SUMMARY
The interplay of transcription, topological tension and chromosome breakage is a subject of intense interest, but, with so many facets to the problem, difficult to test. Here, we vary the orientation of promoters relative to one another in a yeast system that permits sensitive detection of chromosome breaks. Interestingly, convergent transcription that would direct RNA polymerases into one another does not increase chromosome breakage. In contrast, divergent transcription that would create underwound (potentially single-stranded) DNA does cause a marked increase in chromosome breakage. Furthermore, we examine the role that topoisomerases are playing in preventing genome instability at these promoters and find that Top2 is required to prevent instability at converging promoters.
INTRODUCTION
The events surrounding collision of RNA polymerases during transcription are complex and of universal relevance for all living organisms (Garcia-Rubio and Aguilera, 2012; Hobson et al., 2012; Liu and Alberts, 1995; Prescott and Proudfoot, 2002). Much of the early analysis of this phenomenon was from work performed in bacteria where it became clear that not only must the steric nature of two transcription complexes be considered, but also the effect that the complexes have on the DNA they are transcribing. The twin domain model proposed by Liu and Wang (Liu and Wang, 1987) explains how the movement of an RNA polymerase can generate both positive and negative supercoiling in the DNA. In eukaryotes, where linear chromosomes are packaged into nucleosomes, the dynamics of transcription and torsional stress, and the effect of the positioning of transcription units on DNA structure has been an active area of study (Naughton et al., 2013; Teves and Henikoff, 2014). Our interest is in how these dynamics affect an organism physiologically, particularly as it relates to DNA breakage.
In both yeast and mammals, promoter regions represent a nucleosome-depleted region (NDR) where the pre-initiation complex (PIC) assembles between a −1 and a +1 nucleosome before traveling to a downstream transcription start site (TSS). Divergent promoters can occur in a single NDR where two separate PICs form and direct the transcription units away from each other, creating an area of active chromatin and an expansive NDR determined by the distance between TSSs (Rhee and Pugh, 2012; Scruggs et al., 2015). As the RNA polymerase II (RNAP2) machinery departs the TSS and moves into the gene body, further repositioning of nucleosomes would be necessary, but a key question concerns what happens in the underwound region behind the two RNAP2 complexes? One facet of this question is that this promoter orientation is somehow used to regulate gene expression (Wei et al., 2011). Another facet is that this interplay of transcription, torsional stress, and active chromatin can cause genome instability. Determining the latter is especially important, given that it is now clear that expression of small RNAs is tissue and cell stage-specific; that is, a region that does not have divergent protein-coding promoters in one tissue may have divergent promoters in another cellular context (Lu et al., 2005; Lu et al., 2015; Schotte et al., 2011).
Importantly, closely-spaced convergent promoters may also be a source of potential instability. While convergently expressed genes would have their promoter regions separated by the two gene bodies, some convergent promoters may be close enough to occupy the same NDR. Work has shown that as two RNAP2 complexes approach each other from convergent promoters in a head-on collision, the two complexes stall and prevent each other from proceeding (Garcia-Rubio and Aguilera, 2012; Hobson et al., 2012; Saeki and Svejstrup, 2009). However, the consequences of very close convergent promoters in vivo has only recently begun to be studied, as it now appears that antisense transcription can lead to convergent transcription, especially at proposed super-enhancer regions (Lu et al., 2015; Meng et al., 2014).
Recently, two studies have reported that closely positioned transcription units appear to be correlated with chromosomal translocations (Meng et al., 2014; Pefanis et al., 2014). The conclusions of these large-scale, genome-wide studies make it unclear whether it is divergent or convergent transcription that more greatly potentiates the risk of a DSB. Here we sought to develop a simple genetic assay as a starting point to determine if, indeed, closely-spaced convergent or divergent promoters result in genome instability. We compare divergent and convergent transcription chromosomal regions in a yeast system that permits detection of gross chromosomal rearrangements (GCRs) (Chen and Kolodner, 1999). We find that convergent transcription that would direct RNA polymerases into one another does not increase GCR rate; however, divergent transcription that would create negatively supercoiled or underwound DNA (Kouzine et al., 2008; Liu and Wang, 1987; Sinden, 1987) with the potential for single-strandedness does cause a marked increase in GCR rate.
In this simple system, we can further ask what part do topoisomerases play in preventing genome instability at these sites? Three classes of topoisomerase enzymes are conserved between mammals and yeast (Wang, 2002). In S. cerevisiae, the type IA, IB, and II topoisomerases are encoded by the TOP3, TOP1, and TOP2 genes, respectively, and have multiple and overlapping roles in maintaining genome integrity during transcription, DNA replication, and DNA repair (Allen-Soltero et al., 2014; Bailis et al., 1992; Bennett et al., 2000; Brill and Sternglanz, 1988; El Hage et al., 2010; Fasching et al., 2015; Putnam et al., 2009; Yadav et al., 2014). In this study, we find that the Top2 enzyme is essential for preventing GCRs at converging promoters as the rate significantly increases in a top2-1 mutant. This may have implications for chemotherapy treatments that utilize topoisomerase II inhibitors (Libura et al., 2005).
RESULTS AND DISCUSSION
Experimental design
We wished to study the extent that transcription can result in DNA double-strand breaks (DSBs) when the promoters are arranged in either a convergent or divergent orientation. As these events may be rare, we needed to use a genetic assay with a very low background and preferably one that does not rely on a specific type of repair event to be detected, thus maximizing the sensitivity. We therefore chose to construct a version of a gross chromosomal rearrangement (GCR) assay in S. cerevisiae (Chen and Kolodner, 1999). Briefly, the URA3 marker, which encodes a product that confers uracil prototrophy and sensitivity to 5-fluoroorotic acid (5-FOA), and the CAN1 marker, which confers sensitivity to canavanine, were inserted in a non-essential region of chromosome I, 33.6 kb from the left end of the chromosome (Figure 1A). The endogenous URA3 and CAN1 genes on chromosome V were deleted to prevent their interaction with the assay locus. GCR rate is measured by determining the number of cells that become resistant to 5-FOA and canavanine due to simultaneous loss of both markers. Work from other labs has found that the vast majority of all events resulting in resistance to both 5-FOA and canavanine are due to loss of genetic material from the end of the chromosome (Chen and Kolodner, 1999; Yadav et al., 2014). While the final product can vary, most GCRs are likely initiated by a DSB. Our assay was designed to test whether zones of convergent or divergent transcription would increase the GCR rate.
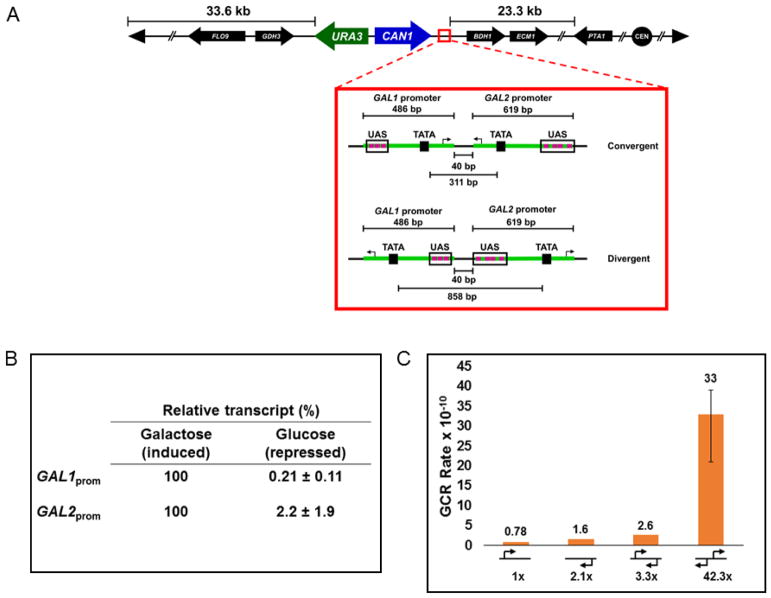
Figure 1. Divergent transcription increases GCR rate.
A. Construction of a gross chromosomal rearrangement (GCR) assay to study convergent and divergent transcription. DNA fragments carrying the URA3 and CAN1 markers and various configurations of the GAL1 and GAL2 promoters were integrated at the BDH2 locus of chromosome I 33.6 kb from the left end of the chromosome and 23.3 kb away from the first essential gene, PTA1. The inset shows specific details of the convergent and divergent configurations immediately downstream of the CAN1 terminator including the size of each promoter region, the distance between the two promoters in the assay, and the distance between the TATA boxes of each promoter. Purple boxes indicate the upstream activation sequences (UAS) and arrows the approximate transcription start sites (TSS). B. Relative percentage of transcript produced by the GAL1 or GAL2 promoters following induction or repression. The promoters are fully active in the presence of galactose (100%) and the relative decrease in transcript abundance when grown in glucose is reported +/− the 95% confidence interval from three biological replicates. C. GCR rate in wild-type strains with the indicated promoter orientations. Rates with error bars were calculated by the method of median, rates without error bars were calculated by Luria-Delbrück fluctuation analysis. Fold-increase relative to value with the GAL1 promoter alone is indicated below the graph.
Immediately downstream of the CAN1 terminator were inserted sequences corresponding to the promoter regions of the well-studied GAL1 and GAL2 genes (Figure 1A) (Johnston and Davis, 1984; Kuras et al., 2003) so that transcription driven by these strong promoters either converges or diverges when grown in medium containing galactose. We verified that the two promoters are active by measuring the relative change in RNA transcript produced by each promoter under inducing (galactose) and repressing (glucose) conditions by RT-qPCR (Figure 1B). When grown in galactose, there is a clear and significant increase in an RNA transcript corresponding to the sequence immediately downstream of each promoter.
The GCR rates were not significantly different when either the GAL1 or GAL2 promoters alone were present (Figure 1C). Also, both of these rates were at the limit of detection of the assay because the rate measured in a strain that was grown in galactose with no promoter sequence present at the assay locus was <7.9 × 10−11 (Table 1).
Table 1.
GCR rate in indicated background with no galactose-induced promoters at the assay locus1
| Genotype | GCR Rate2 × 10−10 |
|---|---|
| Wild-type | < 0.79 |
| top1Δ | 0.80 |
| top3Δ | 320 (130–890) |
| top2-1 | 41 |
Cells were still cultured in galactose, but neither the GAL1 nor GAL2 promoter sequences are present at the GCR testing locus.
Rates determined by method of the median have the 95% confidence interval indicated in parentheses. All other rates were determined by Luria-Delbrück fluctuation analysis (see Methods).
Divergent, but not convergent, transcription increases GCR rate
Recently, work has been undertaken to understand how a cell responds to the collision of two converging RNA polymerase II (RNAP2) complexes. In vitro and in vivo work demonstrates that two RNAP2 complexes cannot pass each other, and a head-on collision results in each complex pausing and disrupting transcription until one or both complexes can be removed (Garcia-Rubio and Aguilera, 2012; Hobson et al., 2012)_ENREF_1. We wished to determine if this type of collision could stimulate genome instability by causing a DSB. The GCR rate increased approximately three-fold when the converging GAL promoters were present at the assay locus (Figure 1B), but this was not significantly different from the rate when a single GAL1 promoter was used (p=0.13). These results suggest that the converging RNAP2 complexes are not resolved by breaking the DNA, but possibly removed by polyubiquination and degradation as has been suggested (Hobson et al., 2012).
Next, we wanted to examine the consequences of having the RNAP2 complexes transcribing away from each other in a divergent orientation. As the two complexes move away from each other, the DNA between them will become underwound. This more open conformation could potentially expose transient regions of ssDNA to damage until the negative supercoiling can be resolved (Kouzine et al., 2008; Liu and Wang, 1987; Sinden, 1987). Interestingly, unlike with convergent transcription, here we measure a 42-fold increase in the GCR rate, indicating that diverging promoters are a significant source of DSBs while converging promoters are not. The stark contrast measured for divergent and convergent transcription in our system likely indicates that the GAL promoters transcribe unidirectionally, because any significant antisense transcription would equalize the two values.
Recent work has also shown that the region between divergent TSSs creates a NDR that becomes larger as the distance between the promoters increases (Scruggs et al., 2015). Therefore, in addition to the generation of non-B form DNA, the region would become more accessible. The divergent transcription developed in our system, initiated by two separate promoters, may also be distinct from the divergent transcription described as sense and antisense transcription from a single promoter (Pefanis et al., 2014; Scruggs et al., 2015; Seila et al., 2009) as, here, a much larger region of DNA may be affected.
Top1 and Top3 do not appear to be involved in preventing GCRs at closely spaced promoters
As we are invoking supercoiling and the twin domain model (Liu and Wang, 1987; Sinden, 1994) to explain increased GCRs with divergent transcription, a logical next step was to determine which, if any, topoisomerases are involved in relieving torsional strain at converging or diverging promoters. Like mammals, S. cerevisiae possesses three classes of topoisomerases, type IB, type IA, and type II, encoded by the TOP1, TOP3, and TOP2 genes, respectively (Wang, 2002). Both type I topoisomerases relieve topological tension by transiently nicking one strand of the DNA while type II enzymes act by generating a transient DSB.
Overall, we found that neither loss of TOP1, which can resolve both positive and negative supercoiling (Thrash et al., 1985; Wang, 2002), nor TOP3, which has a weak activity only on negative supercoils and binds strongly to single-stranded heteroduplex DNA (Wang, 2002), appeared to have an effect on the GCR rate with convergent or divergent transcription. The GCR rates measured with a top1Δ allele did not differ significantly from those measured in a WT background (Figure 2A). Importantly, this does not mean that the Top1 enzyme is not acting at these sites, given that its role in transcription has been firmly established (Brill and Sternglanz, 1988; El Hage et al., 2010; Yadav et al., 2014); rather, this only means that DSBs are not generated in its absence in the context we are studying, perhaps due to compensation from Top2. For the top3Δ allele, there was a marked increase in GCR rate for each substrate with no clear indication of any effect of transcription (Figure S1), suggesting that the background GCR rate in a top3Δ mutant is so high that clear effects from transcription cannot be accurately measured (See the Supplemental Discussion for further details).
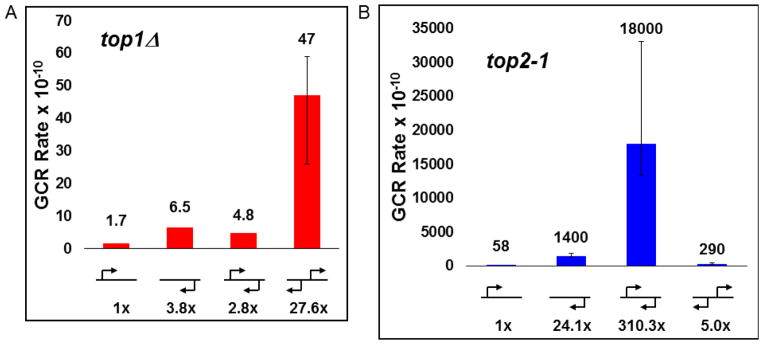
Figure 2. Convergent transcription combined with loss of Top2 activity increases GCR rate.
GCR rate for indicated GAL promoter orientations in A. top1Δ and B. top2-1 strains. All other details as in Figure 1B.
Top2 prevents GCRs at convergent promoters
In contrast to loss of TOP1 and TOP3, a defective Top2 enzyme has a strong effect on genome instability in relation to transcription. Top2 resolves both positive and negative supercoiling through the generation of a DSB (Wang, 2002). TOP2 is an essential gene; therefore, to determine if the Top2 enzyme affects formation of DSBs due to the orientation of transcription, we utilized the temperature sensitive top2-1 allele (Nitiss et al., 1993). top2-1 produces an enzyme that exhibits WT activity at 25°C, is defective at 30°C to the point where cells bearing the mutation are no longer sensitive to the topoisomerase II poison etoposide, and results in cell death at the non-permissive temperature of 37°C. For these experiments, cultures were inoculated from fresh colonies of meiotic segregants grown at 20°C. Cultures were grown to saturation at 30°C. Following plating to selective and non-selective media, cells were incubated at 20°C until colonies formed.
As in our previous results with the top1Δ and top3Δ alleles, there was no significant difference in the GCR rate when either no promoter or only the GAL1 promoter is present at the assay locus (p=0.55) (Table 1, Figure 2B). Comparing WT and top2-1 strains with GAL1 transcription towards the centromere, we measure a 74-fold increase in the GCR rate. These results indicate that a defective Top2 enzyme, alone, can increase genome instability.
Strikingly, when the GCR rate is measured for convergent promoters in a top2-1 strain, we see a significant increase of 310- and 13-fold over the individual GAL1 or GAL2 promoters, respectively, indicating Top2 has a clear role in resolving topological tension, presumably positive supercoiling, between two RNAP2 as they approach each other. The difference in the GCR rate for the GAL1 and GAL2 promoters alone may reflect a convergence with the distant CAN1 promoter, though the 2.0 kb distance and the rate of fire between these promoters likely attenuates that effect compared to the convergent GAL promoters.
Interestingly, the rate measured with convergent transcription is also 62-fold higher than the rate for divergent transcription in a top2-1 background, a clear contrast to the higher rate for divergent transcription in WT and top1Δ backgrounds. Perhaps this is indicative of a redundancy for Top1 to resolve negative supercoiling that Top2 does not resolve; however, the fact that Top1 cannot compensate for defective Top2 in the context of converging promoters is intriguing and may indicate a specific role for Top2.
Future directions
The intent of this short report is to communicate our finding that closely-spaced divergent promoter fragments are able to cause genome instability in WT cells. Convergent promoters do not impart this instability to WT cells as long as the Top2 enzyme is functional (Figure 3A). We wished to report this information with the community quickly, as the role of convergent versus divergent transcription has recently become a point of broad interest (Meng et al., 2014; Pefanis et al., 2014). From our brief but clear initial findings, there are many avenues of research to pursue using this assay system.
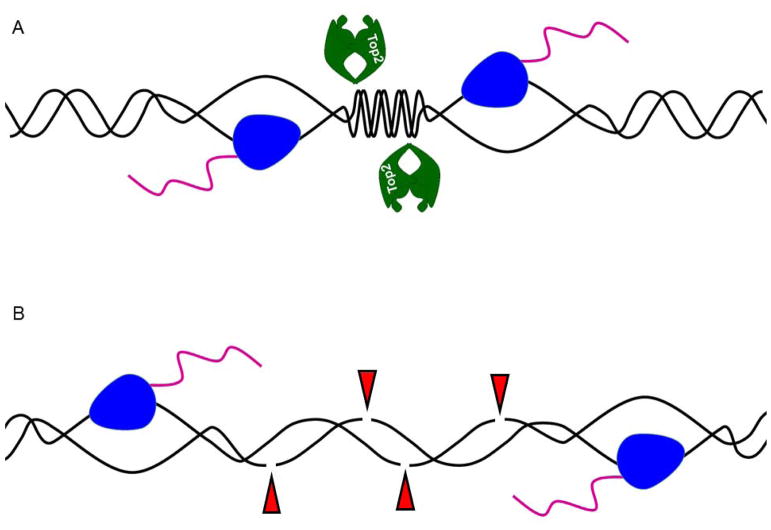
Figure 3. Model for effect of divergent and convergent transcription.
A. As RNAP2 complexes converge, positive supercoiling builds up that is relieved by the type II topoisomerase activity of Top2. B. As RNAP2 complexes move away from each other in a divergent orientation, negative supercoiling is generated between them resulting in localized DNA melting and ssDNA formation, increasing the risk for damage to the DNA by endogenous or exogenous sources.
For example, the distance between the promoter fragments can be varied to make them further apart to determine if this attenuates the GCR effect. This could be especially relevant for the convergent promoters as assembly of the large PIC requires significant space (Murakami et al., 2013). Confirming that the promoters are far enough apart to allow for dual assembly and initial elongation, therefore, may be important. Also, altering the sequence composition between both the divergent and convergent promoters could yield interesting results. Another variation would be to use promoters with a rate of transcription that can be controlled, such as in the pTET system that can be repressed to varying degrees based upon dosage of doxycycline (Kim and Jinks-Robertson, 2011). Combinations of GAL and TET promoters would allow us to determine the effect of a strong and a weak promoter together. These are a small subset of the many important questions that one can address with our system for studying the interplay of transcriptional orientation and genetic instability.
Concluding remarks
The finding that divergent transcription increases GCRs has implications for higher eukaryotes. Firing of divergent promoters leads to a spreading of negative supercoiling as the RNAP2 complexes move away from each other, in turn increasing DNA melting and the potential to form ssDNA in the region (Kouzine et al., 2013; Liu and Wang, 1987; Pefanis et al., 2014; Sinden, 1987) (Figure 3B). Thus, it is possible that divergent transcription opens up a region of DNA, making it more susceptible to DSB formation. While the S. cerevisiae genome is considerably smaller than that of mammals, with genes organized more compactly, and lacking the high number of introns, our work has relevance to the human genome where a significant fraction of genes are transcribed from divergent promoters (Adachi and Lieber, 2002; Takai and Jones, 2004; Trinklein et al., 2004). Divergent configurations result in increased DNA melting and ssDNA, which are necessary for some enzymes that cut or mutate DNA (Bransteitter et al., 2003).
That convergent transcription in wild-type cells does not increase the GCR rate would seem to fit with data showing that two RNAP2 complexes colliding head-on cause each to stall and do not necessarily result in a DSB (Hobson et al., 2012). Our results with the top2-1 mutant indicate, however, that the real danger with opposing RNAP2 transcription units, in terms of DSBs, is the increased local torsional stress, presumably arising even when the two promoters fire at different times so that there is no collision at all. Without a way to mitigate the topological tension, there is a clear increase in the GCR rate with this promoter configuration.
What is a possible cause for breaks measured at convergent promoters in cells with functional topoisomerases then? One possibility is that a convergent arrangement where two promoters are close together will eventually become divergent when the two RNA polymerases progress away from the zone of overlap. While two RNAP2 complexes that initiate simultaneously would collide and stall, it is critical to note that staggered transcription initiation (which is likely to be much more common) would provide adequate opportunity for the RNAP2 complexes to avoid a collision (Lu et al., 2015). This would explain the finding that some convergent configurations can also cause instability, as, at a certain point, the RNAP2 complexes actually move divergently. This latter point may provide unity to conflicting interpretations about convergent versus divergent transcription at preferred zones of mammalian chromosomal translocation.
EXPERIMENTAL PROCEDURES
Construction of yeast strains used in the GCR assay
All S. cerevisiae strains used were isogenic and derived from W303 (Thomas and Rothstein, 1989) corrected to express wild-type RAD5 and are listed in Table S1. Standard yeast genetic and molecular techniques were used (Sambrook et al., 1989; Sherman F., 1986). PCR amplifications were done using Q5 high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA). See supplemental experimental procedures for details of strain construction.
The top1::LEU2, top3::hisG, and top2-1 alleles have been described previously (Brill and Sternglanz, 1988; Gangloff et al., 1994; Thrash et al., 1985; Wallis et al., 1989) and were a kind gift from Dr. Adam Bailis. All strains bearing topoisomerase mutants were maintained as heterozygous diploids with only freshly dissected meiotic segregants being used in each assay.
GCR assay
Single colonies growing on synthetic complete (SC) medium lacking uracil were disbursed in ddH20 and cell count was determined by hemocytometer. Approximately 50 cells were used to inoculate 5 mL of YP-galactose (1% yeast extract, 2% peptone, 2% galactose). Cultures were grown at 30°C for 3 days to saturation. Cells were washed in PBS. A dilution was plated to YP-dextrose to determine viable count and the remaining cells were plated to SC lacking arginine supplemented with 750 mg/L of 5-fluoroorotic acid (Gold Biotechnology, St. Louis, MO), 60 mg/L of L-canavanine (Sigma-Aldrich, St. Louis, MO), and 2x uracil (40 mg/L) and incubated at 30°C for four days. For each particular strain, 10 to 20 individual trials were performed.
Colony counts were used to determine the GCR rate. If greater than 50% of the trials yielded colonies, the method of the median (Lea and Coulson, 1949) was employed. In these cases, a 95% confidence interval was also determined. When fewer than 50% of the trials yield colonies, Luria-Delbrück fluctuation analysis (Luria and Delbruck, 1943) was used. Since a 95% confidence interval cannot be determined using this method, significance was measured by comparing the data from two trials in a Mann-Whitney test (Hammer, 2001).
RT-qPCR
Total RNA was isolated from cells grown in glucose and galactose that carry the GCR substrate with either the GAL1 or GAL2 promoters (NPX220-18B and NPX298-4C, respectively) and used to synthesize cDNA. cDNA produced from each strain under each condition was used as template in a qPCR to measure the abundance of transcript utilizing TaqMan probes corresponding to a sequence immediately downstream of each promoter that is unique to the assay locus and not present at the endogenous GAL1 and GAL2 loci. Data are presented as the percentage of the quantity measured in glucose conditions over galactose conditions and represent three biological replicates each measured in duplicate. For further details, see Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Convergent transcription does not increase genome instability in wild-type cells
Divergent transcription leads to a significant increase in genome instability
Top2 is critical for relieving topological tension at closely-spaced promoters
Direction of transcription affects chromosome instability via topological tension
Acknowledgments
We thank Dr. Adam Bailis for yeast strains and for careful reading of the manuscript. The work was supported by NIH grants to M.R.L. Funding for N.R.P. is provided by a generous memorial endowment established by Barbara Knight through the ARCS Foundation, Inc. Los Angeles founder chapter - John H. Richardson Postdoctoral Fellowship.
Footnotes
Author Contributions N.R.P. performed all experiments. N.R.P. and M.R.L. conceived the experiments, analyzed the data, and wrote the paper.
References
- Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis AM, Arthur L, Rothstein R. Genome rearrangement in top3 mutants of Saccharomyces cerevisiae requires a functional RAD1 excision repair gene. Mol Cell Biol. 1992;12:4988–4993. doi: 10.1128/mcb.12.11.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol Cell. 2015;57:595–606. doi: 10.1016/j.molcel.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio ML, Aguilera A. Topological constraints impair RNA polymerase II transcription and causes instability of plasmid-borne convergent genes. Nucleic Acids Res. 2012;40:1050–1064. doi: 10.1093/nar/gkr840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Paleontologia Electronica. 2001;4:9. [Google Scholar]
- Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst) 2011;10:953–960. doi: 10.1016/j.dnarep.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci U S A. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. Journal of genetics. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Libura J, Slater D, Felix C, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- Liu B, Alberts BM. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science. 1995;267:1131–1137. doi: 10.1126/science.7855590. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu Z, Pannunzio NR, Greisman HA, Casero D, Parekh C, Lieber MR. Convergent BCL6 and lncRNA promoters demarcate the major breakpoint region for BCL6 translocations. Blood. 2015 doi: 10.1182/blood-2015-07-657999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, et al. Convergent Transcription at Intragenic Super-Enhancers Targets AID-Initiated Genomic Instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science. 2013;342:1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL, Liu YX, Hsiung Y. A temperature sensitive topoisomerase II allele confers temperature dependent drug resistance on amsacrine and etoposide: a genetic system for determining the targets of topoisomerase II inhibitors. Cancer Res. 1993;53:89–93. [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci U S A. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schotte D, Akbari Moqadam F, Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R, den Boer ML. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- Scruggs BS, Gilchrist DA, Nechaev S, Muse GW, Burkholder A, Fargo DC, Adelman K. Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Sherman F, FG, Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sinden RR. Supercoiled DNA: Biological Significance. Journal of Chemical Education. 1987;64:294–301. [Google Scholar]
- Sinden RR. DNA Structure and Function. San Diego: Academic Press; 1994. [Google Scholar]
- Takai D, Jones PA. Origins of bidirectional promoters: computational analysis of intergenic distance in the human genome. Mol Biol Evol. 2004;21:463–467. doi: 10.1093/molbev/msh040. [DOI] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash C, Bankier AT, Barrell BG, Sternglanz R. Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc Natl Acad Sci U S A. 1985;82:4374–4378. doi: 10.1073/pnas.82.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N, Aldred SF, Hartman S, Schroeder D, Otillar R, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Jarvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P, Harcy V, Argueso JL, Dominska M, Jinks-Robertson S, Kim N. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet. 2014;10:e1004839. doi: 10.1371/journal.pgen.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.