Abstract
APOA1 is the most abundant protein in HDL. It modulates interactions that affect HDL’s cardioprotective functions, in part via its activation of the enzyme, LCAT. On nascent discoidal HDL, APOA1 comprises 10 α-helical repeats arranged in an anti-parallel stacked-ring structure that encapsulates a lipid bilayer. Previous chemical cross-linking studies suggested that these APOA1 rings can adopt at least two different orientations, or registries, with respect to each other; however, the functional impact of these structural changes is unknown. Here, we placed cysteine residues at locations predicted to form disulfide bonds in each orientation and then measured APOA1’s ability to adopt the two registries during HDL particle formation. We found that most APOA1 oriented with the fifth helix of one molecule across from fifth helix of the other (5/5 helical registry), but a fraction adopted a 5/2 registry. Engineered HDLs that were locked in 5/5 or 5/2 registries by disulfide bonds equally promoted cholesterol efflux from macrophages, indicating functional particles. However, unlike the 5/5 registry or the WT, the 5/2 registry impaired LCAT cholesteryl esterification activity (P < 0.001), despite LCAT binding equally to all particles. Chemical cross-linking studies suggest that full LCAT activity requires a hybrid epitope composed of helices 5–7 on one APOA1 molecule and helices 3–4 on the other. Thus, APOA1 may use a reciprocating thumbwheel-like mechanism to activate HDL-remodeling proteins.
Keywords: apolipoproteins, cholesterol metabolism, lecithin:cholesterol acyltransferase, high density lipoprotein metabolism, high density lipoprotein, proteomics, electron microscopy, surface plasmon resonance, cholesterol/efflux, apolipoprotein A1
Low levels of HDL cholesterol strongly associate with an increased risk of coronary artery disease in epidemiological studies (1). One proposed cardioprotective effect of HDL and its primary protein component, APOA1, is the transport of cellular cholesterol from the artery wall to the liver for catabolism. APOA1 acts as a structural scaffold that maintains lipid packaging and mediates the docking of a variety of accessory proteins (lipases, enzymes, transfer proteins, cell surface proteins), which remodel lipoproteins throughout their lifespan (2). These proteins likely account for other potentially cardioprotective functions of HDL as they have known functions in lipid metabolism (3), anti-inflammation (4), and immune response (5). Thus, it is important to understand the molecular details of how proteins interact with the APOA1 scaffold and how those interactions affect HDL metabolism.
One of the best known HDL modifying factors is LCAT, an APOA1-activated enzyme that drives HDL maturation by catalyzing the fatty acyl esterification of cholesterol in plasma, possibly to prevent backflow of free cholesterol into peripheral cells (6). LCAT acts at the lipid/water interface (7); however, its reactivity increases up to 300-fold in the presence of APOA1 (8). Evidence gathered from studies on natural (9, 10) and engineered APOA1 mutants (11–14), as well as recent hydrogen-deuterium exchange experiments (15), strongly suggests that LCAT interacts directly with APOA1 on HDL. However, the details of how APOA1 stimulates LCAT activation are poorly understood.
APOA1 is a 243 amino acid 28 kDa single polypeptide that lacks glycosylation or disulfide linkages. A consensus structure for lipid-free monomeric APOA1 has recently been published (16); however, its structural transitions during HDL biogenesis are less well-understood. APOA1 contains an N-terminal domain composed of amino acids 1–43 and a C-terminal domain (amino acids 44–243) with 10 amphipathic α-helical repeats (eight 22-mers and two 11-mers) that are frequently punctuated by proline residues (17). In model discoidal HDL particles of 9.6 nm in diameter, there are two molecules of APOA1 per particle (18). Based on decades of data (2) from spectroscopy, electron microscopy (EM), and a crystal structure of an APOA1 deletion mutant (17), APOA1 molecules are thought to orient in stacked anti-parallel rings around the circumference of these discs (19–21) in an arrangement known as the double belt model (22). Computer analysis suggested an intermolecular registry in which helix 5 (specifically G129) of both molecules is directly opposed, giving the highest weighted score of potential intermolecular salt bridges (19). This is called the 5/5 (G129j) registry, and numerous variations of this model have been proposed (15, 23–25). Most recently, a study that combined nuclear magnetic resonance, electron paramagnetic resonance, and transmission EM confirmed many aspects of the double belt model in reconstituted HDL (rHDL) particles made with a shortened construct of APOA1 (26). We previously used chemical cross-linking and MS analyses of rHDL discs containing full-length APOA1 to reveal interactions between the anti-parallel molecules (27). The cross-links supported a 5/5 registry, but a fraction of the particles seemed to contain evidence of an alternate registry where helix 5 in one molecule is adjacent to helix 2 of its antiparallel partner [5/2 (K206 note: this can also be called a 4/4 orientation)]. Additional work found evidence for this alternative registry in rHDL of various sizes (28), spherical HDL (29), and in “real” HDL particles isolated from human plasma (27).
The concept that APOA1 may exist in at least two helical registries is consistent with prior fluorescence energy transfer experiments by Li et al. (30) and raises the intriguing possibility that the shifting of APOA1 helical registries may alter its interactions with HDL remodeling proteins. In the current study, we first confirmed the presence of differing APOA1 helical registries in rHDL, using an orthogonal technique versus our original cross-linking observation. We then engineered three “registry-locked” mutant HDL particles and investigated how APOA1 helical registry affects HDL function in terms of cholesterol efflux and LCAT activation. The results indicate that APOA1 may modulate HDL maturation by forming a discontinuous epitope involving two molecules of APOA1 for optimal LCAT activation.
MATERIALS AND METHODS
APOA1 expression/purification
Cysteine (Cys) residues were introduced by Quick Change™ mutagenesis (Agilent) at K133, G129, Q132, K206, E205, K195, and R177 in the mature human APOA1 cDNA in a pET-30 vector (Novagen, now EMD Millipore) per the manufacturer’s instructions. The sequence was verified by Sanger sequencing at the Cincinnati Children’s Hospital DNA Core. WT APOA1 and mutants containing a tobacco etch virus (TEV) protease cleavable N-terminal histidine (His)-tag were expressed and purified as previously described (31). Briefly, the APOA1-containing vector was transformed into BL-21 Escherichia coli cells (Protein Express, #961-003). Lysogeny broth containing kanamycin (62 mM) was inoculated with single colonies and grown to an OD600 of 0.7–0.8 by shaking at 225 RPM overnight at 37°C. Protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (Gold Biotechnology, #1248165) and shaking for 2 h at 37°C. The cells were pelleted at 8,000 RCF for 10 min at 4°C in a Sorvall RC-5C Plus centrifuge with an SLA 1500 rotor (Thermo Scientific) and the pellet was stored at −20°C until purification. 15N-labeled protein was expressed as previously described (32).
Cell pellets were solubilized in binding buffer [5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl (pH 7.9)] with PMSF, pepstatin, and leupeptin at a final concentration of 1 μM. Cells were lysed by sonication with a tapered probe (Sonic Dismembrator 550; Fisher Scientific) on ice for 10 min at a 50% duty cycle, and cell debris was pelleted at 14,000 RCF for 20 min at 4°C. The supernatant was passed through a 0.45 μm filter (VWR, #28145-505), applied to His-binding columns containing NiO4S·6 H2O (Qiagen, #30230), and eluted with buffer containing 1 M imidazole, 500 mM NaCl, and 20 mM Tris-HCl (pH 7.9). Purification steps were analyzed by sodium dodecyl sulfate-polyacrylamide gradient gel electrophoresis (SDS-PAGGE) (4–15%; BioRad, #456-1084) in reducing conditions [3 mM β-mercaptoethanol (β-ME)], stained with Coomassie Blue (GE Healthcare, #17-0518-01), and eluates containing pure protein were collected and dialyzed against standard Tris buffer (STB) (1 mM Tris-HCl, 15 mM NaCl, 0.1 mM EDTA, 0.02% NaN3) containing 3 mM DTT. His-tags were cleaved by TEV protease at a ratio of 80:1 (mass APOA1/mass TEV) for 2 h at 21°C. The sample was dialyzed against STB without EDTA and reapplied to the His-binding column to capture the cleaved His-tag and recover pure protein. Purification steps were analyzed by SDS-PAGGE and pure protein was collected in the wash buffer [60 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl (pH 7.9)]. Protein was dialyzed against 10 mM ammonium bicarbonate, frozen on dry ice, lyophilized, and stored at −80°C until ready for use.
14N/15N APOA1 preparation
WT (14N) and isotopically labeled (15N) APOA1 were solubilized in STB containing 3 M guanidine HCl and added together at a 1:1 ratio. The protein mixture was stored at 4°C and vigorously vortexed every 20 min for 1 h. Protein was then purified on a Superdex 200 column (10/30; GE Healthcare Life Sciences) at 0.6 ml/min with an STB mobile phase. The fractions were analyzed by SDS-PAGGE for purity.
Generation and characterization of 9.6 nm rHDL
Lyophilized protein was solubilized and denatured in STB containing 3 M guanidine HCl and 3 mM DTT for 1 h at 4°C followed by refolding at 4°C by dialyzing against 3 changes of 4L STB for a minimum of 3 h each. rHDL particles were generated with a modified sodium cholate dialysis as previously described (33). A molar ratio of 80:8:1 POPC (Avanti Polar Lipids, Birmingham, AL), cholesterol (Sigma, St. Louis, MO), and APOA1, respectively, was used to generate 9.6 nm rHDL. The particles were purified away from unreacted protein and lipid by size exclusion chromatography (SEC) with a Superose 6 column (10 × 300 mm; GE Healthcare Life Sciences) in experiment-specific buffer at 0.5 ml/min. The fractions were analyzed by SDS-PAGGE, and pure particles were stored under N2 at 4°C until ready for experiments. Particles were used within 3 days. Particle size and homogeneity was determined by 8–25% nondenaturing gradient gel electrophoresis (NDGGE) (GE Healthcare Life Sciences) with Coomassie Blue staining. Protein concentrations were determined by the modified Markwell-Lowry assay (34). Choline-containing phospholipid (PL) and cholesterol concentrations were determined by colorimetric assay (Wako, #997-01811, and Pointe Scientific, #C7509STD, #C7510, respectively).
Cys-proximity assay
WT and Cys-mutant APOA1 in STB were reduced at a final concentration of 3 mM DTT and analyzed by nonreducing SDS-PAGGE. rHDLs were generated as detailed above, making sure to maintain reducing conditions at each step. To initiate disulfide formation, the particles were incubated at 21°C for 48 h. The extent of dimerization was analyzed by nonreducing SDS-PAGGE.
Circular dichroism
Circular dichroism (CD) measurements were completed on an Aviv 215 spectropolarimeter. Particles were dialyzed at 4°C against 10 mM NaH2PO4 and 100 mM NaF (pH 7.4) for 3 changes of 4L at a minimum of 3 h each. Samples at equal protein concentration (0.18 mg/ml) were analyzed in a 0.01 cm cuvette from 190 to 300 nm at a rate of 1 nm/s at 25°C. The step resolution and bandwidth were 1 nm. Data were analyzed with SELCON3 (35), reference set 4, on DichroWeb (36), using a mean residue weight of 115.3. Results were expressed as the molar ellipticity for three independent experiments.
Cholesterol efflux
HDL particles were assayed for their ability to efflux cholesterol [1,2-3H (N)]cholesterol ([3H]; Perkin Elmer) from labeled RAW 264.7 macrophages. The macrophages were grown to 80% confluency and incubated with medium containing 1.0 μCi/ml [3H]cholesterol ± 0.3 mM 8-bromo-cAMP for 16 h in 10% CO2 at 37°C. Medium-containing rHDL samples (10 μg/ml protein) ± 0.3 mM 8-bromo-cAMP were then incubated with the macrophages in triplicate for 6 h in 10% CO2 at 37°C. Medium alone and medium containing 10 μg/ml of lipid-free APOA1 isolated from plasma were included as controls. Cholesterol efflux was measured by harvesting the medium, passing it through a 0.45 μM filter, and quantifying radiolabeled cholesterol in 100 μl of sample by liquid scintillation counting. Total cell counts were determined by extracting intracellular cholesterol from cells using isopropanol, drying under air, solubilizing in toluene, and counting. The percent efflux was calculated by dividing the number of counts in the medium by the total internalized counts per well (determined from the medium-only-treated cells). ABCA1-dependent cholesterol efflux was calculated as described (37). Percent efflux was normalized to the medium-alone control condition.
LCAT activity assay (8)
The efficiency of LCAT-catalyzed cholesterol esterification on HDL was measured using a 67 kDa recombinant LCAT produced in CHO cells (38). The assay was performed on particles containing 10 μCi of [3H]cholesterol. Particles (356 nM) were incubated with BSA (60 μM) and LCAT (3.0 nM) at 37°C in STB ± 10 mM β-ME for 30 min. Lipids were extracted and separated using instant thin-layer chromatography silica gel plates (iTLC-SG; Agilent) with a mobile phase of 300:60:1 (v/v) of petroleum ether, ethyl ether, and acetic acid. The free cholesterol and cholesteryl ester (CE) bands were excised, and [3H]cholesterol counts were determined using scintillation counting. The fractional esterification rate was calculated as described (8).
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were performed on a Biacore T100 biosensor (GE Healthcare). Two CM5 sensorchips (GE Healthcare) were used, each normalized with a 70% glycerol solution. A mouse anti-human antibody specific to the N-terminus of APOA1 [4H1 (39)] was preimmobilized on flow cells 2–4 of the sensorchip at a flow rate of 10 μl/min for 400 s in PBS with 1.5 μM BSA (pH 7.4) (supplemental Fig. S6a). Flow cell 1 was coupled with a mouse monoclonal antibody against ubiquitin as a reference surface (supplemental Fig. S6b). Before beginning the experiment, nine buffer washes (400 s) were performed to establish a stable baseline. Particles were then captured by the immobilized antibodies at a flow rate of 15 μl/min in the same buffer for 600 s to a density of 50 RU in flow cells 2–4. The particles were washed with buffer for 900 s to establish a stable baseline. LCAT (67 kDa, >99% pure based on SDS-PAGGE) in PBS (pH 7.4) was injected over the flow cells at concentrations of 0.2–55 μM at 25°C with a flow rate of 60 μl/min to collect equilibrium binding data. Each run involved LCAT injections for 120 s, sequentially, from highest to lowest concentration, and then from lowest to highest for duplicate measurements. After each injection, rHDL was washed with buffer for 600 s to ensure complete regeneration of the rHDL baseline before the next injection. Four buffer blanks were included and injected after each quarter of the run. Concentrations above 13.8 μM were excluded from analysis due to evidence of nonspecific binding. Certain concentrations for WT and K133C on sensorchip 2 were not included in the analysis because the traces did not result in a stable equilibrium plateau. Data were collected at a rate of 1 Hz. For equilibrium binding analyses, the steady-state response was taken 4 s before the end of the injection window for each analyte concentration to create a binding isotherm. Equilibrium dissociation constants were determined using GraphPad Prism 7, nonlinear regression (curve fit), and specific binding with Hill slope based on reference channel and blank subtracted response.
Cross-linking rHDL to LCAT
rHDL particles were generated that contained 14N and 15N. 14N/15N rHDLs were incubated with LCAT in PBS at a ratio of 4:1 APOA1:LCAT (mol/mol) for 30 min at 37°C. Interactions were captured by the addition of the bis(sulfosuccinimidyl)suberate (BS3; Thermo Fisher Scientific #21580) cross-linker at a ratio of 100:1 BS3:APOA1 (mol/mol) for 2 h at 4°C and were quenched by addition of Tris-HCl to a final concentration of 25 mM. The rHDL:LCAT complex was isolated using SEC with three Superdex 200 10/30 columns (GE Healthcare) in series containing 10 mM ammonium bicarbonate (pH 7.4) at 0.3 ml/min. Fractions were evaluated using SDS-PAGGE and fractions containing purified rHDL:LCAT complex were pooled and prepared for MS analysis.
Sample preparation for MS
Samples were prepared for analysis using organic solvent delipidation as previously described (40). Lipid was extracted by addition of 2:1 chloroform:methanol (v/v) and protein was resolubilized in 50 mM ammonium bicarbonate (pH 8.1). Disulfide bonds were reduced by addition of DTT to a final concentration of 10 mM and incubated at 42°C for 30 min. Reduced protein was carbamidomethylated with iodoacetamide at a final concentration of 40 mM at 21°C for 30 min in the dark. Samples were digested by addition of sequencing-grade trypsin (Promega, #V5111) at a ratio of 1:20 trypsin:protein (mass/mass) and incubated at 37°C for 16 h. Samples were spiked with additional trypsin and incubated an additional 2 h at 37°C. Peptides were lyophilized to dryness and stored at −20°C until MS analysis.
MS analysis of the rHDL:LCAT complex
As the sample yield of the rHDL:LCAT complex was limited, MS analysis was performed using Nano-LC-MS/MS analyses with a TripleTOF® 5600+ (Sciex, Concord, Canada) coupled to an Eksigent (Dublin, CA) NanoLC-Ultra® nanoflow system with increased sensitivity. Briefly, dried samples were reconstituted in 0.1% formic acid, and 5 μl (∼1 μg of digest) were loaded onto a C18 IntegraFrit™ trap column (New Objective, Inc.) at 2 μl/min in 0.1% formic acid in water for 15 min to desalt and concentrate the samples. For the chromatographic separation, the trap column was switched to align with the analytical column, Acclaim® PepMap100 (Thermo Fisher Scientific). Peptides were eluted at 300 nl/min using a varying mobile phase gradient from 95% phase A (0.1% formic acid in water) to 40% phase B (0.1% formic acid in acetonitrile) for 70 min (1% per minute), then from 40% B to 85% B in 5 min with reequilibration. Effluent was introduced to the mass spectrometer using a NANOSpray® III source (Sciex). The instrument was operated in positive ion mode for 90 min, where each cycle consisted of one TOF-MS scan (0.25 s accumulation time in a m/z 350–1,500 window) followed by 50 information-dependent acquisition mode MS/MS scans on the most intense candidate ions selected from the initially performed TOF-MS scan during each cycle. Each product ion scan had an accumulation time of 0.05 s and a rolling collision energy with a spread of five selected. The .wiff files were converted to Mascot generic files using PeakView® v1.2.0.3 software (AB Sciex). Injection blank gradient runs over 25 min were run between samples to minimize sample carryover.
Identification and analysis of cross-linked peptides
Cross-links were initially identified from the MGF files using SimXL (v. 1.1.2.2) as previously described (41). For positive identification, cross-linked peptides were within 20 ppm of the precursor and fragment ions, had a SimXL score of ≥2.0, and were identified in at least two of three independent experiments. Additionally, all reported cross-links initially identified using SimXL were confirmed using either MassHunter qualitative analysis software (v B.07.00, Agilent) or PeakView (v. 1.2) to manually visualize the MS1 peak pattern.
Statistical methods
Characterization of rHDL particles was performed in triplicate for n ≥ 3 independent particle preparations. Particles were characterized in triplicate for a single preparation of particles for CD and EM. A one-way ANOVA was performed using SigmaPlot (v. 11.0) to assess statistically significant differences for LCAT functional assays (n = 3). Significant differences (P < 0.05) were then analyzed with the Holm-Sidak post hoc test. Error bars represent SD. SPR analysis was performed in duplicate.
RESULTS
Cys-proximity assay, an independent test of APOA1 helical registry hypothesis
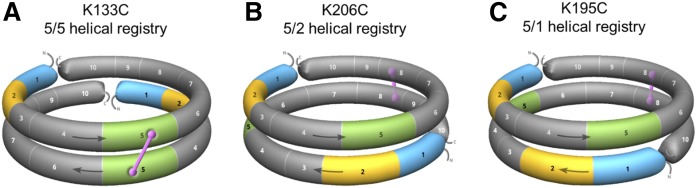
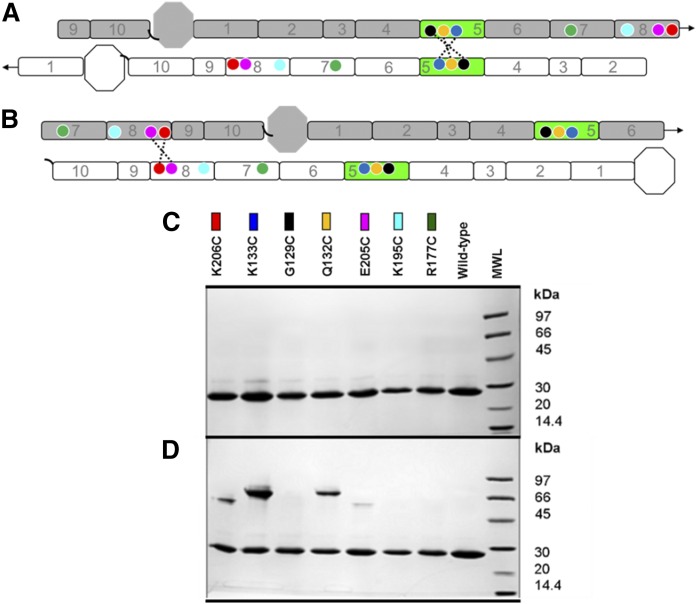
To determine whether APOA1 molecules in discoidal HDL can adopt 5/5 and 5/2 helical registries, we exploited the fact that APOA1 lacks Cys residues. Cys residues were introduced by site-directed mutagenesis in positions predicted to be in close proximity between two APOA1 molecules in both registries. We hypothesized that if the natural helical registry of APOA1 brought the Cys residues into close opposition, disulfide bonds could form under oxidizing conditions. If the natural registry kept them far apart, few disulfide bonds would form. In the case of the 5/5 helical registry, mutations K133C (Fig. 1A), G129C, and Q132C were made. For the 5/2 helical registry, we generated K206C (Fig. 1B) and E205C. We also produced K195C (5/1) (Fig. 1C) and R177C (5/9) to test helical registries that we had not observed experimentally in previous cross-linking studies. 2D diagrams of the 5/5 Cys mutants (Fig. 2A) and 5/2 Cys mutants (Fig. 2B) are also shown. The mutants were expressed, purified, and incorporated into rHDL entirely under reducing conditions (3 mM DTT). Figure 2C shows an SDS-PAGGE analysis of APOA1 mutants with minimal disulfide-mediated dimerization under reducing conditions. After removal of the reducing agent, K133C formed significant amounts of dimer, consistent with a close proximity of the two Cys residues across the intermolecular interface (Fig. 2D). Q132C also formed dimer though to a lesser extent. G129C did not form a dimer; however, models of the 5/5 double-belt suggest that the G129C substitution may point the Cys residues away from each other. K206C and E205C also showed dimer formation, though not to the extent of K133C and G129C [Note: the dimers show differential gel migration depending on the point of linkage as described by Sorci-Thomas et al. (42)]. Both K195C and K177C failed to form dimers, suggesting that the residues were too far apart to form disulfide linkages. In repeated experiments, we occasionally noticed some dimer formation in K195C, though always less than both K133C and K206C. The reason for this is not clear, but may result from protein dynamics given the relatively close proximity of residue 195 to 206 in the sequence. Overall, this data was consistent with the hypothesis that APOA1 can form at least two helical registries on HDL.
Fig. 1.
Cartoons showing the 3D spatial relationships of APOA1 helix (H)5 (green), H2 (yellow), and H1 (blue) registry-locked rHDL preparations. A: In oxidizing conditions, K133C (H5) locks two antiparallel APOA1 monomers into a 5/5 helical registry through a disulfide bond linkage (pink). B: K206C (H8) locks two APOA1 monomers into a 5/2 helical registry. C: K195C (H8) locks APOA1 monomers into a 5/1 helical registry. All other helices of APOA1 are represented in gray. Arrows show the direction of the antiparallel APOA1 helices (N terminus, H1 through H10, C terminus).
Fig. 2.
Design of mutants for the postulated APOA1 registries in discoidal rHDL. A: A 2D representation of the molecular interactions between two antiparallel APOA1 monomers (imagine two APOA1 belts pulled off the edge of a particle and laid flat). Helix 5 (H5, green) of APOA1 (gray) is shown opposite to H5 of its antiparallel APOA1 partner molecule (white). Circles represent the positions of APOA1 residues K133 (dark blue), G129 (black), Q132 (yellow), K206 (red), E205 (pink), K195 (cyan), and R177 (green). The dotted line shows disulfide bonds predicted to form with APOA1 mutant K133C, G129C, and Q132C in the 5/5 registry. Arrows at the end of the molecules show the continuity of the APOA1 disc helices [N terminus (octagon), H1 through H10, C terminus (black, curved line)]. B: A 2D representation of two antiparallel APOA1 monomers with H5 (green) of APOA1 (gray) opposite to H2 of its APOA1 partner molecule (white). Circles represent the same residues indicated above. The dotted line shows the disulfide bond predicted to form in APOA1 mutant K206C and E205C in a 5/2 registry. C: SDS-PAGGE analysis of WT APOA1 and Cys-mutants reconstituted into rHDL particles under reducing conditions (3 mM DTT). Colored rectangles correspond to the positions of the residues on APOA1 diagrams in A and B. D: SDS-PAGGE (nonreducing) analysis of WT APOA1 and Cys-mutant rHDL after incubation at 21°C for 48 h in the absence of a reducing agent.
Generation and characterization of Cys-mutant rHDL particles
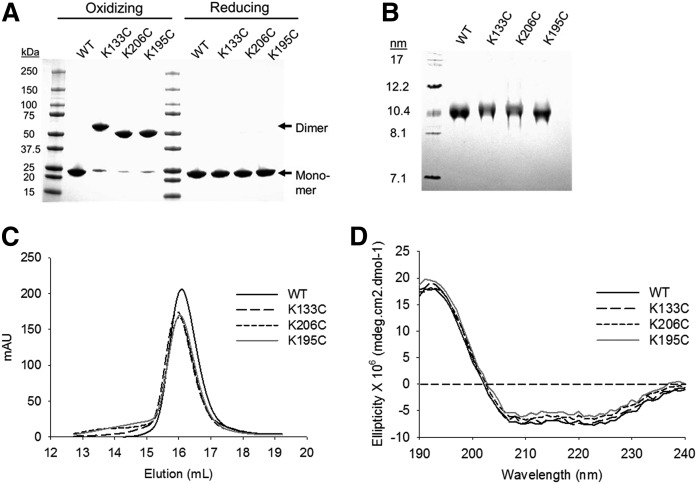
We hypothesized that these helical registries differ with respect to HDL function. To test this, we set out to generate rHDL particles containing APOA1 that was 100% locked in the 5/5, 5/2, or 5/1 registries (Figs. 1, 2). In a serendipitous finding, we noted that the Cys-mutants could be induced to form disulfide-linked dimers when APOA1 was in a lipid-free state. Therefore, we used isolated predimerized mutants to generate rHDL particles via the cholate dialysis method under oxidizing conditions (i.e., disulfide linkage maintained throughout particle formation). In the resulting particles, the Cys-mutants remained primarily dimeric, but could be fully reduced with 3 mM β-ME (Fig. 3A). All particles were of comparable composition in terms of PL to protein ratio (Table 1) and exhibited similar band patterns and hydrodynamic diameters by NDGGE (9.6 ± 0.1 nm; Fig. 3B), gel filtration (Fig. 3C), and negative stain EM (10.0 ± 0.6 nm; Table 1, supplemental Fig. S1). Furthermore, all particles exhibited similar α-helical contents by far-UV CD (Table 1, Fig. 3D).
Fig. 3.
Characterization of Cys-mutant rHDL particles. A: Lipid-free WT APOA1 and Cys-mutants were predimerized in oxidizing conditions. Dimerized protein was then purified from the residual monomer by SEC. Protein purity was analyzed by SDS-PAGGE in oxidizing and reducing (3 mM β-ME) conditions. B: Sodium cholate dialysis was used to generate rHDL particles at a molar ratio of 80:8:1 POPC:cholesterol:APOA1. Native PAGGE was used to determine the purity of the particles in oxidizing conditions. C: SEC was used to determine the size and purity of the particles. Peak intensity is shown in arbitrary units (mAU) based on the elution volume of the particles. D: Far-UV CD spectra of WT and Cys-mutant rHDL particles showing the mean ellipticity of APOA1 based on the 1 nm wavelength increases in the UV spectrum (n ≥ 3 for all characterization experiments).
TABLE 1.
Characterization of rHDL
| rHDL | POPCa | FCa | APOA1b | Particle Diameter (nm)c | Particle Diameter (nm) EMd | Helicity (%)e |
| WT | 102.1 ± 3.9 | 7.7 ± 3.8 | 1.0 | 9.6 | 10.3 ± 1.6 | 82.5 ± 2.2 |
| K133C | 92.6 ± 5.2 | 6.9 ± 2.6 | 1.0 | 9.6 | 10.5 ± 1.2 | 82.6 ± 2.0 |
| K206C | 94.7 ± 8.4 | 6.9 ± 1.8 | 1.0 | 9.5 | 10.2 ± 1.4 | 81.6 ± 3.0 |
| K195C | 94.4 ± 4.9 | 7.5 ± 1.7 | 1.0 | 9.6 | 9.1 ± 1.4 | 83.4 ± 4.1 |
rHDL particles were reconstituted at an initial molar ratio of 80:8:1 POPC:FC:APOA1. Results shown indicate mean ± SD of at least three independent rHDL preparations. Calculations represent scans of three independent samples of rHDL preparations. FC, free cholesterol.
Phospholipid and cholesterol concentrations determined as indicated in the Materials and Methods.
APOA1 concentration was determined using a Markwell Protein Lowry.
Particle size determined using NDGGE by comparison to protein standards.
Particle size determined by EM.
Percent α-helicity determined by CD.
To confirm that the Cys-mutants were in their predicted helical registries on rHDL, the particles were cross-linked with BS3-H12/D12 under oxidizing conditions. Cross-linked peptides were then analyzed by LC-MS/MS. Cross-links consistent with the 5/5 registry were present in WT and K133C rHDL, but not in K206C or K195C particles (supplemental Fig. S2). By contrast, cross-links consistent with the 5/2 orientation were present in the K206C particles, but only minimally in WT or K133C species. Some cross-links consistent with the 5/2 orientation were also identified in K195C particles, again possibly due to the close proximity of residues 206 and 195 in the sequence.
Effect of helical registry on HDL function
Cholesterol efflux.
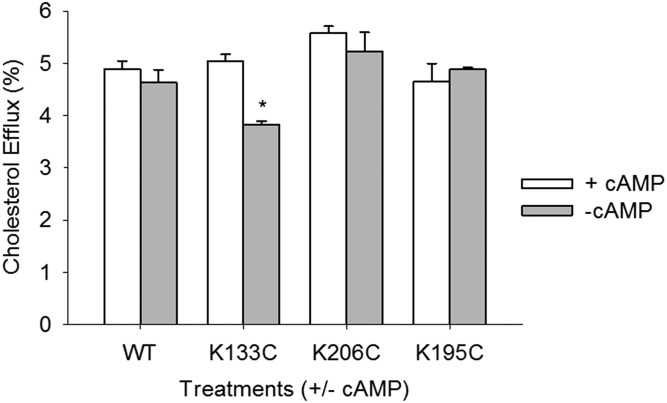
The ability of the registry-locked rHDL species to stimulate cholesterol efflux from cells was evaluated using RAW mouse macrophages treated in the presence or absence of cAMP. In the presence of cAMP, both aqueous diffusion and ABCA1-mediated mechanisms of efflux are active (43). In the absence of cAMP, only non-ABCA1-mediated cholesterol efflux occurs from RAW cells, and is the main mechanism utilized by rHDL. Figure 4 shows that, in the presence of cAMP, all registry-locked particles performed similarly to WT. Without cAMP, all particles again performed similarly with the possible exception of K133C rHDL, which was slightly decreased in this experiment for reasons that are not clear. Overall, the ability of the particles to act as acceptors of cellular cholesterol was not markedly affected by APOA1 helical registry.
Fig. 4.
Cholesterol efflux capacity of WT and Cys-mutant rHDL particles. RAW mouse macrophages were exchange labeled with [3H]free cholesterol and incubated with rHDL generated with WT or Cys-mutant APOA1. Ten micrograms per milliliter (protein) of the various rHDL particles were incubated with the cells for 6 h at 37°C. The cells were treated in the presence or absence cAMP to distinguish between ABCA1-mediated (with cAMP, white bar) and aqueous diffusion (without cAMP, gray bar) cholesterol efflux. Experiments were performed in triplicate and bars represent mean ± SD. The mean was normalized to the medium alone control condition. A two-way ANOVA and Holm-Sidak post hoc test were used to determine cholesterol efflux differences between samples and within treatments with or without cAMP (K133C + cAMP vs. K133C − cAMP; *P < 0.001, t = 6.818).
LCAT activation.
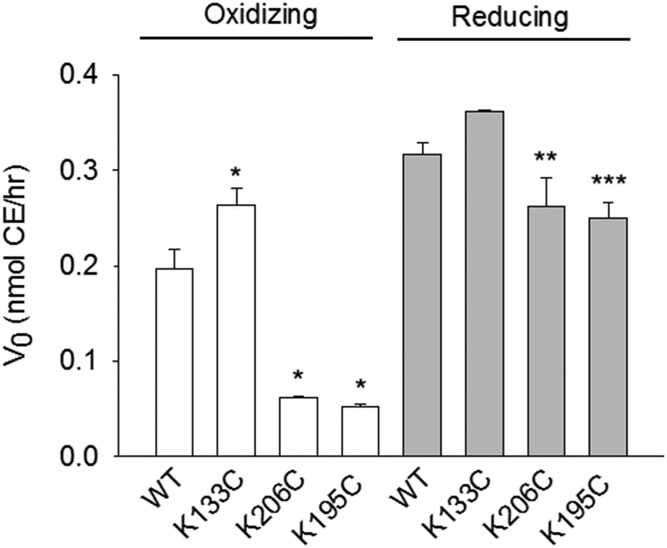
We next assessed the ability of LCAT to convert [3H]cholesterol to [3H]CE using registry-locked rHDL particles. In preliminary work, we generated substrate/velocity curves with WT rHDL from 0 to 1.4 μM (supplemental Fig. S3) and selected a substrate concentration (0.4 μM) in which the reaction rate was on the linear portion of the curve. This concentration was used for subsequent comparisons. Figure 5 and supplemental Table S1 show that WT 9.6 nm rHDL activated LCAT effectively, as widely reported. K133C rHDL stimulated LCAT activity to a slightly higher extent (P < 0.001). However, LCAT activity was clearly attenuated for K206C (P < 0.001) and K195C rHDL (P < 0.001) versus WT. When the disulfide bonds were reduced with DTT prior to LCAT addition, LCAT activation by K133C was nearly reduced to WT-like levels. [Note: the increase in activity of the WT is consistent with previous work showing that another reducing agent, β-ME, enhances LCAT activity (44, 45).] K206C and K195C rHDL recovered a significant amount of activity when the disulfide bond was reduced compared with oxidizing conditions (P < 0.001). This may indicate that the 5/2 and 5/1 registry-locked particles can revert to an LCAT-permissive registry during the course of the experiment. Moreover, this shows that APOA1 helical registry was the factor responsible for differences in LCAT activation rather than the Cys mutations themselves.
Fig. 5.
LCAT activation by WT and Cys-mutant rHDL. rHDL particles (0.4 μM) generated with [3H]free cholesterol were incubated with LCAT (0.02 μM) in oxidizing (disulfide “locked,” white bars) and reducing (“unlocked,” 10 mM β-ME, gray bars) conditions as described in the Materials and Methods. The amount of labeled CE, as separated by thin-layer chromatography, was quantitated by scintillation counting. Bars represent mean ± SD of triplicate measurements for each condition. A one-way ANOVA and Holm-Sidak post hoc test were used to determine differences in the reaction velocity (V0) of CE production between WT and Cys-mutant rHDL (K133C, K206C, K195C vs. WT; *P < 0.001, F = 189.740, DF = 3; K206C reducing vs. K206C oxidizing, **P < 0.001, t = −11.823, DF = 4; K195C reducing vs. K195C oxidizing, ***P < 0.001, t = −22.023, DF = 4).
Effect of APOA1 helical registry on LCAT binding affinity
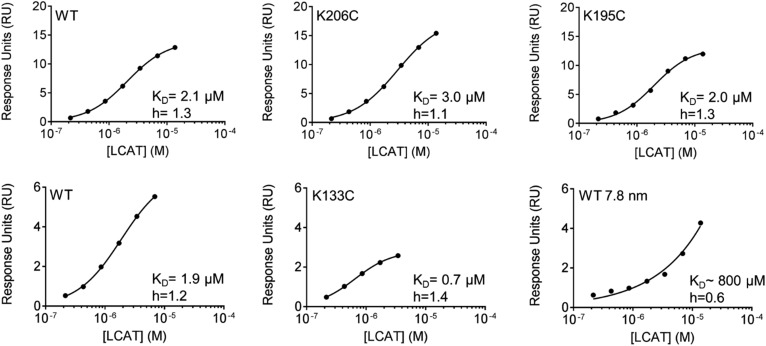
LCAT-catalyzed CE production is the result of a multistep process where LCAT must first bind to the rHDL particle and then catalyze the transfer of a fatty acid from PL to cholesterol. We investigated whether the APOA1 helical registry could affect the first step in this process, the binding of LCAT to Cys-mutant rHDL, using SPR. We measured the binding of varying concentrations of LCAT to immobilized rHDL substrates to determine equilibrium binding constants. Figure 6 shows that LCAT bound to WT rHDL with a KD of 1.9–2.1 μM, consistent with previous reports (46). K133C rHDL exhibited a similar KD of 0.7 μM. Interestingly, K206C (5/2) and K195C (5/1) also exhibited comparable KD values (3 μM and 2 μM, respectively). To demonstrate a biologically relevant difference in KD, we included a smaller (7.8 nm) rHDL particle generated with WT APOA1 that has been shown to be a poor LCAT activator (47). As is clear from the curve in Fig. 6, we observed no indication of binding saturation to the 7.8 nm particle over the concentration range studied for the 9.6 nm particles. This precluded an accurate KD measurement, but the software extrapolated a value of ∼800 μM, many orders of magnitude higher than the WT 9.6 nm particle. Additionally, Hill coefficients indicated mild positive cooperativity in all of the 9.6 nm rHDL, regardless of APOA1 registry.
Fig. 6.
LCAT affinity for WT and Cys-mutant rHDL assessed by SPR. The 4H1, a mouse anti-APOA1 monoclonal antibody recognizing amino acids 2–8 of APOA1, was immobilized on flow cells 2–4 of a CM5 sensorchip. Mouse anti-ubiquitin monoclonal antibody was immobilized on flow cell 1 to serve as a reference channel. WT and Cys-mutant rHDL particles (2.5 mM) were coupled to 4H1 in flow cells 2–4. WT APOA1 7.8 nm rHDL particles served as a negative control. Particles were bound to the chip, washed, and then LCAT was flowed over for 120 s at concentrations from 0.2 to 55 μM in duplicate, including four buffer blanks. LCAT was washed from rHDL particles for 600 s before injecting a new concentration. Equilibrium dissociation constants (KD) were determined using GraphPad Prism 7, nonlinear regression (curve fit), specific-binding with Hill slope based on reference channel, and blank subtracted response units (RU). Binding curves are plotted on a semi-log scale. Top panel: WT, K206C, and K195C were captured on sensorchip 1. Bottom panel: WT, K133C, and WT 7.8 nm were captured on sensorchip 2.
As a complementary approach, we evaluated the ability of LCAT to bind rHDL using chemical cross-linking. rHDL and LCAT were coincubated in the presence of BS3, an amine to amine homobifunctional cross-linking agent. When evaluated by NDGGE, we noted the appearance of a band that was larger than all Cys-mutant particles (supplemental Fig. S4). (Note: proteins cross-linked with amine-to-amine cross-linkers appear at a decreased size when evaluated with NDGGE, due to increased net negative charge.) The band representing the LCAT/APOA1 complex was present at a similar intensity (“excised band”) for WT and all registry-locked rHDL based on densitometry. MS analysis of the excised band identified peptides from both LCAT and APOA1 (not shown). Overall, this experiment is consistent with the SPR results, indicating that LCAT binds similarly to 9.6 nm rHDL, irrespective of APOA1 registry.
Cross-linking LCAT to rHDL
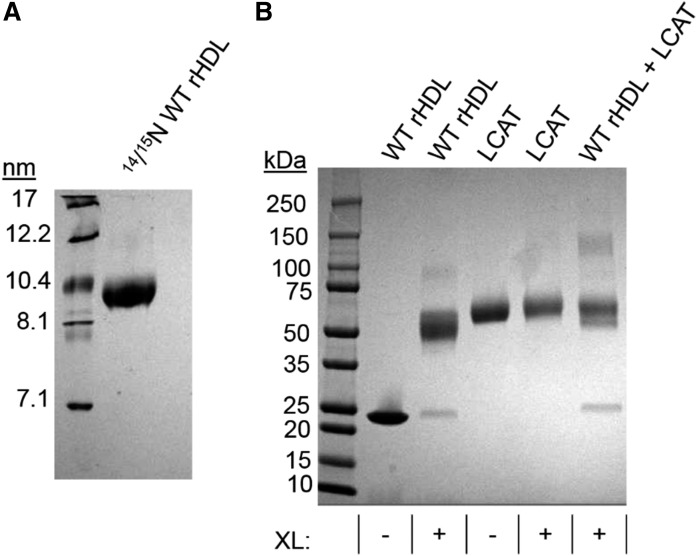
To understand the specific regions of APOA1 that interact with LCAT, we produced 9.6 nm WT rHDL particles containing a 1:1 mixture of 14N and 15N APOA1 (Fig. 7A). Particles were incubated with LCAT and cross-linked with BS3. SDS-PAGGE analysis revealed a cross-linker-dependent band that was consistent with the molecular mass of two APOA1 molecules and one LCAT molecule in complex (∼123 kDa) (Fig. 7B). We then isolated the LCAT/APOA1 complex with SEC and performed a peptide analysis by MS (Table 2). LCAT residues S108 and K240 (48) were found cross-linked to APOA1 at residues located in helices 4, 5, and 7.
Fig. 7.
Identification of LCAT interaction sites on rHDL containing WT and isotopically labeled APOA1 with chemical cross-linking. A 4:1 ratio of rHDL and LCAT was incubated with BS3, a lysine-to-lysine cross-linker, at a 100:1 molar ratio of cross-linker to APOA1 for 16 h at 4°C. A: NDGGE analysis of 1:1 14N/15N APOA1 rHDL prepared by sodium cholate dialysis. B: SDS-PAGGE analysis of rHDL incubated with LCAT, with (+) or without (−) BS3 cross-linking (XL). Cross-linked complexes were purified by SEC with a 50 mM ammonium bicarbonate mobile phase at 0.6 ml/min and prepared for MS analysis.
TABLE 2.
Identified BS3 cross-links between APOA1-containing rHDL and LCAT
| Cross-link | Identified Peptide Cross-links | Modificationa | Peptide Mass (Da)b | Number of Identificationsc | Experiment | ||
| ApoA1 | LCAT | ApoA1 | LCAT | ||||
| K182 | K240 | 178-LEALKENGGAR-188 | 239-LKEEQR-244 | XL | 2,097.5 ± 0.5 | 28 | 1,2,3 |
| K140 | K240 | 134-LHELQEKLSPLGEEMR-149 | 239-LKEEQR-244 | XL, H | 2,859.2 ± 41.1 | 15 | 1,2,3 |
| K118 | K240 | 117-QKVEPLR-123 | 239-LKEEQR-244 | XL | 1,809.0 ± 0.0 | 13 | 1,2 |
| K118 | S108 | 117-QKVEPLR-123 | 106-TYSVEYLDSSK-116 | XL | 2,298.4 ± 0.4 | 6 | 1,2 |
BS3 cross-linking capability: KK-, KS-, KN-term, cross-linked residues are in bold.
Chemical modifications: XL = 1 complete cross-link between APOA1 and LCAT (BS3 adds 138.06808 Da), H = hydrolyzed cross-link (BS3 adds 156.07864 Da).
Experimentally derived mass of cross-linked peptides.
Number of cross-links uniquely identified in at least two of three independent cross-linking experiments.
DISCUSSION
This work resulted in two major findings. First, molecules of APOA1 on HDL particles can adopt at least two (but possibly more) helical registries. Second, these registries can impact the functionality of the HDL particles, at least with respect to LCAT. Each of these aspects will be discussed in more detail below.
APOA1 helical registry in HDL
Our Cys-proximity experiments support the hypothesis proposed by Silva et al. (27) that APOA1 can adopt at least the 5/5 and 5/2 helical registries in rHDL particles. Subsequently, we also identified cross-links consistent with these registries in reconstituted spherical particles (29) and those isolated from human plasma (28). Our impression is that the 5/5 helical registry tends to dominate, accounting for the majority of the particles in a given preparation based on cross-linking and the Cys-proximity assay. A similar conclusion was reached by Li, Jones, and Segrest (49) using disulfide linkage formation as a readout for APOA1 helical registry. In our hands, the 5/2 helical registry is likely the next most common, but we cannot preclude the possibility of other registries. Indeed, other laboratories have found evidence for alternative registries (Y. He and J. Heinecke, personal communication).
What is the genesis of these registries? One possibility is that the differential registries are created at the time of particle formation and are static for the lifetime of the particle. The two orientations might arise from different conformations of lipid-free APOA1 at the time of lipid interaction. Another possibility is that these registries can interconvert in a dynamic equilibrium after the particle is fully assembled. The LCAT activity studies reported here may support this idea. Particles that were fully locked into the 5/2 orientation activated LCAT poorly, yet those same particles regained significant activity once the disulfide bond was reduced, suggesting that some may have reverted to the permissive 5/5 orientation. We can envision two general ways that APOA1 molecules could rotate with respect to each other on an HDL particle. First, the entire length of both molecules could simply slide past the other. This seems unlikely given the high number of salt bridges predicted to occur between the two molecules of APOA1 on a disc (19), particularly because the strength of these interactions should be increased at the lipid/water interface (50). A more likely scenario is that one molecule “walks” along the other like an inchworm. Part of the protein may disassociate from the other APOA1 molecule and this localized region could propagate by breaking interactions in front of it and forming the new shifted interactions behind it. The energetics driving this are not clear. Segrest et al. (19) performed an analysis in which different APOA1 registries were compared on the basis of salt-bridge formation. The 5/5 orientation, also the same orientation observed in the Borhani crystal structure of truncated APOA1 (17), exhibited the lowest weighted salt bridge score (about −20), indicating the most possible salt bridges. Other orientations, such as 5/4, exhibited comparable, though slightly less favorable, scores. Of note, the 5/2 orientation was not remarkable with a score of about −10. Thus, the 5/2 registry may be stabilized by other forces, such as hydrophobic interactions between the proteins.
Functional impact of registry shifts
The concept of APOA1 molecules shifting registries has significant implications for the functionality and metabolism of APOA1 and HDL. Given that APOA1 forms extended antiparallel belts around particles, it is difficult to imagine that most HDL accessory proteins dock only to a single copy of APOA1. For example, LCAT is a relatively large enzyme (67 kDa) with respect to an APOA1 double belt (∼56 kDa) or even an entire rHDL disc (∼250 kDa including the lipid). Our work here shows that LCAT cross-links to residues in helices 4, 5, and 7, consistent with the notion that it interacts with a hybrid epitope created by both APOA1 molecules on the particle edge when in the 5/5 helical registry.
To investigate this further, we surveyed the literature for instances of APOA1 mutations that did not affect cholesterol efflux (and thereby likely did not cause global perturbations in particle structure), but did impact LCAT activation (supplemental Table S2, supplemental Fig. S5). Natural and engineered point mutations in APOA1 that fit these criteria were concentrated in helices 4, 6, and 7, consistent with a hybrid LCAT interaction site. Unfortunately, several mutations in helix 3 have not been assayed for LCAT activation or ability to stimulate cholesterol efflux (51). Hazen and colleagues reported hydrogen-deuterium exchange experiments showing that the presence of LCAT alters hydrogen exchange across APOA1 residues 159–170 (helices 6/7, the “solar flare”) (15). Helices 3/4 on the opposing APOA1 molecule were not evaluated due to lack of sequence coverage in that region (S. L. Hazen, personal communication). However, monoclonal antibodies generated against the 3/4 region of APOA1 are known to decrease LCAT activation (52). One antibody showed decreased LCAT activation despite overlapping with two regions of APOA1 (one in the 3/4 region, one unknown), and the authors suggested that this antibody may bind to a discontinuous epitope of APOA1. We propose that this unknown region could encompass residues 159–170.
Other HDL proteins have been shown to interact in a similar region of APOA1. Haptoglobin, an acute phase protein, interacts with APOA1 in the helix 6/7 region (53); however, the 3/4 region was not considered for analysis. Additionally, APOA2, the second most abundant protein on HDL, was shown to interact with APOA1 in the 3/4 region, as well as in helices 7–9 (54). Considering that haptoglobin and APOA2 are known to inhibit LCAT activation, steric hindrance of the LCAT activation site on APOA1 may be the cause (8). The 6/7 region of APOA1 is more solvent exposed compared with the 3/4 region, suggesting that LCAT interfacial activation may be mediated by the 6/7 region, and the more hydrophobic 3/4 region may play a role in lipid substrate shuttling (15, 55, 56). For instance, aside from LCAT, the 3/4 region of APOA1 has been shown to be important for ABCG1- (57) and SR-B1-mediated (58) cholesterol mobilization. The conformation of the 3/4 region changes as the size and cholesterol content of HDL increases and inhibits cholesterol mobilization (59). Therefore, it is possible that, as LCAT esterifies cholesterol and particles become spherical, LCAT activation could be modulated by a conformational change in the 3/4 region that could also affect the 6/7 region of APOA1 due to weakened protein interactions (60). All of this evidence predicts that disruption of this intermolecular epitope, such as by altering the registry, should impact LCAT activation.
Indeed, this proved to be the case. rHDL that contained APOA1 molecules locked into the 5/2 registry were impaired in LCAT activation compared with WT and the 5/5 orientation. This is despite the fact that helices 3/4 and 6/7, in terms of sequence, were identical in both cases. The only difference was the shifted APOA1 registry, strongly implying that it is important to the overall LCAT reaction. Interestingly, APOA1 registry did not affect LCAT binding to the particles. This may indicate that initial LCAT binding is mediated primarily by the lipid interface, as suggested by Jonas and colleagues (61), or that only one strand of APOA1 is critical for the initial binding event, perhaps the helix 6/7 region, as recent work by Gu et al. (46) points to the solar flare region as responsible for LCAT binding. However, our data show that APOA1 registry plays a role in subsequent reaction steps, such as the hydrolysis of PL or the esterification of cholesterol. Alternatively, APOA1 registry might impact either the extraction of lipid substrates from the particle lipid bilayer or the movement of lipid products from the active site of the enzyme back into the particle core. Further mechanistic work is needed to delineate these details.
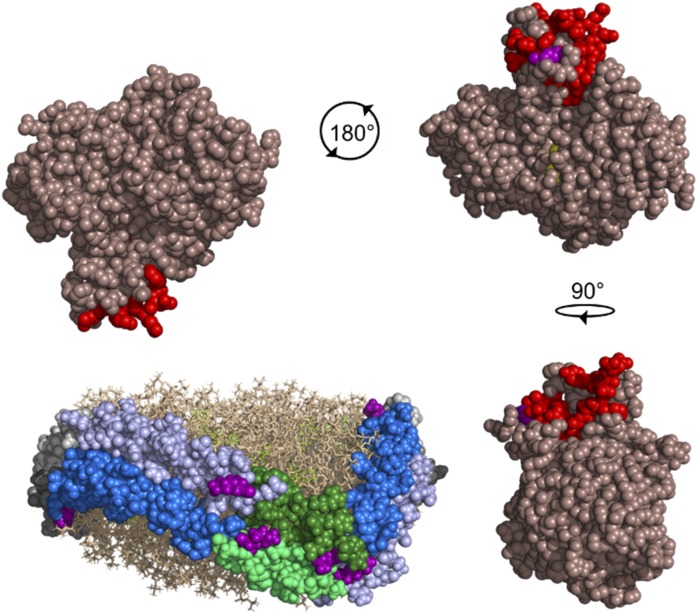
It is of interest to consider the consequences of the reciprocating nature of APOA1 in light of the concept of hybrid intermolecular epitopes. Because of the antiparallel orientation of two APOA1 molecules, one can see that two LCAT interaction epitopes are created per particle, each separated by the central helix 5 sequence (Fig. 8). It is compelling that these sites are in the opposite orientation with respect to the “top” and “bottom” PL face. This may indicate that LCAT can interact in two orientations, perhaps to access PL on both faces of the particle. In the case of more complicated particles in which APOA1 is arranged in a trefoil or quatrefoil arrangement (29), there would be additional LCAT interaction sites (equaling the number of APOA1 molecules present). These might allow LCAT to access each lune of PL in the particle, thus consuming the PL substrate in a uniform manner across the particle. Hill coefficients >1 for LCAT binding to rHDL may suggest that binding of additional LCAT molecules is facilitated after one site is occupied. Additionally, we have shown that other apolipoproteins in the A class have similar symmetrical and reciprocating structures. For example, our crystal structure of a human APOA4 dimer (32) bears a remarkable resemblance to the crystal structure of the human APOA1 dimer (62) and both are expected to maintain this general architecture in lipidated particles. Thus, we predict that APOA4, also known to activate LCAT (63), may feature LCAT interaction domains that are situated symmetrically about a central shared helical domain. This is despite the fact that there is no clearly conserved sequence that is known to activate LCAT across different proteins (60). Perhaps specific 3D structural properties or specific charge distributions are required for LCAT to efficiently complete its reaction cycle. It is also interesting to note that Bibow et al. (26) have described a zipper-like pattern of cation-π interactions between helices 6 and 4 of APOA1 when oriented in a right to right helical orientation.
Fig. 8.
Postulated model of LCAT interacting with the 6/4 region of APOA1 molecules in the 5/5 helical registry. The Piper et al. (48) crystal structure of LCAT (dark salmon) is shown with the lid region highlighted in red and the catalytic triad in yellow. LCAT residue S108 (purple) involved in a cross-link to APOA1 is visible near the lid region. The rHDL particle is depicted with two anti-parallel molecules of APOA1 (molecule A, light gray; molecule B, dark gray) with helix (H)6 (blue), H4 (light blue), and H5 (molecule A, dark green; molecule B, light green) highlighted on both molecules. Phospholipids (tan) and cholesterol (yellow-green) are shown. Residues (purple) cross-linked in helices 4, 5, and 7 of APOA1 to LCAT are shown. In a 5/5 helical registry, there are two locations where helices 6 and 4 are opposing, meaning two possible sites for LCAT activation to occur. LCAT is depicted rotating 180° and 90° from the starting location to interact with H6 on molecule B and H4 on molecule A, and H6 on molecule A and H4 on molecule B, respectively. The figure was generated with PyMOL, version 2.0, Schrodinger, LLC.
Our results raise the possibility that the helical registry of APOA1 on HDL particles could be a thumbwheel-like mechanism by which LCAT activity is regulated. Particles in the 5/5 orientation may facilitate LCAT activity (Fig. 8) until some stimulus promotes an APOA1 shift to the nonpermissive 5/2 orientation. Factors that might promote APOA1 registry shifting could include particle lipid composition and diameter, the presence or absence of other modifying proteins, such as APOA2, or a combination of these factors. It is even possible that the 5/2 orientation may facilitate interactions with other HDL-modifying factors that are not active in the 5/5. We are working to develop ways to quantitate APOA1 registry so that the impact on HDL composition can be assessed, as well as the effect on other enzymatic activities.
In summary, we have demonstrated that APOA1 molecules can exist in at least two different molecular registries on HDL particles. APOA1 helical registry did not affect the particles’ ability to promote cholesterol efflux or the ability of LCAT to bind to the particles. However, the ability of LCAT to complete the cholesterol esterification reaction was highly sensitive to APOA1 registry. Further work is needed to determine whether APOA1 registries are important in vivo and whether they can mediate the docking and activity of different sets of HDL remodeling factors.
Supplementary Material
Acknowledgments
The authors acknowledge the Plant-Microbe Genomics Facility at Ohio State University in Columbus, OH for assisting with the SPR studies.
Footnotes
Abbreviations:
- β-ME
- β-mercaptoethanol
- BS3
- bis(sulfosuccinimidyl)suberate
- CD
- circular dichroism
- CE
- cholesteryl ester
- Cys
- cysteine
- EM
- electron microscopy
- His
- histidine
- NDGGE
- nondenaturing gradient gel electrophoresis
- PL
- phospholipid
- rHDL
- reconstituted high density lipoprotein
- SDS-PAGGE
- sodium dodecyl sulfate-polyacrylamide gradient gel electrophoresis
- SEC
- size exclusion chromatography
- SPR
- surface plasmon resonance
- STB
- standard Tris buffer
- TEV
- tobacco etch virus
- 5/2
- helix 5 of one APOA1 molecule opposing helix 2 of another
- 5/5
- helix 5 of one APOA1 molecule opposing helix 5 of the other
This work was supported by American Heart Association Grant 15PRE25220021 (predoctoral fellowship to A.L.C.) and National Institutes of Health Grants R01 HL127649 (to W.S.D.) and P01 HL12803 (to W.S.D. and J.P.S.). The mass spectrometry data was acquired in the University of Cincinnati Proteomics Laboratory under the direction of Ken Greis on a mass spectrometer funded in part through National Institutes of Health Grant RR027015 (S10 shared instrumentation grant; Gries). Electron microscopy image collection was performed in part through the use of the VUCell Imaging Shared Resource, which is supported by National Institutes of Health Grants CA68485, DK20593, DK58404, DK59637, and EY08126. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no financial conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Gordon T., Castelli W. P., Hjortland M. C., Kannel W. B., and Dawber T. R.. 1977. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 62: 707–714. [DOI] [PubMed] [Google Scholar]
- 2.Shah A. S., Tan L., Long J. L., and Davidson W. S.. 2013. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 54: 2575–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu X., Trigatti B., Xu S., Acton S., Babitt J., and Krieger M.. 1998. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J. Biol. Chem. 273: 26338–26348. [DOI] [PubMed] [Google Scholar]
- 4.Puranik R., Bao S., Nobecourt E., Nicholls S. J., Dusting G. J., Barter P. J., Celermajer D. S., and Rye K. A.. 2008. Low dose apolipoprotein A-I rescues carotid arteries from inflammation in vivo. Atherosclerosis. 196: 240–247. [DOI] [PubMed] [Google Scholar]
- 5.Navab M., Imes S. S., Hama S. Y., Hough G. P., Ross L. A., Bork R. W., Valente A. J., Berliner J. A., Drinkwater D. C., and Laks H.. 1991. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J. Clin. Invest. 88: 2039–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soutar A. K., Garner C. W., Baker H. N., Sparrow J. T., Jackson R. L., Gotto A. M., and Smith L. C.. 1975. Effect of the human plasma apolipoproteins and phosphatidylcholine acyl donor on the activity of lecithin: cholesterol acyltransferase. Biochemistry. 14: 3057–3064. [DOI] [PubMed] [Google Scholar]
- 7.Jonas A. 1991. Lecithin-cholesterol acyltransferase in the metabolism of high-density lipoproteins. Biochim. Biophys. Acta. 1084: 205–220. [DOI] [PubMed] [Google Scholar]
- 8.Jonas A., Sweeny S. A., and Herbert P. N.. 1984. Discoidal complexes of A and C apolipoproteins with lipids and their reactions with lecithin: cholesterol acyltransferase. J. Biol. Chem. 259: 6369–6375. [PubMed] [Google Scholar]
- 9.Zannis V. I., Chroni A., and Krieger M.. 2006. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. (Berl.). 84: 276–294. [DOI] [PubMed] [Google Scholar]
- 10.Fotakis P., Kuivenhoven J. A., Dafnis E., Kardassis D., and Zannis V. I.. 2015. The effect of natural LCAT mutations on the biogenesis of HDL. Biochemistry. 54: 3348–3359. [DOI] [PubMed] [Google Scholar]
- 11.Chroni A., Duka A., Kan H. Y., Liu T., and Zannis V. I.. 2005. Point mutations in apolipoprotein A-I mimic the phenotype observed in patients with classical lecithin:cholesterol acyltransferase deficiency. Biochemistry. 44: 14353–14366. [DOI] [PubMed] [Google Scholar]
- 12.Sviridov D., Hoang A., Sawyer W. H., and Fidge N. H.. 2000. Identification of a sequence of apolipoprotein A-I associated with the activation of lecithin:cholesterol acyltransferase. J. Biol. Chem. 275: 19707–19712. [DOI] [PubMed] [Google Scholar]
- 13.Cho K. H., Durbin D. M., and Jonas A.. 2001. Role of individual amino acids of apolipoprotein A-I in the activation of lecithin:cholesterol acyltransferase and in HDL rearrangements. J. Lipid Res. 42: 379–389. [PubMed] [Google Scholar]
- 14.Alexander E. T., Bhat S., Thomas M. J., Weinberg R. B., Cook V. R., Bharadwaj M. S., and Sorci-Thomas M.. 2005. Apolipoprotein A-I helix 6 negatively charged residues attenuate lecithin-cholesterol acyltransferase (LCAT) reactivity. Biochemistry. 44: 5409–5419. [DOI] [PubMed] [Google Scholar]
- 15.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M. 3rd, Smith J. D., Gogonea V., and Hazen S. L.. 2007. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14: 861–868. [DOI] [PubMed] [Google Scholar]
- 16.Melchior J. T., Walker R. G., Cooke A. L., Morris J., Castleberry M., Thompson T. B., Jones M. K., Song H. D., Rye K. A., Oda M. N., et al. . 2017. A consensus model of human apolipoprotein A-I in its monomeric and lipid-free state. Nat. Struct. Mol. Biol. 24: 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borhani D. W., Rogers D. P., Engler J. A., and Brouillette C. G.. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonas A., Wald J. H., Toohill K. L., Krul E. S., and Kezdy K. E.. 1990. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J. Biol. Chem. 265: 22123–22129. [PubMed] [Google Scholar]
- 19.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., and Harvey S. C.. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758. [DOI] [PubMed] [Google Scholar]
- 20.Panagotopulos S. E., Horace E. M., Maiorano J. N., and Davidson W. S.. 2001. Apolipoprotein A-I adopts a belt-like orientation in reconstituted high density lipoproteins. J. Biol. Chem. 276: 42965–42970. [DOI] [PubMed] [Google Scholar]
- 21.Koppaka V., Silvestro L., Engler J. A., Brouillette C. G., and Axelsen P. H.. 1999. The structure of human lipoprotein A-I. Evidence for the “belt” model. J. Biol. Chem. 274: 14541–14544. [DOI] [PubMed] [Google Scholar]
- 22.Segrest J. P., Harvey S. C., and Zannis V.. 2000. Detailed molecular model of apolipoprotein A-I on the surface of high-density lipoproteins and its functional implications. Trends Cardiovasc. Med. 10: 246–252. [DOI] [PubMed] [Google Scholar]
- 23.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., and Thomas M. J.. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46: 7811–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., and Thomas M. J.. 2005. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J. Biol. Chem. 280: 33015–33025. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., et al. . 2009. Double superhelix model of high density lipoprotein. J. Biol. Chem. 284: 36605–36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bibow S., Polyhach Y., Eichmann C., Chi C. N., Kowal J., Albiez S., McLeod R. A., Stahlberg H., Jeschke G., Guntert P., et al. . 2017. Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat. Struct. Mol. Biol. 24: 187–193. [DOI] [PubMed] [Google Scholar]
- 27.Silva R. A., Hilliard G. M., Li L., Segrest J. P., and Davidson W. S.. 2005. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 44: 8600–8607. [DOI] [PubMed] [Google Scholar]
- 28.Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., and Davidson W. S.. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., and Davidson W. S.. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Lyles D. S., Thomas M. J., Pan W., and Sorci-Thomas M. G.. 2000. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J. Biol. Chem. 275: 37048–37054. [DOI] [PubMed] [Google Scholar]
- 31.Tubb M. R., Smith L. E., and Davidson W. S.. 2009. Purification of recombinant apolipoproteins A-I and A-IV and efficient affinity tag cleavage by tobacco etch virus protease. J. Lipid Res. 50: 1497–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker R. G., Deng X., Melchior J. T., Morris J., Tso P., Jones M. K., Segrest J. P., Thompson T. B., and Davidson W. S.. 2014. The structure of human apolipoprotein A-IV as revealed by stable isotope-assisted cross-linking, molecular dynamics, and small angle x-ray scattering. J. Biol. Chem. 289: 5596–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonomo E. A., and Swaney J. B.. 1988. A rapid method for the synthesis of protein-lipid complexes using adsorption chromatography. J. Lipid Res. 29: 380–384. [PubMed] [Google Scholar]
- 34.Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J.. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- 35.Sreerama N., and Woody R. W.. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287: 252–260. [DOI] [PubMed] [Google Scholar]
- 36.Whitmore L., and Wallace B. A.. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32: W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith L. E., and Davidson W. S.. 2010. The role of hydrophobic and negatively charged surface patches of lipid-free apolipoprotein A-I in lipid binding and ABCA1-mediated cholesterol efflux. Biochim. Biophys. Acta. 1801: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rousset X., Vaisman B., Auerbach B., Krause B. R., Homan R., Stonik J., Csako G., Shamburek R., and Remaley A. T.. 2010. Effect of recombinant human lecithin cholesterol acyltransferase infusion on lipoprotein metabolism in mice. J. Pharmacol. Exp. Ther. 335: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leblond L., and Marcel Y. L.. 1991. The amphipathic alpha-helical repeats of apolipoprotein A-I are responsible for binding of high density lipoproteins to HepG2 cells. J. Biol. Chem. 266: 6058–6067. [PubMed] [Google Scholar]
- 40.Davidson W. S., and Hilliard G. M.. 2003. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass specrometry study. J. Biol. Chem. 278: 27199–27207. [DOI] [PubMed] [Google Scholar]
- 41.Melchior J. T., Walker R. G., Morris J., Jones M. K., Segrest J. P., Lima D. B., Carvalho P. C., Gozzo F. C., Castleberry M., Thompson T. B., et al. . 2016. An evaluation of the crystal structure of C-terminal truncated apolipoprotein A-I in solution reveals structural dynamics related to lipid binding. J. Biol. Chem. 291: 5439–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorci-Thomas M., Kearns M. W., and Lee J. P.. 1993. Apolipoprotein A-I domains involved in lecithin-cholesterol acyltransferase activation. Structure:function relationships. J. Biol. Chem. 268: 21403–21409. [PubMed] [Google Scholar]
- 43.Oram J. F., Lawn R. M., Garvin M. R., and Wade D. P.. 2000. ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J. Biol. Chem. 275: 34508–34511. [DOI] [PubMed] [Google Scholar]
- 44.Jauhiainen M., and Dolphin P. J.. 1986. Human plasma lecithin-cholesterol acyltransferase. An elucidation of the catalytic mechanism. J. Biol. Chem. 261: 7032–7043. [PubMed] [Google Scholar]
- 45.Verdery R. B., and Gatt S.. 1981. Assay for lecithin: cholesterol acyltransferase. Methods Enzymol. 72: 375–384. [DOI] [PubMed] [Google Scholar]
- 46.Gu X., Wu Z., Huang Y., Wagner M. A., Baleanu-Gogonea C., Mehl R. A., Buffa J. A., DiDonato A. J., Hazen L. B., Fox P. L., et al. . 2016. A systematic investigation of structure/function requirements for the apolipoprotein A-I/lecithin cholesterol acyltransferase interaction loop of high-density lipoprotein. J. Biol. Chem. 291: 6386–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cavigiolio G., Geier E. G., Shao B., Heinecke J. W., and Oda M. N.. 2010. Exchange of apolipoprotein A-I between lipid-associated and lipid-free states: a potential target for oxidative generation of dysfunctional high density lipoproteins. J. Biol. Chem. 285: 18847–18857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piper D. E., Romanow W. G., Gunawardane R. N., Fordstrom P., Masterman S., Pan O., Thibault S. T., Zhang R., Meininger D., Schwarz M., et al. . 2015. The high-resolution crystal structure of human LCAT. J. Lipid Res. 56: 1711–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L., Li S., Jones M. K., and Segrest J. P.. 2012. Rotational and hinge dynamics of discoidal high density lipoproteins probed by interchain disulfide bond formation. Biochim. Biophys. Acta. 1821: 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davidson W. S., Hazlett T., Mantulin W. W., and Jonas A.. 1996. The role of apolipoprotein AI domains in lipid binding. Proc. Natl. Acad. Sci. USA. 93: 13605–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogonea V. 2016. Structural insights into high density lipoprotein: old models and new facts. Front. Pharmacol. 6: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banka C. L., Bonnet D. J., Black A. S., Smith R. S., and Curtiss L. K.. 1991. Localization of an apolipoprotein A-I epitope critical for activation of lecithin-cholesterol acyltransferase. J. Biol. Chem. 266: 23886–23892. [PubMed] [Google Scholar]
- 53.Spagnuolo M. S., Cigliano L., D’Andrea L. D., Pedone C., and Abrescia P.. 2005. Assignment of the binding site for haptoglobin on apolipoprotein A-I. J. Biol. Chem. 280: 1193–1198. [DOI] [PubMed] [Google Scholar]
- 54.Gauthamadasa K., Vaitinadin N. S., Dressman J. L., Macha S., Homan R., Greis K. D., and Silva R. A.. 2012. Apolipoprotein A-II-mediated conformational changes of apolipoprotein A-I in discoidal high density lipoproteins. J. Biol. Chem. 287: 7615–7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao B., Bergt C., Fu X., Green P., Voss J. C., Oda M. N., Oram J. F., and Heinecke J. W.. 2005. Tyrosine 192 in apolipoprotein A-I is the major site of nitration and chlorination by myeloperoxidase, but only chlorination markedly impairs ABCA1-dependent cholesterol transport. J. Biol. Chem. 280: 5983–5993. [DOI] [PubMed] [Google Scholar]
- 56.Maiorano J. N., and Davidson W. S.. 2000. The orientation of helix 4 in apolipoprotein A-I-containing reconstituted high density lipoproteins. J. Biol. Chem. 275: 17374–17380. [DOI] [PubMed] [Google Scholar]
- 57.Daniil G., Zannis V. I., and Chroni A.. 2013. Effect of apoA-I mutations in the capacity of reconstituted HDL to promote ABCG1-mediated cholesterol efflux. PLoS One. 8: e67993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chroni A., Kan H. Y., Kypreos K. E., Gorshkova I. N., Shkodrani A., and Zannis V. I.. 2004. Substitutions of glutamate 110 and 111 in the middle helix 4 of human apolipoprotein A-I (apoA-I) by alanine affect the structure and in vitro functions of apoA-I and induce severe hypertriglyceridemia in apoA-I-deficient mice. Biochemistry. 43: 10442–10457. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez M. C., Toledo J. D., Tricerri M. A., and Garda H. A.. 2008. The central type Y amphipathic alpha-helices of apolipoprotein AI are involved in the mobilization of intracellular cholesterol depots. Arch. Biochem. Biophys. 473: 34–41. [DOI] [PubMed] [Google Scholar]
- 60.Bashtovyy D., Jones M. K., Anantharamaiah G. M., and Segrest J. P.. 2011. Sequence conservation of apolipoprotein A-I affords novel insights into HDL structure-function. J. Lipid Res. 52: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kosek A. B., Durbin D., and Jonas A.. 1999. Binding affinity and reactivity of lecithin cholesterol acyltransferase with native lipoproteins. Biochem. Biophys. Res. Commun. 258: 548–551. [DOI] [PubMed] [Google Scholar]
- 62.Mei X., and Atkinson D.. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emmanuel F., Steinmetz A., Rosseneu M., Brasseur R., Gosselet N., Attenot F., Cuine S., Seguret S., Latta M., and Fruchart J. C.. 1994. Identification of specific amphipathic alpha-helical sequence of human apolipoprotein A-IV involved in lecithin:cholesterol acyltransferase activation. J. Biol. Chem. 269: 29883–29890. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.