over five data sets and species, and concluded that the interest of Bayesian procedures is to complement regular association analysis. Anyway, Bayesian regression is of interest for three reasons: first, the Bayesian analysis has interesting properties of automatically accounting for multiple test, structure, unbiasedness, false discovery rate and power (Wakefield 2009; Fernando and Garrick 2013); second, genomic evaluation routinely generates marker estimates and these may be used for GWAS; third, complex models used in genetic evaluation can be considered, for instance multiple trait disease all-or-none traits (Parker Gaddis et al. 2014).
The analysis that we propose can be seen as an approximation to a mixture analysis such as BayesCPi. For a given marker, we ask the question: “is this marker worth being included in the model?” whereas we pretend that all the other markers are included in the model. Implicitly, the prior is of a normal distribution with a known variance for loci not being tested and a mixture of a point mass at zero and a normal distribution for the locus being tested. In a mixture model (BayesCPi and similar ones), all markers are scrutinized simultaneously, and the strength of evidence compared against the probability value that a marker should be included in the model (usually labeled as π). This is probably why the actual numbers for the BF are so different across both methods.
We stress that the SNP-BLUP or GBLUP estimation is run only once, and its results are used to construct BFs for different groups of markers (consecutive or not), if desired. This BF combining information from several markers is quite different from estimating the effect of segments of alleles forming haplotypes, where a haplotype can be seen as a multiallelic marker, and where a different complete estimation must be run for each segment length.
In Bayesian regression models there is a lack of unique criterion to define “relevance” of the association and of corresponding well-defined thresholds; see (Legarra et al. 2015) for a description.The numerical values depend strongly on the assumed prior for marker effects (as can be seen in Figure 1). Thus, two researchers fitting, say, BayesCPi and BayesA may obtain different results. The most popular procedure for genomic evaluation and Bayesian regression is SNP-BLUP or its equivalent GBLUP, both of which assume multivariate normality of marker effects. Most often, a reasonable assumption (point estimate) on the variance of marker effects exists, by a transformation of previous estimates of genetic variance (obtained by pedigree analysis or, using the same data set, by genomic REML or similar methods). Using this point estimate underestimates noise linked to estimation of variance components. Here, we present for the first time a closed-form method to estimate BFs for association analysis based on GBLUP results, and we advocate its use. The statistical properties of the BF have been extensively discussed in the statistics literature, but for mapping causal variants it has two very few relevant properties: the BF can show evidence against and for the null hypothesis, and as data cumulates, the Bayes Factor favors the true hypothesis.
Our results from real data sets show that all methods point to the right marker (the one in stronger LD with the unobserved, but known, QTL). Classical regression analysis is significant and BayesCPi yields a “strong” BF signal. However, the BF observed from SNP-BLUP is 1.07 for the truly associated marker, which is very small support.
Evidence from BF increases when we extend the BF to gather evidence from several markers. A multi – SNP test captures the divergence of the posterior distribution from the 0 vector, and takes into account the posterior dependencies, due to LD, between marker estimates. This is similar to the idea of using the amount of variance explained by each genomic segment (Pérez-Enciso and Varona 2000; Hayes et al. 2010; Nagamine et al. 2012; Fernando and Garrick 2013). The inconvenience of these methods is mostly computational: they require to do either Restricted Maximum Likelihood (Nagamine et al. 2012) or MCMC (Hayes et al. 2010; Fernando and Garrick 2013) to estimate variance components, and that only the Restricted Maximum Likelihood estimation has an associated statistical test (Likelihood ratio test), for which consensual threshold exist (such as 0.05 genome-wide corrected by Bonferroni) whereas the MCMC methods use ad hoc thresholds that are less consensual. Our proposal does not require MCMC or Restricted Maximum Likelihood, but establishing a threshold for the BF is still ambiguous. An approximate method pools information from estimates of marker effects (Wang et al. 2012), but this does not consider not the error in the estimation of marker effects, neither their a posteriori correlation in presence of LD. Our proposal is exact, given a point estimate of variance components but does not necessarily require Restricted Maximum Likelihood or MCMC.
The interpretation of the BF in this study is as follows. There are two models, in the first (null) model all markers have 0 effect, whereas in the second (alternative) model at least one of the markers has an effect. In other words, the BF is a contrast between the region contributing, or not, to the genetic variance. When markers’ evidence is pooled across contiguous markers, the evidence for either of the two competing models increases.
Strangely, in our study including more markers in multiple marker EMMAX does not reinforce evidence, contrary to the BF. This is contrary to results of Chen et al. (2017). The reason is possibly due to the not-too-strong linkage disequilibrium in our data set, for which p-values do not cumulate information across multiple markers. It would seem that, in our data set, it is more difficult to disprove several null hypotheses (null hypothesis in EMMAX: all markers are zero) than to prove an alternative hypothesis (alternative hypothesis in BF: some marker is different from zero).
Conclusions
We present a Bayesian method (the BF) that complements existing EMMAX methods for QTL detection using marker estimates from SNP-BLUP or (SS)GBLUP from a commonly accepted prior (multivariate normality combined with prior estimates of the genetic variance) and commonly accepted, and used, methods (SNP-BLUP and SSGBLUP). Computations are reasonable and pooling information from several markers is straightforward. Based on our real data set, single marker EMMAX is better to claim significance, whereas multiple marker BF gives a better perspective of influence of LD on the result. This is likely to be data dependent.
Acknowledgments
INRA SelGen metaprogram is acknowledged for financing in its project EpiSel. Project partly supported by the Toulouse Midi-Pyrenees Bioinformatics platform. Luis Varona acknowledges the CGL2016-80155 project of Ministerio de Economía y Competitividad of Spain. Editor and reviewers of preliminary and current submissions made very useful comments and pointed out the Bernal Rubio et al. study.
Appendix
EMMAX tests from GBLUP and SNP-BLUP results
Most of this development is in (Gualdrón Duarte et al. 2014; Bernal Rubio et al. 2016; Chen et al. 2017).
SNP-BLUP:
The procedure is easier to be presented from the SNP-BLUP method point of view. In this method, the multivariate prior distribution of marker effect is, for a random locus,
and is a variance component that usually (but not necessarily) is assumed related to genetic variance in the form (Fernando et al. 2007; VanRaden 2008). For several loci, i.e., loci effects are assumed uncorrelated a priori. A linear model for SNP-BLUP is where Z is a matrix of coded genotypes. In SNP-BLUP, the posterior distribution of a can be obtained by Markov Chain MonteCarlo (MCMC) (Legarra and Misztal 2008) or from the inverse of the left hand side of Henderson’s Mixed Model Equations:
| (A1) |
In both cases, it is possible to obtain (a) the estimate of the marker effects is , and (b) two measures of incertitude of , the (frequentist) sampling variance, i.e. and the (Bayesian) posterior variance, i.e. . For instance, if the inverse of the right hand side of (A1) is computed:
then (Bayesian, conditional on data) and (frequentist, over repeated sampling of y). Matrix contains a posteriori covariances of marker effects, which reflect allelic frequencies (i.e., a rare SNP is more difficult to estimate) and linkage disequilibrium across markers (two markers in strong LD will have correlated estimates a posteriori, and any of them will be less accurate than a marker not in LD with any other). If estimates are obtained by MCMC, can be estimated as the covariance matrix of the samples of the posterior distribution, . The R package RRBLUP (Endelman 2011) produces , and our software GS3 (Legarra et al. 2011) can produce parts of .
GBLUP:
the equivalence between GBLUP and SNP-BLUP implies that marker solutions () can be backsolved for individual solutions ( (VanRaden 2008; Strandén and Garrick 2009). Proof is as follows. If individual effects are the sum of marker effects then u and a follow a joint degenerate multivariate normal distribution such that . Under the usual assumptions and (VanRaden 2008), and it can be shown (if G is invertible) that
The estimation error of a from GBLUP estimation is more cumbersome to obtain. Assume that the model for GBLUP is , and mixed model equations are:
| (A2) |
With the inverse of the left hand side equations . Gualdrón-Duarte et al. (2014) showed that:
where is the element of the inverse of the left hand side matrix corresponding to u; this inverse can be computed by inversion or, again, by MCMC. Finally, the posterior variance of a is
This allows computing estimates of marker effects and their errors for very complex models, something difficult to do with standard GWAS or Bayesian Regressions.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6241928.
Communicating editor: G. de los Campos
Literature Cited
- Aguilar I., Misztal I., Johnson D., Legarra A., Tsuruta S., et al. , 2010. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score1. J. Dairy Sci. 93: 743–752. 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- Andersson L. S., Larhammar M., Memic F., Wootz H., Schwochow D., et al. , 2012. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488: 642–646. 10.1038/nature11399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal Rubio Y. L., Gualdrón Duarte J. L., Bates R. O., Ernst C. W., Nonneman D., et al. , 2016. Meta-analysis of genome-wide association from genomic prediction models. Anim. Genet. 47: 36–48. 10.1111/age.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brard S., Ricard A., 2015. Should we use the single nucleotide polymorphism linked to in genomic evaluation of French trotter? J. Anim. Sci. 93: 4651–4659. 10.2527/jas.2015-9224 [DOI] [PubMed] [Google Scholar]
- Casiró S., Velez-Irizarry D., Ernst C. W., Raney N. E., Bates R. O., et al. , 2017. Genome-wide association study in an F2 Duroc x Pietrain resource population for economically important meat quality and carcass traits. J. Anim. Sci. 95: 545–558. 10.2527/jas.2016.1003 [DOI] [PubMed] [Google Scholar]
- Chen C., Steibel J. P., Tempelman R. J., 2017. Genome-Wide Association Analyses Based on Broadly Different Specifications for Prior Distributions, Genomic Windows, and Estimation Methods. Genetics 206: 1791–1806. 10.1534/genetics.117.202259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen O. F., 2012. Compatibility of pedigree-based and marker-based relationship matrices for single-step genetic evaluation. Genet. Sel. Evol. 44: 37 10.1186/1297-9686-44-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikmen S., Cole J. B., Null D. J., Hansen P. J., 2013. Genome-wide association mapping for identification of quantitative trait loci for rectal temperature during heat stress in Holstein cattle. PLoS One 8: e69202 10.1371/journal.pone.0069202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. 10.3835/plantgenome2011.08.0024 [DOI] [Google Scholar]
- Fernando R. L., Habier D., Stricker C., Dekkers J. C. M., Totir L. R., 2007. Genomic selection. Acta Agric. Scand. A 57: 192–195. [Google Scholar]
- Fernando R. L., Garrick D., 2013. Bayesian methods applied to GWAS. Genome-Wide Assoc. Stud. Genomic Predict.: 237–274. 10.1007/978-1-62703-447-0_10 [DOI] [PubMed] [Google Scholar]
- Gualdrón Duarte J. L., Cantet R. J., Bates R. O., Ernst C. W., Raney N. E., et al. , 2014. Rapid screening for phenotype-genotype associations by linear transformations of genomic evaluations. BMC Bioinformatics 15: 246 10.1186/1471-2105-15-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Pryce J., Chamberlain A. J., Bowman P. J., Goddard M. E., 2010. Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits. PLoS Genet. 6: e1001139 10.1371/journal.pgen.1001139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath S. C., 1997. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am. J. Hum. Genet. 61: 748–760. 10.1086/515506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarquín D., Crossa J., Lacaze X., Cheyron P. D., Daucourt J., et al. , 2014. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor. Appl. Genet. 127: 595–607. 10.1007/s00122-013-2243-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Jannink J.-L., 2012. Multiple-Trait Genomic Selection Methods Increase Genetic Value Prediction Accuracy. Genetics 192: 1513–1522. 10.1534/genetics.112.144246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Zaitlen N. A., Kong S., Freimer N. B., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795. 10.1080/01621459.1995.10476572 [DOI] [Google Scholar]
- Kennedy B., Quinton M., Van Arendonk J., 1992. Estimation of effects of single genes on quantitative traits. J. Anim. Sci. 70: 2000–2012. 10.2527/1992.7072000x [DOI] [PubMed] [Google Scholar]
- Kruglyak L., 1999. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22: 139–144. 10.1038/9642 [DOI] [PubMed] [Google Scholar]
- Legarra A., Misztal I., 2008. Technical note: Computing strategies in genome-wide selection. J. Dairy Sci. 91: 360–366. 10.3168/jds.2007-0403 [DOI] [PubMed] [Google Scholar]
- Legarra A., Ricardi A., Filangi O., 2011 GS3: Genomic Selection, Gibbs Sampling, Gauss-Seidel (and BayesCp). Available at: http://genoweb.toulouse.inra.fr/∼alegarra.
- Legarra A., Croiseau P., Sanchez M. P., Teyssèdre S., Sallé G., et al. , 2015. A comparison of methods for whole-genome QTL mapping using dense markers in four livestock species. Genet. Sel. Evol. 47: 6 10.1186/s12711-015-0087-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legarra A., Vitezica Z. G., 2015. Genetic evaluation with major genes and polygenic inheritance when some animals are not genotyped using gene content multiple-trait BLUP. Genet. Sel. Evol. 47: 89 10.1186/s12711-015-0165-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco D. L., Tsuruta S., Fragomeni B. O., Masuda Y., Aguilar I., et al. , 2015. Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus. J. Anim. Sci. 93: 2653–2662. 10.2527/jas.2014-8836 [DOI] [PubMed] [Google Scholar]
- Maier R., Moser G., Chen G.-B., Ripke S., Absher D., et al. , 2015. Joint Analysis of Psychiatric Disorders Increases Accuracy of Risk Prediction for Schizophrenia, Bipolar Disorder, and Major Depressive Disorder. Am. J. Hum. Genet. 96: 283–294. 10.1016/j.ajhg.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Strabel T., Auvray B., Druet T., et al. , 2002. BLUPF90 and related programs (BGF90), pp. 28–07 in: 7th World Congress on Genetics Applied to Livestock Production, CD-ROM Communication, Montpellier, France. [Google Scholar]
- Moser G., Lee S. H., Hayes B. J., Goddard M. E., Wray N. R., et al. , 2015. Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model. PLoS Genet. 11: e1004969 10.1371/journal.pgen.1004969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y., Pong-Wong R., Navarro P., Vitart V., Hayward C., et al. , 2012. Localising Loci underlying Complex Trait Variation Using Regional Genomic Relationship Mapping. PLoS One 7: e46501 10.1371/journal.pone.0046501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker Gaddis K. L., Cole J. B., Clay J. S., Maltecca C., 2014. Genomic selection for producer-recorded health event data in US dairy cattle. J. Dairy Sci. 97: 3190–3199. 10.3168/jds.2013-7543 [DOI] [PubMed] [Google Scholar]
- Pérez-Enciso M., Varona L., 2000. Quantitative trait loci mapping in F2 crosses between outbred lines. Genetics 155: 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard A., 2015. Does heterozygosity at the DMRT3 gene make French trotters better racers? Genet. Sel. Evol. 47: 10 10.1186/s12711-015-0095-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandén I., Garrick D. J., 2009. Technical note: Derivation of equivalent computing algorithms for genomic predictions and reliabilities of animal merit. J. Dairy Sci. 92: 2971–2975. 10.3168/jds.2008-1929 [DOI] [PubMed] [Google Scholar]
- Teyssèdre S., Elsen J.-M., Ricard A., 2012. Statistical distributions of test statistics used for quantitative trait association mapping in structured populations. Genet. Sel. Evol. 44: 32 10.1186/1297-9686-44-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta S., Misztal I., Aguilar I., Lawlor T., 2011. Multiple-trait genomic evaluation of linear type traits using genomic and phenotypic data in US Holsteins. J. Dairy Sci. 94: 4198–4204. 10.3168/jds.2011-4256 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 91: 4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Varona L., García-Cortés L. A., Pérez-Enciso M., 2001. Bayes factors for detection of quantitative trait loci. Genet. Sel. Evol. 33: 133–152. 10.1186/1297-9686-33-2-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varona L., 2010. Understanding the use of Bayes factor for testing candidate genes. J. Anim. Breed. Genet. 127: 16–25. 10.1111/j.1439-0388.2009.00826.x [DOI] [PubMed] [Google Scholar]
- Wakefield J., 2009. Bayes factors for genome‐wide association studies: comparison with P‐values. Genet. Epidemiol. 33: 79–86. 10.1002/gepi.20359 [DOI] [PubMed] [Google Scholar]
- Wakefield J., 2012. Commentary: Genome-wide significance thresholds via Bayes factors. Int. J. Epidemiol. 41: 286–291. 10.1093/ije/dyr241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Misztal I., Aguilar I., Legarra A., Muir W., 2012. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 94: 73–83. 10.1017/S0016672312000274 [DOI] [PubMed] [Google Scholar]
- Wang T., Chen Y.-P. P., Bowman P. J., Goddard M. E., Hayes B. J., 2016. A hybrid expectation maximisation and MCMC sampling algorithm to implement Bayesian mixture model based genomic prediction and QTL mapping. BMC Genomics 17: 744 10.1186/s12864-016-3082-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6241928.