Abstract
Purpose:
A number of studies have tested the hypothesis that breast cancer patients with low-activity CYP2D6 genotypes achieve inferior benefit from tamoxifen treatment, putatively due to lack of metabolic activation to endoxifen. Studies have provided conflicting data, and meta-analyses suggest a small but significant increase in cancer recurrence, necessitating additional studies to allow for accurate effect assessment. We conducted a retrospective pharmacogenomic analysis of a prospectively collected community-based cohort of patients with estrogen receptor-positive breast cancer to test for associations between low-activity CYP2D6 genotype and disease outcome in 500 patients treated with adjuvant tamoxifen monotherapy and 500 who did not receive any systemic adjuvant therapy.
Methods:
Tumor-derived DNA was genotyped for common, functionally consequential CYP2D6 polymorphisms (*2, *3, *4, *6, *10, *41 and copy number variants) and assigned a CYP2D6 activity score (AS) ranging from none (0) to full (2). Patients with poor metabolizer (PM, AS=0) phenotype were compared to patients with AS>0 and in secondary analyses AS was analyzed quantitatively. Clinical outcome of interest was recurrence free survival (RFS) and analyses using long-rank test were adjusted for relevant clinical covariates (nodal status, tumor size, etc.).
Results:
CYP2D6 AS was not associated with RFS in tamoxifen treated patients in univariate analyses (p>0.2). In adjusted analyses, increasing AS was associated with inferior RFS (Hazard ratio (HR)=1.43, 95% confidence interval: 1.00–2.04, p=0.05). In patients that did not receive tamoxifen treatment, increasing CYP2D6 AS, and AS>0, were associated with superior RFS (each p=0.0015).
Conclusions:
This population-based study does not support the hypothesis that patients with diminished CYP2D6 activity achieve inferior tamoxifen benefit. These contradictory findings suggest that the association between CYP2D6 genotype and tamoxifen treatment efficacy is null or near null, and unlikely to be useful in clinical practice.
Keywords: Pharmacogenetic, CYP2D6, tamoxifen, prognostic, predictive
Introduction
The selective estrogen receptor modulator (SERM) tamoxifen, and third generation aromatase inhibitors (AI) including anastrozole, letrozole, and exemestane have played a substantial role in decreasing breast cancer mortality, especially when used in the adjuvant setting[1]. Approximately 60–70% of newly diagnosed breast cancers are estrogen receptor (ER)-positive, but only 60% of these will respond to therapy[2]. It is not currently possible to identify which patients with ER-positive cancers will respond to anti-estrogens nor is it possible to determine whether a particular treatment (tamoxifen or an AI) will be more effective for an individual patient.
Tamoxifen is an estrogen receptor antagonist in breast cancer cells, accounting for its favorable anti-neoplastic effect. There are data suggesting that the effectiveness of tamoxifen can be partially attributed to its metabolic activation to more potent anti-estrogenic metabolites including 4-OH-tamoxifen (4-OH-tam) and 4-OH-N- desmethyl-tamoxifen, also called endoxifen[3]. This bioactivation is mediated primarily by the cytochrome P450 (CYP) family member 2D6 (CYP2D6), which shows large phenotypical variations due to genetic polymorphisms[4]. Though hundreds of polymorphisms have been identified, the majority of variation in metabolic activity can be accounted for by a relatively small number of no function (*3, *4, *6) or diminished function (*10, *41) alleles[5].
Low-activity polymorphisms in CYP2D6 are associated with decreased plasma concentrations of endoxifen[6,7,3,8]. We and others have hypothesized that tamoxifen efficacy would be diminished in patients who have lower endoxifen concentration[9,10] or carry low-activity CYP2D6 polymorphisms[11–14], however, these associations have not been established[15,16]. These inconsistent results can be attributed to a number of factors including differences in patient, tumor, or treatment characteristics or incomplete genotyping analysis[17,18]. Alternatively, another explanation for the varying results is that the underlying hypothesis is null or near null[19,20]. Given the potential clinical significance of a predictive biomarker for tamoxifen efficacy, it is necessary to conduct additional studies in large cohorts of tamoxifen treated patients.
Here we report the results from a retrospective pharmacogenetic analysis of a large prospectively collected patient cohort. Specifically, our objective was to test for an association between CYP2D6 phenotype and benefit of tamoxifen[21], utilizing DNA from tumors collected from patients treated with surgery and adjuvant tamoxifen or with surgery only (n=500 for each group). Our prespecified hypothesis was that patients who had low CYP2D6 metabolic activity, based on CYP2D6 genotype, would have worse treatment outcomes in the tamoxifen treated cohort but similar outcomes in the surgery only group.
Methods:
Patient Cohort
This secondary analysis was performed using patients from two breast cancer databases and corresponding biobanks maintained by the Breast Center at Baylor College of Medicine (BCM, Houston, TX) that have been previously described in detail[22]. Briefly, the PPG/P01 database and biobank, funded by the National Cancer Institute (Bethesda, MD), collected tissue and data on disease, adjuvant treatment and outcomes from community physicians for patients with early breast cancer diagnosed between 1970 and 1999. Clinical characteristics and outcomes were similar to data from the Surveillance, Epidemiology and End Results Registry for the same period. The second database and biobank, maintained as part of a Breast Cancer Specialized Program of Research Excellence (SPORE) grant funded by the National Cancer Institute, collected similar tissue and clinical data, with follow-up information coming from tumor registries for patients with early breast cancer who were diagnosed and treated between 1984 and 1999 from community hospitals throughout the United States. Comparison to SEER data for early breast cancer from more or less the same time period suggests that death has been very reliably ascertained, while disease recurrence was slightly underascertained. This is expected, given that data derived from hospital tumor registries not MD offices. The effective sample size slightly reduced; however, there is no reason to think there is a difference in completeness by genotype.
Selection of patients from these databases for the BCM Breast Tumor DNA Bank-v1 has been described previously[22]. Briefly, Caucasian patients from either database with ER+ tumors (>3 fmol/mg protein) that received surgery and tamoxifen (“treated”, n=500) or no systemic treatment (“untreated”, n=500), had complete patient and tumor information, and sufficient banked tumor material, were selected. Treatment within these observational registries was in accordance with standard clinical practice. Duration of tamoxifen therapy reflects community practice during the time period and patient/physician preference. No patients were treated with adjuvant chemotherapy. A total of n=213, and n=787 samples came from the P01 and SPORE banks, respectively.
Genotyping and CYP2D6 Phenotype Assignment
Fresh, whole-tumor specimens were flash frozen and maintained in the biobank. These specimens thawed for approximately three days during a tropical storm that flooded the biobank, prior to being refrozen. DNA was isolated using Puregene® DNA Purification Kit (Qiagen) in the BCM Genetics Core. The DNA samples were genotyped for CYP2D6 gene variants using the Taqman® Allelic Discrimination assays (Applied Biosystems, Inc., Foster City, CA) as described previously[7]. The CYP2D6 gene variants determined include: *2 (rs1135840), *3 (rs35742686), *4 (rs3892097), *6 (rs5030655), *10 (rs1065852), *41 (rs28371725), Assays were run in a Step-One Plus instrument (Applied Biosystems, Inc. Foster City, CA). Detailed information on CYP2D6 allele nomenclature can be found at http://www.cypalleles.ki.se/cyp2d6.htm. Call rate for each allele genotyped was >99%; random selection and re-genotyping of approximately 10% of the samples yielded concordance >99%. Hardy-Weinberg Equilibrium (HWE) was assessed for each polymorphism via exact tests by using the HWE function in the R package ‘genetics’. Each patient was assigned a predicted CYP2D6 phenotypic activity score (AS) based on the method recommended by PharmGKB by adding the AS assigned to each of the patient’s two alleles (*3, *4, *4xN, *6, *6xN = 0; *10, *41 = 0.5; *1, *2, *10xN, *41xN = 1; *1xN, *2xN = 2). Each patient’s AS was then transformed into a predicted CYP2D6 metabolizer activity phenotype (poor metabolizer (PM)=0, intermediate (IM)=0.5, extensive (EM)=1.0–2.0, and ultra-rapid (UM)>2.0).[23,24]
Statistical Analyses
This analysis had a pre-specified primary outcome and method of quantifying CYP2D6 activity, and was calculated to have 80% power to detect a hazard ratio (HR) of 2.5 assuming 6% of cases were PM and a sample size of n=500 in each group. The primary endpoint for all analyses was recurrence free survival, defined as the period of time following surgery until first recurrence or death, or censoring due to loss of followup. Overall survival (OS), the time from diagnosis to death or censoring due to loss of follow-up, was used for secondary analyses. Due to the sparseness of very long-term follow-up data, all patients and analyses were censored at 150 months (12.5 years) of follow up. Survival curves were estimated using Kaplan-Meier method. Clinical characteristics and tendency to be treated with tamoxifen differed between patients obtained from the P01 and SPORE databases, therefore, analyses were stratified by database.
Clinical characteristics, genotype frequencies, and outcomes were compared between tamoxifen treated and tamoxifen untreated patients using Chi square or Wilcoxon Rank-Sum tests, as appropriate. Cox proportional hazards analysis was used to identify clinical factors (age, progesterone receptor (PR) status, nodal status, tumor size, database) significantly associated with outcome. Genotype data was defined in two ways, in the primary analysis, PM patients (AS=0) were compared to all other patients (AS>0) and in secondary analyses the AS (0–3) was analyzed as a continuous variable. Associations between CYP2D6 PM status (AS=0) and prognostic clinical variables (age, nodal status, tumor size) were analyzed separately in the tamoxifen treated and untreated cohorts using chi square and Fisher’s exact tests, as appropriate. Statistical significance of a relationship between genotype and treatment outcomes were assessed using the log-rank test independently in the tamoxifen treated and untreated cohorts. Schoenfeld residuals were inspected and the proportional hazards assumption was tested using the Kolmogorov-type supremum test on 1000 simulated patterns. Variables that violated the proportional hazards assumption (database in all models and PR status in the untreated and combined model) were used as stratifiers in subsequent models. Multivariable models were constructed including significant clinical variables and CYP2D6 genotype to test for independent contribution of CYP2D6. All statistical analyses were performed using SAS v9.3 with two-tailed tests and a standard significance threshold of p<0.05.
Results
Patient Characteristics
After exclusion of patients missing genetic or clinical information, 476 patients who received adjuvant tamoxifen and 481 patients who did not receive any adjuvant treatment were evaluable for pharmacogenetic analyses (Figure 1). The demographic, disease, and treatment characteristics of the patients are reported in Table 1. All tumors were ER+ and 77% were PR+. In general, this patient population has favorable prognostic features such as small tumors (48% <2 cm) and low rates of metastasis (66% node negative). There are significant differences between patients who received tamoxifen treatment and those who did not in several of the patient and tumor characteristics including age, tumor size, and nodal status. This expected finding reflects the nature of the non-randomized and community-based cohorts; retrospective population based analysis; patients with more aggressive tumors were more likely to receive additional adjuvant treatment, as decided by their treating physicians. The median follow-up for patients was 121 and 124 months for tamoxifen-treated and untreated patients, respectively.
Fig 1.
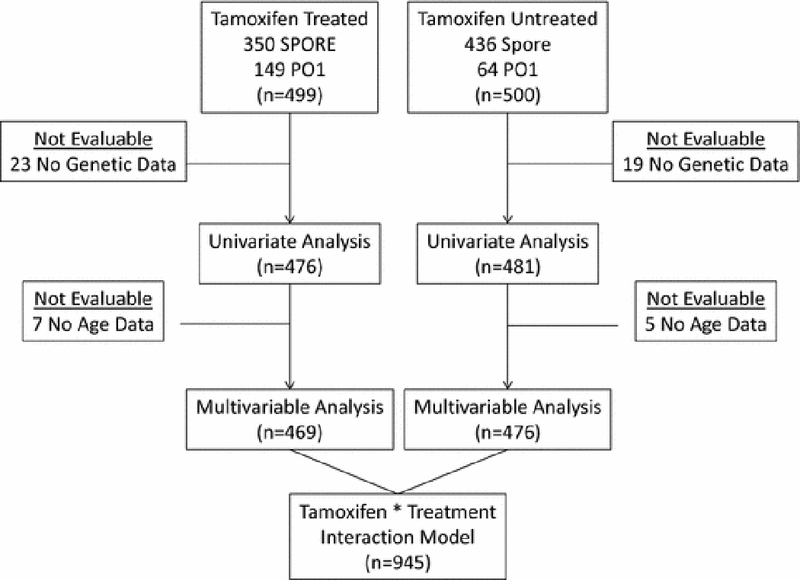
Consort diagram depicting the patient flow from initial selection from the SPORE or P01 databases into the final analysis.
Table 1:
Summary of patient, tumor, genetic, and outcomes data in tamoxifen treated and untreated cohorts.
| Tamoxifen Treated (n=476) | Tamoxifen Untreated (n=481) | Tamoxifen treated vs. untreated p-value | ||||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Source Bank | P01 | 148 | 31.1 | 59 | 12.3 | <0.0001 |
| SPORE | 328 | 68.9 | 422 | 87.7 | ||
| Agea | Age <50 | 29 | 6.2 | 74 | 15.5 | <0.0001 |
| Age > 50yrs | 440 | 93.8 | 402 | 84.5 | ||
| Median | 67.0 | 66.0 | 0.024 | |||
| PR | Negative: < 5 fmol/mg | 103 | 21.6 | 117 | 24.3 | 0.32 |
| Positive: > 5 fmol/mg | 373 | 78.4 | 364 | 75.7 | ||
| Tumor size | < 2 cm | 189 | 39.7 | 272 | 56.6 | <0.0001 |
| >2 and < 5 cm | 264 | 55.5 | 191 | 39.7 | ||
| >5 cm | 23 | 4.8 | 18 | 3.7 | ||
| Nodal Status | Negative | 241 | 50.6 | 388 | 80.7 | <0.0001 |
| 1–3 Positive Nodes | 163 | 34.2 | 66 | 13.7 | ||
| 4–9 Positive Nodes | 51 | 10.7 | 20 | 4.2 | ||
| >10 Positive Nodes | 21 | 4.4 | 7 | 1.5 | ||
| CYP2D6 Phenotype | Poor Metabolizer | 28 | 5.9 | 30 | 6.2 | 0.0008 |
| Intermediate Metabolizer | 15 | 3.2 | 23 | 4.8 | ||
| Extensive Metabolizer | 430 | 90.3 | 406 | 84.4 | ||
| Ultra-Rapid Metabolizer | 3 | 0.6 | 22 | 4.6 | ||
| Months of follow-upb | Median | 110.7 | 115.0 | N/A | ||
5 patients from tamoxifen treated and 7 from untreated groups missing age information
Truncated at 150 months
Association between Clinical Variables and Treatment Outcome
Differences between patient cohorts from the two DNA banks was highly significantly associated with RFS (Table 2) and OS (data not shown) in both the tamoxifen treated and untreated cohorts, therefore, all analyses were stratified according to the two cohorts. As expected, age, tumor size, and nodal status were independently associated with RFS in the treated and untreated cohorts (all univariate p<0.05). PR status was not associated with outcome (p=0.32).
Table 2:
Associations with Recurrence Free Survival in Tamoxifen Treated and Untreated Patients in Univariate and Multivariable Analyses.
| Model/Variable | HR | 95% Cl | p-value | HR | 95% Cl | p-value |
|---|---|---|---|---|---|---|
| Univariate Analysis | Tamoxifen Treated (n=476) | Tamoxifen Untreated (n=481) | ||||
| CYP2D6 non-PM Status (AS>0) | 0.68 | 0.33–1.40 | 0.29 | 0.44 | 0.22–0.89 | 0.023 |
| CYP2D6 AS (continuous) | 1.16 | 0.84–1.62 | 0.37 | 0.72 | 0.52–1.00 | 0.051 |
| Age (continuous) | 0.98 | 0.96–1.00 | 0.07 | 0.98 | 0.97–1.00 | 0.044 |
| Age (>50 vs. <50) | 0.67 | 0.34–1.32 | 0.24 | 0.71 | 0.41–1.23 | 0.22 |
| PR status (Positive vs. negative) | 0.83 | 0.53–1.31 | 0.43 | 0.66 | 0.41–1.09 | 0.10 |
| Tumor Size (vs. <2 cm) | ||||||
| >2 and <5 cm | 2.01 | 1.28–3.14 | 0.0003 | 2.43 | 1.50–3.94 | 0.0002 |
| >5 cm | 4.03 | 1.95–8.35 | 4.12 | 1.70–9.99 | ||
| Nodes (vs. negative) | ||||||
| 1–3 | 1.29 | 0.80–2.07 | <0.0001 | 1.85 | 1.04–3.28 | 0.010 |
| 4–9 | 3.77 | 2.24–6.36 | 1.47 | 0.46–4.70 | ||
| >10 | 6.52 | 3.39–12.54 | 4.41 | 1.60–12.20 | ||
| Database (P01 vs. SPORE)a | 2.06 | 1.40–3.05 | 0.0003 | 4.46 | 2.70–7.36 | <0.0001 |
| CYP2D6 PM Status Multivariable Modelb | Tamoxifen Treated | (n=469)d | Tamoxifen Untreated (n=476)d | |||
| Age (continuous) | 0.98 | 0.96–1.00 | 0.059 | 0.99 | 0.97–1.01 | 0.19 |
| Tumor Size | ||||||
| >2 and <5 cm | 1.77 | 1.12–2.79 | 0.034 | 1.84 | 1.10–3.09 | 0.014 |
| >5 cm | 2.07 | 0.96–4.46 | 3.20 | 1.27–8.06 | ||
| Nodes | ||||||
| 1–3 | 1.12 | 0.69–1.83 | <0.0001 | 1.33 | 0.72–2.47 | 0.44 |
| 4–9 | 3.11 | 1.82–5.34 | 1.19 | 0.36–3.93 | ||
| >10 | 5.14 | 2.49–10.62 | 2.26 | 0.76–6.74 | ||
| CYP2D6 non-PM Status | 1.11 | 0.50–2.44 | 0.80 | 0.41 | 0.20–0.84 | 0.015 |
| CYP2D6 AS Multivariable Modelc | 1.43 | 1.00–2.04 | 0.050 | 0.66 | 0.47–0.92 | 0.015 |
Abbreviations: AS: Activity score, Cl: Confidence Interval, HR: Hazard Ratio, PM: Poor Metabolizer
Violated proportional hazards assumption
Analysis stratified by database (P01 and SPORE)
HR, 95% Cl, and p-values reported are for the covariates in the final multivariable that includes CYP2D6 non- PM status. Covariate values for the models with CYP2D6 AS were not meaningfully different.
Tamoxifen treated models were stratified by database, untreated and overall models were stratified by database and PR status.
12 Patients missing age data were excluded from multivariable analyses
Genotyping Results
The genotype counts for tamoxifen treated and untreated patients included in the analysis are reported in Supplementary Table 1. All minor allele frequencies were similar to expected frequencies in a predominantly Caucasian cohort[23]. Of note, the common no-activity CYP2D6*4 and diminished activity *41 alleles were within expected Hardy-Weinberg proportions. The CYP2D6*2 allele was not within the expected Hardy- Weinberg proportions, however, this is irrelevant as the *2 allele is categorized as metabolically normal (AS=1), similar to wild-type *1[24]. CYP2D6 diplotype was translated into a predicted activity phenotype for each patient (Supplementary Table 1).
Association between CYP2D6 and Prognostic Clinical Variables
CYP2D6 poor metabolizer status (AS=0) was not associated with age or tumor size in either the tamoxifen treated or untreated cohorts (all p>0.05, data not shown). A nominal association with nodal status was detected in the tamoxifen treated patients, in which patients with CYP2D6 PM status were more likely to have 10 or more positive nodes (5/28=17.9%) than patients with AS>0 (16/449=3.6%) (p=0.015). A similar association was not detected in the tamoxifen untreated patients (p=0.42), however, the association maintained significance when the treated and untreated cohorts were combined (p=0.026, Supplementary Table 2).
Association between CYP2D6 and Treatment Outcome in Tamoxifen Treated Patients
In the primary analysis there was no association between CYP2D6 non-PM status (AS>0) and RFS in tamoxifen treated patients (HR=0.68, 95% Confidence Interval: 0.33–1.40, p=0.29, Table 2, Figure 2 (left). A Cox-based survival curve assuming average clinical variables (1–3 nodes, tumor size of 2–5 cm, and 66.5 years of age) is depicted in Supplementary Figure 1 (left). Similarly, in a secondary analysis of AS, as a continuous variable, there was no association with RFS (HR=1.16, 95% CI: 0.84–1.62, p=0.37, Table 2). After adjusting for relevant clinical covariates (age, tumor size, positive nodes), CYP2D6 non-PM status (p=0.80) was not associated with RFS, however, there was a borderline significant association of worse RFS as CYP2D6 AS increased (HR=1.43, 95% CI: 1.00–2.04, p=0.05). CYP2D6 non-PM status (p=0.28) and AS (p=0.57) were not associated with OS in tamoxifen treated patients (data not shown).
Fig 2.
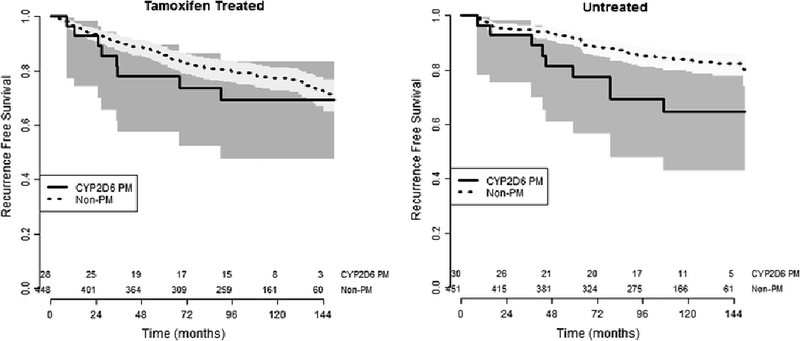
Recurrence free survival curves stratified by CYP2D6 PM Status including 95% confidence intervals (shaded areas) and number at risk (along X-axis). In tamoxifen treated patients (left) there was no association between CYP2D6 genotype and recurrence free survival. In the tamoxifen untreated cohort (right) the patients with CYP2D6 non-poor metabolizer phenotype had significantly better recurrence free survival ((HR=0.44, 95% CI: 0.22–0.89, p=0.023) than patients with poor metabolizer phenotype.
Association between CYP2D6 and Treatment Outcomes in Tamoxifen Untreated Patients
A parallel analysis was performed in the cohort of patients that did not receive adjuvant systemic treatment. In the univariate analysis CYP2D6 non-PM status was associated with superior RFS (HR=0.44, 95% CI: 0.220.89, p=0.023, Table 2, Figure 2 (right). A Cox-based survival curve assuming average clinical variables (1–3 nodes, tumor size of 2–5 cm, and 66.5 years of age) is depicted in Supplementary Figure 1 (right). In a secondary analysis of AS as a continuous variable, increasing AS was nearly significantly associated with improved RFS (HR=0.72, 95% CI: 0.51–1.00, p=0.051). In the multivariable model of RFS, nodal status did not maintain significance (p=0.44). In adjusted analyses patients with CYP2D6 non-PM status had superior RFS compared to patients with PM phenotype (HR=0.41, 95% CI: 0.20–0.84, p=0.015) and similar results were found when analyzing CYP2D6 AS as a continuous variable (HR=0.66, 95% CI: 0.47–0.92, p=0.015). CYP2D6 non-PM status (p=0.83) and AS (p=0.74) were not associated with OS in patients not receiving adjuvant treatment (data not shown).
Discussion
A number of studies have tested the hypothesis that patients with breast cancer who carry low-activity CYP2D6 genotypes have inferior tamoxifen treatment outcomes. A recent meta-analysis detected a small, but statistically significant, increase in tumor recurrence for patients with diminished CYP2D6 activity, particularly for those who carry two non-functional copies of CYP2D6 (PMs, AS=0)[16]. However, this meta-analysis relied on data from several independent studies, and there is concern that publication bias as well as exclusion of several large studies[14,12] may be artificially inflating meta-analysis estimates away from the null hypothesis[25–27]. Therefore, it is important that additional large, well-conducted analyses testing the CYP2D6/tamoxifen hypothesis are published, regardless of their findings. The current study utilized two large breast cancer registries and biobanks with long-term follow up to test for an association between CYP2D6 genotype and recurrence free survival in two subcohorts, one which received adjuvant tamoxifen treatment and the other that received no adjuvant treatment. All patients had ER+ tumors, did not receive adjuvant chemotherapy, and CYP2D6 allelic coverage was relatively comprehensive, three factors that have been identified as limitations of many of the previous retrospective studies[17]. In this analysis, there was no decrease in tamoxifen effectiveness for patients with CYP2D6 PM phenotype, though there was evidence of an association in the opposite direction when CYP2D6 activity score quantitatively was analyzed with adjustment for other important clinical characteristics. Additionally, in patients who did not receive adjuvant treatment, higher CYP2D6 metabolic activity was associated with superior outcomes.
After adjustment for clinical characteristics, we found that patients with low CYP2D6 activity have superior tamoxifen treatment outcomes. These data contradict the hypothesis that extent of metabolic activation of tamoxifen to endoxifen is a biomarker for therapeutic effectiveness and are consistent with two previous studies[28,29]. Analyses of the CYP2D6-tamoxifen hypothesis with the highest strength of evidence, Conducted in large prospective clinical trials, have yielded similarly conflicting results[14,12,13]. The potential biases and limitations of all studies to date has been discussed [30–33], but the overall equivocal results suggest that a true association, if one exists, is likely marginal and only detectable in the most highly selected cohorts. This conclusion is supported by the results of the meta-analysis from the International Tamoxifen Pharmacogenetics Consortium, which only detected an association with recurrence free survival in a carefully selected subcohort of the overall analysis population[16], a filtering process that itself was debated by the research community[26].
This study, unexpectedly, detected an improvement in RFS in patients with higher CYP2D6 activity in the cohort who did not receive adjuvant tamoxifen treatment. Inclusion of an untreated cohort in pharmacogenetic studies is necessary to differentiate between true pharmacogenetic effects that are predictive of treatment outcome and prognostic genetic effects[34–36]. If the tamoxifen/CYP2D6 hypothesis were true, one would expect to see patients with higher CYP2D6 activity have superior in outcomes in the tamoxifen treated cohort and similar outcomes in the tamoxifen untreated outcomes. In contrast, our results indicate that patients with higher CYP2D6 activity have superior outcomes in the tamoxifen untreated cohort and similar, or perhaps inferior, outcomes in the tamoxifen treated cohort. Contrary to the hypothesis, these results suggest that increased CYP2D6 activity may be a prognostic factor associated with superior treatment outcomes in patients not receiving systemic treatment. If true, this adds an additional layer of complexity to previous studies of the CYP2D6/tamoxifen hypothesis, which did not include a tamoxifen untreated control group. However, skepticism is warranted, as a plausible biological rationale for a prognostic effect of systemic CYP2D6 activity on ER+ breast cancer prognosis is not readily available. The physiological role of CYP2D6 is not well defined as few high-affinity endogenous substrates have been identified. CYP2D6 is responsible for O-demethylation of pinoline[37] and of 6-methoxytryptamine to serotonin[38], which may account for the well-established association between CYP2D6 activity and personality[39,40]. It is unlikely, though possible, that these physiological differences are related to prognosis of ER+ breast cancer. CYP2D6 has very weak affinity for testosterone[41], suggesting a possible relationship with ER+ breast cancer occurrence or prognosis, however, associations of CYP2D6 polymorphisms with occurrence of ER+ breast cancer have not been detected in very large genome-wide screens[42].
Genotyping for this analysis was performed using DNA isolated from whole-tumor specimens, and not from peripheral blood. Several studies have confirmed a near perfect concordance between CYP2D6 genotypes obtained from tumor and matched germline DNA[43–45] [46,11] and these are in contrast with a single study reporting some discordance between CYP2D6M genotypes, potentially due to somatic loss of heterozygosity (LOH)[47], Tumor LOH has been hypothesized to explain large deviations from HWE in the BIG 1–98 analysis[12,30]. In the present study, CYP2D6M was well within expected Hardy-Weinberg proportions, further refuting the hypothesis that tumor genotyping causes meaningful misclassification. Deviations from HWE seen in BIG 1–98 are more likely result from a well-known consequence of population ad mixture [48], similar to the deviations from HWE detected in the multi-center studies included in the ITPC, regardless of whether the genotyping was performed in DNA derived from blood ortumor[49]. Deviation from Hardy-Weinberg proportions for the *2 allele, and potential misclassification of *1 and *2, would have no effect on this analysis as both alleles are fully functional alleles with assigned AS=1, based on CPIC recommendations[24,5].
Other limitations of this analysis are also worth mentioning. The use of patients from non-trial-based breast cancer biobanks is subject to biases inherent in retrospective analyses[50], including under-ascertainment of recurrence, and several important data elements were not available for some or all patients including menopausal status (available for most), tamoxifen treatment duration and/or adherence, and concomitant administration of CYP2D6 inhibitors. Each of these variables has been hypothesized to be an important consideration in analyses of this pharmacogenetic association[17,16,51]. Given these limitations, it is critical that our current findings are interpreted in the context of the dozens of previously published studies. The inconsistency of these findings, spanning the full range of effect from protective, null, to enhanced risk, are consistent with random sampling from a distribution with a modest effect, at most. The marginal association, detectable only in carefully selected patient populations, and the relative infrequency of the PM phenotype (frequency«6% in Caucasian cohorts), further support recommendations of ASCO[52] and the NCCN[53] against genotyping CYP2D6 to guide tamoxifen treatment, despite confirmation that doing so is feasible and safe[54–58,15].
In conclusion, in this large, retrospective analysis, patients who received tamoxifen treatment with low-activity CYP2D6 genotype had similar, or perhaps slightly better, treatment outcomes compared with patients with normal or slightly diminished CYP2D6 activity. In a parallel analysis, patients with low CYP2D6 activity genotype who did not receive tamoxifen treatment had inferior treatment outcomes. These findings contradict the underlying hypothesis that low activity CYP2D6 genotype is associated with inferior tamoxifen benefit. These findings further suggest that the true association between CYP2D6 activity and tamoxifen effectiveness, if one exists, is unlikely to be clinically meaningful.
Supplementary Material
Supp Fig 1 Recurrence Free Survival in Tamoxifen Treated and Untreated Patients by CYP2D6 PM-Status. Cox-based survival curves assuming 1-3 nodes, tumor size 2-5 cm, and age 66.5 years. Recurrence free survival (RFS) was not associated with CYP2D6 PM-status in the tamoxifen treated patients (left). In the untreated patients (right), CYP2D6 non-PM status was associated with superior RFS (p=0.023). Shading indicates 95% confidence intervals of the RFS estimate.
Acknowledgements:
We would like to acknowledge the National Cancer Institute for the funding to collect and maintain these biobanks and databases through the following grants: the Specialized Program of Research Excellence (SPORE) (2P50_CA058183 and P50_CA186784) and the MOPP from (P01_CA30195). This study was supported in part by the Breast Cancer Research Foundation (BCRF) (N003173 to JMR) and the National Institutes of Health (GM099143 to JMR).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
This work was presented in part at the 2010 San Antonio Breast Cancer Symposium.
References
- 1.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ (2005) Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med 353 (17):1784–1792. doi:10.1056/NEJMoa050518 [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists’ Collaborative G (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. The Lancet 378 (9793):771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA (2003) Active Tamoxifen Metabolite Plasma Concentrations After Coadministration of Tamoxifen and the Selective Serotonin Reuptake Inhibitor Paroxetine. Journal of the National Cancer Institute 95 (23):1758–1764. doi:10.1093/jnci/djg108 [DOI] [PubMed] [Google Scholar]
- 4.Ingelman-Sundberg M (2005) Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J 5 (1):6–13. doi:10.1038/sj.tpj.6500285 [DOI] [PubMed] [Google Scholar]
- 5.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, Callaghan JT, Kharasch ED, Skaar TC, Clinical Pharmacogenetics Implementation C (2012) Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clinical pharmacology and therapeutics 91 (2):321–326. doi:10.1038/clpt.2011.287; 10.1038/clpt.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, Hillman G, Hayes DF, Stearns V, Flockhart DA (2006) Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clinical pharmacology and therapeutics 80 (1):61–74 [DOI] [PubMed] [Google Scholar]
- 7.Borges S, Desta Z, Jin Y, Faouzi A, Robarge JD, Philip S, Nguyen A, Stearns V, Hayes D, Rae JM, Skaar TC, Flockhart DA, Li L (2010) Composite Functional Genetic and Comedication CYP2D6 Activity Score in Predicting Tamoxifen Drug Exposure Among Breast Cancer Patients. The Journal of Clinical Pharmacology 50 (4):450–458. doi:10.1177/0091270009359182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee K-H, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA (2005) CYP2D6 Genotype, Antidepressant Use, and Tamoxifen Metabolism During Adjuvant Breast Cancer Treatment. JNatlCancer Inst 97 (1):30–39. doi:10.1093/jnci/dji005 [DOI] [PubMed] [Google Scholar]
- 9.Saladores P, Murdter T, Eccles D, Chowbay B, Zgheib NK, Winter S, Ganchev B, Eccles B, Gerty S, Tfayli A, Lim JS, Yap YS, Ng rC , Wong NS, Dent R, Habbal MZ, Schaeffeler E, Eichelbaum M, Schroth W, Schwab M, Brauch H (2015) Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. The pharmacogenomics journal 15 (1):84–94. doi:10.1038/tpj.2014.34 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madlensky L, Natarajan L, Tchu S, Pu M, Mortimer J, Flatt SW, Nikoloff DM, Hillman G, Fontecha MR, Lawrence HJ, Parker BA, Wu AHB, Pierce JP (2011) Tamoxifen Metabolite Concentrations, CYP2D6 Genotype, and Breast Cancer Outcomes. Clinical pharmacology and therapeutics 89 (5):718–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN (2005) Pharmacogenetics of Tamoxifen Biotransformation Is Associated With Clinical Outcomes of Efficacy and Hot Flashes. Journal of Clinical Oncology 23 (36):9312–9318. doi:10.1200/JCO.2005.03.3266 [DOI] [PubMed] [Google Scholar]
- 12.Regan MM, Leyland-Jones B, Bouzyk M, Pagani O, Tang W, Kammler R, Dell’orto P, Biasi MO, Thurlimann B, Lyng MB, Ditzel HJ, Neven P, Debled M, Maibach R, Price KN, Gelber RD, Coates AS, Goldhirsch A, Rae JM, Viale G, Breast International Group 1–98 Collaborative G (2012) CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1–98 trial. Journal of the National Cancer Institute 104 (6):441–451. doi:10.1093/jnci/djs125; 10.1093/jnci/djs125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goetz MP, Suman VJ, Hoskin TL, Gnant M, Filipits M, Safgren SL, Kuffel M, Jakesz R, Rudas M, Greil R, Dietze O, Lang A, Offner F, Reynolds CA, Weinshilboum RM, Ames MM, Ingle JN (2013) CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clinical cancer research : an official journal of the American Association for Cancer Research 19 (2):500–507. doi:10.1158/1078-0432.CCR-12-2153; 10.1158/1078–0432.CCR-12–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, Haynes BP, Salter J, Sestak I, Cuzick J, Dowsett M, trialists A (2012) CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. Journal of the National Cancer Institute 104 (6):452–460. doi:10.1093/jnci/djs126; 10.1093/jnci/djs126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertz DL, Rae JM (2016) One step at a time: CYP2D6 guided tamoxifen treatment awaits convincing evidence of clinical validity. Pharmacogenomics 17 (8):823–826. doi:10.2217/pgs-2016-0059 [DOI] [PubMed] [Google Scholar]
- 16.Province MA, Goetz MP, Brauch H, Flockhart DA, Hebert JM, Whaley R, Suman VJ, Schroth W, Winters, Zembutsu H, Mushiroda T, Newman WG, Lee MTM, Ambrosone CB, Beckmann MW, Choi JY, Dieudonne AS, Fasching PA, Ferraldeschi R, Gong L, Haschke-Becher E, Howell A, Jordan LB, Hamann U, Kiyotani K, Krippl P, Lambrechts D, Latif A, Langsenlehner U, Lorizio W, Neven P, Nguyen AT, Park BW, Purdie CA, Ouinlan P, Renner W, Schmidt M, Schwab M, Shin JG, Stingl JC, Wegman P, Wingren S, Wu AHB, Ziv E, Zirpoli G, Thompson AM, Jordan VC, Nakamura Y, Altman RB, Ames MM, Weinshilboum RM, Eichelbaum M, Ingle JN, Klein TE (2013) CYP2D6 Genotype and Adjuvant Tamoxifen: Meta-Analysis of Heterogeneous Study Populations. Clinical pharmacology and therapeutics doi: 10.1038/clpt.2013.186 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertz DL, McLeod HL, Irvin WJ Jr., (2012) Tamoxifen and CYP2D6: a contradiction of data. Oncologist 17 (5):620–630. doi: 10.1634/theoncologist.2011-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyotani K, Mushiroda T, Hosono N, Tsunoda T, Kubo M, Aki F, Okazaki Y, Hirata K, Takatsuka Y, Okazaki M, Ohsumi S, Yamakawa T, Sasa M, Nakamura Y, Zembutsu H (2010) Lessons for pharmacogenomics studies: association study between CYP2D6 genotype and tamoxifen response. Pharmacogenet Genomics 20 (9): 565–568 [DOI] [PubMed] [Google Scholar]
- 19.Lash TL, Lien EA, Sorensen HT, Hamilton-Dutoit S (2009) Genotype-guided tamoxifen therapy: time to pause for reflection? The Lancet Oncology 10 (8):825–833. doi:10.1016/S1470-2045(09)70030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cronin-Fenton DP, Damkier P, Lash TL (2014) Metabolism and transport of tamoxifen in relation to its effectiveness: new perspectives on an ongoing controversy. Future oncology (London, England) 10 (1): 107–122. doi:10.2217/fon.13.168 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lum DWK, Perei P, Hingorani AD, Holmes MV (2013) CYP2D6 Genotype and Tamoxifen Response for Breast Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 8 (10):e76648. doi: 10.1371/journal.pone.0076648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathiraja TN, Shetty PB, Jelinek J, He R, Hartmaier R, Margossian AL, Hilsenbeck SG, Issa J-PJ, Oesterreich S (2011) Progesterone receptor isoform-specific promoter méthylation —Association of PRA promoter méthylation with worse outcome in breast cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research 17 (12):4177–4186. doi: 10.1158/1078-0432.CCR-10-2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE (2012) Pharmacogenomics knowledge for personalized medicine. Clinical pharmacology and therapeutics 92 (4):414–417. doi: 10.1038/clpt.2012.96; 10.1038/clpt.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS (2008) The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clinical pharmacology and therapeutics 83 (2):234–242. doi: 6100406 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Ahern TP, Hertz DL, Damkier P, Ejlertsen B, Hamilton-Dutoit SJ, Rae JM, Regan MM, Thompson AM, Lash TL, Cronin-Fenton DP (2017) Cytochrome P-450 2D6 (CYP2D6) Genotype and Breast Cancer Recurrence in Tamoxifen-Treated Patients: Evaluating the Importance of Loss of Heterozygosity. American journal of epidemiology 185 (2):75–85. doi:10.1093/aje/kww178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry DA (2014) CYP2D6 genotype and adjuvant tamoxifen. Clinical pharmacology and therapeutics 96 (2): 138–140. doi:10.1038/clpt.2014.96 [doi] [DOI] [PubMed] [Google Scholar]
- 27.Zeng Z, Liu Y, Liu Z, You J, Chen Z, Wang J, Peng Q, Xie L, Li R, Li S, Qin X (2013) CYP2D6 polymorphisms influence tamoxifen treatment outcomes in breast cancer patients: a meta-analysis. Cancer Chemother Pharmacol 72 (2):287–303. doi:10.1007/s00280-013-2195-9 [DOI] [PubMed] [Google Scholar]
- 28.Wegman P, Vainikka L, Stal O, Nordenskjold B, Skoog L, Rutqvist L-E, Wingren S (2005) Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Research 7 (3):R284–R290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, MacLeod SL, Kadlubar FF, Ambrosone CB (2005) Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. vol 91 Springer Netherlands. doi:10.1007/s10549-004-7751-x [DOI] [PubMed] [Google Scholar]
- 30.Nakamura Y, Ratain MJ, Cox NJ, McLeod HL, Kroetz DL, Flockhart DA (2012) Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1–98 trial. Journal of the National Cancer Institute 104 (16):1264; author reply 1266–1268. doi:10.1093/jnci/djs304; 10.1093/jnci/djs304 [DOI] [PubMed] [Google Scholar]
- 31.Ratain MJ, Nakamura Y, Cox NJ (2013) CYP2D6 genotype and tamoxifen activity: understanding interstudy variability in methodological quality. Clin Pharmacol Ther 94 (2):185–187. doi:10.1038/clpt.2013.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rae JM, Leyland-Jones B, Regan M, Thompson AM (2015) RE: Loss of Heterozygosity at the CYP2D6 Locus in Breast Cancer: Implications for Germline Pharmacogenetic Studies. Journal of the National Cancer Institute 107 (5). doi:10.1093/jnci/djv065 [DOI] [PubMed] [Google Scholar]
- 33.Rae JM, Leyland-Jones B, Regan M (2014) Response. J Natl Cancer Inst 106 (5). doi:10.1093/jnci/dju064 [DOI] [PubMed] [Google Scholar]
- 34.Henry NL, Hayes DF (2006) Uses and abuses of tumor markers in the diagnosis, monitoring, and treatment of primary and metastatic breast cancer. Oncologist 11 (6):541–552. doi:10.1634/theoncologist.11-6-541 [DOI] [PubMed] [Google Scholar]
- 35.Gasparini G, Pozza F, Harris AL (1993) Evaluating the potential usefulness of new prognostic and predictive indicators in node-negative breast cancer patients. J Natl Cancer Inst 85 (15):1206–1219 [DOI] [PubMed] [Google Scholar]
- 36.Chen JJ, Lin WJ, Chen HC (2013) Pharmacogenomic biomarkers for personalized medicine. Pharmacogenomics 14 (8):969–980. doi:10.2217/pgs.13.75 [DOI] [PubMed] [Google Scholar]
- 37.Yu AM, Idle JR, Herraiz T, Kupfer A, Gonzalez FJ (2003) Screening for endogenous substrates reveals that CYP2D6 is a 5-methoxyindolethylamine O-demethylase. Pharmacogenetics 13 (6):307–319. doi:10.1097/01.fpc.0000054094.48725.b7 [DOI] [PubMed] [Google Scholar]
- 38.Yu AM, Idle JR, Byrd LG, Krausz KW, Kupfer A, Gonzalez FJ (2003) Regeneration of serotonin from 5- methoxytryptamine by polymorphic human CYP2D6. Pharmacogenetics 13 (3):173–181. doi:10.1097/01.fpc.0000054066.98065.7b [DOI] [PubMed] [Google Scholar]
- 39.Bertilsson L, Alm C, De Las Carreras C, Widen J, Edman G, Schalling D (1989) Debrisoquine hydroxylation polymorphism and personality. Lancet 1 (8637):555. [DOI] [PubMed] [Google Scholar]
- 40.Penas-Lledo EM, Llerena A (2014) CYP2D6 variation, behaviour and psychopathology: implications for pharmacogenomics-guided clinical trials. Br J Clin Pharmacol 77 (4):673–683. doi:10.1111/bcp.12227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geier M, Braun A, Fladischer P, Stepniak P, Rudroff F, Hametner C, Mihovilovic MD, Glieder A (2013) Double site saturation mutagenesis of the human cytochrome P450 2D6 results in regioselective steroid hydroxylation. The FEBS journal 280 (13):3094–3108. doi:10.1111/febs.12270 [DOI] [PubMed] [Google Scholar]
- 42.Ghoussaini M, French JD, Michailidou K, Nord S, Beesley J, Canisus S, Hillman KM, Kaufmann S, Sivakumaran H, Moradi Marjaneh M, Lee JS, Dennis J, Bolla MK, Wang Q, Dicks E, Milne RL, Hopper JL, Southey MC, Schmidt MK, Broeks A, Muir K, Lophatananon A, Fasching PA, Beckmann MW, Fletcher O, Johnson N, Sawyer EJ, Tomlinson I, Burwinkel B, Marme F, Guenel P, Truong T, Bojesen SE, Flyger H, Benitez J, Gonzalez-Neira A, Alonso MR, Pita G, Neuhausen SL, Anton-Culver H, Brenner H, Arndt V, Meindl A, Schmutzler RK, Brauch H, Hamann U, Tessier DC, Vincent D, Nevanlinna H, Khan S, Matsuo K, Ito H, Dork T, Bogdanova NV, Lindblom A, Margolin S, Mannermaa A, Kosma VM, Wu AH, Van Den Berg D, Lambrechts D, Floris G, Chang-Claude J, Rudolph A, Radice P, Barile M, Couch FJ, Hallberg E, Giles GG, Haiman CA, Le Marchand L, Goldberg MS, Teo SH, Yip CH, Borresen-Dale AL, Zheng W, Cai Q, Winqvist R, Pylkas K, Andrulis IL, Devilee P, Tollenaar RA, Garcia-Closas M, Figueroa J, Hall P, Czene K, Brand JS, Darabi H, Eriksson M, Hooning MJ, Koppert LB, Li J, Shu XO, Zheng Y, Cox A, Cross SS, Shah M, Rhenius V, Choi JY, Kang D, Hartman M, Chia kS , Kabisch M, Torres D, Luccarini C, Conroy DM, Jakubowska A, Lubinski J, Sangrajrang S, Brennan P, Olswold C, Slager S, Shen CY, Hou MF, Swerdlow A, Schoemaker MJ, Simard J, Pharoah PD, Kristensen V, Chenevix-Trench G, Easton DF, Dunning AM, Edwards SL (2016) Evidence that the 5p12 Variant rs10941679 Confers Susceptibility to Estrogen-Receptor-Positive Breast Cancer through FGF10 and MRPS30 Regulation. Am J Hum Genet 99 (4):903–911. doi:10.1016/j.ajhg.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rae JM, Cordero KE, Scheys JO, Lippman ME, Flockhart DA, Johnson MD (2003) Genotyping for polymorphic drug metabolizing enzymes from paraffin-embedded and immunohistochemically stained tumor samples. Pharmacogenetics 13 (8):501–507. doi:10.1097/01.fpc.0000054114.14659.77 [doi] [DOI] [PubMed] [Google Scholar]
- 44.Rae JM, Regan MM, Thibert JN, Gersch C, Thomas D, Leyland-Jones B, Viale G, Pusztai L, Hayes DF, Skaar T, Van Poznak C (2013) Concordance between CYP2D6 genotypes obtained from tumor-derived and germline DNA. J Natl Cancer Inst 105 (17):1332–1334. doi:10.1093/jnci/djt204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson A, Johnson A, Quinlan P, Hillman G, Fontecha M, Bray S, Purdie C, Jordan L, Ferraldeschi R, Latif A, Hadfield K, Clarke R, Ashcroft L, Evans D, Howell A, Nikoloff M, Lawrence J, Newman W (2010) Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Springer Netherlands. doi:10.1007/s10549-010-1139-x [DOI] [PubMed] [Google Scholar]
- 46.Ahern TP, Christensen M, Cronin-Fenton DP, Lunetta KL, Rosenberg CL, Sorensen HT, Lash TL, Hamilton-Dutoit S (2010) Concordance of metabolic enzyme genotypes assayed from paraffin-embedded, formalin-fixed breast tumors and normal lymphatic tissue. Clinical epidemiology 2:241 −246. doi:10.2147/CLEP.S13811 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goetz MP, Sun JX, Suman VJ, Silva GO, Perou CM, Nakamura Y, Cox NJ, Stephens PJ, Miller VA, Ross JS, Chen D, Safgren SL, Kuffel MJ, Ames MM, Kalari KR, Gomez HL, Gonzalez-Angulo AM, Burgues O, Brauch HB, Ingle JN, Ratain MJ, Yelensky R (2014) Loss of Heterozygosity at the CYP2D6 Locus in Breast Cancer: Implications for Germline Pharmacogenetic Studies. Journal of the National Cancer Institute 107 (2):10. doi:10.1093/jnci/dju401 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng HW, Chen WM, Recker RR (2001) Population admixture: detection by Hardy-Weinberg test and its quantitative effects on linkage-disequilibrium methods for localizing genes underlying complex traits. Genetics 157 (2):885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kidwell KM, Hertz DL, Leyland-Jones B, Regan MM, Dowsett M, Rae JM (2016) Abstract P6–09-02: Analysis of the International tamoxifen pharmacogenomics consortium (ITPC) dataset shows that genotyping DNA derived from tumor does not introduce CYP2D6 genotyping error or mask an association with tamoxifen efficacy. Cancer Research 76 (4 Supplement):P6–09–02 [Google Scholar]
- 50.Lash TL, Cole SR (2009) Immortal person-time in studies of cancer outcomes. J Clin Oncol 27 (23):e55–56. doi:10.1200/jco.2009.24.1877 [DOI] [PubMed] [Google Scholar]
- 51.Rae JM, Sikora MJ, Henry NL, Li L, Kim S, Oesterreich S, Skaar TC, Nguyen AT, Desta Z, Storniolo AM, Flockhart DA, Hayes DF, Stearns V (2009) Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J 9 (4):258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, Hammond EH, Kuderer NM, Liu MC, Mennel RG, Van Poznak C, Bast RC, Hayes DF, American Society of Clinical O (2016) Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 34 (10):1134–1150. doi:10.1200/JCO.2015.65.2289 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Referenced from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Breast Cancer V.3.2014. © National Comprehensive Cancer Network, Inc 2014. All rights reserved. Accessed [August 22, 2014]. To view the most recent and complete version of the guideline, go online to www.nccn.org.NATIONAL COMPREHENSIVE CANCER NETWORK®, NCCN®, NCCN GUIDELINES®, and all other NCCN Content are trademarks owned by the National Comprehensive Cancer Network, Inc.
- 54.Barginear MF, Jaremko M, Peter I, Yu C, Kasai Y, Kemeny M, Raptis G, Desnick RJ (2011) Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clinical pharmacology and therapeutics 90 (4):605–611. doi:10.1038/clpt.2011.153 [doi] [DOI] [PubMed] [Google Scholar]
- 55.Kiyotani K, Mushiroda T, Imamura CK, Tanigawara Y, Hosono N, Kubo M, Sasa M, Nakamura Y, Zembutsu H (2012) Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast cancer research and treatment 131 (1): 137–145. doi: 10.1007/s10549-011-1777-7 [doi] [DOI] [PubMed] [Google Scholar]
- 56.Martinez de Duenas E, Ochoa Aranda E, Blancas Lopez-Barajas I, Ferrer Magdalena T, Bandres Moya F, Chicharro Garcia LM, Gomez Capilla JA, Zafra Ceres M, de Haro T, Romero Llorens R, Ferrer Albiach C, Ferriols Lisart R, Chover Lara D, Lopez Rodriguez A, Munarriz Ferrandis J, Olmos Anton S (2014) Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast (Edinburgh, Scotland) 23 (4):400–406. doi:10.1016/j.breast.2014.02.008 [doi] [DOI] [PubMed] [Google Scholar]
- 57.Hertz DL, Deal A, Ibrahim JG, Walko CM, Weck KE, Anderson S, Magrinat G, Olajide O, Moore S, Raab R, Carrizosa DR, Corso S, Schwartz G, Graham M, Peppercorn JM, Jones DR, Desta Z, Flockhart DA, Evans JP, McLeod HL, Carey LA, Irvin WJ Jr., (2016) Tamoxifen Dose Escalation in Patients With Diminished CYP2D6 Activity Normalizes Endoxifen Concentrations Without Increasing Toxicity. Oncologist 21 (7):795–803. doi: 10.1634/theoncologist.2015-0480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hertz DL, Rae JM (2016) Individualized Tamoxifen Dose Escalation: Confirmation of Feasibility, Ouestion of Utility. Clin Cancer Res 22 (13):3121–3123. doi: 10.1158/1078-0432.ccr-16-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Fig 1 Recurrence Free Survival in Tamoxifen Treated and Untreated Patients by CYP2D6 PM-Status. Cox-based survival curves assuming 1-3 nodes, tumor size 2-5 cm, and age 66.5 years. Recurrence free survival (RFS) was not associated with CYP2D6 PM-status in the tamoxifen treated patients (left). In the untreated patients (right), CYP2D6 non-PM status was associated with superior RFS (p=0.023). Shading indicates 95% confidence intervals of the RFS estimate.


