Abstract
BioID is a unique method to screen for physiologically relevant protein interactions that occur in living cells. This technique harnesses a promiscuous biotin ligase to biotinylate proteins based on proximity. The ligase is fused to a protein of interest and expressed in cells, where it biotinylates proximal endogenous proteins. Because it is a rare protein modification in nature, biotinylation of these endogenous proteins by BioID fusion proteins enables their selective isolation and identification with standard biotin-affinity capture. Proteins identified by BioID are candidate interactors for the protein of interest. BioID can be applied to insoluble proteins, can identify weak and/or transient interactions, and is amenable to temporal regulation. Initially applied to mammalian cells, BioID has potential application in a variety of cell types from diverse species.
Keywords: BioID, biotinylation, proximity-dependent labelling, protein-protein interaction
INTRODUCTION
This unit describes the design and execution of the BioID method to screen for candidate protein interactions. This method has several theoretical advantages over alternative methods to screen for protein interactions. Based on its mechanism of action, BioID is effective when applied to insoluble and membrane-associated proteins, two classes of proteins that may be refractory to screening with conventional approaches. Additional advantages include the ability to identify weak and/or transient interactions, the ability to screen for interactions in a relatively natural cellular setting, and temporal inducibility of biotin labeling. The overall process of BioID can be broken down into two stages: (1) generation and characterization of a BioID fusion protein in a mammalian (or other) expression vector for stable expression in a cell line (Basic Protocol 1) and (2) use of that cell line for a large-scale BioID pull-down to identify protein candidates by mass spectrometry (Basic Protocol 2). Note that the protocols here are specific to the application of BioID in common mammalian cell lines. With modifications, BioID has been shown to be effective in a wide variety of cell types and species.
STRATEGIC PLANNING
Design of the BioID Fusion Protein
The fundamental component of this system is the BioID fusion protein. If the purpose of BioID is to screen for physiologically relevant protein interactions for a protein of interest, then the ideal fusion protein will represent a functional replacement of the original protein. Thus, care should be taken in deciding where in the protein the biotin ligase will be incorporated. Prior evidence of successful fusions with similarly sized proteins, such as GFP, can be a good starting point to guide this decision. Posttranslational modifications to either the N- or C-terminus must be considered as well. For example, C-terminal prenylation or an N-terminal signal peptide should not be disrupted. This may complicate the molecular cloning process of generating the expression plasmid for the fusion protein, but is worth the effort. If there is little information as to how N- or C-terminal fusions might affect the protein of interest, it is recommended to try both in parallel, at least to the point of validating each fusion protein for proper targeting.
BioID Options
The original ligase for BioID was based on the Escherichia coli biotin ligase (Roux, Kim, Raida,&Burke, 2012). A second-generation biotin ligase, called BioID2, is derived from the Aquifex aeolicus biotin ligase (Kim et al., 2016). BioID2, which naturally lacks a DNA-binding domain, is approximately one-third smaller than BioID, enabling enhanced targeting and localization of the fusion protein. Compared to BioID, BioID2 requires less biotin to accomplish similar biotinylation, a trait potentially beneficial in systems where biotin supplementation may be difficult. To extend the labeling radius of BioID, estimated to be about 10 to 15 nm (Kim et al., 2014), an extended flexible linker can be used (Kim et al., 2016a). This increased labeling range may be desired when the protein of interest is significantly larger than the labeling radius of BioID2 alone, and/or when the goal is to map the constituency of a larger protein complex or discrete subcellular region.
Functional Validation of the Fusion Protein
Another consideration is how to ensure that the fusion protein is functional. In many cases, this is not possible unless the function of the protein is well known, some form of knockout/knockdown system exists, and there is a phenotypic outcome to rescue. However, it is typically possible to at least compare the localization of the fusion protein to a more minimally tagged, or preferably endogenous, protein by immunofluorescence microscopy. Attention should be paid to expression levels, as overexpression of proteins can often lead to inappropriate localization. In most cases, low levels of BioID fusion proteins, at or below the level of the endogenous protein, are sufficient for the identification of candidates. Thus, when validating by transient transfection, attention should be paid to cells expressing lower levels of the fusion protein, as this is the desired, and often likely outcome for the cells that stably express the fusion protein.
Choice of Cells and Expression Method
Once the BioID fusion protein has been designed and an expression plasmid generated, it is necessary to consider the cell type in which to express the BioID fusion protein. This is obviously a situation-specific decision; however, there are a few concepts to consider. Preferably, the expression will be similar in all cells within the population to be used for large-scale BioID pull-down. Thus, traditional transient transfection is ill-advised for more than preliminary functional validation. A population of cells stably expressing similar amounts of the fusion protein is far more useful. This can be accomplished with standard random integration or various viral expression systems. Since it is usually sufficient to screen for BioID candidates, and more likely to be physiologically relevant, an extremely low level of protein expression is ideal. Inducible expression of the fusion protein can be utilized, especially if expression of the bait is toxic. However, inducible expression is not necessary in most instances, since biotinylation itself is induced by the addition of excess biotin.
Establishing Controls
Incorporation of a BioID-only control that expresses the biotin ligase without a fused protein of interest has become standard practice to help interpret the potentially lengthy list of proteins identified by mass spectrometry in a BioID experiment. The BioID-only control identifies proteins that may be biotinylated randomly due to sheer abundance and/or due to an unnatural affinity to the BioID ligase itself. This experimental condition also controls for those proteins that specifically or nonspecifically adhere to the avidin/streptavidin beads used for biotin-affinity purification. Proteins that are detected at similar levels for the BioID-only and BioID-fusion samples should be considered nonspecific to the protein of interest. With this BioID-only control, an additional control that lacks expression of any BioID ligase is unnecessary. Alternative controls should be considered if the fusion protein lies outside of the cytoplasm or nucleus (e.g., mitochondrial matrix) such that a minimal targeting motif is used to localize the ligase to that same compartment as the BioID-protein of interest.
BASIC PROTOCOL 1
GENERATION OF CELLS EXPRESSING A BioID FUSION PROTEIN
This protocol describes the creation and characterization of a BioID fusion protein expression plasmid followed by its stable expression in mammalian cells. Techniques such as PCR cloning and generation of stably expressing cells will not be described, as they vary dramatically depending on the situation. The methods needed to test the targeting of the fusion protein and its expression and activity in cells prior to a large-scale BioID pull-down are described.
Materials
Expression plasmid for BioID fusion protein (see Internet Resources)
Gene for protein of interest
Cells of choice for transfection and appropriate medium
1 mM (20×) biotin (see recipe)
Fixative, e.g., paraformaldehyde (PFA)
Triton X-100
Streptavidin-Alexa Fluor (Invitrogen)
DNA labeling reagent (e.g., Hoechst, DAPI)
Phosphate-buffered saline (PBS; APPENDIX 2E)
SDS-PAGE sample buffer (see recipe)
BSA blocking buffer (see recipe)
Streptavidin-HRP (AB7403, Abcam)
ABS blocking buffer (see recipe)
Enhanced chemiluminescence (ECL) reagent (from commercial source, or see recipe)
30% H2O2
Antibodies specific to BioID fusion protein (e.g., anti-myc/HA) or chicken anti-BioID (BID-CP-100, BioFront)
Secondary antibodies to detect chosen primary antibody (Alexa Fluor form and HRP-conjugated form)
6-well plates
Glass coverslips
Sonicator
SDS-PAGE electrophoresis unit (Mini-PROTEAN II Electrophoresis Cell, Bio-Rad)
Semi-dry transfer cell (Trans-Blot, Bio-Rad)
Additional reagents and equipment for PCR cloning (Elion, Marina, & Yu, 2007), transfection (Kingston, 2003), SDS-PAGE separation (UNIT 10.1; Gallagher, 2012), protein transfer (UNIT 10.7; Goldman, Ursitti, Mozdzanowski, & Speicher, 2015), and immunoblot detection (UNIT 10.10; Ni, Xu, & Gallagher, 2017)
Validation of BioID fusion protein
-
1Prepare the expression plasmid for the BioID fusion protein.This typically requires the use of PCR cloning (see Elion et al., 2007) to generate an expression vector in which a protein of interest is fused in-frame with the BioID ligase. This requires plasmid DNA that is of sufficient quality for cellular transfection.To obtain mammalian expression plasmids for BioID, see the Internet Resources section.
-
2
For each experimental condition (e.g., control, BioID fusion protein), plate two wells of cells in a standard 6-well plate, one with a single glass coverslip and the other without.
-
3Express the fusion protein in the cells of choice by transient transfection (see Kingston, 2003). Process mock or nontransfected cells in parallel. Apply biotin (50 µM final concentration) to cells at time of transfection. If transfection protocol requires replacement of medium shortly after transfection, add biotin at that time.Lipofectamine 2000/3000 (Life Technologies) reagents work well with the manufacturer’s suggested protocol.The presence of excess biotin promotes biotinylation by the BioID fusion protein (usually over a period of 18 to 24 hr). This permits simultaneous analysis of expression, targeting, and function of the BioID fusion protein by immunofluorescence microscopy and immunoblot.
-
4Process the cells on coverslips for analysis by immunofluorescence microscopy 1 day following transient transfection.It is recommended to use PFA fixation (3% PFA in PBS) for 10 min, followed by Triton X-100 permeabilization (0.4% Triton X-100 in PBS) for 15 min. Use of methanol fixation will result in a strong mitochondrial signal observed with fluorescently labeled streptavidin (Streptavidin–Alexa Fluor). Use the labeled streptavidin at 1:1000 with the Alexa Fluor secondary antibody and DNA labeling reagent (e.g., Hoechst, DAPI). For more information on immunofluorescence staining, see Donaldson (2001).
-
5
Process the cells for immunoblot analysis approximately 24 hr following transient transfection and addition of biotin. To do this, briefly rinse cells in room temperature PBS to remove serum proteins prior to lysis in SDS-PAGE sample buffer using a volume appropriate for the number of cells being lysed (e.g., 200 µl for 1 × 106 cells). Sonicate to shear DNA and heat to 96°C for 5 min to denature proteins.
-
6Following whole-cell lysis, SDS-PAGE separation (UNIT 10.1; Gallagher, 2012), and protein transfer (UNIT 10.7; Goldman et al., 2015), agitate membrane in BSA blocking buffer for 30 min at room temperature.BSA is preferred for blocking to eliminate any free biotin that is potentially present in milk or serum. Free biotin will compete with biotinylated proteins for binding to streptavidin-HRP.
-
7Agitate membrane in streptavidin-HRP at 1:40,000 in BSA blocking buffer for 40 min at room temperature.It may be necessary to optimize concentration depending on the source of streptavidin-HRP. This incubation can be performed overnight at 4°C for convenience.
-
8
Quickly wash membrane in PBS two to three times to wash away unbound streptavidin-HRP.
-
9Agitate membrane in ABS blocking buffer for 5 min.This step is critical to reduce background signal on membrane.
-
10
Quickly wash membrane in PBS two to three times.
-
11
Agitate membrane in PBS for 5 min.
-
12
Add enhanced chemiluminescence (ECL) reagent to observe biotinylated proteins.
-
13Following successful analysis of biotinylated proteins, quench HRP signal on membrane by agitating for 20 min in 30% H2O2.Removal of streptavidin-HRP with standard membrane stripping methods is futile given the strength of the biotin-streptavidin interaction. The quenching permanently inactivates the HRP, allowing further probing with additional antibodies. Consider reapplication of ECL to the membrane and visualization to confirm success of quenching.
-
14
Wash membrane with PBS two to three times to remove residual hydrogen peroxide.
-
15Proceed to immunoblot membrane with antibodies specific to the BioID fusion protein (e.g., anti-myc/HA and corresponding HRP-conjugated secondary antibody) to confirm its expression and migration by SDS-PAGE.Use standard protocol (see UNIT 10.10; Ni et al., 2017) for this, beginning with blocking in ABS or equivalent (BSA blocking not required). As an alternative to HA/myc tag detection of the BioID fusion protein, an anti-BirA antibody is effective for immunofluorescence and immunoblot (chicken anti-BioID, BID-CP-100, BioFront).
Generation of cells stably expressing BioID fusion protein
-
16Initiate generation of stable cell lines by stable transfection or viral transduction.This is a highly variable process depending on the strategy and cell type.
-
17If subcloning of cells is performed, screen subclones first by immunofluorescence. Perform immunoblot analysis on subclones that pass the immunofluorescence screening. If viral infection is utilized, screen population of infected cells by immunofluorescence and immunoblot.It is important to add 50 µM biotin to the cells as in step 3 to monitor the biotinylation function of the BioID fusion protein.
-
18
Freeze down multiple vials of stably expressing cells for future BioID experiments and store in liquid nitrogen.
BASIC PROTOCOL 2
BioID PULL-DOWN TO IDENTIFY CANDIDATE PROTEINS
This protocol describes the utilization of cells stably expressing a BioID fusion protein (along with BioID-only control cells) to perform large-scale BioID pull-down experiments. The purpose of these experiments is to isolate sufficient amounts of proteins biotinylated by the BioID fusion protein to be identified by mass spectrometry. The starting material for these experiments may vary depending on a number of factors. These include the efficiency of biotinylation by the BioID fusion protein and the number of desired candidate proteins. This protocol describes the analysis of four confluent 10-cm plates of cells/condition (4 × 107 cells); however, comparable results can be obtained with two confluent 10-cm plates of cells/condition. In this case, the volume of Triton X-100 added (step 7), lysis diluent (step 10), and streptavidin beads (step 15) is halved. The protocol ends immediately prior to analysis by mass spectrometry, a service typically performed by a core facility.
Materials
Four 10-cm dishes of cells for each experimental condition (cells expressing BioID constructs or control cells)
Complete medium
1 mM (20×) biotin (see recipe)
Phosphate-buffered saline (PBS)
Lysis buffer (see recipe)
20% Triton X-100
50 mM Tris·Cl, pH 7.4 (APPENDIX 2)
Streptavidin Sepharose High Performance Beads (GE Healthcare)
Wash buffer (see recipe)
1 mM biotin in 50 mM ammonium bicarbonate (NH4HCO3), made freshly
1× SDS-PAGE sample buffer (see recipe)
DNase/RNase-free tubes, 15-ml conical and 2-ml microcentrifuge
Tube cap opener
Sonicator (Branson Sonifier-250 or equivalent)
Rotator
Day 1: Perform cell lysis
Lysis and wash steps are performed at room temperature. To reduce keratin contamination, use DNase/RNase-free tubes that have not previously been opened, wear gloves, and use a tube cap opener.
-
1
Begin with four 10-cm dishes for each experimental condition (cells expressing BioID fusion protein or BioID-only constructs).
-
2
When cells reach approximately 80% confluency, change medium to fresh complete medium containing 50 µM biotin (1×).
-
3Incubate cells for 24 hr.Incubation time and conditions may vary depending on the goals of the experiment (e.g., cell cycle stage-specific labeling). In general, 16 to 18 hr of biotin results in a maximal labeling, whereas after 6 hr of biotin there is a considerably reduced level of biotinylated proteins (approximate 25%). Pilot studies with immunofluorescence and immunoblot analysis should be performed prior to large-scale experiments.
-
4Remove medium completely by aspiration and rinse the cells twice at room temperature with 5 ml/dish of PBS.This step is important in that it removes residual free-biotin from the medium.
-
5Add 540 µl of lysis buffer/dish and scrape cells gently to harvest the cells. Perform this step at room temperature.The purpose of this harsh lysis is to try to disrupt all protein interactions and completely denature/solubilize the proteins.
-
6
Transfer lysed cells to a 15-ml conical tube.
-
7
Add 120 µl of 20% Triton X-100 (final concentration 1%) and mix by trituration. Keep tube on ice during subsequent sonication.
-
8
Position the sonicator probe tip in the sample just above the tube bottom.
-
9Apply sonication for two sessions with 60 pulses using a Branson Sonifier 250 (or equivalent) at 30% duty cycle and an output level of 4. Let the tube sit on ice for 2 min between each session to prevent overheating.If the sample is still viscous and cloudy after sonication, apply an additional period of sonication. Sonication also functions to shear DNA.
-
10Add 2.52 ml of prechilled lysis buffer, and mix well.This dilution provides more favorable conditions for affinity capture.
-
11Apply one session of sonication (60 pulses at 30% duty cycle and an output level of 4, using a Branson Sonifier 250 or equivalent).This step helps solubilize any precipitated proteins that may be present due to reduced detergent concentrations from the previous step and assists in mixing the sample.
-
12
Aliquot the sample evenly into three prechilled 2-ml tubes (~1.6 ml each).
-
13
Spin down 10 min at 16,500 × g, 4°C.
Perform affinity purification of biotinylated proteins
-
14
During the centrifugation in step 13, add 1 ml of room temperature lysis buffer to one 2-ml tube.
-
15Mix the stock of streptavidin beads well with gentle tapping to resuspend and add 100 µl of beads to each tube prepared in step 14.This step functions to equilibrate the beads in the binding buffer. Earlier protocols utilized paramagnetic streptavidin Dynabeads, which interfere with mass spectrometric analysis.
-
16Centrifuge the equilibrated beads for 2 min at 1000 × g, then remove the supernatant gently by pipetting.This step removes the buffer in which the beads equilibrated.
-
17After sample centrifugation (step 13), carefully transfer 1 ml of supernatant to the tubes prepared in step 16. Do not disturb the small insoluble pellet on the tube wall when removing the supernatant.Step 17 should be performed quickly to prevent the beads from drying out.
-
18
Resuspend the samples and beads with gentle pipetting and transfer to a pre-chilled 15-ml conical tube. Add the rest of the sample from the 2-ml tubes, being careful not to disturb the pellets on the tube walls.
-
19
Incubate the tube on a rotator at 4°C overnight.
Day 2: Bead washing
-
20
Spin the tubes for 5 min at 1000 × g to collect streptavidin beads.
-
21
Remove the supernatant gently by pipetting. Try not to disturb beads.
-
22Add 1 ml of wash buffer to each tube, resuspend beads gently by flicking the tube, and transfer to a 1.5-ml microcentrifuge tube.Avoid pipetting the beads excessively, as they can bind to the pipet tips, leading to sample loss.
-
23
Place tubes on a rotator for 8 min at room temperature.
-
24
Centrifuge for 2 min at 1000 × g and remove supernatant.
-
25
Repeat steps 21 to 24 three more times (four washes total).
-
26
Resuspend with 1 ml wash buffer.
-
27Save 100 µl (10% of total) of resupended beads for further analysis by western blot.The remaining 900 µl are the beads destined for mass spectrometry analysis.
-
28
Centrifuge to collect beads as described in step 24 (both 900 µl and 100 µl tubes).
-
29
Remove the supernatant completely. Do not disrupt beads at the tube bottom.
-
30Add 50 µl of 1 mM biotin in 50 mM ammonium bicarbonate to the tube that contained 900 µl of sample (destined for mass spectrometry analysis) and resuspend gently with flicking.Biotin binds all remaining unbound streptavidin to stabilize the protein and prevent tryptic-released peptides that are biotinylated from binding the beads. The final wash buffer may vary depending on analysis method. Before mass spectrometry analysis, freeze the sample quickly in liquid nitrogen. Frozen samples can be stored at −80°C until biotinylation has been confirmed by immunoblot.
-
31
For the 100-µl sample, after removing supernatant, resuspend beads in 50 mM Tris·Cl, pH 7.4 to remove urea.
-
32
Collect beads by centrifugation (2 min, 1000×g) again and remove the supernatant.
-
33Add 100 µl of 1× SDS-PAGE sample buffer. Heat samples at 98°C for 5 min.Samples can be stored at −20°C for further analysis by immunoblot as described in Basic Protocol 1.Prior to sending samples for analysis by mass spectrometry, it is recommended to perform immunoblot analysis (UNIT 10.10; Ni et al., 2017) of 15 µl from the protein in the SDSPAGE sample buffer. This should follow the same protocol in Basic Protocol I to check for evidence of biotinylated proteins from BioID fusion proteins. If immunoblot shows clear evidence of biotinylation in control and BioID fusion protein samples, proceed to mass spectrometry (Chapter 16).
REAGENTS AND SOLUTIONS
Use Milli-Q purified water or equivalent in all recipes. For common stock solutions, see APPENDIX 2E.
ABS blocking buffer
10% (v/v) adult bovine serum
1% (w/v) Triton X-100
Bring up to final volume with 1× PBS (APPENDIX 2E)
Store up to 4 weeks at 4°C
Biotin, 1 mM (20×)
Dissolve 12.2 mg biotin (Sigma, cat. no. B4501) in 50 ml of serum-free DMEM (or standard tissue culture medium). Pipetting may be required to dissolve biotin completely. Sterilize by passing through a 0.22-µmsyringe-driven filter unit (Millex). Dispense into sterile 50-ml tube; cap tightly. Store up to 8 weeks at 4°C.
BSA blocking buffer
1% bovine serum albumin, fraction V
0.2% (w/v) Triton X-100
Bring up to final volume with 1× PBS (APPENDIX 2E)
Store up to 4 weeks at 4°C
ECL reagent
Stock solutions:
250 mM luminol; 90 mM coumaric acid; 1 M Tris·Cl, pH 8.5 (APPENDIX 2E); 30% hydrogen peroxide
Store luminol and coumaric acid stock solutions in 500-µl aliquots at −20°C, others at 4°C
Solution 1 (wrap in tin foil to protect from light):
1 ml 1 M Tris·Cl, pH 8.5 (APPENDIX 2E)
45 µl coumaric acid
100 µl luminol
8.9 ml H2O
Solution 2:
1 ml 1 M Tris·Cl, pH 8.5 (APPENDIX 2E)
6 µl 30% hydrogen peroxide
9 ml H2O
Store solutions 1 and 2 in the dark at 4°C for up to 1 month
Mix equal volumes of solutions 1 and 2 together and immediately add to membrane for 1 min prior to exposure to film or ChemiDoc detection
Lysis buffer
8 M urea in 50 mM Tris·Cl, pH 7.4 (APPENDIX 2E)
1× protease inhibitor (Halt Protease Inhibitor Cocktail, EDTA-Free, Thermo Scientific)
1 mM dithiothreitol (DTT)
Prepare fresh
This lysis buffer composition differs from that described in the initial publication of BioID (Roux et al., 2012).
SDS-PAGE sample buffer
50 mM Tris·Cl, pH 6.8 (APPENDIX 2E)
12% sucrose
2% SDS
0.004% bromophenol blue
20 mM dithiothreitol (DTT)
Prepare fresh
Wash buffer
50 mM Tris·Cl, pH 7.4 (APPENDIX 2E) containing:
8 M urea
Prepare fresh
COMMENTARY
Background Information
BioID is a system to screen for protein interactions as they occur in living cells. The fundamental concept of BioID is derived from another method called DamID in which a prokaryotic Dam methylase is fused to a protein of interest to monitor DNA-protein interactions in eukaryotes (van Steensel & Henikoff, 2000). For BioID, a prokaryotic biotin ligase called BirA from E. coli has been mutated (R118G) to promote promiscuous biotinylation (Choi-Rhee, Schulman, & Cronan, 2004; Cronan, 2005; Roux et al., 2012). This mutant ligase (designated BioID) is fused to a protein of interest to screen for protein interactions. The wild-type BirA catalyzes a two-step reaction: first, the generation of reactive biotinyl-AMP from biotin and ATP, and second, the attachment of that biotinyl-AMP (bioAMP) to a specific lysine on a subunit of the acetyl-CoA carboxylase. BioID is capable of generating the reactive bioAMP, albeit with reduced affinity for biotin, but the mutant also has a significantly reduced affinity for the reactive bioAMP intermediate (Kwon & Beckett, 2000). This results in the premature release of the reactive bioAMP, which will covalently react with adjacent primary amines (e.g., lysine). As biotin is a relatively rare protein modification and is capable of being selectively isolated, BioID permits the identification of those proteins that were proximate to the fusion protein (Fig. 19.23.1). BioID is a complementary alternative to existing methods such as yeast-2-hybrid (Y2H) or affinity complex purification. Y2H is a yeast-based genetic assay in which both the specific bait and the variable prey are exogenous fusion proteins expressed in specific compartments of the yeast cells. The interaction between bait and prey generates a selectable readout permitting identification of the prey based on cDNA sequence analysis. Advantages of Y2H include the ability to generate cDNA libraries of prey from diverse cell types (e.g., spermatocytes) in which expression of exogenous fusion proteins may not be practical. Furthermore, with classical Y2H, a positive candidate implies that a direct interaction has occurred between the bait and prey. The main disadvantage of Y2H is its generally high rate of false positive and negative results that can result from a number of factors such as the testing of interactions in an inappropriate cellular environment and the absence of post-translational modifications and/or accessory proteins. In contrast, affinity complex purification is based on the isolation of protein complexes derived from live cells. This is frequently accomplished by introducing a fusion protein containing one or more highly specific tags, either an epitope for antibody-based capture and/or a protein domain that imparts a specific binding property (e.g., protein-A, calmodulin binding peptide). The basis for affinity complex purification is the solubilization of stable protein complexes that can be captured and selectively purified for identification of constituents by mass spectrometry. In principle, this approach also permits discrimination of multiple complexes that can be separated by various properties such as size. The main advantage of this method is that isolated complexes contain endogenous proteins that are interacting within their normal cellular context. Disadvantages include loss of weak interactions and difficulty detecting transient interactions. These effects may be exacerbated by intrinsic insolubility of the complexes. Additional complications may arise during the unavoidable admixture of proteins from different cellular compartments that occurs during cell lysis. These complications can be further compounded by the inability to apply stringent wash conditions (which may cause complex dissociation). Taken together, these effects may result in a considerable background level of irrelevant proteins. Of more practical concern, it is generally not possible to derive information concerning protein proximity when employing traditional complex purification.
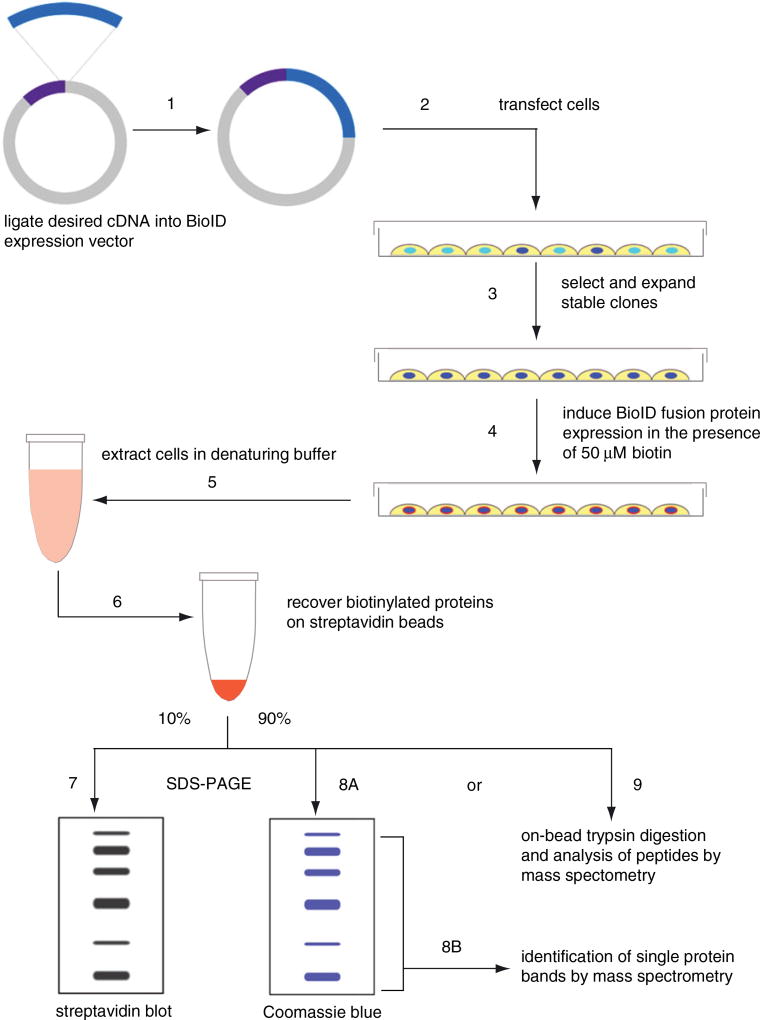
Figure 19.23.1.
Outline of the BioID method. The following steps outline the BioID process. (1) Ligate bait cDNA and BioID in-frame and in an appropriate expression vector. (2) Express BioID fusion protein in cells to (3) generate stably expressing cells. (4) Induce biotinylation with excess biotin prior to (5) cell lysis and protein denaturation. (6) Perform biotin-affinity capture of biotinylated proteins. (7) Reserve 10% of the sample for streptavidin immunoblot analysis. Use the remainder of the sample for mass spectrometry either by (8A) SDS-PAGE separation with (8B) identification of proteins in isolated bands or (9) analysis of peptides released by on-bead digestion.
BioID has its own intrinsic advantages and limitations. BioID fusion proteins label adjacent proteins over a period of time in a natural cellular setting. All of the proteins are denatured and solubilized, and only those that are biotinylated are selectively captured. This makes the method generally insensitive to protein solubility or aggregation/filament formation, with obvious applicability to membrane proteins and cytoskeletal constituents, a major advantage for BioID as compared to complex purification or Y2H. Its mechanism of action also permits the identification of weak or transient interactions that are generally lost with complex purification. The ability to regulate the biotinylation process, essentially a built-in inducibility, is accomplished with the necessary addition of excess biotin to the cell culture medium. This feature can potentially be used to provide low-resolution kinetic data; however, the optimal times for biotinylation are over 12 hr, which precludes short labeling periods that capture a snapshot of protein associations. BioID does excel at generating a history of protein associations over time that would enable identification of transient or even possibly low-frequency associations. Limitations of BioID include the need to express an exogenous fusion protein, although the levels of that protein need not be great. The addition of biotin ligase, a protein somewhat larger than GFP, may impair normal targeting, stability, or function; however, the smaller biotin ligase, BioID2, may be the better option if faced with these issues. Another limitation is that the identity of a candidate protein does not immediately imply a direct or indirect interaction with the bait, but could reflect a close proximity [currently estimated to be ~10 to 15 nm (Kim et al., 2014)]. It is possible that false negatives may occur if candidates lack accessible primary amines for biotinylation. The permanent addition of biotin to primary amines may alter protein function by modifying charge and removing accessibility of those sites for alternative modifications. Negative consequences from expression of a BioID fusion protein have not been observed, but must be considered possible.
Critical Parameters and Troubleshooting
Troubleshooting is most likely required during two distinct stages. The first is the initial creation of the BioID fusion protein expression plasmid, where difficulties with molecular cloning can slow the process considerably; however, this is not specific to BioID. The second area where difficulties are likely to be encountered is the establishment of cells stably expressing a low level of the fusion protein. If random integration of episomal DNA is the method utilized to generate these cells, multiple rounds of subcloning may be needed to obtain a relatively pure population of cells expressing the desired level of the fusion protein. A retroviral or lentiviral approach is often optimal to rapidly achieve stable low-level expression of BioID fusion protein in mammalian cell lines.
The subcellular locations in which BioID has successfully been applied include the centrosome, nucleus, cytoplasm, Golgi, ER, endosome, lysosome, mitochondrial matrix, cell-cell junctions, and flagella (Kim & Roux, 2016). Other cellular compartments remain untested. The efficiency of BioID in the ER appears lower than that in the other compartments, perhaps due to availability of biotin or ATP. However, the efficiency in the ER lumen is sufficient to yield more than 80 candidate proteins from a standard BioID pull-down (D.I.K. and K.J.R., unpub. observ.).
BioID constructs have now been successfully expressed in many different cell lines and have also been expressed in several different unicellular organisms including Saccharomyces cerevisiae, Trypanasoma brucei, Toxoplasma gondii, and Dictyostelium discoideum (Batsios, Meyer, & Graf, 2016; Morriswood et al., 2013; Opitz et al., 2017). BioID has been used in mouse tumor xenograft studies and via viral transduction in the mouse brain (Chan et al., 2014; Uezu et al., 2016). Aprotein-complementation-based split-BioID has been generated in which the ligase is only active when two proteins, each fused to a complementary fragment of the ligase, are brought together by an interaction (De Munter et al., 2017).
To extend the labeling radius of BioID a linker can be added between the BioID enzyme and the bait protein (Kim et al., 2016a). This may extend the range of biotinylation and/or provide the enzyme with increased accessibility to label adjacent proteins. It is worth noting that depending on restriction site usage within the multiple cloning site of a BioID expression plasmid, a linker of variable length may be unintentionally generated.
This protocol does not provide highly specific details for identification of proteins by mass spectrometry. The original BioID experiments utilized on-bead tryptic digestion to release peptides for analysis by 1-D LC-MS/MS (Roux et al., 2012). Advantages of on-bead digestion include circumventing the difficulty in removing biotinylated proteins/peptides from the streptavidin matrix without also removing the streptavidin itself, which may interfere with the mass spectrometry. An additional benefit from this approach is elimination of the SDS-PAGE separation of proteins, a common source of keratin contamination and mass-spectrometry interference from residual SDS.
Anticipated Results
The first major milestone in the process of BioID is the generation of cells that stably express the BioID fusion protein. Ideally, this population of expressing cells should be as close as possible to 100% of the total cells. As that number drops, there will be an increase in the relative amount of background proteins at the expense of candidates unique to the fusion protein. The percentage of cells expressing is determined by fluorescence microscopy (see Fig. 19.23.2A, for example). At this point it is worth confirming that the fusion protein is properly targeted. Sometimes, especially with fusion proteins in which BioID is in the ER lumen, the targeting and levels of biotinylation are not consistent until the protein is stably expressed. This may be due to excessive levels of fusion protein generated by transient transfection. Another outcome to anticipate at this stage in the process is that these stably expressing cells actually make a fusion protein of the expected molecular weight and that endogenous proteins are indeed being biotinylated. This is accomplished by immunoblot analysis of the fusion protein and streptavidin-HRP detection of the biotinylated proteins (see Fig. 19.23.2B, for example). At this point it is also useful to try to evaluate the level of expression of the fusion protein as compared to its endogenous counterpart, if one exists. In most cases, the lowest level of expression needed to detect biotinylation of endogenous proteins is optimal.
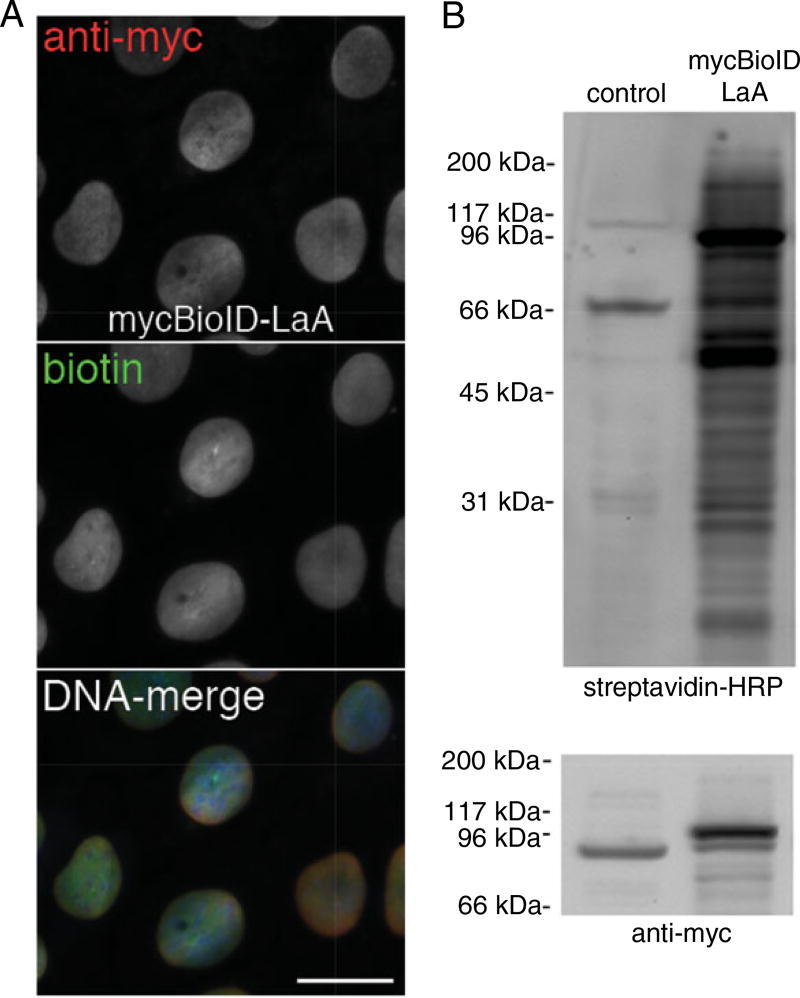
Figure 19.23.2.
Analysis of cells stably expressing a BioID-fusion protein. U2OS cells stably expressing BioID-lamin A (LaA) were analyzed prior to large-scale pull-down experiments. (A) By fluorescence microscopy it can be observed that all of the cells express similar levels of the myc-tagged BioID-LaA protein (red) targeted to the nuclear envelope. Following 24 hr incubation with excess biotin, the vast majority of the biotin signal, detected with streptavidin–Alexa Fluor 488 (green), co-localizes with the fusion protein. DNA is detected with Hoechst dye (blue). Scale bar is 15 µm. (B) Following SDS-PAGE separation, the protein constituents of BioID-LaA and control U2OS cells were probed with both streptavidin-HRP (top) and anti-myc (bottom). As compared to the few naturally biotinylated proteins in the control sample, there is extensive biotinylation of endogenous proteins in the BioID-LaA sample.
The ultimate outcome is the identification of proteins that interact with and/or are proximate to the BioID fusion protein. For a full-scale BioID pull-down, the results will likely arrive as a list of proteins that were identified by mass spectrometry. This list should contain not only the names of the proteins but the peptide sequences and number of times each peptide was identified. This provides some information as to the relative abundance of that protein in the sample. In parallel, there will be results from the BioID-only control cells. This list usually contains the five naturally biotinylated carboxylases (pyruvate carboxylase, PC; methylcrotonoyl-CoA carboxylase subunit alpha, MCC1; acetyl-CoA carboxylase 1, ACACA; acetyl-CoA carboxylase 1, ACACB; propionyl-CoA carboxylase alpha chain, PCCA), contaminating keratins and ribosomal proteins, and histones, as well as proteins that appear to be preferentially biotinylated by BioID-only including AHNAK, FLNA, PARP1, EEF1A1, PRKDC, TOP1, and PKM which are typically abundant in control pull-downs. These are to be subtracted from the mass spectrometry results from the BioID fusion protein samples. These background proteins can be subtracted based on their relative abundance depending on the MS approaches and analysis used. It is cost-effective to coordinate experiments so that a single control is used for several BioID pull-downs, as long as the control pull-down is performed in parallel under the same conditions and in the same cellular background as the BioID fusion proteins. BioID typically results in a relatively large list of candidates, from 50 to several hundred, depending on the experimental conditions and fusion protein. These can be ranked by abundance and/or based on what is already known about those proteins. It should be noted that the abundance, or lack thereof, does not necessarily imply biological relevance, and the user will have to make decisions as how to best utilize the results.
Time Considerations
The process of generating the BioID fusion protein expression plasmid by PCR cloning and validation of the fusion protein by transient transfection can be accomplished in as little as 1 week. Generation and selection of stably expressing cells could take as little as 1 week if viral expression is utilized or 3 to 4 weeks if relying on random integration following transient transfection and selection. A large-scale BioID pull-down can be accomplished in under 1 week. This includes the time required for the generation of sufficient cell numbers (typically 4 × 107 cells). Identification of candidates by mass spectrometry typically takes 1 to 2 weeks. Thus, a standard BioID experiment can be accomplished in 1 to 2 months from start to finish. However, this may take significantly longer if any difficulties are encountered. These delays would most likely occur during PCR cloning and generation of stable cell lines.
Acknowledgments
This work was supported by Sanford Research, R01GM102203 and P20GM103620 from the National Institutes of Health (K.J.R., D.I.K., D.G.M.) and the Agency for Science, Technology and Research (A*STAR, Singapore) (B.B.). The authors acknowledge Pete Vitiello, Morgan Greenfield, and K.C. Birendra for their helpful discussions.
Footnotes
Internet Resources
https://www.addgene.org/Kyle_Roux
BioID and BioID2 plasmids can be obtained from the non-profit plasmid repository Addgene.
https://www.sanfordresearch.org/researchcenters/childrenshealth/rouxlab/bioidresources/
Up-to-date information on BioID.
Literature Cited
- Batsios P, Meyer I, Graf R. Proximity-dependent biotin identification (BioID) in Dictyostelium amoebae. Methods in Enzymology. 2016;569:23–42. doi: 10.1016/bs.mie.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Chan P, Srikumar T, Dingar D, Kalkat M, Penn L, Raught B. BioID data of c-MYC interacting protein partners in culutured cells and xenograft tumors. Data Brief. 2014;1:76–78. doi: 10.1016/j.dib.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, Van C, Bradley PJ. Novel components of the Toxoplasma inner membrane complex revealed by BioID. MBio. 2015;6(1):e02357–14. doi: 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Science. 2004;13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE. Targeted and proximity-dependent promiscuous protein biotinylation by a mutant Escherichia coli biotin protein ligase. Journal of Nutritional Biochemistry. 2005;16:416–418. doi: 10.1016/j.jnutbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- De Munter S, Gornemann J, Derua R, Lesage B, Qian J, Heroes E, Bollen M. Split-BioID: A proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Letters. 2017;591(2):415–424. doi: 10.1002/1873-3468.12548. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Immunofluorescence staining. Current Protocols in Cell Biology. 2001;00:4.3.1–4.3.6. doi: 10.1002/0471143030.cb0403s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA, Marina P, Yu L. Constructing recombinant DNA molecules by PCR. Current Protocols in Molecular Biology. 2007;78:3.17.1–3.17.12. doi: 10.1002/0471142727.mb0317s78. [DOI] [PubMed] [Google Scholar]
- Gallagher SR. One-dimensional SDS gel electrophoresis of proteins. Current Protocols in Protein Science. 2012;68:10.1:10.1.1–10.1.44. doi: 10.1002/0471140864.ps1001s68. [DOI] [PubMed] [Google Scholar]
- Goldman A, Ursitti JA, Mozdzanowski J, Speicher DW. Electroblotting from polyacrylamide gels. Current Protocols in Protein Science. 2015;82:10.7.1–10.7.16. doi: 10.1002/0471140864.ps1007s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(24):E2453–2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Jensen SC, Noble KA, Birendra KC, Roux KH, Motamedchaboki K, Roux KJ. An improved smaller biotin ligase for BioID proximity labeling. Molecular Biology of the Cell. 2016;27(8):1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Roux KJ. Filling the void: Proximity-based labeling of proteins in living cells. Trends in Cell Biology. 2016;26(11):804–817. doi: 10.1016/j.tcb.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE. Introduction of DNA into mammalian cells. Current Protocols in Molecular Biology. 2003:9.0.1–9.0.5. doi: 10.1002/j.1934-3647.1990.tb00198.x. [DOI] [PubMed] [Google Scholar]
- Kwon K, Beckett D. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Science. 2000;9:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Warren G. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryotic Cell. 2013;12:356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni D, Xu P, Gallagher S. Immunoblotting and immunodetection. Current Protocols in Protein Science. 2017;88:10.10.1–10.10.37. doi: 10.1002/cpps.32. [DOI] [PubMed] [Google Scholar]
- Opitz N, Schmitt K, Hofer-Pretz V, Neumann B, Krebber H, Braus GH, Valerius O. Capturing the Asc1p/RACK1 microenvironment at the head region of the 40S ribosome with quantitative BioID in yeast. Molecular and Cell Proteomics. 2017 doi: 10.1074/mcp.M116.066654. pii: mcp.M116.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. Journal of Cell Biology. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, Soderling SH. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353(6304):1123–1129. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Aponte A, Tietgens AJ, Gucek M, Fredriksson K, Anderson JM. The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. Journal of Biological Chemistry. 2013;288:13775–13788. doi: 10.1074/jbc.M113.466193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nature Biotechnology. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]