Abstract
BACKGROUND:
Interferon- (IFN-) is an adjuvant to chemotherapy and radiotherapy for hepatocellular carcinoma (HCC), but some HCC patients do not respond to treatment with IFN-.
METHODS:
We performed loss-of-function and gain-of-function experiments to examine the role of ISG15 in the IFN- sensitivity of LH86, HLCZ01, SMMC7721, and Huh7 cell lines and tumor samples.
RESULTS:
The overexpression of ISG15 reduced apoptosis in Huh7 and LH86 cells in the presence of IFN-, whereas the shRNA-mediated knock down of ISG15 expression increased apoptosis in both Huh7 and LH86 cells. We identified a putative miR-370 target site in the 3’-UTR in the ISG15 mRNA, and the level of miR-370 expression in HCC cell lines reflected the level of IFN--induced apoptosis exhibited by each. Both HCC cell lines and tumor samples had significantly lower levels of miR-370 than the control cells and tissues ( 0.05). The overexpression of miR-370 in IFN--treated LH86 and Huh7 cells increased apoptosis and reduced the volume of LH86- and Huh7-derived xenograft tumors in mice treated with IFN- compared with the control tumors.
CONCLUSIONS:
Our findings suggest that miR-370 functions as an HCC tumor suppressor and regulator of IFN- sensitivity and that miR-370 might be a useful prognostic marker for HCC patients.
Keywords: Hepatocellular carcinoma cells, interferon-α, ISG15, miR-370, prognostic marker
1. Introduction
Hepatocellular carcinoma (HCC) ranks as the second and sixth leading world-wide cause of cancer-related death in men and women, respectively [1]. About 80% to 90% of HCC patients have advanced disease at the time of diagnosis, and are thus ineligible for surgical intervention [2]. Therefore, the development of effective chemotherapy strategies for HCC is a critical research objective. Studies of melanoma and cancer of the breast, stomach, pancreas and bladder have reported the beneficial effects of using IFN- as an adjuvant to chemotherapy and radiation treatments following tumor resection [3, 4, 5, 6], and multiple cell culture, animal and clinical studies have shown that the progression and recurrence of HCC were inhibited by treatment with IFN- [7, 8].
The majority of HCC cases are chronically infected with the hepatitis B virus (HBV) or hepatitis C virus (HCV) [9]. Although the induction of microRNA (miRNA) expression by type I IFNs contributes to the regulation of cellular antiviral defenses [10], altered miRNA expression has also been reported in various types of cancer in humans [11]. One such miRNA is miR-370, which has been shown to suppress HBV replication [12]. Aberrant miR-370 expression has been reported in esophageal squamous cell carcinoma [13], laryngeal squamous cell carcinoma [14], gastric carcinoma [15], and prostate cancer [16], as well as in cholangiocarcinoma cell lines [17, 18]. Although miR-370 serves as a tumor suppressor in malignant cholangiocytes [19] and laryngeal carcinoma cells [14], the overexpression of miR-370 contributes to the progression of gastric carcinoma [15], prostate cancer [16], and acute myeloid leukemia [20]. The biological mechanisms through which miR-370 is involved in tumorigenesis in this wide range of tissue types remains unclear.
The expression of IFN-stimulated gene 15 (ISG15) and ISG15-mediated conjugation have been implicated in malignancies of the breast [21], prostate [22], bladder cancer [23], oral cavity [24], esophagus [25], stomach [26], pancreas [27], colon [28], and liver [29], but how ISG15 influences tumorigenesis is unclear. Although ISG15, a ubiquitin-like protein, forms conjugates with various cellular proteins via a pathway similar to that used by ubiquitin, conjugation by ISG15 does not induce proteasome-dependent degradation of the conjugated protein [30, 31], and the biological consequences of ISG15 conjugation are unknown. The expression of ISG15 is induced by IFN- in HCV-infected hepatoma cells [32]. Appropriately regulated ISG15 expression is associated with apoptosis in various cell systems, whereas the perturbation of ISG15 regulation is correlated by cell proliferation and migration [33].
In our previous study, we found that ISG15 is a novel prognostic biomarker for HCC in patients with chronic HBV infection [34]. In our current study, we performed ISG15 loss-of-function and gain-of-function experiments to examine its role in the sensitivity of various HCC cell lines to treatment with IFN-. We also investigated whether miRNA might target the 3’ untranslated region (UTR) of ISG15, and examined whether the function of ISG15 in IFN-induced apoptosis in HCC cells was influenced by the candidate miRNA. Our results suggested that miR-370 mediates the role of ISG15 in the antitumor effects of IFN- in HCC cells.
2. Materials and methods
2.1. Tissues, cell lines and antibodies
The Hunan Provincial Cancer Hospital Review Board approved the protocol for the analysis of HCC tumor and noncancerous liver tissue specimens. The HCC tumor tissues and adjacent noncancerous tissue samples were collected at the Hunan Provincial Tumor Hospital (Changsha, China). Informed written consent was obtained from all patients prior to collection. The human HCC cell lines, HLCZ01, LH86, LO2, Huh7 and SMMC7721 were obtained from the Translational Medicine Research Center at Hunan University, and were grown in Dulbecco Modified Eagle Medium (DMEM, Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum at a temperature of 37C in an atmosphere of 5% CO. Recombinant human IFN- was obtained from Kexing Biotech (Beijing, China) and rabbit anti-poly (ADP-ribose) polymerase (PARP) polyclonal antibodies were purchased from Cell Signaling Technologies (Danvers, MA, USA). The rabbit anti-ISG12a polyclonal antibodies, rabbit anti-ISG15 polyclonal antibodies, and mouse anti--actin monoclonal antibody were purchased from Abcam (ab171919; Cambridge, UK), Santa Cruz Biotechnology (sc-166755; Dallas, TX, USA), and Sigma-Aldrich (AC-74; St. Louis, MO, USA), respectively.
2.2. Plasmids, oligonucleotides and reagents
Transfections for the knock down of ISG15 were performed using the siPORT NeoFx reagent (ThermoFisher, Waltham, MA, USA). The anti-ISG15 siRNA and randomized negative control RNA oligonucleotide were purchased from Santa Cruz Biotechnology. The pre-miR miRNA precursor molecule and anti-miR miRNA inhibitor for miRNA-370 (henceforth referred to as miR-370 and anti-miR-370, respectively) and the corresponding negative control RNAs were purchased from Ambion (ThermoFisher). The pFLAG-ISG15 plasmid was constructed by inserting the ISG15 cDNA into the multiple cloning site of the pFLAG-CMV-2 plasmid (Sigma-Aldrich), which allowed the expression of FLAG-tagged ISG15 protein in transfected cells. For siRNA knock down of ISG15 expression, the pSilencer-shISG15 plasmid was constructed based on the pSilencer system (ThermoFisher), which expressed a 21-nt short hairpin RNA with a 9-nt loop. The ISG15 shRNA-coding sequences used to construct pSilencer-ISG15 were as follows: 5’-GATCCCCGCACCTACGAGGTACGGCTTTCAAGA GAAGCCGTACCTCGTAGGTGCTTTTTA-3’ (sense); 5’-AGCTTAAAAAGCACCTACGAGGTACGGCTTCTCTTGAAAGCCGTACCTCGTAGGTGCGGG-3’ (antisense).
2.3. Cell viability assay
We examined the IFN- sensitivity of LH86, HLCZ01, SMMC7721 and Huh7 cells. Ninety-six well plates were seeded with approximately 8 10 cells/per well in 100 L of medium for 24 h at 37C. After aspirating the medium, 100 L of fresh medium containing 125, 250, 500, 1,000, 2,000 or 4,000 IU/mL IFN- was added. After incubating the cells for an additional 24, 48 or 72 h, 20 mL of 5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to each well. After a 4-h incubation, the medium with MTT was aspirated, and 100 L of DMSO was added to each well to dissolve insoluble formazan, which was measured at 570 nm in a plate reader.
2.4. Western blotting
Cells were washed twice with phosphate-buffered saline (PBS), and lysed in buffer containing 137 mM NaCl, 2 mM EDTA, 20 mM Tris-HCl (pH 7.4), 10% glycerin, 1% Triton X-100 and a protein inhibitor cocktail (Roche Diagnostics) for 30 min on ice. The lysate was centrifuged at 14,000 g for 20 min at 4C, and the supernatants were collected. Samples were subjected to SDS-PAGE in a 10% acrylamide gel and the resolved protein bands were transferred electrophoretically onto a nitrocellulose membrane. The membranes were blocked for 1.5 h at room temperature in Tris-buffered saline (TBS) containing 5% nonfat dry milk and 0.1% Tween-20, after which the membranes were probed with primary antibodies at 4C overnight. After washing in TBS, the membranes were incubated in TBS containing horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Sigma-Aldrich). Bound primary antibody was visualized using an enhanced chemiluminescence (ECL) detection kit (GE Healthcare, Chicago, IL, USA), and the membranes were scanned using an LAS-4000 luminescence imager (Fujifilm, Tokyo, Japan). After stripping the membranes in 20 mM glycine-HCl (pH 2.3), protein loading normalization was performed by probing with anti--actin antibody, incubation with HRP-anti-mouse antibody and visualized using ECL.
2.5. Flow cytometry
Apoptosis in the LH86 and Huh7 cell lines was assessed using flow cytometry after transfection. Vehicle controls were added to maintain equivalent transfectant volumes, and 2,000 IU/mL IFN- was used for all of the IFN- treatments. After transfection for 48 h, apoptosis was also evaluated based on annexin V (AV) binding of extracellular phosphatidylserine, a marker of early-stage apoptosis, and intracellular staining with propidium iodide (PI), an indicator of late-stage apoptosis, using the Dead Cell Apoptosis kit (ThermoFisher), according to the manufacturer’s instructions. The cells were analyzed and the levels of FITC and PI fluorescence were calculated using a FACS-Canto flow cytometer (BD Biosciences, San Jose, CA, USA) and Cell Quest software (BD Biosciences).
2.6. miRNA target prediction
To investigate the mechanisms involved in the repression of ISG15 in IFN- resistant cells, we performed an in silico analysis of the human ISG15 mRNA (Genbank accession no. NM_005101.3) using PicTar (http://pictar.mdc-berlin.de) to identify potential miRNA binding sites. The PicTar computational utility provides alignments of 3’ UTR sequences and predicted miRNA target sites with links to various public databases.
2.7. Relative quantification of miRNA
Relative quantification of the level of miR-370 in human tumor tissues; the LH86, HLCZ01, L02, SMMC-7721, and Huh7 cell lines; and LH86- and Huh7-derived xenograft tumors was performed using qRT-PCR. Total RNA was isolated from tissues using the MagMAX mirVana Total RNA Isolation Kit (ThermoFisher), and miRNA was isolated from cultured cells using the TaqMan MicroRNA Cells-to-C Kit (ThermoFisher). The miR-370 level was measured using the Taqman Advanced miRNA Assay for human miR-370 (cat. no. A25576; ThermoFisher, Waltham, MA, USA), according to the manufacturer’s instructions. Real-time PCR was performed using the TaqMan Fast Advanced Master Mix.
2.8. Fluorescence microscopy
Apoptosis in the LH86 and Huh7 cell lines was assessed using fluorescence microscopy after transfection with the following: IFN- only; miR-370 with or without IFN-; anti-miR-370 siRNA with or without IFN-; ISG15 with IFN-; and ISG15 with IFN- and miR-370. Vehicle controls were added to maintain equivalent transfectant volumes and 2,000 IU/mL IFN- was used for all of the IFN- treatments. After transfection for 48 h, the cells were fixed for 5 min at room temperature in 4% paraformaldehyde dissolved in PBS, and stained for 30 min using 0.5 g/mL 4,6 diamidino-2 phenylindole (DAPI; Sigma-Aldrich) in PBS. After washing with PBS, the cells were mounted in 90% glycerol in PBS. The cells were viewed using a Nikon Eclipse TE2000-U inverted fluorescence microscope (Nikon, Tokyo, Japan). Apoptotic nuclei were identified based on the presence of condensed chromatin and nuclear fragmentation and the percentage apoptosis was calculated relative to the control cells.
2.9. Tumor xenografting
Our animal protocols were approved by the Hunan Provincial Cancer Hospital Review Board (Changsha, China) and the Hunan Province Animal Care and Use Committee, and all of the animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, USA). All animals were housed under standard conditions in a pathogen-free, temperature and light-controlled facility. The LH86 and Huh7 cell lines were infected with adenovirus expressing miR-370 (Ad-miR-370; Vector Biolabs, Malvern, PA, USA) or control adenovirus expressing green fluorescent protein (Ad-GFP; Vector Biolabs). After antibiotic selection for 10 days, 5 10 cells in 200 L PBS were injected subcutaneously into the lower right flank of 6-week old male NOD/SCID mice to induce xenograft tumor formation. Three days after the LH86 and Huh7 cells were implanted, IFN- treatment was initiated by intraperitoneal injection of 5 10 U/kg every 3 days. Tumor volume (TV) was calculated using the following formula: TV 0.5 width length. The mice were sacrificed 42 days after HCC cell implantation.
2.10. Statistical analysis
All statistical analyses were performed using SPSS software, version 17.0 (IBM, Armonk, NY, USA). Student -tests, log-rank sum, Wilcoxon’s matched-pairs analysis, Mann-Whitney U tests, and chi-squared analysis were used to determine statistically significant differences between data sets with the level of statistical significance set at 0.05.
3. Results
3.1. IFN- induced apoptosis and ISG15 expression in human HCC cell lines
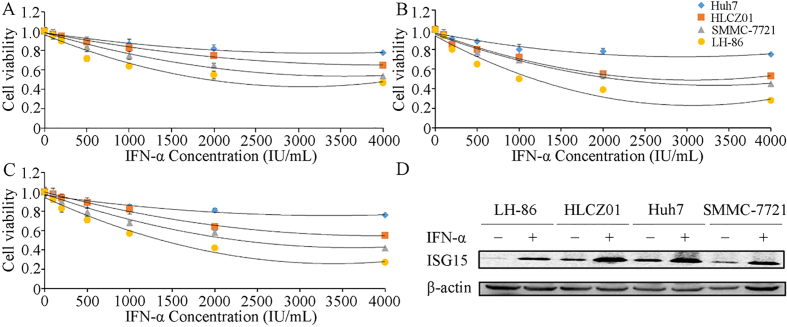
The expression of ISG15 is associated with the tumor grade, metastasis and survival in HCC patients [29]. Therefore, we evaluated ISG15 expression and apoptosis in LH86, HLCZ01, SMMC-7721 and Huh7 cells after treatment with various concentrations of IFN-. The MTT assays showed that among the cell lines tested, LH86 cells were most sensitive to IFN- at all concentrations and time points, whereas Huh7 cells were least sensitive to IFN-. The threshold of maximum sensitivity for all of the cell lines tested occurred at approximately 2,000 IU/mL IFN- for the 48- and 72-h treatments (Fig. 1A–C). Western blot analysis showed that treatment with 2,000 IU/mL IFN- for 48 h induced expression of ISG15 protein in all of the HCC cell lines (Fig. 1D). These results confirmed that IFN- induces ISG15 expression in HCC cells, and that the sensitivity to IFN--induced apoptosis varies between different HCC cell lines.
Figure 1.
Effects of IFN- on human HCC cell lines. (A) Huh7, HLCZ01, SMMC7721, and LH86 cells were treated with 125, 250, 500, 1,000, 2,000 or 4,000 IU/mL IFN- for (A) 24 h, (B) 48 h, (C) and 72 h, and cell viability was assessed by MTT assay. (D) Expression of the ISG15 and ISG12a proteins was detected by western blotting using -actin as a loading control. The data presented are representative of 3 independent experiments.
3.2. ISG15 regulates the sensitivity of HCC cells to IFN- treatment
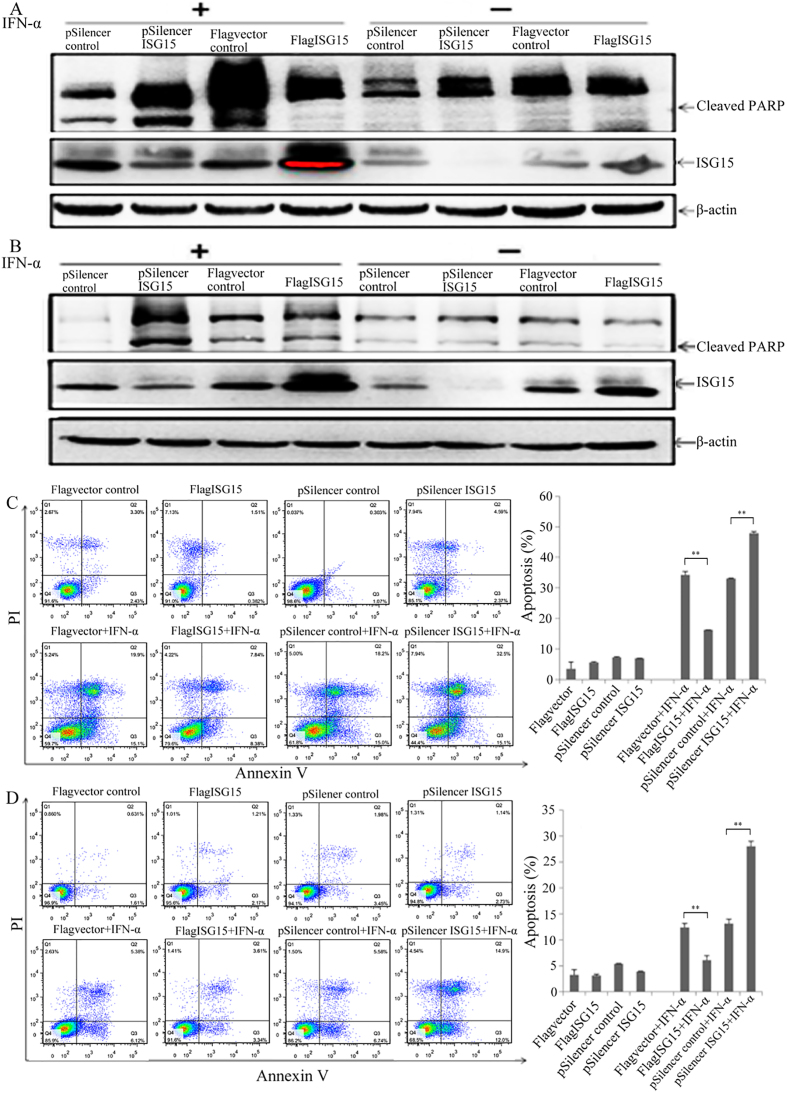
To investigate the role of ISG15 expression in IFN--induced apoptosis, we examined whether changes in ISG15 expression influenced PARP cleavage and/or apoptosis in IFN--treated LH86 and Huh7 cells. Western blot analysis showed that in the absence of IFN- overexpression of ISG15 did not significantly affect PARP cleavage (Fig. 2A and B) or apoptosis (Fig. 2C and D) in either cell line, compared to the vector control, nor did the shRNA-mediated knock down of ISG15 mRNA expression (Fig. 2A–D). In the absence of ISG15 overexpression, treatment with 2,000 IU/mL IFN- increased both PARP cleavage and apoptosis in LH86 cells, compared to the LH86 control without IFN- and no ISG15 overexpression (Fig. 2A and C). The overexpression of ISG15 in IFN--treated LH86 cells reduced apoptosis (Fig. 2C) while reducing PARP cleavage (Fig. 2A), relative to the IFN--treated vector control. In the Huh7 cells treated with IFN- alone (pSilencer control), the level of cleaved PARP was not noticeably different from Huh7 cells without IFN- (Fig. 2B). The overexpression of ISG15 in Huh7 cells treated with IFN- did not affect PARP cleavage (Fig. 2B), but ISG15 overexpression did reduce apoptosis (Fig. 2D), relative to the IFN--treated Huh7 vector control. The shRNA-mediated knock down of ISG15 mRNA expression increased both PARP cleavage and apoptosis in both LH86 and Huh7 cells treated with IFN- (Fig. 2A–D), compared with pSilencer control cells treated with IFN-. These results suggested that the mechanism underlying sensitivity to IFN--induced apoptosis in HCC cells involves the regulation of ISG15 expression. In addition, the magnitude of the changes in apoptosis in response to ISG15 overexpression and shRNA-mediated ISG15 knock down was consistently greater in LH86 cells than in Huh7 cells, which showed that factors other than ISG15 expression contributed to the differences in IFN- sensitivity observed between the different HCC cell lines.
Figure 2.
ISG15 mediates IFN- sensitivity in HCC cells. Western blotting was used to detect PARP cleavage and ISG15 protein expression in (A) LH86 and (B) Huh7 cells after treatment with 2,000 IU/mL IFN- for 48 h. Apoptosis in (C) LH86 and (D) Huh7 cells was assessed using flow cytometry after treatment with 2,000 IU/mL IFN- for 48 h. The data presented are representative of 3 independent experiments.
3.3. ISG15 expression is regulated by interaction between miR-370 and ISG15 mRNA
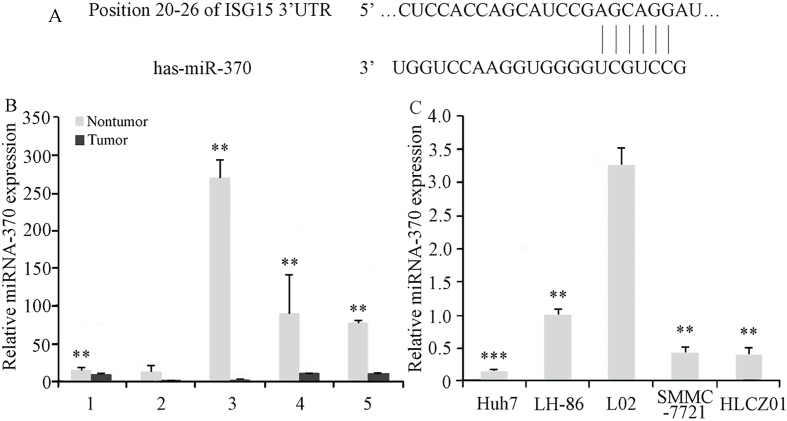
To investigate the factors involved in the regulation of ISG15 expression in HCC cells, we analyzed the ISG15 mRNA sequence to identify possible miRNA target sites. Our sequence analysis revealed a high level of complementarity between human miR-370 and a 7-bp sequence in the 3’-UTR of human ISG15 mRNA (Fig. 3A). Our analysis of miR-370 expression showed that the level of miR-370 was significantly lower in 4 of the 5 HCC tumor samples tested, compared to those in adjacent noncancerous tissues (Fig. 3B). The levels of miR-370 expression in the HCC cell lines, HLCZ01, LH86, Huh7, and SMMC7721, were also reduced, compared with the level of miR-370 in the irradiation immortalized, normal human liver cell line, LO2 (Fig. 3C). These results suggested that reduced miR-370 expression contributes to HCC progression.
Figure 3.
Identification of miR-370 target sequence in ISG15 mRNA, and analysis of miR-370 expression in HCC tumor tissues and cell lines. (A) The 3-UTR of ISG15 mRNA contains one predicted miR-370 binding site, as indicated in the alignment of the seed region of miR-370 with ISG15 3-UTR sequence. Relative quantification of the level of miR-370 in (B) HCC tumor samples (** 0.01 compared to adjacent noncancerous tissues) and (C) HCC cell lines (** 0.05 and * 0.05 compared with LO2 and LH86 cells, respectively) was performed using qRT-PCR, and the results were normalized based on U6 expression. The data presented are representative of 3 independent experiments.
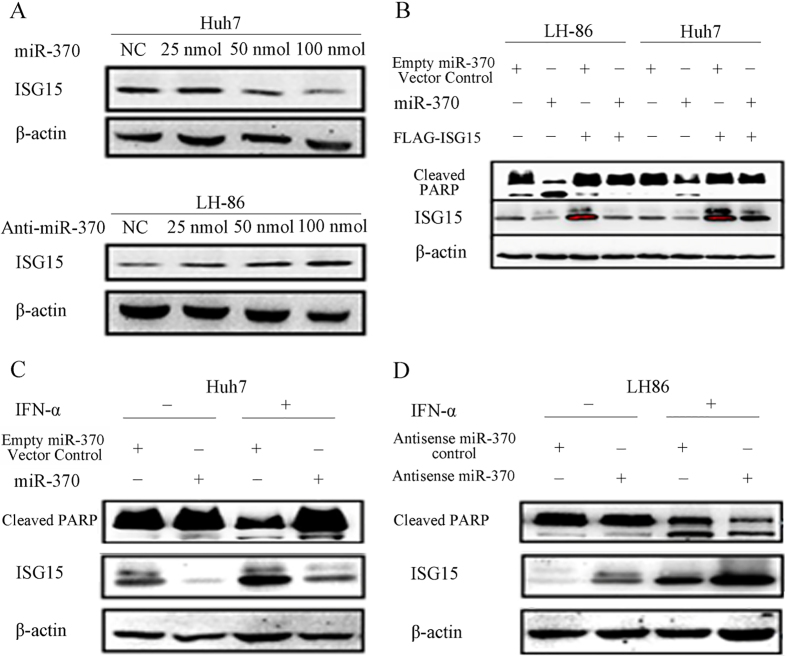
Western blot analysis showed that transfection with miR-370 reduced ISG15 protein expression in Huh7 and LH86 cells without IFN- treatment (Fig. 4A and B) and in IFN- treated Huh7 cells (Fig. 4C), whereas tranfection of LH86 cells with anti-miR-370 increased ISG15 expression in LH86 cells with or without IFN- treatment (Fig. 4A and D). In the absence of IFN-, transfection with miR-370 also increased PARP cleavage in both LH86 and Huh7 cells, and the overexpression of exogenous FLAG-ISG15 reversed the effect of miR-370 on PARP cleavage.
Figure 4.
Effects of miR-370 on ISG15 expression and PARP cleavage. (A) Huh7 and LH86 cells were transfected with 25, 50 or 100 nmol miR-370 or anti-miR-370 siRNA for 24 h before assessing the levels of ISG15 and actin (loading control) proteins by western blotting. (B) After transfection with 100 nmol miR-370 and/or pFLAG-ISG15 for 24 h, LH86 and Huh7 cells were subjected to western blotting to assess the levels of cleaved PARP, ISG15, and actin proteins. (C and D) After treatment with or without 2,000 IU/mL IFN- for 48 h, Huh7 and LH86 cells were transfected with 100 nmol miR-370 or anti-miR-370 siRNA for 24 h, and the levels of cleaved PARP, ISG15, and actin proteins were assessed by western blotting. The data presented are representative of 3 independent experiments.
The combined results of our bioinformatic (Table S1) and western blot analyses suggested that miR-370 downregulates ISG15 expression by targeting the 3’-UTR of the ISG15 mRNA.
3.4. Targeting of ISG15 by miR-370 modulates the IFN- sensitivity in HCC cells
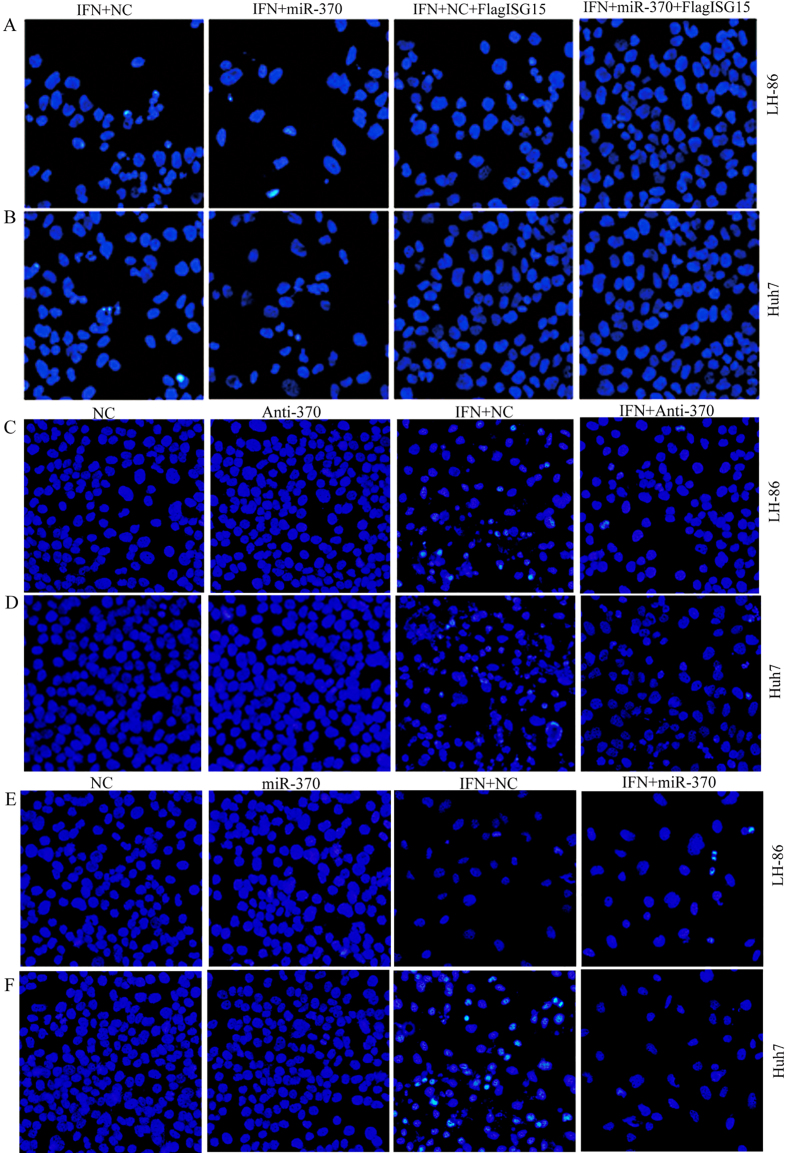
The role of miR-370-mediated regulation of ISG15 expression in its sensitivity to IFN--induced apoptosis in HCC cells was investigated using DAPI staining and fluorescence microscopy. Transfection with miR-370 increased the sensitivity to IFN--induced apoptosis in both LH86 and Huh7 cells (Fig. 5A and B). The overexpression of exogenous FLAG-ISG15 reduced the sensitivity to IFN-, compared to the IFN--treated controls, as evidenced by reduced apoptosis in both LH86 and Huh7 cells, with the Huh7 cells exhibiting lower sensitivity to IFN--induced apoptosis than the LH86 cells. The overexpression of exogenous FLAG-ISG15 in LH86 and Huh7 cells transfected with miR-370 reversed the enhancing effect of miR-370 on IFN--induced apoptosis. Knock down of endogenous miR-370 expression by transfection with anti-miR-370 reduced the sensitivity of LH86 cells to IFN--induced apoptosis compared to LH86 cells treated with IFN- alone, mimicking the resistance of Huh7 cells to IFN--induced apoptosis (Fig. 5C and F). These results suggested that miR-370 modulates IFN--induced apoptosis in HCC cells through the suppression of ISG15 expression.
Figure 5.
The sensitivity of cancer cells to IFN- induced apoptosis is modulated through the miR-370 mediated regulation of ISG15 expression. After treatment with IFN- for 48 h, (A) LH86 and (B) Huh7 cells were transfected with miR-370 and/or pFLAG-ISG15 for 24 h, (C) LH86 cells and (D) Huh7 cells treated with IFN- for 48 h and/or transfected with anti-miR-370 for 24 h, and (E) LH86 cells and (F) Huh7 cells treated with IFN for 48 h or/and transfected with miR-370 for 24 h. Apoptosis was assessed based on the presence of condensed chromatin and nuclear fragmentation using DAPI staining and fluorescence microscopy. The data presented are representative of 3 independent experiments.
3.5. miR-370 modulates IFN--mediated tumor immunity in HCC in vivo
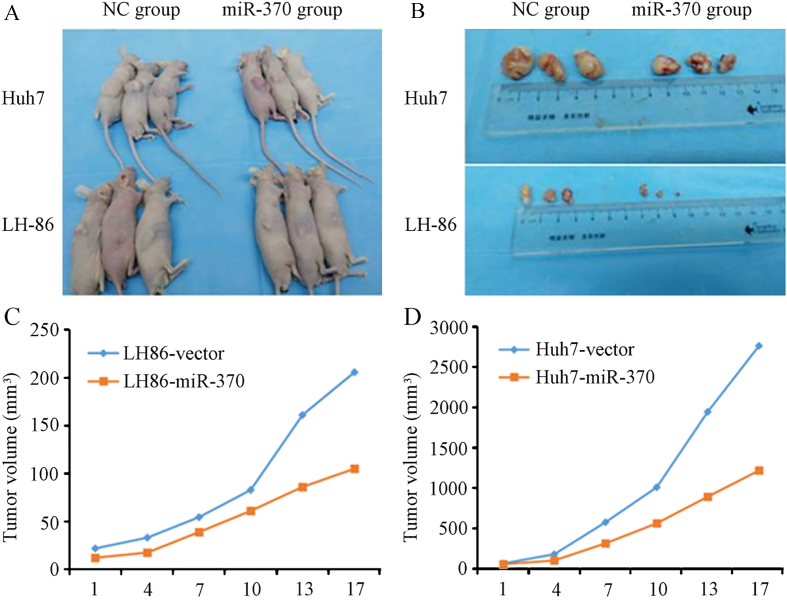
We performed HCC xenograft experiments in immunodeficient NOD/SCID mice to determine whether the effects of miR-370 on ISG15-mediated IFN- sensitivity in LH86 and Huh7 cells could be confirmed in vivo using an animal model of HCC. The LH86 and Huh7 cells were transduced with Ad-miR-370 before implantation into the right flank of mice (Fig. 6A), and the recipient mice were treated with IFN-. On day 17 post-implantation, the tumors overexpressing miR-370 were approximately one-half the size of the tumors with the control virus (Fig. 6B and D). These results suggested that miR-370 is an important factor for IFN--mediated HCC tumor immunity in vivo.
Figure 6.
MiR-370 modulates the sensitivity of cancer cells to IFN- induced apoptosis in vivo. (A) Xenograft tumors were induced in NOD/SCID mice by implantation of Huh7 or LH86 cells infected with an adenovirus expressing miR-370, and the mice were treated with IFN- every 3 days thereafter. (B) The mice were sacrificed 42 days after HCC cell implantation and the tumor volume was calculated. The volumes of the (C) LH86 derived and (D) Huh7 derived tumors were compared with the control virus. The data presented are representative of 3 independent experiments.
4. Discussion
The pleiotropic cytokine IFN- induces the expression of multiple proapoptotic genes, and IFN- has been shown to exhibit direct effects on the growth and proliferation of malignant HCC tumor cells [35, 36]. IFN- also inhibits angiogenesis in HCC, which contributes further to the suppression of tumor progression [37]. Treatment with IFN- improves the antitumor response and prognosis in HCC patients receiving chemotherapeutic agents including 5-fluorouracil, cisplatin, methotrexate and doxorubicin [38, 39, 40], and combination therapy using antiviral drugs and IFN- are the first-line treatments for HBV and HCV infections, which contribute to the majority of HCC cases. However, a significant portion of HCC patients do not respond well to IFN- adjuvant therapy. Therefore, elucidating the biological mechanism(s) underlying hepatocyte sensitivity to IFN- is an important research objective.
We found that IFN- increased ISG15 expression in various HCC cell lines, including LH86, HLCZ01, SMMC7721 and Huh7 cells (Fig. 1). These findings are consistent with those of Zhu et al. based on their analysis of ISG15 expression in Huh7 and FCA1 cells treated with IFN- [32]. Our results also showed that ISG15 overexpression reduced apoptosis in Huh7 and LH86 cells in the presence of IFN-, but not in the absence of IFN-, whereas the shRNA mediated knock down of ISG15 expression increased apoptosis in both Huh7 and LH86 cells (Fig. 2). These results differ somewhat from those of Wan et al., who found that, although IFN- induced ISG15 expression in HepG2 cells, treatment with IFN- alone did not induce apoptosis, which suggests that HepG2 cells are less sensitive to IFN- than Huh7 cells, and exhibit the lowest sensitivity to IFN- among the cell lines tested in our study [41]. Wan et al. also reported that ISG15 overexpression increased apoptosis in HepG2 cells in the absence of IFN- [41]. In our experiments, overexpressing ISG15 did not increase apoptosis in Huh7 and LH86 cells in the absence of IFN- (Fig. 2C and D), compared to the rate of apoptosis in cells without ISG15 overexpression and no IFN- treatment, which suggests that the mechanism linking ISG15 expression to apoptotic pathways in HepG2 cells is not dependent on the expression of other IFN- stimulated genes.
Our bioinformatic analysis suggested that miR-370 downregulated ISG15 expression in Huh7 and LH86 cells (Fig. 4) through a putative interaction between miR-370 and the 3’-UTR of ISG15 mRNA (Fig. 3). Xu et al. showed that miR-370 expression gradually decreased over the course of HCC tumorigenesis in rats, and they also showed that reduced expression of miR-370 in tumor tissue samples was associated with tumor progression, angiogenesis, microsatellite tumors, capsular invasion, and poor prognosis in HCC patients [41]. We observed significantly lower levels of miR-370 in all the HCC cell lines examined and in most of the HCC tumor samples, compared with LO2 cells (normal hepatocytes) and noncancerous adjacent tissues, respectively (Fig. 3). We also showed that miR-370 expression reduced the expression of ISG15 and increased apoptosis in Huh7 and LH86 cells treated with IFN- (Fig. 5). In mice treated with IFN-, xenograft tumors derived from Huh7 or LH86 cells overexpressing miR-370 were smaller than those derived from Huh7 or LH86 cells without miR-370 overexpression (Fig. 6). In animal experiments, Li et al. showed that the knock down of ISG15 expression inhibited the progression of xenograft tumors derived from HepG2 and 97L cells without IFN- treatment [29], which suggested that ISG15-induced apoptosis might also involve factors other than IFN--induced genes.
Li et al. have also shown that increased ISG15 expression was associated with tumor progression, metastasis and poor survival in HCC patients without IFN- treatment [29]. We did not examine the effects of miR-370 on xenograft tumors without IFN- treatment. However, in our cell culture experiments, we found that the level of miR-370 expression in HCC cell lines reflected the level of IFN- induced apoptosis (Fig. 3). We also observed that the overexpression of miR-370 reduced ISG15 expression both in the presence and absence of IFN- (Fig. 4). Combined with our results, the findings of Li et al. suggest that other factors contribute to the miR-370 mediated regulation of ISG15 expression in HCC cells, and our findings are consistent with those of previous studies regarding a potential role for the reduced expression of miR-370 in the tumorigenesis of HCC. In addition, our data showed that HCC cells exhibiting reduced miR-370 expression produced larger xenograft tumors (Figs 3 and 6), suggesting that miR-370 expression might correlate with reduced sensitivity to IFN-.
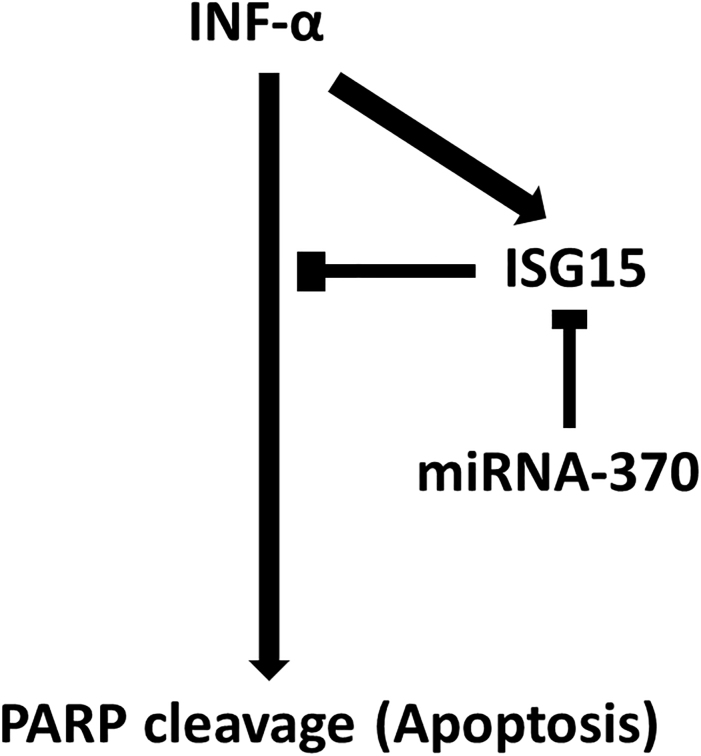
In conclusion, our results showed that miR-370 downregulates IFN- activity, reducing ISG15 expression in Huh7 and LH86 cells through a putative miR-370 target sequence in the 3’-UTR of ISG15 mRNA, which in turn leads to recovered IFN- activation and enhanced apoptosis (Fig. 7). All of the HCC cell lines and most of the HCC tumor samples that we tested had significantly lower levels of miR-370 than the control cells and tissues. Both the knock down of ISG15 expression and the overexpression of miR-370 increased apoptosis in the HCC cell lines treated with IFN- and the overexpression of miR-370 reduced the volume of LH86 and Huh7 derived xenograft tumors in mice treated with IFN-, compared to control tumors.
Figure 7.
Scheme of the study findings. INF- induced apoptosis and concomitantly upregulated ISG15, which is a negative regulator of its apoptotic activity. miRNA-370 leads to silencing of ISG15 via a 7-bp sequence in the 3’-UTR and interrupts apoptosis downregulation.
Our findings suggest that the miR-370 functions as a tumor suppressor in HCC cells, and might be a useful prognostic marker for HCC, especially in patients with IFN- resistant tumors. Therefore, large-scale studies of miR-370 expression particularly in HCC tumor samples from patients are required to confirm our findings.
Acknowledgments
This work was supported by a grant from the Changsha City Third Science and Technology Project (grant number K1403380-31), and the National Natural Science Foundation of China (grant number 816701030).
Supplementary data
Table S1.
The bioinformatic results of miR-370 downregulates ISG15 expression by targeting the 3’-UTR of the ISG15
| Predicted consequential pairing of | Seed | Site-type | 3’pairing | Local AU | Position | TA | SPS | Context | Context score | Conserved | Per | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| target region (top) and miRNA (bottom) | match | contribution | contribution | contribution | contribution | contribution | contribution | score | percentile | branch length | ||
| Position 3–9 of ISG15 has-miR-650 | 5’ NNNNNNNNNNNNNNG 3’ CAGGACUCUCGCG | 7mer-1A | 0.074 | NA | NA | NA | NA | NA | NA | Too close to ORF | 0.073 | NA |
| Position 3–9 of ISG15 has-miR-4443 | 5’ NNNNNNNNNNNNNNG 3’ UUUUGGGU | 7mer-m8 | 0.120 | NA | NA | NA | NA | NA | NA | Too close to ORF | 0.073 | NA |
| Position 3–9 of ISG15 has-miR-3612 | 5’NNNNNNNNNNNNNNG 3’AGGUAAAGAGUUCU | 7mer-1A | 0.074 | NA | NA | NA | NA | NA | NA | Too close to ORF | 0.073 | NA |
| Position 6–12 of ISG15 has-miR-3615 | 5’NNNNNNNNNNNGGGC 3’ACUCCAGUGUAACG | 7mer-1A | 0.074 | NA | NA | NA | NA | NA | NA | Too close to ORF | 0.073 | NA |
| Position 7–13 of ISG15 has-miR-4456 | 5’NNNNNNNNNNGGGCC 3’UUUUCCUU | 7mer-m8 | 0.120 | NA | NA | NA | NA | NA | NA | Too close to ORF | 0.343 | NA |
| Position 8–15 of ISG15 has-miR-138 | 5’ NNNNNNNNNGG 3’GCCGGACUAA | 8mer | 0.247 | NA | NA | NA | NA | NA | NA | Too close to ORF | 0.343 | 0.1 |
| Position 17–23 of ISG15 has-miR-423-3p | 5’GGCCUCCA 3’UGACUCCCCGGAGUC | 7mer-1A | 0.074 | 0.006 | 0.034 | 0.048 | 0.08 | 0.085 | 0.26 | 71 | 0.013 | NA |
| Position 20–26 of ISG15 has-miR-370 | 5’CUCCACCAG 3’UGGUCCAAG | 7mer-1A | 0.074 | 0.002 | 0.042 | 0.048 | 0.02 | 0.063 | 0.13 | 59 | 0.013 | NA |
| Position 24–30 of ISG15 has-miR-640 | 5’ACCAGCAUCCGAGCA 3’UCUCCGUCCAAGGA | 7mer-1A | 0.074 | 0.002 | 0.053 | 0.048 | 0.015 | 0.022 | 0.11 | 52 | 0.368 | NA |
| Position 27–33 of ISG15 has-miR-4780 | 5’AGCAUCCGAGCAGGA 3’CGAUCCCUAGUCCG | 7mer-m8 | 0.120 | 0.007 | 0.048 | 0.058 | 0.01 | 0.031 | 0.18 | 76 | 0.073 | NA |
| Position 33–40 of ISG15 has-miR-1180 | 5’CGAGCAGGAUCAA 3’UGUGUGGGUGCG | 8mer | 0.247 | 0.003 | 0.043 | 0.109 | 0.137 | 0.114 | 0.56 | 97 | 0.013 | NA |
| Position 34–40 of ISG15 has-miR-4664-3p | 5’GAGCAGGAUCAAGGG 3’CUGCCCCGAGUGUCU | 7mer-1A | 0.074 | 0.002 | 0.013 | 0.048 | 0.088 | 0.044 | 0.24 | 67 | 0.013 | NA |
| Position 54–61 of ISG15 has-miR-4753-3p | 5’AAUAAAGGCUGUUGU 3’UGUGUUCCGAUUUC | 8mer | 0.247 | 0.060 | 0.008 | 0.114 | 0.054 | 0.078 | 0.30 | 99 | 0.406 | NA |
Conflict of interest
The authors have no conflicts of interest.
References
- [1]. Torre L., Bray F., Siegel R., Ferlay J., Lortet-Tieulent J. and Jemal A., Global cancer statistics, 2012, CA: A Cancer Journal for Clinicians 65 (2015), 87. [DOI] [PubMed] [Google Scholar]
- [2]. Fan S.T., Mau Lo C., Poon R.T., Yeung C., Leung Liu C., Yuen W.K., Ming Lam C., Ng K.K. and Ching Chan S., Continuous improvement of survival outcomes of resection of hepatocellular carcinoma: a 20-year experience, Ann Surg 253 (2011), 745–58. [DOI] [PubMed] [Google Scholar]
- [3]. Durante M., Reppingen N. and Held K., Immunologically augmented cancer treatment using modern radiotherapy, Trends in Molecular Medicine 19 (2013), 565. [DOI] [PubMed] [Google Scholar]
- [4]. Barker C.A. and Postow M.A., Combinations of Radiotherapy and Immunotherapy for Melanoma: A Review of Clinical Outcomes, International Journal of Radiation Oncology, Biology, Physics 88 (2014), 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Rocha F., Hashimoto Y., Traverso L., Dorer R., Kozarek R., Helton W. and Picozzi V., Interferon-based Adjuvant Chemoradiation for Resected Pancreatic Head Cancer: Long-term Follow-up of the Virginia Mason Protocol, Annals of Surgery 263 (2016), 376. [DOI] [PubMed] [Google Scholar]
- [6]. Lamm D., Brausi M., O’Donnell M.A. and Witjes J.A., Interferon alfa in the treatment paradigm for non-muscle-invasive bladder cancer, Urol Oncol 32 (2014), 35 e21–30. [DOI] [PubMed] [Google Scholar]
- [7]. Sun H., Tang Z., Wang L., Qin L., Ma Z., Ye Q., Zhang B., Qian Y., Wu Z. and Fan J., Postoperative interferon alpha treatment postponed recurrence and improved overall survival in patients after curative resection of HBV-related hepatocellular carcinoma: a randomized clinical trial, Journal of Cancer Research and Clinical Oncology 132 (2006), 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Mazzaferro V., Romito R., Schiavo M., Mariani L., Camerini T., Bhoori S., Capussotti L., Calise F., Pellicci R. and Belli G., Prevention of hepatocellular carcinoma recurrence with alpha-interferon after liver resection in HCV cirrhosis, Hepatology (Baltimore, Md.) 44 (2006), 1543. [DOI] [PubMed] [Google Scholar]
- [9]. Miamen A.G., Dong H. and Roberts L.R., Immunotherapeutic approaches to hepatocellular carcinoma treatment, Liver Cancer 1 (2012), 226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Pedersen I.M., Cheng G., Wieland S., Volinia S., Croce C.M., Chisari F.V. and David M., Interferon modulation of cellular microRNAs as an antiviral mechanism, Nature 449 (2007), 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Nana-Sinkam S.P. and Croce C.M., Clinical applications for microRNAs in cancer, Clin Pharmacol Ther 93 (2013), 98–104. [DOI] [PubMed] [Google Scholar]
- [12]. Fan H., Lv P., Lv J., Zhao X., Liu M., Zhang G. and Tang H., miR-370 suppresses HBV gene expression and replication by targeting nuclear factor IA, Journal of Medical Virology (2016). [DOI] [PubMed] [Google Scholar]
- [13]. Zhang C., Wang C., Chen X., Yang C., Li K., Wang J., Dai J., Hu Z., Zhou X. and Chen L., Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma, Clinical Chemistry 56 (2010), 1871. [DOI] [PubMed] [Google Scholar]
- [14]. Yungang W., Xiaoyu L., Pang T., Wenming L. and Pan X., miR-370 targeted FoxM1 functions as a tumor suppressor in laryngeal squamous cell carcinoma (LSCC), Biomed Pharmacother 68 (2014), 149–154. [DOI] [PubMed] [Google Scholar]
- [15]. Fan C., Liu S., Zhao Y., Han Y., Yang L., Tao G., Li Q. and Zhang L., Upregulation of miR-370 contributes to the progression of gastric carcinoma via suppression of FOXO1, Biomed Pharmacother 67 (2013), 521–526. [DOI] [PubMed] [Google Scholar]
- [16]. Wu Z., Sun H., Zeng W., He J. and Mao X., Upregulation of MircoRNA-370 induces proliferation in human prostate cancer cells by downregulating the transcription factor FOXO1, PLoS One 7 (2012), e45825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Meng F., Wehbe-Janek H., Henson R., Smith H. and Patel T., Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes, Oncogene 27 (2008), 378. [DOI] [PubMed] [Google Scholar]
- [18]. Olaru A.V., Ghiaur G., Yamanaka S., Luvsanjav D., An F., Popescu I., Alexandrescu S., Allen S., Pawlik T.M. and Torbenson M., A microRNA downregulated in human cholangiocarcinoma controls cell cycle through multiple targets involved in the G1/S checkpoint, Hepatology (Baltimore, Md.) 54 (2011), 2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Tracy C.M., Epstein A.E., Darbar D., Dimarco J.P., Dunbar S.B., Estes N.A., 3rd, Ferguson T.B., Jr., Hammill S.C., Karasik P.E., Link M.S., Marine J.E., Schoenfeld M.H., Shanker A.J., Silka M.J., Stevenson L.W., Stevenson W.G. and Varosy P.D., 2012 ACCF/AHA/HRS focused update of the 2008 guidelines for device-based therapy of cardiac rhythm abnormalities: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, J Am Coll Cardiol 60 (2012), 1297–313. [DOI] [PubMed] [Google Scholar]
- [20]. Dixon-McIver A., East P., Mein C.A., Cazier J.B., Molloy G., Chaplin T., Andrew Lister T., Young B.D. and Debernardi S., Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia, PLoS One 3 (2008), e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Hadjivasiliou A., ISG15 implicated in cytoskeleton disruption and promotion of breast cancer, Expert Rev Proteomics 9 (2012), 7. [PubMed] [Google Scholar]
- [22]. Wood L.M., Pan Z.K., Seavey M.M., Muthukumaran G. and Paterson Y., The ubiquitin-like protein, ISG15, is a novel tumor-associated antigen for cancer immunotherapy, Cancer Immunol Immunother 61 (2012), 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Andersen J.B., Aaboe M., Borden E.C., Goloubeva O.G., Hassel B.A. and Orntoft T.F., Stage-associated overexpression of the ubiquitin-like protein, ISG15, in bladder cancer, Br J Cancer 94 (2006), 1465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Vincent-Chong V.K., Ismail S.M., Rahman Z.A., Sharifah N.A., Anwar A., Pradeep P.J., Ramanathan A., Karen-Ng L.P., Kallarakkal T.G., Mustafa W.M., Abraham M.T., Tay K.K. and Zain R.B., Genome-wide analysis of oral squamous cell carcinomas revealed over expression of ISG15, Nestin and WNT11, Oral Dis 18 (2012), 469–76. [DOI] [PubMed] [Google Scholar]
- [25]. Tao J., Hua P., Wen J., Hu Y., Yang H. and Xie X., Prognostic value of ISG15 mRNA level in drinkers with esophageal squamous cell cancers, International Journal of Clinical and Experimental Pathology 8 (2015), 10975. [PMC free article] [PubMed] [Google Scholar]
- [26]. Yang S., Jeung H.-C., Jeong H.J., Choi Y.H., Kim J.E., Jung J.-J., Rha S.Y., Yang W.I. and Chung H.C., Identification of genes with correlated patterns of variations in DNA copy number and gene expression level in gastric cancer, Genomics 4 (2007), 451–459. [DOI] [PubMed] [Google Scholar]
- [27]. Sainz B., Martín B., Tatari M., Heeschen C. and Guerra S., ISG15 Is a Critical Microenvironmental Factor for Pancreatic Cancer Stem Cells, Cancer Research (2014). [DOI] [PubMed] [Google Scholar]
- [28]. Desai S.D., Haas A.L., Wood L.M., Tsai Y.-C., Pestka S., Rubin E.H. and Saleem A., Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway, Cancer Research 66 (2006), 921–928. [DOI] [PubMed] [Google Scholar]
- [29]. Li C., Wang J., Zhang H., Zhu M., Chen F., Hu Y., Liu H. and Zhu H., Interferon-stimulated Gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma, Oncotarget 5 (2014), 8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Giannakopoulos N.V., Luo J.K., Papov V., Zou W., Lenschow D.J., Jacobs B.S., Borden E.C., Li J., Virgin H.W. and Zhang D.E., Proteomic identification of proteins conjugated to ISG15 in mouse and human cells, Biochem Biophys Res Commun 336 (2005), 496–506. [DOI] [PubMed] [Google Scholar]
- [31]. Zuo C., Sheng X., Ma M., Xia M. and Ouyang L., ISG15 in the tumorigenesis and treatment of cancer: An emerging role in malignancies of the digestive system, Oncotarget 7 (2016), 74393–74409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Zhu H., Zhao H., Collins C., Eckenrode S., Run Q., McIndoe R., Crawford J., Nelson D., She J. and Liu C., Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line, Hepatology (Baltimore, Md.) 37 (2003), 1180. [DOI] [PubMed] [Google Scholar]
- [33]. Cajee U.F., Hull R. and Ntwasa M., Modification by ubiquitin-like proteins: significance in apoptosis and autophagy pathways, Int J Mol Sci 13 (2012), 11804–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Qiu X., Hong Y., Yang D., Xia M., Zhu H., Li Q., Xie H., Wu Q., Liu C. and Zuo C., ISG15 as a novel prognostic biomarker for hepatitis B virus-related hepatocellular carcinoma, Int J Clin Exp Med 8 (2015), 17140–50. [PMC free article] [PubMed] [Google Scholar]
- [35]. Herzer K., Hofmann T., Teufel A., Schimanski C., Moehler M., Kanzler S., Schulze-Bergkamen H. and Galle P., IFN-alpha-induced apoptosis in hepatocellular carcinoma involves promyelocytic leukemia protein and TRAIL independently of p53, Cancer research 69 (2009), 855. [DOI] [PubMed] [Google Scholar]
- [36]. Jiang W., Zhang C., Tian Z. and Zhang J., hIFN-alpha gene modification augments human natural killer cell line anti-human hepatocellular carcinoma function, Gene Ther 20 (2013), 1062–1069. [DOI] [PubMed] [Google Scholar]
- [37]. Wang L., Wu W., Sun H., Wu X., Qin L., Liu Y., Liu K. and Tang Z., Mechanism of interferon alpha on inhibition of metastasis and angiogenesis of hepatocellular carcinoma after curative resection in nude mice, Journal of Gastrointestinal Surgery: Official Journal of the Society for Surgery of the Alimentary Tract 7 (2003), 587. [DOI] [PubMed] [Google Scholar]
- [38]. Leung T., Patt Y., Lau W., Ho S., Yu S., Chan A., Mok T., Yeo W., Liew C. and Leung N., Complete pathological remission is possible with systemic combination chemotherapy for inoperable hepatocellular carcinoma, Clinical cancer Research: An Official Journal of the American Association for Cancer Research 5 (1999), 1676. [PubMed] [Google Scholar]
- [39]. Urabe T., Kaneko S., Matsushita E., Unoura M. and Kobayashi K., Clinical pilot study of intrahepatic arterial chemotherapy with methotrexate, 5-fluorouracil, cisplatin and subcutaneous interferon-alpha-2b for patients with locally advanced hepatocellular carcinoma, Oncology 55 (1998), 39. [DOI] [PubMed] [Google Scholar]
- [40]. Kaneko S., Urabe T. and Kobayashi K., Combination chemotherapy for advanced hepatocellular carcinoma complicated by major portal vein thrombosis, Oncology 62 (2002), 69. [DOI] [PubMed] [Google Scholar]
- [41]. Xu W.P., Yi M., Li Q.Q., Zhou W.P., Cong W.M., Yang Y., Ning B.F., Yin C., Huang Z.W., Wang J., Qian H., Jiang C.F., Chen Y.X., Xia C.Y., Wang H.Y., Zhang X. and Xie W.F., Perturbation of MicroRNA-370/Lin-28 homolog A/nuclear factor kappa B regulatory circuit contributes to the development of hepatocellular carcinoma, Hepatology 58 (2013), 1977–91. [DOI] [PubMed] [Google Scholar]