Abstract
Lipidomics, the mass spectrometry-based comprehensive analysis of lipids, has attracted attention as an analytical approach to provide novel insight into lipid metabolism and to search for biomarkers. However, an ideal method for both comprehensive and quantitative analysis of lipids has not been fully developed. Here, we have proposed a practical methodology for widely targeted quantitative lipidome analysis using supercritical fluid chromatography fast-scanning triple-quadrupole mass spectrometry (SFC/QqQMS) and theoretically calculated a comprehensive lipid multiple reaction monitoring (MRM) library. Lipid classes can be separated by SFC with a normal-phase diethylamine-bonded silica column with high resolution, high throughput, and good repeatability. Structural isomers of phospholipids can be monitored by mass spectrometric separation with fatty acyl-based MRM transitions. SFC/QqQMS analysis with an internal standard-dilution method offers quantitative information for both lipid class and individual lipid molecular species in the same lipid class. Additionally, data acquired using this method has advantages, including reduction of misidentification and acceleration of data analysis. Using the SFC/QqQMS system, alteration of plasma lipid levels in myocardial infarction-prone rabbits to the supplementation of EPA was first observed. Our developed SFC/QqQMS method represents a potentially useful tool for in-depth studies focused on complex lipid metabolism and biomarker discovery.—Takeda, H., Y. Izumi, M. Takahashi, T. Paxton, S. Tamura, T. Koike, Y. Yu, N. Kato, K. Nagase, M. Shiomi, and T. Bamba.
Keywords: quantitative analysis, multiple reaction monitoring, phospholipids, sphingolipids, neutral lipids, omega-3 fatty acids
Imbalance of lipid metabolites in vivo is believed to affect the onset of serious diseases such as dyslipidemia and resultant atherosclerosis (1, 2). In recent years, cohort studies using metabolomics have also helped discover reliable biomarkers (3, 4). Accordingly, the development of a novel lipidomic analytical system that enables the acquisition of comprehensive and quantitative information of individual lipid molecular species is needed for identification and validation of potential biomarkers across multiple specimens and different equipment and/or facilities (5). Recently, chromatographic and/or tandem mass spectrometric analytical approaches for lipidomics have been proposed (6). Features of representative lipidomic methodologies are summarized in supplemental Table S1.
The shotgun lipidomics approach, using direct infusion tandem mass spectrometry (DI/MS/MS), is often used to rapidly quantitate lipid molecular species in a short time (7–9). Each lipid level can be quantitated using mass spectrometry because the ion-suppression and/or ion-enhancement effects of the biological matrix can be normalized by adding the appropriate internal standards of each lipid class (e.g., chemically synthesized lipid standards not detected in vivo or stable isotope-labeled lipid standards). However, it is difficult to identify some isomeric compounds due to coelution, and low abundant lipid molecular species are poorly detected because of strong ionization suppression. In-source fragmentation caused by DI/MS/MS analysis can also confound lipid identification and quantification.
LC/MS/MS techniques have been developed to identify and quantitate lipid molecular species. The advantages of LC/MS/MS methods over DI/MS/MS are as follows: i) improvement of detection sensitivity, since ESI mass spectrometry (ESI-MS) is a concentration-sensitive device (10); ii) decrease in the effects of the biological matrix by liquid chromatography (LC) separation; and iii) consequently, an increase in the number of lipids detected. Reverse-phase LC (RPLC) is a widely used separation technique of lipid molecular species based on the hydrophobic interaction between nonpolar side chains of C18 particles and hydrophobic fatty acyl chains of lipids. RPLC can enhance the chromatographic resolution of a wide range of lipids including some isomers (11, 12). However, biological matrix effects cannot be normalized in all detected peaks, because it is not possible to prepare appropriate internal standards corresponding to all the detected peaks. In contrast, normal-phase LC MS/MS (NPLC/MS/MS) can be used to separate each lipid class according to the polar head group (13–15). In principle, the NPLC/MS/MS approach can lead to reliable identification and quantification of each lipid class and some of individual lipid species by the addition of appropriate internal standards. However, the mobile phase of NPLC (e.g., hexane, isopropanol, and chloroform) causes a reduction in ionization efficiency. Thus, the sensitivity of lipid molecular species is decreased. Instead, hydrophilic interaction chromatography MS/MS (HILIC/MS/MS)-based lipidome analysis has been applied to overcome this limitation (16–21). Separation behavior of HILIC for lipidomics is similar to that of NPLC. The sensitivity of HILIC/MS/MS is higher than that of NPLC/MS/MS because HILIC/MS/MS uses ESI-compatible solvents (e.g., acetonitrile, methanol, and water) as the mobile phase. However, since HILIC methods have insufficient retention and separation for nonpolar lipids [e.g., cholesteryl ester (CE), diacylglycerol (DAG), and triacylglycerol (TAG)] (18), HILIC methods are mainly applied to polar lipids such as glycerophospholipids and sphingolipids (16, 17, 19–21). In order to carry out HILIC/MS/MS-based lipidome analysis with high repeatability of retention times (RTs) and peak areas, long equilibration times are also required to form a water-rich layer on the surface of the polar stationary phase (17–21).
A supercritical fluid (SCF), kept beyond its critical temperature and pressure, is defined as a high-density and noncondensable fluid. SCF chromatography (SFC) is a chromatographic separation technique that uses an SCF as a mobile phase. Supercritical carbon dioxide (SCCO2) is the most frequently used mobile phase for SFC, because carbon dioxide can easily be converted to its supercritical state (critical temperature, 31.1°C; critical pressure, 7.38 MPa) and exhibits favorable properties such as being nonflammable, chemically inert, relatively nontoxic, easy to handle, and inexpensive. Moreover, like n–hexane, SCCO2 has relatively low polarity, and the polarity of the mobile phase in SFC can be changed considerably by adding a polar organic solvent such as methanol as a modifier. In previous work, we have presented that SFC/MS/MS with a reverse-phase column can be used for the analysis of global lipids without ESI-incompatible solvents (22–24). Very recently, Lísa and Holčapek reported that SFC coupled with a quadrupole TOF mass spectrometer (SFC/QTOFMS) system with a normal-phase silica-based column has been applied to perform high-throughput lipidome analysis (25). Lísa and Holčapek also revealed that lipid class separation using SFC would be superior to conventional lipid class separation methods (i.e., NPLC and HILIC) in terms of short analysis time and chromatographic resolution for a wide variety of lipids. However, SFC separation efficiency of individual lipid classes, chromatographic and mass spectrometric separation of individual lipid species, including positional or structural isomers, and quantitative evaluation have not been fully investigated. It is necessary to further investigate chromatographic and mass spectrometric separation for quantification of a wide range of lipids.
Multiple reaction monitoring (MRM) using triple-quadrupole mass spectrometry (QqQMS) has highly sensitive and selective quantitative performance, allowing for reliable monitoring of low-abundance lipids. Furthermore, mass spectrometric separation of structural isomers [e.g., phosphatidylcholine (PC) 16:0–20:4 and PC 18:2–18:2] can be provided using fatty acyl-based MRM transitions. In lipidome analysis, fragmentation was induced regularly based on the lipid class and FA side chain. A rapid development of fast-scanning QqQMS has also contributed to conduct multitargeted MRM transition settings (i.e., large-scale MRM mode) for simultaneous analysis of several hundred lipids (namely, widely targeted lipidomics) (26, 27).
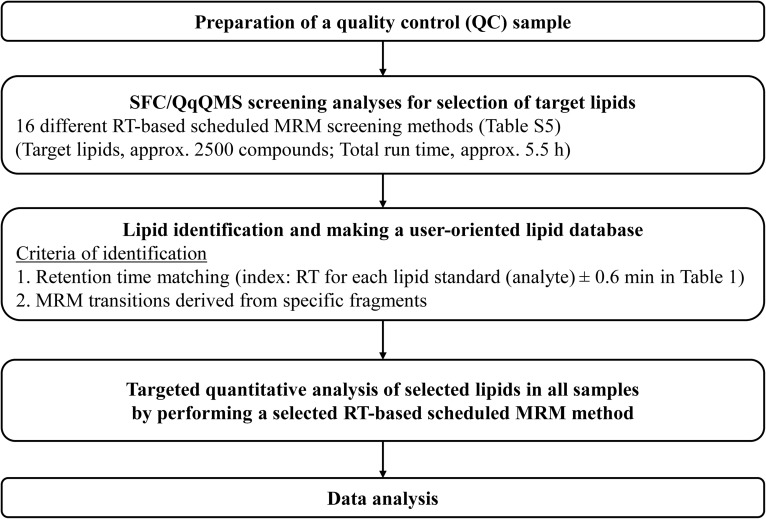
In the present study, we aimed to develop an advanced methodology for widely targeted quantitative analysis of individual lipid molecular species of 22 lipid classes, including lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), lysophosphatidylglycerol (LPG), lysophosphatidic acid (LPA), lysophosphatidylinositol (LPI), lysophosphatidylserine (LPS), PC, alkyl-acyl PC, alkenyl-acyl PC, phosphatidylethanolamine (PE), alkenyl-acyl PE, phosphatidylglycerol (PG), phosphatidic acid (PA), phosphatidylinositol (PI), phosphatidylserine (PS), SM, ceramide (Cer), CE, monoacylglycerol (MAG), DAG, TAG, and FFA, in complex biological samples using SFC coupled with fast-scanning QqQMS (SFC/QqQMS) and theoretically calculated a comprehensive lipid MRM library (Fig. 1). Based on this analytical method, we investigated the effect of administration of EPA on the plasma lipid compositions using myocardial infarction-prone Watanabe heritable hyperlipidemic (WHHLMI) rabbits (28, 29).
Fig. 1.
Easy and user-oriented method development workflow for widely targeted quantitative lipidome analysis using SFC/QqQMS system with in-house lipid MRM library.
MATERIALS AND METHODS
Chemicals and reagents
Ammonium acetate was obtained from Sigma-Aldrich Co. (St. Louis, MO). Distilled water (LC/MS grade) and methanol (LC/MS grade) were purchased from Kanto Chemical Co., Ltd (Tokyo, Japan). HPLC-grade chloroform was obtained from Kishida Chemical (Osaka, Japan). All synthetic lipid standards were purchased from Avanti Polar Lipids Inc. (Alabaster, AL) except for FA 17:0 (Sigma-Aldrich Co.). Carbon dioxide (99.5% grade, Yoshida Sanso Co., Ltd., Fukuoka, Japan) was used as the SFC mobile phase. Supplemental Table S2 lists the FA composition used to construct an in-house lipid MRM library.
Animals
All animal procedures were approved by the Kobe University Animal Care and Use Committee and were conducted in accordance with the Regulations for Animal Experimentation of Kobe University, Act on Welfare and Management of Animals (Law No. 105, 1973, revised in 2006), Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain (Notification No. 88, 2006), and Fundamental Guidelines for the Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the Jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology (Notice No. 71, 2006). Twelve WHHLMI rabbits (10.2 ± 0.4 months old) were used. The rabbits were housed individually in metal cages in a room with constant temperature (22 ± 2°C) and a constant lighting cycle (12 h light/dark). EPA was suspended using a 0.5% carboxymethyl cellulose suspension, and 300 mg per weight (kg) of EPA was administered to six WHHLMI rabbits orally every day. Another set of six WHHLMI rabbits (placebo group) was administered only a 0.5% carboxymethyl cellulose suspension. After administration, the rabbits were fed standard rabbit chow (120 g per day; CR-3, Clea, Tokyo, Japan) and water ad libitum.
Rabbit plasma sample preparation
Blood was collected from the auricular central artery of the 12 WHHLMI rabbits, who had undergone a period of fasting of more than 12 h. Lipid extraction from rabbit plasma was performed using Bligh and Dyer’s method with minor modifications (30). Before lipid extraction, a dodecanoyl- or heptadecanoyl-based synthetic internal standard mixture was added to samples for quantification of the endogenous lipid molecular species (31). The concentrations of internal standard mixture spiked into the plasma were as follows: 1.6 μM (SM d18:1–17:0 and Cer d18:1–17:0); 8 μM (LPC 17:0); 16 μM (MAG 17:0, DAG 12:0–12:0, and TAG 17:0–17:0–17:0); 130 μM (PE 17:0–17:0); 160 μM (LPE 17:1, PC 17:0–17:0, CE 17:0, and FFA 17:0); 320 μM (PG 17:0–17:0); 450 μM (PA 17:0–17:0); 1200 μM (LPG 17:1); 1,300 μM (LPI 17:1 and PS 17:0–17:0); and 1,600 μM (LPA 17:0 and LPS 17:1). In short, 20 μl of plasma was mixed with 30 μl of internal standard mixture and 1,000 μl of methanol/chloroform/water (10:5:3, vol/vol/vol) and vortexed at the maximum setting for 1 min. The plasma sample was further extracted by ultrasonication for 5 min before centrifugation at 16,000 g for 5 min at 4°C. The supernatant (700 μl) was transferred to clean tubes. After adding 195 μl of chloroform and 195 μl of water, phase separation of aqueous and organic layers was performed by centrifugation at 16,000 g for 3 min at 4°C. Aliquots (100 μl) of the organic layer were transferred to clean tubes. The plasma lipid extract (100 μl) for WHHLMI rabbit was diluted to a final volume of 400 μl with a solution of 2:1 (vol/vol) methanol:chloroform for SFC/QqQMS analysis. A quality control (QC) sample (240 μl) was prepared by mixing equal amounts (10 μl each) of 24 plasma extracts.
Flow injection conditions
An SFC/QqQMS system was composed of an ACQUITY Ultra-Performance Convergence Chromatography (UPC2) system and a Xevo TQ-S micro triple-quadrupole mass spectrometer with an ESI ion source (Waters, Milford, MA). In order to enhance ionization efficiency, HPLC 515 pump (Waters) was used as a make-up pump. The SFC and QqQMS systems and data acquisition were controlled by MassLynx software version 4.1 (Waters). Standard solutions (1 μM) of each synthetic lipid were prepared in methanol/chloroform (2/1, vol/vol). MRM transition settings for each lipid class were performed using flow injection. The SFC flow injection conditions were as follows: injection volume, 2 μl; mobile phase (A), SCCO2; mobile phase (B) (modifier) and make-up pump solvent; methanol with 0.1% (wt/vol) ammonium acetate; flow rate of mobile phase, 1.0 ml min–1; flow rate of make-up pump, 0.1 ml min–1; modifier condition; 40% B (isocratic condition); temperature of column manager, 40°C; temperature of sample manager, 10°C; active back pressure regulator (ABPR), 1,500 pounds per square inch (psi); analytical time, 1 min. QqQMS analytical conditions were as follows: capillary voltage, 3.0 kV; desolvation temperature, 500°C; cone gas flow rate, 50 lh–1; desolvation gas flow rate, 1,000 lh–1. MRM conditions, including the cone voltage and collision energy of each lipid, were automatically optimized with the aid of the MassLynx QuanOptimize using standard solutions.
SFC/QqQMS conditions
The SFC conditions were as follows: injection volume, 1 μl; mobile phase (A), SCCO2; mobile phase (B) (modifier) and make-up pump solvent; methanol/water (95/5, vol/vol) with 0.1% (wt/vol) ammonium acetate; flow rate of mobile phase, 1.0 ml·min–1; flow rate of make-up pump, 0.2 ml·min–1; modifier gradient; 1% (B) (1 min), 1–65% (B) (11 min), 65% (B) (6 min), 65–1% (B) (0.1 min), 1% (B) (1.9 min); temperature of column manager, 50°C; ABPR, 1,500 psi; analytical time, 20 min; columns, ACQUITY UPC2TM ethylene bridged hybrid (BEH), ACQUITY UPC2TM 2-ethylpyridine (2-EP), ACQUITY UPC2TM Torus 2-picolylamine (2-PIC), ACQUITY UPC2TM Torus 1-aminoanthracene (1-AA), ACQUITY UPC2TM Torus DIOL, and ACQUITY UPC2TM Torus diethylamine (DEA) [each, 100 × 3.0 mm inner diameter (i.d.), particle size: sub-1.7 μm, Waters]. The final MS analysis conditions were as follows: capillary voltage, 3.0 kV; desolvation temperature, 500°C; cone gas flow rate, 50 lh–1; and desolvation gas flow rate, 1,000 lh–1. The MRM parameters per one time period were as follows: limit on number of MRM transitions, 150; dwell time, 1 ms; MS inter-scan and interchannel delay, 2 ms; and polarity switch interscan, 15 ms.
Method validation
For the creation of calibration curves of each lipid, their standard solutions were prepared at the following concentrations: 0, 1, 5, 10, 50, 100, 500, 1,000, 5,000, 10,000, 50,000, and 100,000 nM, each with the internal standard of 22 nM (SM d18:1–17:0 and Cer d18:1–17:0); 110 nM (LPC 17:0); 220 nM (MAG 17:0, DAG 12:0–12:0, and TAG 17:0–17:0–17:0); 1,800 nM (PE 17:0–17:0); 2,200 nM (LPE 17:1, PC 17:0–17:0, CE 17:0, and FFA 17:0); 4,400 nM (PG 17:0–17:0); 6,200 nM (PA 17:0–17:0); 16,000 nM (LPG 17:1); 18,000 nM (LPI 17:1 and PS 17:0–17:0); and 22,000 nM (LPA 17:0 and LPS 17:1).
Spike-and-recovery test
A standard spike-and-recovery test was carried out in optimal analytical conditions by adding the same amount of synthetic lipid standard as the actual amount in the representative plasma lipid extract for the WHHLMI rabbit (placebo group).
Data analysis for quantitation
Identification and quantification of lipid molecular species were performed using MassLynx software version 4.1. The quantitative values were calculated using the ratio of the chromatographic peak area of each analyte to that of the internal standard of its representative lipid class.
Biochemical analysis for total cholesterol
Total cholesterol level was measured using a Cholesterol E-Test Assay Kit (Wako Pure Chemical Industries, Ltd.) following the manufacturer’s instruction.
Statistical analysis
Statistical significance between baseline and 5 weeks after administration of EPA was determined using paired t-test (*P < 0.05, **P < 0.01, and ***P < 0.001). The difference of lipid profiles was examined by volcano plot which determined the relationship between fold-change and statistical significance.
RESULTS AND DISCUSSION
Overview of SFC/QqQMS system
A schematic diagram of the Waters SFC/QqQMS system used in this study is shown in supplemental Fig. S1. Recently, a modern analytical SFC system, namely, the Waters ACQUITY UPC2, has been developed. The new SFC system benefits from a novel back pressure regulator design (i.e., ABPR) and is largely based on ultra-high-performance LC technology, including higher upper pressure limits and reduced void volume. Improved quantitative performance (accuracy, repeatability, robustness, etc.) using columns packed with sub-2 μm particles have broadened the bioanalytical applications (32). A new fast-scanning QqQMS, namely, the Waters Xevo TQ-S micro, has also been developed, allowing for increased scan speed [maximum scan speed: previous QqQMS, approximately (approx.) 10,000 u/s; and Xevo TQ-S micro, 20,000 u/s] and large-scale MRM mode (minimum dwell time: previous QqQMS, approx. 5–10 ms; and Xevo TQ-S micro, 1 ms). Therefore, it is possible to conduct multitargeted MRM transition settings for simultaneous analysis of several hundred compounds.
In SFC/MS, SCCO2 vaporizes at the inlet of the ESI-MS interface, and a small amount of organic solvent used as the modifier and make-up solvent is introduced into the ESI-MS. From a theoretical point of view, ionization/desolvation efficiency in SFC/MS should be better than that in RPLC/MS, which uses an aqueous organic solvent (e.g., water/methanol). In our previous investigation, we conformed that SFC/QqQMS showed much higher sensitivity than LC/QqQMS when the analytical conditions were fully optimized for SFC/QqQMS (33).
Optimization of MRM transition settings
Identification of the precursor ion and elucidation of the fragmentation patterns of each of the 22 lipid classes (LPC, LPE, LPG, LPA, LPI, LPS, PC, alkyl-acyl PC, alkenyl-acyl PC, PE, alkenyl-acyl PE, PG, PA, PI, PS, SM, Cer, CE, MAG, DAG, TAG, and FFA) were carried out by SFC flow injection analysis in both positive and negative ion modes. Precursor ions of each lipid were clearly observed in the positive ion mode (mainly [M+H]+ or [M+NH4]+) and/or in the negative ion mode (mainly [M–H]– or [M+CH3COO]–) by the addition of ammonium acetate to the modifier and make-up solvent. In this study, the MS/MS spectra of the individual lipid precursor ion, which produce signature product ions specific for each molecular substructure, were acquired by collision-induced dissociation (CID) with argon. Among the 22 lipid classes, only FFA was difficult to dissociate in QqQMS using CID. Based on the most abundant adduct precursor ion and the selected product ions for each lipid class, we optimized the MS cone voltage and collision energy. Supplemental Table S3 shows the optimization results of the MS cone voltage, collision energy, and MRM transition setting for each lipid class.
Effect of modifier and make-up solvent on SFC lipid class separation and QqQMS detection
The effect of mobile phase composition in SFC on peak shapes of targeted lipids was first evaluated by SFC analysis with a silica-based BEH column as the reference column because of the lack of functional groups. In the case of using 100% methanol with 0.1% (wt/vol) ammonium acetate, peak shapes of the polar lipids such as LPA, LPS, PA, and PS were broadened. On the other hand, their peak shapes were improved by the addition of 5% water to the modifier solvent (supplemental Fig. S2). It is known that use of a mixture of 5% water in methanol as the polar modifier led to an improvement in peak shape for hydrophilic compounds (e.g., flavonoid glycosides) because of increased analyte solubility and enhanced solvating power of the mobile phase (34). Thus, it is suggested that similar improvement in peak shape was observed in the above-mentioned relative polar lipid species. Using this water-containing modifier solvent, we then optimized the flow rate of the make-up solvent. The coupling with QqQMS consists of a double T with polyetheretherketone tubing with fixed dimensions. The first T enables the addition of make-up solvent to the column effluent to enhance ionization. In the second T, the flow is split between the ABPR and MS source in a ratio of approx. 9:1 (supplemental Fig. S1). In the initial solvent condition in SFC with the BEH column, the mobile phase consists of SCCO2, and weakly retaining lipids such as CE are eluted under this condition. In order to enhance the ionization efficiency for CE, we investigated the flow rate of the make-up pump (supplemental Fig. S3A). As a result, the flow rate of the make-up pump was set to 0.2 ml·min–1.
Column screening for high-resolution SFC separation of lipid classes and their positional isomers
A previous study described that SFC/QTOFMS system using a normal-phase silica-based BEH column (100 × 3.0 mm i.d., particle size: sub-1.7 μm, Waters) identified a total of 436 molecular species in porcine brain extract (25). However, lysophospholipid positional isomers [e.g., LPC 16:0 (sn-1) and LPC 16:0 (sn-2)] and diacyl phospholipid structural isomers (e.g., PC 16:0–22:6 and PC18:2–20:4) were not identified in the previous methodology. To investigate the separation efficiency of each lipid class and some lipid isomers, including positional isomers (i.e., MAG, DAG, and lysophospholipids), we used six different normal-phase columns including BEH, 2-EP, 2-PIC, 1-AA, DIOL, and DEA, which were developed based on 1.7 μm BEH particle technologies. First, we set the optimal SFC condition under the maximum system pressure limit (6,000 psi) in this SFC system. The initial modifier concentration was set to 1% in the first 1 min. From this isocratic condition, the peak shape of weakly retaining lipids such as CE was improved in all SFC conditions using each of the six different columns (supplemental Fig. S3B). Afterward, other lipid classes were eluted with gradient conditions. In SFC conditions using the DEA column, LPA had the strongest retention. Therefore, the modifier percentage was increased from 1% to 65%. Next, we checked the effect of column oven temperature on lipid class separation and system pressure. By raising the column oven temperature, the retention of each lipid class became slightly stronger, and system pressure decreased in all SFC conditions using each of the six different columns (supplemental Fig. S4A); however, lipid class separation did not improve dramatically. In this study, the optimal column oven temperature was set to 50°C. Subsequently, we investigated for the optimal ABPR condition. Separation of each lipid class and its peak shape showed the same behavior under each ABPR (1,500; 1,750; and 2,000 psi). Thus, we decided to set it to 1,500 psi, the minimum condition of ABPR. Finally, we investigated the best flow rate conditions (supplemental Fig. S4B). In the case of 1.0 ml·min–1 flow rate in the SFC using the DEA column, LPA was eluted within 20 min, and the system pressure was kept within the upper limit of the instrument (6,000 psi).
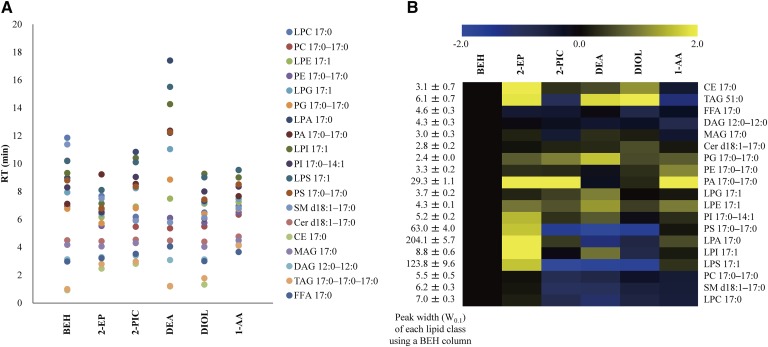
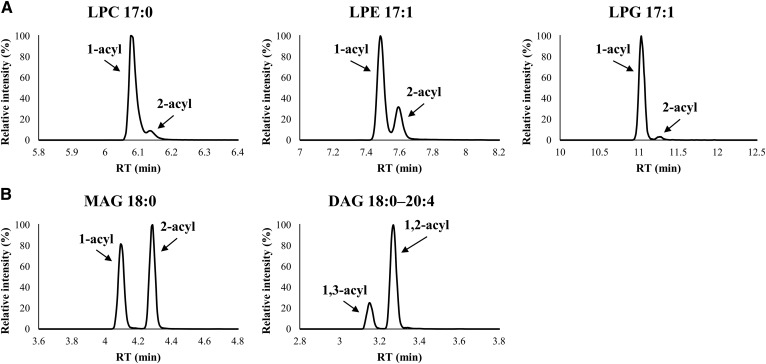
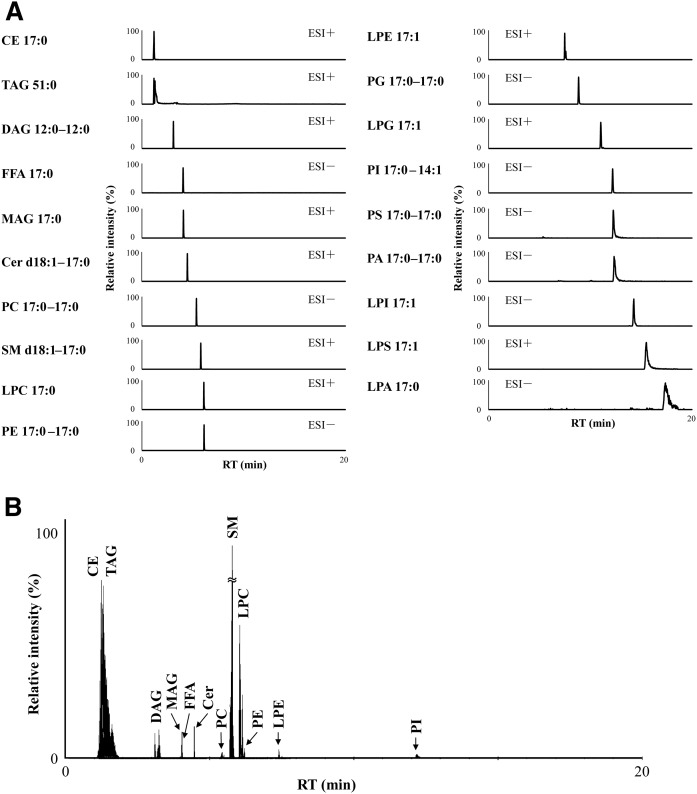
We examined the separation behavior of lipid classes using six different SFC columns under the same SFC condition (supplemental Table S4). Figure 2A shows the visualization of retention behavior of each lipid class. In the case of using the three types of columns (2-EP, 2-PIC, and 1-AA) with a benzene ring as a functional group, more hydrophobic lipids, such as CE and TAG, had a relatively stronger retention than the other columns (supplemental Table S4). In contrast, among screening columns, SFC separation with the DEA column had the best hydrophilic interaction with acidic polar lipids such as PI, PS, PA, LPI, LPS, and LPA. Furthermore, peak width for these acidic polar lipids was improved to a greater extent in SFC separation using the DEA column than using the BEH column (Fig. 2B). One of the most plausible reasons for this result is that the DEA stationary phase, which has strongly basic properties, contributed to the retention and separation of acidic polar lipids. By using the DEA column, baseline separation of positional isomers such as lysophospholipids (e.g., LPC, LPE, and LPG) and neutral lipids (e.g., MAG and DAG) were also obtained (Fig. 3). Lysophospholipids are known as bioactive molecules, precursors, and intermediates in the metabolism of glycerophospholipids and sphingolipids. However, the difference in function between 1-acyl-2-lysophospholipid and 2-acyl-1-lysophospholipid in vivo is not well known at the moment (35). Similarly, the differences in the roles of 1-acyl-2-MAG and 2-acyl-1-MAG or 1,2-acyl-DAG and 1,3-acyl-DAG have not been elucidated. Thus, our analytical method will be useful in future research for elucidation of the function and roles of these molecules. Based on these observations, we decided to use the DEA column for widely targeted lipidome analysis. SFC with the DEA column achieved good separation of internal standard mixture of 19 lipid classes within 20 min (Fig. 4A). In SFC separation using the DEA column, the minimum peak width for DAGs was less than 5 s (supplemental Fig. S5). In order to maintain 10 data points across a peak, MRM parameters, including dwell time, interscan and interchannel delay, and polarity switch interscan, were optimized (see Materials and Methods). High-performance chromatographic separation of lipid classes not only avoids ion-enhancement and/or ion-suppression effects among lipid classes, but also leads to improved compound identification and quantitative accuracy.
Fig. 2.
Column screening results using SFC/QqQMS with six different normal-phase columns including BEH, 2-EP, 2-PIC, DEA, DIOL, and 1-AA. A: Separation behavior of each lipid class. The data show the mean value of five replicated experiments. B: Ratio of W0.1 of each column to that of BEH column in each lipid class. Data were log2-transformed, and the mean value of five replicated experimental data was presented. W0.1, peak width of 10% peak height (second).
Fig. 3.
SFC/QqQMS chromatograms of positional isomers of using a DEA column. A: Lysophospholipids (LPC, LPE, and LPG). B: Neutral lipids (MAG and DAG).
Fig. 4.
High-resolution SFC separation of each lipid class using a DEA column. A: SFC/QqQMS (MRM) chromatograms of internal standard mixture of 19 lipid classes. B: SFC/MRM chromatograms of lipids in the WHHLMI rabbit plasma.
Discrimination of diacyl phospholipid structural isomers by fast-scanning QqQMS
Discrimination of diacyl phospholipid structural isomers with different FA side chains (e.g., PC 16:0–20:4 and PC 18:2–18:2) was subsequently investigated based on MS/MS separation. We validated the fragmentation patterns of each of the 22 lipid classes and determined the MRM transition(s) for identification and quantification of each lipid molecule (supplemental Table S3). The optimized MRM transitions for phospholipids including PC, PE, PG, PA, PI, and PS were set to the MS/MS fragments derived from their FA side chains in the negative ion mode. Based on the MRM transitions derived from FA side chains, PC 16:0–20:4 (840.6 > 255.3 and 840.6 > 303.2) and PC 18:2–18:2 (840.6 > 279.3) could be monitored individually.
Identification of individual TAG molecular species could not be achieved by MRM transitions derived from neutral loss of FA side chains. Since TAG comprises three fatty acyl tails at the sn-1, -2, and -3 positions of the glycerol backbone, the coeluted TAG structural isomers cannot be monitored individually. For example, TAG 16:0–18:1–20:4 (898.8 > 577.5, 898.8 > 599.5, and 898.8 > 625.5) and TAG 16:0–18:2–20:3 (898.8 > 575.5, 898.8 > 601.5, and 898.8 > 625.5) have the same MRM transition as 898.8 > 625.5. Thus, individual TAG levels cannot be obtained by the lipid class-based separation method of SFC with the DEA column. In the present study, we quantified TAG molecular species by summing lipid levels of these structural isomers using the same precursor-to-product ion transition (i.e., TAG 54:5, 898.8 > 898.8) (supplemental Table S3). In order to obtain detailed molecular information of TAG with FA side chain information, reverse-phase-based separation in LC/MS/MS or SFC/MS/MS is recommended (36).
Widely targeted quantitative lipidomics platform using SFC/QqQMS with theoretically calculated comprehensive lipid MRM library
The SFC/QqQMS analytical system was optimized by the investigation of SFC column screening, SFC separation conditions, and MRM parameter settings. In addition, according to the MS/MS fragment patterns of each lipid class and major FA composition (supplemental Table S2), we conducted widely targeted MRM transition settings (supplemental Table S3). In general, MRM assays using QqQMS are selected for quantitative analysis of targeted compounds, including minor components. However, this approach has limitations, which are inherent to the targeted acquisition. In other words, MRM-based methods are a problem in terms of comprehensive and simultaneous lipid measurements. In the present study, an in-house lipid MRM library is utilized to perform comprehensive lipids measurements using fast-scanning QqQMS. We targeted 22 lipid classes with a variety of 23 FA side chains; thus, targeted lipid molecular species totaled approx. 2,500 compounds (supplemental Table S5). Based on the structurally representative MS/MS fragments derived from each lipid, we selected approx. 5,000 MRM transitions for the screening of lipids contained in biological samples used for research (supplemental Table S5). Our strategy of a widely targeted quantitative lipidomics methodology was as follows: i) preparation of a mixture consisting of small aliquots from each sample extract containing the lipid class-specific internal standards (QC sample); ii) selection of target lipids by complete screening of the QC sample using 16 different RT-based scheduled MRM analyses in which the QqQMS is programmed to monitor specific MRM transitions only during the expected RT window; and iii) targeted quantitative analysis of selected lipids in all samples by performing a selected RT-based scheduled MRM method (Fig. 1 and supplemental Fig. S6).
Method validation
We validated the method using lipids standard solutions. The calibration curves were made using the ratio of the chromatographic peak area of each lipid class to that of the corresponding internal standard. The result of the analytical validation is summarized in Table 1. From these results, each lipid class displayed good RT repeatability [relative SDs (RSDs) < 3.8%] and peak areas (RSDs < 13.0%). As shown in supplemental Table S6, our SFC/QqQMS method using the DEA column also gave excellent repeatability for RTs (RSDs < 5.4%) on five different days (interday variation). The SFC/QqQMS method showed linearity with correlation coefficients (R2) over 0.9922. The limits of detections (LODs) for the 22 lipid classes were in the range of 5–1,000 fmol. By our SFC/QqQMS method, the LOD of PC and PE standards were found to be 50 and 10 fmol, respectively, and these sensitivities were 20- and 100-fold higher, respectively, than those in the SFC/QTOFMS full-scan method (supplemental Table S7). QTOFMS allows nontargeted lipidome analysis; however, the sensitivity of each lipid class was lower than that in the QqQMS-based MRM assay. The use of QC samples and in-house lipid MRM library also allowed for efficient selection of target lipid molecules. Thus, our SFC/QqQMS-based approach could also achieve comprehensive and simultaneous analysis of lipids.
TABLE 1.
Analytical validation of SFC/QqQMS for determination of lipids
| Lipid class | Ion mode | Analyte | Internal standard | RT for each analyte | RSD (%, n = 5)a | Linear range | R2 value | Range of RTsd | Spike-and-recovery teste | ||
| (min) | RRTb | RPAc | (nM) | (min) | Analyte | Recovery (%, n = 5) | |||||
| LPC | ESI+ | LPC 18:0 | LPC 17:0 | 6.12 | 0.09 | 2.7 | 50–50,000 | 0.9998 | 6.03–6.30 | LPC 18:0 | 71.9 ± 4.9 |
| LPE | ESI+ | LPE 18:0 | LPE 17:1 | 7.45 | 0.07 | 1.73 | 50–100,000 | 0.9987 | 7.32–7.56 | LPE 18:0 | 95.9 ± 3.5 |
| LPG | ESI+ | LPG 18:0 | LPG 17:1 | 10.87 | 0.08 | 8.18 | 50–100,000 | 0.9940 | – | – | – |
| LPA | ESI− | LPA 18:0 | LPA 17:0 | 17.43 | 0.57 | 8.97 | 1,000–100,000 | 0.9942 | – | – | – |
| LPI | ESI− | LPI 18:0 | LPI 17:1 | 14.02 | 0.09 | 3.5 | 5–50,000 | 0.9966 | – | – | – |
| LPS | ESI+ | LPS 16:0 | LPS 17:1 | 15.36 | 0.07 | 2.27 | 100–100,000 | 0.9999 | – | – | – |
| PC | ESI− | PC 18:0–18:2 | PC 17:0–17:0 | 5.46 | 0.08 | 4.35 | 50–50,000 | 0.9922 | 5.33–5.51 | PC 16:0-20:4 | 77.7 ± 3.5 |
| Alkyl-acyl PC (e) | ESI+ | PC 16:0e–18:1 | LPC 17:0 | 5.40 | 0.08 | 2.44 | 5–10,000 | 0.9993 | – | – | – |
| Alkenyl-acyl PC (p) | ESI+ | PC 18:0p–20:4 | LPC 17:0 | 5.45 | 0.1 | 3.65 | 50–50,000 | 0.9977 | 5.33–5.45 | PC 18:0p-20:4 | 78.0 ± 10.4 |
| PE | ESI− | PE 18:0–20:4 | PE 17:0–17:0 | 6.32 | 0.11 | 3.09 | 10–50,000 | 0.9929 | 5.80–6.32 | PE 18:0-20:4 | 101.9 ± 17.0 |
| Alkenyl-acyl PE (p) | ESI+ | PE 18:0p–20:4 | LPE 17:1 | 6.22 | 0.09 | 9.91 | 10–100,000 | 0.9976 | 6.05–6.33 | PE 18:0p-20:4 | 95.8 ± 17.3 |
| PG | ESI− | PG 16:0–18:1 | PG 17:0–17:0 | 8.99 | 0.08 | 1.72 | 5–100,000 | 0.9975 | – | – | – |
| PA | ESI− | PA 16:0–18:1 | PA 17:0–17:0 | 12.43 | 0.82 | 4.17 | 10–100,000 | 0.9985 | – | – | – |
| PI | ESI− | PI 18:0–20:4 | PS 17:0–17:0 | 12.06 | 0.1 | 2.26 | 100–50,000 | 0.9994 | 11.84–11.98 | PI 18:0-20:4 | 103.5 ± 7.6 |
| PS | ESI− | PS 18:0–18:1 | PS 17:0–17:0 | 12.23 | 0.09 | 1.87 | 50–100,000 | 0.9959 | – | – | – |
| SM | ESI+ | SM d18:1–18:0 | SM d18:1–17:0 | 5.83 | 0.09 | 2.56 | 10–10,000 | 0.9950 | 5.74–5.92 | SM d18:1-18:0 | 64.9 ± 3.0 |
| Cer | ESI+ | Cer d18:1–18:0 | Cer d18:1–17:0 | 4.48 | 0.12 | 12.95 | 10–100,000 | 0.9977 | 4.48–4.51 | Cer d18:1-18:0 | 91.0 ± 14.2 |
| CE | ESI+ | CE 16:0 | CE 17:0 | 1.12 | 1.81 | 11.32 | 50–50,000 | 0.9994 | 1.12–1.30 | CE 16:0 | 67.2 ± 5.7 |
| MAG | ESI+ | MAG 16:0 | MAG 17:0 | 3.97 | 0.18 | 3.49 | 500–100,000 | 0.9974 | 4.02–4.15 | MAG 16:0 | 98.4 ± 1.8 |
| DAG | ESI+ | DAG 18:0–20:4 | DAG 12:0–12:0 | 3.25 | 0.14 | 8.36 | 5–10,000 | 0.9976 | 3.14–3.40 | DAG 16:0-16:0 | 86.6 ± 9.1 |
| TAG | ESI+ | TAG 54:2 | TAG 51:0 | 1.61 | 3.76 | 2.86 | 500–100,000 | 0.9992 | 0.97–1.76 | TAG 54:2 | 72.8 ± 5.0 |
| FFA | ESI− | FFA 18:0 | FFA 17:0 | 4.02 | 0.11 | 1.18 | 500–100,000 | 0.9988 | 3.97–4.40 | FFA 18:0 | 91.8 ± 7.1 |
Amounts per injection used for validation of quantitative repeatability were 10 pmol.
Relative RT (analyte/internal standard).
Relative peak area (analyte/internal standard).
Range of RTs observed by the analysis of individual lipid molecular species in the all WHHLMI rabbit plasma samples in this study.
Representative WHHLMI rabbit plasma (placebo group) were used to spike-and-recovery test samples.
Previous studies demonstrated that the ionization efficiency of lipid molecular species was mainly dependent on their polar head groups and slightly dependent on their FA side chains (7, 37). Furthermore, there were no significant differences in the ionization efficiency of lipid molecular species differing in FA chain lengths and number of double bonds (31). Therefore, the lipid class-based separation improves the quantification of individual lipid species as the internal standard elutes together with the endogenous species. In order to investigate the quantitative accuracy, a standard spike-and-recovery test was carried out. We analyzed each lipid molecular species in WHHLMI rabbit plasma using the SFC/QqQMS system. Among commercially available chemically synthesized lipid standards, 14 standards of lipid molecular species contained in WHHLMI rabbit plasma were picked up for evaluation of the spike-and-recovery test (Table 1). Afterward, standard spike-and-recovery tests were carried out by adding the same amount of synthetic lipid standard as calculated for lipid levels in the plasma lipid extract. The calculated recoveries ranged from 64.9% to 103.5%, which were within the generally accepted range for quantification (Table 1). These results indicated that this lipidomics method using SFC/QqQMS could simultaneously achieve quantitative analysis with good repeatability and accuracy.
Widely targeted and quantitative lipidome analysis of myocardial infarction-prone animal model by SFC/QqQMS system
Dietary n-3 FAs are essential nutrients to prevent serious diseases, such as atherosclerosis. Many reports have described the association between n-3 FAs and prevention of atherosclerosis from the viewpoint of supplementation (38, 39). For example, administration of n-3 PUFAs such as EPA and docosahexaenoic acid decreased macrophage accumulation and expression of inflammatory molecules in apolipoprotein E KO mice. It was suggested that suppressive effects of n-3 PUFAs against plaque destabilization occurred (39). In the present study, we investigated the alteration of plasma lipid compositions by administration of EPA using the constructed analytical method. We used 12 WHHLMI rabbits (6 rabbits, placebo group; and 6 rabbits, EPA administration group). Supplemental Table S8 shows the age, sex, weight, and total plasma cholesterol levels in both placebo and EPA administration groups. No significant difference in body weight was observed, whereas total cholesterol levels significantly decreased (approx. 24%) by administration of EPA.
First, we performed lipid screening with the QC sample, which was derived from a mixture of equal amounts of the 24 plasma extracts. A total of 413 lipid molecular species were identified in plasma extracts of WHHLMI rabbits (supplemental Table S9). Based on the results of lipid screening, we selected MRM transitions and performed quantitative analysis in the 24 plasma extracts by the SFC/MS/MS with the selected RT-based scheduled MRM. Fig. 4B shows MRM chromatograms of the target lipid molecular species in WHHLMI rabbit plasma. From these results, in SFC separation using the DEA column, lipid species with the same FA composition but different head groups were fully resolved, but entire classes had some overlap (Table 1).
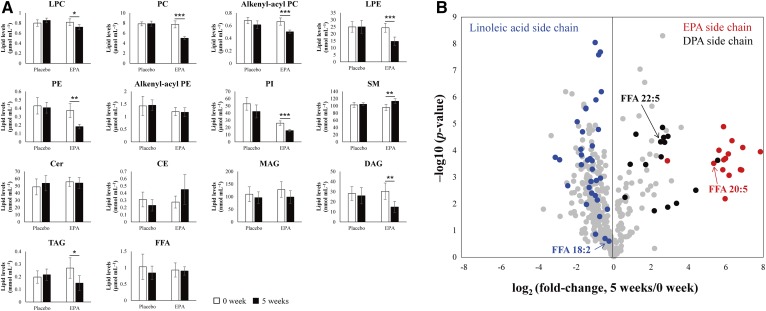
By using our novel analytical system, the alteration of each lipid class as well as individual lipid molecular species, including structural isomers of phospholipids by administration of EPA, could be quantitatively obtained. Figure 5A shows the plasma concentration of each lipid class in both the placebo and the EPA administration groups. The quantitative levels of plasma LPC, LPE, PC, alkenyl-acyl PC, PE, PI, DAG, and TAG were significantly decreased by administration of the EPA. To determine individual lipid levels, the quantitative lipid profile data of the EPA administration group between 0 and 5 weeks were compared using a volcano plot (Fig. 5B). Lipid molecular species with an EPA side chain (20:5, n-3) were dramatically increased (approx. more than 41-fold), and, accordingly, lipid molecular species with a docosapentaenoic acid (DPA) (22:5, n-3) side chain were also increased (approx. more than 5.7-fold) by administration of EPA and extension of the acyl chain by elongase-2 (40). On the other hand, high levels of lipid molecular species with a linoleic acid (18:2, n-6) side chain in WHHLMI rabbit plasma were decreased based on the supplementation of EPA. Therefore, a decrease in some lipid classes (i.e., LPC, LPE, PC, alkenyl-acyl PC, PE, PI, DAG, and TAG) was mainly due to a decrease in lipids with a linoleic acid side chain. This phenomenon may suggest that balance of the n-3 and -6 FAs changes with EPA supplementation.
Fig. 5.
Response of alteration of plasma lipid levels in WHHLMI to the supplementation of EPA. A: Plasma concentration of each lipid class in the placebo group and the EPA administration group. Error bars indicate the SDs of biological replicates (n = 6). Statistical significance was determined using paired t-test. * P < 0.05; ** P < 0.01; *** P < 0.001. B: Individual plasma lipid profiles of before (0 weeks) and after (5 weeks) administration of EPA by volcano plot. Red, black, and blue closed circles indicate the lipid molecular species with EPA side chain, DPA side chain, and linoleic acid side chain, respectively.
CONCLUSIONS
In the present study, we have developed a practical methodology for widely targeted quantitative lipidome analysis using SFC/QqQMS and theoretically calculated a comprehensive lipid MRM library. Lipid classes can be separated by SFC using the DEA column with high resolution, high throughput, and good repeatability. The lipid class-based separation and multitargeted MRM assay also improve the quantification of individual lipid species as the internal standard elutes together with the endogenous species. Our SFC/QqQMS-based analytical platform enabled us to quantitatively capture the changes of individual lipid classes as well as individual lipid molecular species, including positional and structural isomers in WHHLMI rabbit plasma. Quantitative analysis of comprehensive lipids in various biological samples (e.g., blood, cell, and organ) will enable comparison of measurements on different days, using different equipment, and/or facilities. Information of endogenous amounts of lipids will also develop the biological function of lipid molecular species. Taking all of the present results into account, our developed analytical system is a promising new tool for in-depth studies on complex lipid metabolism, drug administration testing, and biomarker discovery.
Supplementary Material
Footnotes
Abbreviations:
- 1-AA
- 1-aminoanthracene
- 2-EP
- 2-ethylpyridine
- 2-PIC
- 2-picolylamine
- ABPR
- active back pressure regulator
- approx.
- approximately
- BEH
- ethylene bridged hybrid
- CE
- cholesteryl ester
- Cer
- ceramide
- DAG
- diacylglycerol
- DEA
- diethylamine
- DI
- direct infusion
- HILIC
- hydrophilic interaction chromatography
- i.d.
- inner diameter
- LC
- liquid chromatography
- LPA
- lysophosphatidic acid
- LPC
- lysophosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- LPG
- lysophosphatidylglycerol
- LPI
- lysophosphatidylinositol
- LPS
- lysophosphatidylserine
- MAG
- monoacylglycerol
- MRM
- multiple reaction monitoring
- MS/MS
- tandem mass spectrometry
- NPLC
- normal-phase liquid chromatography
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- psi
- pounds per square inch
- QC
- quality control
- QqQMS
- triple-quadrupole mass spectrometry
- QTOFMS
- quadrupole TOF mass spectrometry
- RPLC
- reverse-phase liquid chromatography
- RSD
- relative SD
- RT
- retention time
- SCCO2
- supercritical carbon dioxide
- SCF
- supercritical fluid
- SFC
- supercritical fluid chromatography
- TAG
- triacylglycerol
- UPC2
- UltraPerformance Convergence Chromatography
- WHHLMI
- myocardial infarction-prone Watanabe heritable hyperlipidemic
This study was partly supported by the Japan Agency for Medical Research and Development AMED-CREST Programs 18gm0910010h0203, JPMJCR1395, and JP18gm0910013 (Y.I. and T.B.); a grant from the Japan Science and Technology Agency ALCA Program of the (Y.I. and T.B.); Ministry of Education, Culture, Sports, Science, and Technology of Japan Grant-in-Aid for Scientific Research on Innovative Areas 17H06304 (Y.I. and T.B.); Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (C) 26505007 (T.B.); JSPS Grant-in-Aid for Scientific Research (C) 15K06557 (Y.I.); and grants from Takeda Science Foundation (T.B.), Yakult Bio-Science Foundation (T.B.), and Kieikai Research Foundation (T.B.). The study represents a portion of the dissertation submitted by H.T. to Kyushu University in partial fulfillment of the requirement for his PhD degree. The authors declare no competing financial interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Ekroos K., Jänis M., Tarasov K., Hurme R., and Laaksonen R.. 2010. Lipidomics: a tool for studies of atherosclerosis. Curr. Atheroscler. Rep. 12: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda H., Koike T., Izumi Y., Yamada T., Yoshida M., Shiomi M., Fukusaki E., and Bamba T.. 2015. Lipidomic analysis of plasma lipoprotein fractions in myocardial infarction-prone rabbits. J. Biosci. Bioeng. 120: 476–482. [DOI] [PubMed] [Google Scholar]
- 3.Kettunen J., Tukiainen T., Sarin A. P., Ortega-Alonso A., Tikkanen E., Lyytikäinen L. P., Kangas A. J., Soininen P., Würtz P., Silander K., et al. 2012. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 44: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee E. P., Ho J. E., Chen M. H., Shen D., Cheng S., Larson M. G., Ghorbani A., Shi X., Helenius I. T., O’Donnell C. J., et al. 2013. A genome-wide association study of the human metabolism in a community-based cohort. Cell Metab. 18: 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowden J. A., Heckert A., Ulmer C. Z., Jones C. M., Koelmel J. P., Abdullah L., Ahonen L., Alnouti Y., Armando A., Asara J. M., et al. 2017. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using standard reference material 1950 metabolites in frozen human plasma. J. Lipid Res. 58: 2275–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cajka T., and Fiehn O.. 2014. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 61: 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X., and Gross R. W.. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 8.Ståhlman M., Ejsing C. S., Tarasov K., Perman J., Borén J., and Ekroos K.. 2009. High-throughput shotgun lipidomics by quadrupole time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2664–2672. [DOI] [PubMed] [Google Scholar]
- 9.Heiskanen L. A., Suoniemi M., Ta H. X., Tarasov K., and Ekroos K.. 2013. Long-term performance and stability of molecular shotgun lipidomic analysis of human plasma samples. Anal. Chem. 85: 8757–8763. [DOI] [PubMed] [Google Scholar]
- 10.Hopfgartner G., Bean K., Henion J., and Henry R.. 1993. Ion spray mass spectrometric detection for liquid chromatography: a concentration- or a mass-flow-sensitive device? J. Chromatogr. A. 647: 51–61. [Google Scholar]
- 11.Yamada T., Uchikata T., Sakamoto S., Yokoi Y., Fukusaki E., and Bamba T.. 2013. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J. Chromatogr. A. 1292: 211–218. [DOI] [PubMed] [Google Scholar]
- 12.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., and Arita M.. 2015. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 12: 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson A. Å., Michelsen P., Larsen Å., and Odham G.. 1996. Normal-phase liquid chromatography class separation and species determination of phospholipids utilizing electrospray mass spectrometry/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 10: 775–780. [Google Scholar]
- 14.Uran S., Larsen Å., Jacobsen P. B., and Skotland T.. 2001. Analysis of phospholipid species in human blood using normal-phase liquid chromatography coupled with electrospray ionization ion-trap tandem mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 758: 265–275. [DOI] [PubMed] [Google Scholar]
- 15.Pang L. Q., Liang Q. L., Wang Y. M., Ping L., and Luo G. A.. 2008. Simultaneous determination and quantification of seven major phospholipid classes in human blood using normal-phase liquid chromatography coupled with electrospray mass spectrometry and the application in diabetes nephropathy. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 869: 118–125. [DOI] [PubMed] [Google Scholar]
- 16.Schwalbe-Herrmann M., Willmann J., and Leibfritz D.. 2010. Separation of phospholipid classes by hydrophilic interaction chromatography detected by electrospray ionization mass spectrometry. J. Chromatogr. A. 1217: 5179–5183. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y. Y., Xiong Y., and Curtis J. M.. 2011. Measurement of phospholipids by hydrophilic interaction liquid chromatography coupled to tandem mass spectrometry: the determination of choline containing compounds in foods. J. Chromatogr. A. 1218: 5470–5479. [DOI] [PubMed] [Google Scholar]
- 18.Cífková E., Holčapek M., Lísa M., Ovčačíková M., Lyčka A., Lynen F., and Sandra P.. 2012. Nontargeted quantitation of lipid classes using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry with single internal standard and response factor approach. Anal. Chem. 84: 10064–10070. [DOI] [PubMed] [Google Scholar]
- 19.Zhu C., Dane A., Spijksma G., Wang M., van der Greef J., Luo G., Hankemeier T., and Vreeken R. J.. 2012. A efficient hydrophilic interaction liquid chromatography separation of 7 phospholipid classes based on a diol column. J. Chromatogr. A. 1220: 26–34. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki Y., Kamide Y., Hirai M. Y., and Saito K.. 2013. Plant lipidomics based on hydrophilic interaction chromatography coupled to ion trap time-of-flight mass spectrometry. Metabolomics. 9: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonomura K., Kudoh S., Sato T., and Matsuda F.. 2015. Plasma lipid analysis by hydrophilic interaction liquid chromatography coupled with electrospray ionization tandem mass spectrometry. J. Sep. Sci. 38: 2033–2037. [DOI] [PubMed] [Google Scholar]
- 22.Bamba T., Shimonishi N., Matsubara A., Hirata K., Nakazawa Y., Kobayashi A., and Fukusaki E.. 2008. High throughput and exhaustive analysis of diverse lipids by using supercritical fluid chromatography-mass spectrometry for metabolomics. J. Biosci. Bioeng. 105: 460–469. [DOI] [PubMed] [Google Scholar]
- 23.Uchikata T., Matsubara A., Nishiumi S., Yoshida M., Fukusaki E., and Bamba T.. 2012. Development of oxidized phosphatidylcholine isomer profiling method using supercritical fluid chromatography/mass spectrometry. J. Chromatogr. A. 1250: 205–211. [DOI] [PubMed] [Google Scholar]
- 24.Yamada T., Uchikata T., Sakamoto S., Yokoi Y., Nishiumi S., Yoshida M., Fukusaki E., and Bamba T.. 2013. Supercritical fluid chromatography/Orbitrap mass spectrometry based lipidomics platform coupled with automated lipid identification software for accurate lipid profiling. J. Chromatogr. A. 1301: 237–242. [DOI] [PubMed] [Google Scholar]
- 25.Lísa M., and Holčapek M.. 2015. High-throughput and comprehensive lipidomic analysis using ultrahigh-performance supercritical fluid chromatography-mass spectrometry. Anal. Chem. 87: 7187–7195. [DOI] [PubMed] [Google Scholar]
- 26.Tsugawa H., Arita M., Kanazawa M., Ogiwara A., Bamba T., and Fukusaki E.. 2013. MRMPROBS: a data assessment and metabolite identification tool for large-scale multiple reaction monitoring based widely targeted metabolomics. Anal. Chem. 85: 5191–5199. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J., and Yin Y.. 2016. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst. 141: 6362–6373. [DOI] [PubMed] [Google Scholar]
- 28.Shiomi M., and Ito T.. 2009. The Watanabe heritable hyperlipidemic (WHHL) rabbit, its characteristics and history of development: a tribute to the late Dr. Yoshio Watanabe. Atherosclerosis. 207: 1–7. [DOI] [PubMed] [Google Scholar]
- 29.Shiomi M., Ito T., Yamada S., Kawashima S., and Fan J.. 2003. Development of an animal model for spontaneous myocardial infarction (WHHLMI rabbit). Arterioscler. Thromb. Vasc. Biol. 23: 1239–1244. [DOI] [PubMed] [Google Scholar]
- 30.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 31.Jung H. R., Sylvänne T., Koistinen K. M., Tarasov K., Kauhanen D., and Ekroos K.. 2011. High throughput quantitative molecular lipidomics. Biochim. Biophys. Acta. 1811: 925–934. [DOI] [PubMed] [Google Scholar]
- 32.Nováková L., Perrenoud A. G.-G., Francois I., West C., Lesellier E., and Guillarme D.. 2014. Modern analytical supercritical fluid chromatography using columns packed with sub-2 μm particles: A tutorial. Anal. Chim. Acta. 824: 18–35. [DOI] [PubMed] [Google Scholar]
- 33.Fujito Y., Hayakawa Y., Izumi Y., and Bamba T.. 2017. Importance of optimizing chromatographic conditions and mass spectrometric parameters for supercritical fluid chromatography/mass spectrometry. J. Chromatogr. A. 1508: 138–147. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Regalado E. L., Mergelsberg I., and Welch C. J.. 2013. Extending the range of supercritical fluid chromatography by use of water-rich modifiers. Org. Biomol. Chem. 11: 4925–4929. [DOI] [PubMed] [Google Scholar]
- 35.Koistinen K. M., Suoniemi M., Simolin H., and Ekroos K.. 2015. Quantitative lysophospholipidomics in human plasma and skin by LC-MS/MS. Anal. Bioanal. Chem. 407: 5091–5099. [DOI] [PubMed] [Google Scholar]
- 36.Lee J. W., Nagai T., Gotoh N., Fukusaki E., and Bamba T.. 2014. Profiling of regioisomeric triacylglycerols in edible oils by supercritical fluid chromatography/tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 966: 193–199. [DOI] [PubMed] [Google Scholar]
- 37.Han X., and Gross R. W.. 1994. Electrospray ionization mass spectrometric analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA. 91: 10635–10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connor W. E. 2000. Importance of n-3 fatty acids in health and disease. Am. J. Clin. Nutr. 71: 171S–175S. [DOI] [PubMed] [Google Scholar]
- 39.Takashima A., Fukuda D., Tanaka K., Higashikuni Y., Hirata Y., Nishimoto S., Yagi S., Yamada H., Soeki T., Wakatsuki T., et al. 2016. Combination of n-3 polyunsaturated fatty acids reduces atherogenesis in apolipoprotein E-deficient mice by inhibiting macrophage activation. Atherosclerosis. 254: 142–150. [DOI] [PubMed] [Google Scholar]
- 40.Gregory M. K., Gibson R. A., Cook-Johnson R. J., Cleland L. G., and James M. J.. 2011. Elongase reactions as control points in long-chain polyunsaturated fatty acid synthesis. PLoS One. 6: e29662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.