Abstract
Macroautophagy, initially described as a non-selective nutrient recycling process, is essential for the removal of multiple cellular components. In the past three decades, selective autophagy has been characterized as a highly regulated and specific degradation pathway for removal of unwanted cytosolic components and damaged and/or superfluous organelles. Here, we discuss different types of selective autophagy, emphasizing the role of ligand receptors and scaffold proteins in providing cargo specificity, and highlight unanswered questions in the field.
Keywords: mitophagy, receptor, stress, vacuole, yeast
Selective autophagy overview
Autophagy is a highly conserved pathway in eukaryotes involving the cellular recycling of multiple cytoplasmic components during standard physiological conditions and in response to different types of stress including starvation. Macroautophagy (hereafter autophagy) can be either non-selective or selective and involves the sequestration of cytoplasm within double-membrane vesicles termed autophagosomes. Upon maturation, autophagosomes fuse with the vacuole (in yeast and plants) or endosomes and lysosomes (in metazoans) leading to degradation of their cargo by resident hydrolases1, 2. Cargo degradation produces molecular building blocks such as amino acids, which are subsequently recycled back into the cytoplasm for reuse1, 3. Whereas non-selective autophagy, a cellular response to nutrient deprivation, typically involves random uptake of cytoplasm into phagophores, the precursors to autophagosomes, selective autophagy is responsible for specifically removing certain components such as protein aggregates and damaged or superfluous organelles4. Different studies have reported the selective autophagic degradation of several organelles including mitochondria5, peroxisomes6, lysosomes7, endoplasmic reticulum (ER) and the nucleus8 under various conditions. Furthermore, autophagy selectively degrades aggregation-prone misfolded proteins and protein microaggregates implicated in the pathology of various neurodegenerative diseases9. In this review, we address the principal mechanisms of selective autophagy in yeast and mammals, with an emphasis on mitophagy, which is to date the best described type of selective autophagy.
Cytoplasm-to-vacuole targeting (Cvt) pathway
The Cvt pathway is a biosynthetic autophagy-related process specific to yeast, in which vacuolar enzymes are transported from the cytoplasm into the vacuole utilizing the autophagic machinery. Among the enzymes that utilize the Cvt pathway are Ape1, Ape4 and Ams110. Ape1 is first synthesized in the cytoplasm as an inactive proenzyme (prApe1). Following oligomerization, prApe1 is selectively recognized by the non-core autophagy-related (Atg) protein Atg19 that functions as a receptor for Ams1, prApe1 and Ape411, 12. Once the prApe1-Atg19 or Cvt complex is formed, Atg19 binds to the scaffold protein Atg11, which in turn recruits the Cvt complex to the perivacuolar location termed the phagophore-assembly site (PAS) where autophagosomes and Cvt vesicles are formed in yeasts13, 14; interaction of Atg19 with Atg11 is facilitated by Hrr25-dependent phosphorylation of the receptor15. After reaching the PAS, Atg19 interacts with the ubiquitin-like protein Atg813. During autophagy and the Cvt pathway, Atg8 is covalently conjugated through its C terminus to phosphatidylethanolamine (PE); thus, Atg8‒PE is present on both the inner and outer membrane of forming autophagosomes16 (Fig. 1a). Atg8 has been implicated in phagophore expansion and autophagosome size regulation17. Thus, Atg19 binding to Atg8 tethers the Cvt complex to the Atg8‒PE-conjugated sequestering vesicles. Once fully matured, Cvt vesicles fuse with the vacuole and deliver prApe1, which is then processed into its active form by resident hydrolases.
Figure 1.
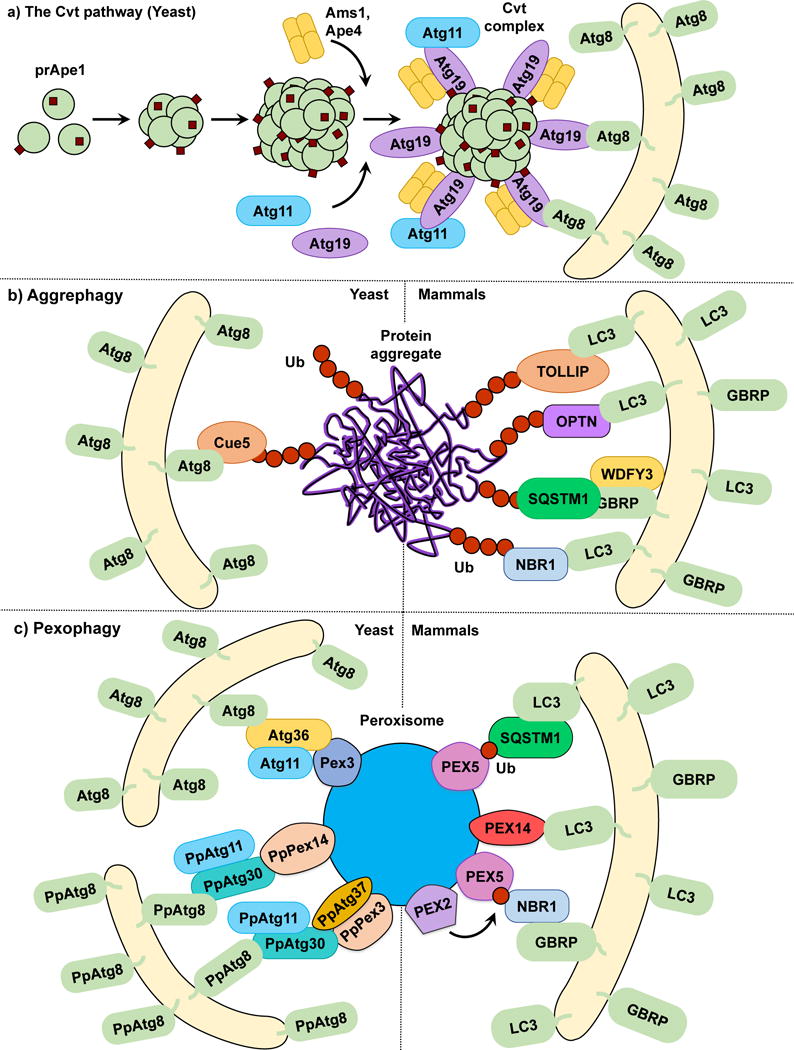
The Cvt pathway, aggrephagy and pexophagy. (a) In the yeast Cvt pathway prApe1, Ape4 and Ams1 are synthesized in the cytoplasm. prApe1 oligomerizes into dodecamers and subsequently higher order structures that are recognized by the receptor Atg19, which in turn binds the scaffold protein Atg11 forming the Cvt Complex. Ams1 and Apr4 also oligomerize and bind Atg19. Atg11 brings the Cvt Complex to the PAS where Atg19 binds Atg8‒PE, tethering the Cvt complex to the phagophore. (b) In both yeast and mammalian aggrephagy, protein aggregates are ubiquitinated and subsequently recognized by cargo receptors. In yeast, Cue5 links the ubiquitinated aggregates to Atg8‒PE. During mammalian aggrephagy, TOLLIP, SQSTM1, NBR1 and OPTN tether the ubiquitinated aggregates to the phagophore by binding LC3/GABARAP family members. WDFY3 has been described as a scaffold for SQSTM1-dependent degradation. (c) In S. cerevisiae pexophagy, Atg36 functions as a receptor linking peroxisomes to the phagophore by binding Pex3 and Atg8‒PE. In P. pastoris pexophagy, PpAtg30 acts as a receptor by linking PpPex3 and PpPex14 to PpAtg8‒PE. Atg11 functions as a scaffold for both S. cerevisiae and P. pastoris. The current model of mammalian pexophagy involves the E3-ubiquitin ligase PEX2-mediated mono-ubiquitination of PEX5, which in turn is recognized by receptors SQSTM1 and NBR1, tethering peroxisomes to the phagophore. PEX14 has also been reported to link peroxisomes to the phagophore by directly binding LC3 family members.
Using the Cvt pathway as a model for selective autophagy we can propose that although the core autophagy machinery directs phagophore membrane expansion and vesicle formation, cargo selectivity is achieved by a ligand receptor and a scaffold protein, roles taken by Atg19 and Atg11, respectively, in the Cvt pathway. Atg19 has a paralog, Atg34 (also phosphorylated by Hrr25), which functions as an Ams1 receptor during nitrogen starvation18. Other types of selective autophagy in yeast, such as mitophagy19 and pexophagy20, 21, also rely on Atg11 as a scaffold for cargo delivery to the PAS. However, a counterpart to Atg11 has yet to be discovered in mammals. Similarly, most types of selective autophagy require the binding of the cargo receptor to the core autophagy machinery. In the Cvt pathway, this process is illustrated by Atg19 binding to Atg8 through a specific WXXL motif found on the Atg19 C terminus, similar to that seen in SQSTM1/p6222, 23. This interaction is evolutionarily conserved as several proteins in yeasts and more complex eukaryotes contain Atg8-interacting motifs (AIM) or LC3-interacting regions (LIRs), respectively. The AIM or LIR provide selective binding to yeast Atg8 or one of the members of the LC3/GABARAP family of Atg8 mammalian homologs24. Recently, a specific type of LIR called GABARAP-interacting motif (GIM) has been proposed, showing enhanced specificity to GABARAP versus LC3 family members25. Multiple examples of scaffold and receptor proteins will be showcased as we discuss different types of selective autophagy (Table 1).
Table 1.
Selective autophagy ligands, receptor and scaffolds in yeast and mammals.
| Process | Organism | Ligand | Receptor | Scaffold |
|---|---|---|---|---|
| Cvt Pathway | Yeast | prApe1, Ams1 | Atg19 | Atg11 |
| Aggrephagy | Yeast | Protein aggregates (Ub) | Cue5 | – |
| Mammals | SQSTM1/p62, NBR1, TOLLIP | WDFY3/ALFY | ||
| Pexophagy | Yeast | Peroxisomes (Pex3, PpPex14, PpPex3) | Atg36, PpAtg30 | Atg11, PpAtg17 |
| Mammals | Peroxisomes (Ub) | SQSTM1/p62, NBR1 | – | |
| Mitophagy | Yeast | Mitochondria | Atg32 | Atg11 |
| Mammals | SQSTM1/p62, BNIP3L/Nix, OPTN, FUNDC1, PHB2 | – | ||
| Reticulophagy | Yeast | Endoplasmic reticulum | Atg39, Atg40 | Atg11 |
| Mammals | RETREG1/FAM134B | – | ||
| Nucleophagy | Yeast | Nucleus, nuclear Nvj1 portions (PMN) | Atg40, Vac8 (PMN) | – |
| Lysophagy | Mammals | Lysosomes (Ub) | SQSTM1/p62 | – |
| Xenophagy | Mammals | Bacteria (Ub), viruses | SQSTM1/p62, CALCOCO2/NDP52, OPTN | – |
| Lipophagy | Mammals | Lipid droplets | – | – |
| Yeast | – | – | ||
| Ferritinophagy | Mammals | Ferritin | NCOA4 | – |
| Glycophagy | Mammals | Glycogen | STBD1 | – |
Aggrephagy
The selective degradation of protein aggregates by autophagy is known as aggrephagy. Multiple aggregation-prone proteins such as amyloid-β26, HTT (huntingtin)27 and SNCA/α-synuclein28 are autophagy substrates. In yeast, Cue5 is a cargo receptor for the clearance of aggregation-prone poly-glutamine (polyQ)-containing proteins. Cue5 possesses a ubiquitin-binding CUE domain and an AIM, mediating the interaction between the ubiquitinated cargo and Atg829. Overexpression of TOLLIP, a Cue5 human homolog that also has a CUE domain, leads to the degradation of polyQ protein aggregates in human cell lines30 (Fig. 1b). Ubiquitination of substrates has been demonstrated as a key mediator in the recognition and degradation of these proteins by selective autophagy31. At least three additional mammalian cargo receptors, SQSTM116, 23, NBR132 and OPTN33, act as ubiquitin binding proteins that mediate the interaction between ubiquitinated proteins and the core autophagy machinery. All three receptors possess LIRs and ubiquitin-binding domains, thus working as a bridge between the LC3/GABARAP family members and the ubiquitinated substrates16, 23–29, 31–34.
The nucleocytoplasmic shuttling protein WDFY3/ALFY has been proposed as a scaffold in aggrephagy35. While unable to directly interact with ubiquitinated substrates, WDFY3 binds the core autophagy protein ATG5, the cargo receptor SQSTM136, GABARAP subfamily members37, and phosphatidylinositol-3-phosphate38, a prominent lipid in the regulation of autophagosome membrane formation. WDFY3 depletion hinders the clearance of aggregated polyQ proteins. The latter observation, in conjunction with its high number of interacting partners, suggests that WDFY3 is an important scaffold protein in the SQSTM1-dependent degradation of ubiquitinated aggregates by selective autophagy.
Ubiquitination plays an important role not only in substrate recognition and degradation by the ubiquitin-proteasome system (UPS), but also by selective autophagy, raising a set of questions regarding the hierarchy between these two degradation pathways. It has been proposed that protein aggregates that cannot be degraded by the UPS (e.g., due to size) may be cleared by autophagy39, 40. At the same time, the Lys residues used for linkage, as well as the length and the nature of the ubiquitin chains, have been proposed as a mechanism to select which degradation pathway is chosen39. However, a recent paper by Lu et al. emphasizes the role of receptor oligomerization over the type of ubiquitination in selecting a degradation pathway41. This finding agrees with data showing the importance of Cue5 and SQSTM1 oligomerization in their association with the phagophore39, 42. Thus, both autophagy and the UPS provide dynamic alternatives to different cellular challenges.
Pexophagy
Pexophagy is the selective removal of peroxisomes. Pexophagy has been mostly studied as a pathway for the removal of superfluous organelles in various fungi43. Incubating these fungi in oleic acid or methanol leads to peroxisome proliferation; following a shift to a preferred carbon source such as glucose, the excess peroxisomes are rapidly degraded through pexophagy43. Similar to other types of selective autophagy, cargo selectivity is provided by receptor proteins; in yeast this role is taken by PpAtg30 in Pichia pastoris20 and Atg3644 in S. cerevisiae. Both Atg36 and PpAtg30 tether peroxisomes targeted for degradation to nascent phagophore membranes by linking Atg8 to peroxisomal membrane proteins, with Atg36 binding Pex3, and PpAtg30 binding both PpPex3 and PpPex1420, 21. Phosphorylatable variants of the classical AIMs have been reported for both Atg36 and PpAtg30; however, disruption of these AIMs only delays pexophagy rather than abrogating it45. As previously mentioned, Atg11 is required for pexophagy46. PpAtg37 is an integral peroxisomal membrane protein specifically required for pexophagy in P. pastoris. During pexophagy, PpAtg37 is necessary for phagophore formation, as PpAtg37 null cells fail to recruit PpAtg11 to peroxisomes47.
In contrast to yeast, no pexophagy-specific cargo receptor has been described in mammals. Rather, mammalian pexophagy relies on the ubiquitination of peroxisomal proteins and their recognition by SQSTM131 and NBR148. Initially, it was reported that PEX3 overexpression leads to peroxisome ubiquitination and pexophagy induction49. However, PEX3 ubiquitination does not prevent pexophagy and this study did not determine the specific peroxisomal proteins targeted for ubiquitination. Subsequently, two studies identified PEX5 mono-ubiquitination as the cargo signal for peroxisome degradation50, 51. PEX5 is a cytosolic protein that shuttles between the peroxisomal membrane and the cytosol in a ubiquitin-dependent manner52. The accumulation of mono-ubiquitinated PEX5 in the peroxisomal membrane, which was unable to shuttle back to the cytosol, triggers pexophagy50. Furthermore, in response to reactive oxygen species (ROS), PEX5 is phosphorylated and subsequently mono-ubiquitinated, which leads to pexophagy induction in a SQSTM1-dependent manner51. A recent study has indicated that the peroxisomal E3-ubiquitin ligase PEX2 is responsible for PEX5 ubiquitination53. These data suggest a model in which mammalian pexophagy is dependent on the membrane accumulation of ubiquitinated peroxisomal proteins such as PEX5 that are recognized by the ubiquitin-binding receptors SQSTM1 and NBR1, and which in turn link the target peroxisomes to LC3/GABARAP-bound sequestration membranes (Fig. 1c). However, this simple model fails to answer several questions. From a mechanistic perspective, how does PEX5 ubiquitination at a specific site determine whether the protein shuttles into the peroxisome or is directed to proteasomal degradation? Are there distinct mechanisms involving ROS and amino acid starvation-induced pexophagy? Regarding this last point, other studies have reported that the peroxisomal membrane protein PEX14, which acts as a docking factor for PEX5, can directly interact with LC3-II under starvation conditions, outcompeting PEX554. This opens the possibility of different pathways being involved under different pexophagy-inducing stimuli. Finally, the human Atg37 ortholog ACBD5 has also been reported as an essential pexophagy factor47. It will be interesting to determine the role of ACBD5 in pexophagy and its connection to possible undiscovered mammalian pexophagy receptors.
Mitophagy
Mitophagy is a critical quality control process that eliminates damaged and/or superfluous mitochondria through their selective autophagic degradation55, 56. Deficiencies in mitophagy have been linked to the development of several pathologies, including neurodegenerative disorders57 such as Parkinson disease (PD).
Mitochondria have multiple metabolic functions and also influence cell fate by regulating apoptosis. Consequently, mitochondrial damage leads to loss of metabolic homeostasis. Additionally, disruption of oxidative phosphorylation (OxPhos) in damaged mitochondria leads to excessive ROS generation58. Mitochondria are high-maintenance organelles, and non-functioning/superfluous mitochondria become an energetic burden. Therefore, the regulation of mitochondrial quality and quantity is of paramount importance. Although mitochondria harbour some internal quality control machinery59, the major contribution towards maintaining mitochondrial integrity comes from mitophagy, which functions in concert with the UPS to ensure mitochondrial homeostasis60.
In fungi, mitophagy can be triggered by nitrogen starvation61–63 or post-log phase growth in a non-fermentable medium. In yeast, selectivity is provided by the outer mitochondrial membrane (OMM) receptor Atg3262, which links targeted mitochondria to the autophagic machinery19, 64. The cytosolic N terminus of Atg32 interacts with Atg1165. Ectopic targeting of the Atg32 N terminus to peroxisomes leads to pexophagy, underscoring the function and sufficiency of Atg32 as an autophagy receptor66. The C terminus of Atg32 faces the intermembrane space, and its proteolytic processing by Yme1 may be required for efficient mitophagy56. The interaction between Atg32 and Atg11 promotes the recruitment of mitochondria to the PAS for sequestration. Atg32 also orchestrates the subsequent expansion of the phagophore around the mitochondria through its interaction with Atg8 via the AIM in its cytosolic domain55, 66. However, mutating the Atg32 AIM causes only a partial mitophagy defect, suggesting that the Atg32-Atg8 interaction increases mitophagy efficiency, but remains auxiliary62, 66, 67.
The expression of Atg32 can be influenced by oxidative stress and nutritional status. In P. pastoris, the Ume6-Sin3-Rpd3 complex, positively regulated by TOR, suppresses ATG32 transcription61. During starvation, TOR is inactivated, promoting the synthesis of Atg32 and starvation-induced mitophagy. However, the upregulation of Atg32 expression is not by itself sufficient to induce mitophagy. Atg32 is activated by phosphorylation at residues Ser114 and Ser119 in its cytosolic domain, facilitating its interaction with Atg1167. Casein kinase 2 (CK2) has been proposed as the Atg32 Ser114 kinase68 as CK2 phosphorylates Atg32 in vitro but fails to phosphorylate Atg32S114A. Similarly, CK2 temperature-sensitive mutants fail to phosphorylate Atg3268. However, CK2 is a multitasking kinase and its activation is independent of mitophagy-inducing stimuli62. Therefore, other signaling pathways may contribute to the temporal selectivity of CK2-mediated phosphorylation of Atg32. Two mitogen-activated protein kinase (MAPK) pathways have been implicated in mitophagy regulation in yeast69. Hog1 is a MAPK in the Ssk1-Pbs2 pathway and Atg32 phosphorylation is suppressed in hog1Δ cells. However, Hog1 does not phosphorylate Atg32 in vitro, suggesting an indirect regulation67. The Slt2 pathway plays a role in mitochondrial recruitment to the PAS69. Although further investigation is required to identify the signaling circuit regulating Atg32 phosphorylation, cooperative expression and activation of Atg32 highlights the multiple levels of regulation involved in mitophagy induction.
Because the dimensions of intact mitochondria are larger than that of autophagosomes, sequestration of damaged mitochondria might be facilitated by mitochondrial fission62, 65. In S. cerevisiae, mitochondrial fission is mediated by several factors including Dnm1 and Fis165. Deletion of either DNM1 or FIS1 significantly suppresses mitophagy63, 70. Dnm1 interacts with Atg11, allowing the former to be recruited to mitochondria targeted for degradation63. The proteins associated with the ER-mitochondrial encounter structure (ERMES) may play a role in modulating mitochondrial fission during mitophagy65. Nevertheless, the exact mechanism of mitophagy-associated mitochondrial fission is unclear, and yet unidentified fission factors may be involved.
Mitophagy in mammals is mechanistically more complex than in yeast and is induced by cellular and developmental cues. In mammalian cells the loss of mitochondrial membrane potential is a potent inducer of mitophagy5, 71. However, while the use of chemicals that target the electron transport chain or act as protonophores is a convenient and efficient way to study mitophagy, the acute dissipation of mitochondrial membrane potential precludes the study of subtle regulatory phenomenon72. Furthermore, such severe mitochondrial damage might not be representative of the true pathophysiological triggers.
In mammals, mitophagy plays important physiological roles in development and cellular differentiation. Erythrocyte development requires the selective degradation of mitochondria in reticulocytes73 and embryonic development in some organisms involves selective degradation of paternal mitochondria in the zygote74. Hypoxia, which disrupts mitochondrial respiration, is another stimulus that promotes mitophagy in mammalian cells55.
The PINK1-PRKN/PARK2/parkin pathway is the most extensively characterized mechanism effecting mitochondrial quality control in most mammalian cells. PINK1 is a Ser/Thr kinase with a C-terminal kinase domain and N-terminal mitochondrial targeting sequence55, and PRKN/PARK2 is an E3-ubiquitin ligase75. Loss of mitochondrial integrity is usually accompanied by mitochondrial depolarization. PINK1, which requires the mitochondrial membrane potential for its inner mitochondrial membrane (IMM) import, acts as a depolarization sensor76. In healthy mitochondria, PINK1 is imported into the matrix where it is cleaved by proteases and subsequently released back into the cytosol for degradation through the N-end rule pathway77, 78. In compromised mitochondria, the loss of membrane potential prevents translocation, and PINK1 is stabilized on the OMM, leading to its activation by autophosphorylation72, 76, 79. Active PINK1 phosphorylates several substrates including ubiquitin, MFN1 (mitofusin 1), MFN2 and PRKN/PARK276, 80. Unphosphorylated PRKN/PARK2 is autoinhibited76, 80; PINK1-mediated phosphorylation of PRKN/PARK276 leads to activation. PINK1 also phosphorylates available ubiquitin attached to OMM proteins at Ser65 generating phospho-ubiquitin81, 82, which acts as a PRKN/PARK2 substrate77. PRKN/PARK2 subsequently links phospho-ubiquitin chains to OMM proteins, which possibly results in a feed-forward amplification loop recruiting more PRKN/PARK276. The phosphorylation of MFN2 by PINK1 might also play a role in PRKN/PARK2 recruitment83, possibly acting along with phospho-ubiquitin at the OMM. However, the role of MFN2 in PRKN/PARK2 recruitment is controversial84.
The classic model for mitophagy involves the recognition of polyubiquitinated mitochondria by autophagy receptors SQSTM1 and OPTN which bind LC376, 85. This interaction tethers damaged mitochondria to the expanding phagophore and promotes their subsequent sequestration within autophagosomes (Fig. 2a). Recent progress in the field suggests a complementary model whereby PINK1-mediated phosphorylation of ubiquitin, independent of PRKN/PARK2 activity, is sufficient to recruit the autophagy receptors CALCOCO2/NDP52 and OPTN and induce low-amplitude mitophagy86. In this model CALCOCO2 and OPTN can successfully recruit ULK1 and facilitate mitophagy initiation upstream of LC3 binding86. The importance of PRKN/PARK2-mediated ubiquitination is indicated by the fact that overexpression of the mitochondrial deubiquitinase USP30 inhibits mitophagy by promoting deubiquitination of PRKN/PARK2 substrates87.
Figure 2.
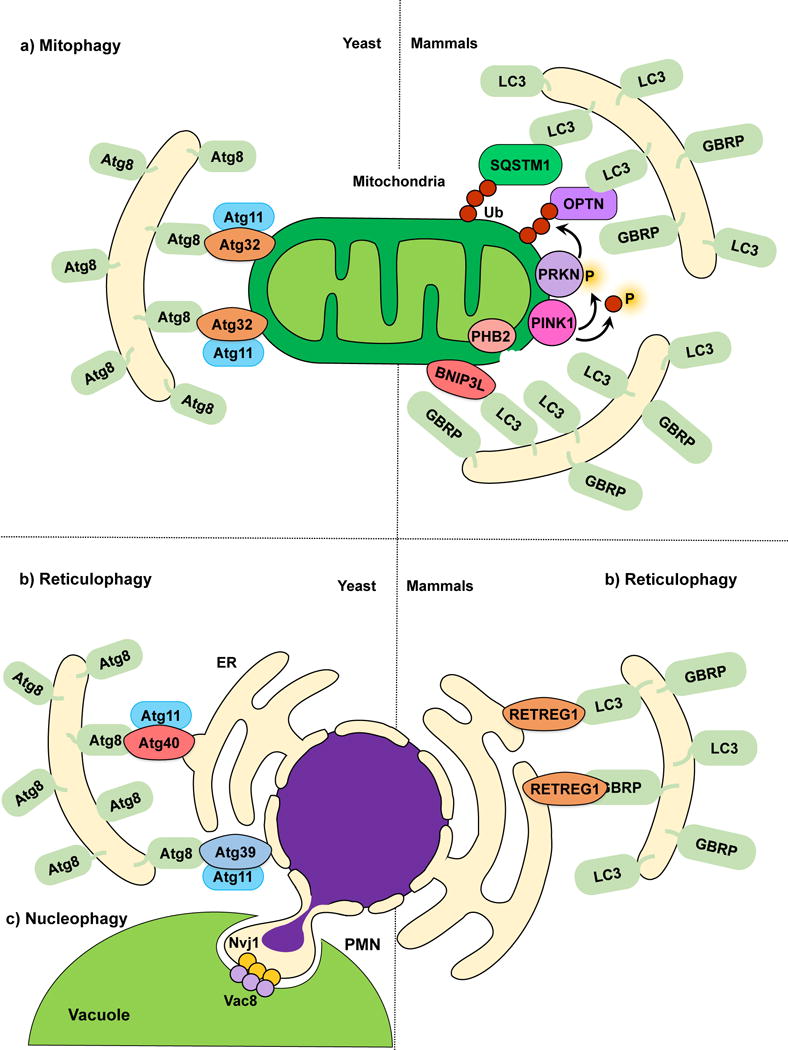
Mitophagy, reticulophagy and nucleophagy. (a) The yeast mitophagy receptor Atg32 links mitochondria to the phagophore by directly binding Atg8‒PE; Atg11 functions as a scaffold. Several cargo receptors (not all shown) have been described for mammalian mitophagy. Mitochondria depolarization leads to PINK1 activation and phosphorylation of ubiquitin and PRKN, and OMM disruption exposes PHB2. Receptors link mitochondria targeted for degradation to the phagophore. (b) In yeast reticulophagy, Atg39 and Atg40 have been proposed as receptor proteins. Atg39 mediates the degradation of the perinuclear ER, and Atg40 mediates cytoplasmic ER degradation. Both Atg39 and Atg40 link their respective ER sites to Atg8‒PE-conjugated membranes for sequestration. Atg11 has been proposed as a scaffold protein for both Atg39 and Atg40-mediated reticulophagy. During mammalian reticulophagy, RETREG1/FAM134B tethers the cytoplasmic ER to LC3/GABARAP family members for membrane sequestration and degradation. (c) Because Atg39 specifically localizes to the perinuclear ER, Atg39-mediated degradation is also considered nucleophagy. During piecemeal microautophagy of the nucleus (PMN), the nuclear envelope protein Nvj1 and vacuolar membrane protein Vac8 form nuclear-vacuolar junctions, which pinch off and engulf part of the nucleus inside the vacuole.
Polyubiquitination also acts as a signal that promotes VCP/p97-mediated extraction of OMM proteins and their subsequent proteasomal degradation88, causing disruption of the OMM88. Recent findings suggest that OMM disintegration serves to expose the IMM protein PHB2 (prohibitin 2), which possesses a LIR and functions as a mitophagy receptor74. PHB2 promotes mitophagy in a PINK1-PRKN/PARK2-dependent manner, and the selective removal of paternal mitochondria in C. elegans embryos requires PHB2 function74. The PINK1-PRKN/PARK2-dependent generation of mitochondria-derived vesicles (MDVs)89, 90 is an alternative pathway to conventional PRKN/PARK2-dependent mitophagy. Limited and localized mitochondrial damage promotes MDV formation to ensure the selective removal of damaged portions of a mitochondrion instead of the entire organelle91. It is possible that the PINK1-PRKN/PARK2 pathway switches between MDV formation and mitophagy depending on the extent of mitochondrial damage55.
PINK1 and PRKN/PARK2 are also involved in regulating the arrest of mitochondrial motility following mitochondrial damage92. Mitochondria are transported by the kinesin KIF5 on microtubules. KIF5 binds mitochondria through the adaptor TRAK1-TRAK2 and the OMM protein RHOT1/Miro193. Following mitochondrial damage, RHOT1 is one of the earliest proteins to be degraded via PRKN/PARK2-mediated ubiquitination94, a process which also requires the interaction of RHOT1 with the LRRK2 kinase95. The removal of RHOT1 halts mitochondrial motility and quarantines damaged mitochondria for degradation95. In cells that harbour mutations in PINK1, PRKN/PARK2 or LRRK2, RHOT1 degradation is inhibited, leading to continued motility of damaged mitochondria and delayed mitophagy96.
Not all mammalian cell types express PARK2/PRKN and several mitochondria-localized mitophagy receptors exist in mammalian cells. BNIP3L/Nix is one such mitophagy receptor and is involved in the selective elimination of mitochondria during the differentiation of reticulocytes into erythrocytes73, 91. BNIP3L localizes to the OMM and contains a LIR near its cytosolic N-terminus94, the activity of which may be regulated by phosphorylation25. However, mutations in the BNIP3L LIR only lead to a partial loss in mitophagy96. Another short motif has recently been reported to be indispensable for BNIP3L function97. Although the exact mechanism by which BNIP3L mediates mitophagy remains unknown, reports suggest that BNIP3L may promote mitochondrial depolarization79 leading to PINK1-PRKN/PARK2 recruitment to mitochondria, and activating mitophagy79. BNIP3L might also work in concert with the related protein BNIP398, which possesses a LIR99.
BNIP3L is also involved in hypoxia-induced mitophagy99, as is the LIR-containing OMM protein FUNDC1100. Mutations in the FUNDC1 LIR lead to loss of function101. Similar to Atg32, FUNDC1 is regulated by reversible phosphorylation. Under normal conditions, FUNDC1 is phosphorylated by SRC kinase and CK2102, including the modification of one site in its LIR. Hypoxia promotes the dephosphorylation of these residues involving the phosphatase PGAM5102. Hypoxia-induced mitophagy is particularly relevant to the pathobiology of tumors, and elucidating the role of BNIP3L and FUNDC1 in these contexts might be an important step towards therapeutic intervention103, 104.
In mammals, mitochondrial dynamics are regulated by the fission-promoting GTPase DNM1L/Drp1 and the profusion factors MFN1-MFN2 and OPA1101, 105. Mitophagy induction is accompanied by a decrease in mitochondrial fusion and an increase in mitochondrial fission to facilitate degradation of damaged mitochondria98. PINK1 activation promotes PRKN/PARK2-mediated degradation of MFN1-MFN2, consistent with the idea of reduced fusion80. The mitophagy receptor FUNDC1 is also involved in regulating mitochondrial dynamics during mitophagy. Whereas FUNDC1 binds to and recruits OPA1 to mitochondria under normal conditions, upon mitochondrial damage it preferentially recruits DNM1L, promoting fission106. Like ERMES in yeast, mitochondria-associated membranes are sites of ER-mitochondria contact in mammals, and have also been proposed to modulate mitophagy-related mitochondrial fission107, although the mechanism remains unclear.
Whereas most selective autophagy receptors are proteins, recent evidence suggests that mitophagy may also be orchestrated by lipid receptors107. Cardiolipin, a lipid unique to the mitochondria, may act as a mitophagy receptor in mammalian cortical neurons. Rotenone-induced mitochondrial damage causes a dramatic translocation of cardiolipin from the inner to the outer mitochondrial membrane108, where it interacts with the LC3 N terminus. Inhibition of cardiolipin synthesis or translocation reduces the efficiency of mitophagy in these neurons108. Cardiolipin was also recently reported to modulate mitophagy in S. cerevisiae33, and ceramide has also been implicated as a mitophagy receptor in certain cancer cell lines109.
Reticulophagy
Reticulophagy describes the degradation of the ER by selective autophagy. Perturbation of ER function results in the accumulation of misfolded proteins and ER stress, which in turn triggers the unfolded protein response (UPR) and ER-associated degradation, in order to recover cellular homeostasis109. Autophagy is also activated by ER stress110 as a means to control ER size and to counterbalance the ER expansion after the UPR111, 112. Other stimuli such as rapamycin treatment and nutrient starvation also activate reticulophagy113, 114. Similar to other selective autophagy pathways, cargo receptors have been described for selective ER degradation. In yeast, starvation-induced reticulophagy depends on Atg39 and Atg40, predicted transmembrane proteins that localize to the perinuclear and cytoplasmic ER, respectively. Consistent with their role as cargo receptors, Atg39 and Atg40 contain AIMs, and interact with both Atg8 and Atg11114. In mammals, RETREG1/FAM134B is a reticulophagy cargo receptor protein, as well as an Atg40 functional homolog113. Similar to Atg40, RETREG1 localizes to the cytoplasmic ER and interacts with LC3 and GABARAP family members through its LIR (Fig. 2b). Consistent with the reported role of reticulophagy in controlling ER size, RETREG1 overexpression increases ER fragmentation, whereas silencing of this protein results in ER expansion.
Nucleophagy
Nucleophagy has been described as the partial or bulk degradation of the nucleus by the vacuole/lysosome. Nucleophagy is closely related to reticulophagy, given that Atg39 localizes to, and mediates the degradation of, the perinuclear ER and nuclear envelope in yeast113. However, to date no Atg39 functional homolog has been described in mammals and it is still unclear how nucleophagy occurs in more complex eukaryotes. However, some studies have suggested selective autophagic degradation of chromatin115 and the nuclear lamina115 could play a role in preventing tumorigenesis.
Other types of nucleophagy termed piecemeal microautophagy of the nucleus (PMN) or micronucleophagy116, and late nucleophagy117 have been described in S. cerevisiae. During PMN the outer nuclear envelope protein Nvj1 interacts with the vacuolar membrane protein Vac8, forming nuclear-vacuolar junctions that pinch off parts of the nucleus, which are later engulfed and degraded by the vacuole117 (Fig. 2c). PMN is activated soon after nutrient starvation and depends on the core autophagic machinery118. In contrast, late nucleophagy occurs after prolonged starvation and is independent of Nvj1, Vac8 and some but not all core autophagy machinery117. Further studies will be required to understand the individual roles of Atg39-induced nucleophagy, PMN and late nucleophagy during nitrogen starvation.
Lysophagy
Lysophagy is the selective degradation of damaged lysosomes by autophagy. Leakage of lysosomal enzymes into the cytosol due to lysosomal membrane rupture leads to lysosomal cell death117. Therefore, the removal of damaged lysosomes is necessary to maintain cellular homeostasis. LGALS3 (galectin 3) binds to glycoproteins exposed upon lysosomal membrane damage and colocalizes with LC3, working as a key lysophagy marker119. Even though the specific mechanisms behind lysophagy are yet to be discovered, two independent reports have suggested a model in which damaged lysosomes are selectively degraded in a ubiquitin-SQSTM1-LC3-dependent manner7, 119 (Fig. 3a). Thus, lysosome degradation appears analogous to other types of organelle-selective autophagy such as mitophagy and pexophagy. Still, many questions regarding the specific ubiquitination targets and their regulation remain. specific physiological conditions in which lysophagy is triggered will need to be determined.
Figure 3.
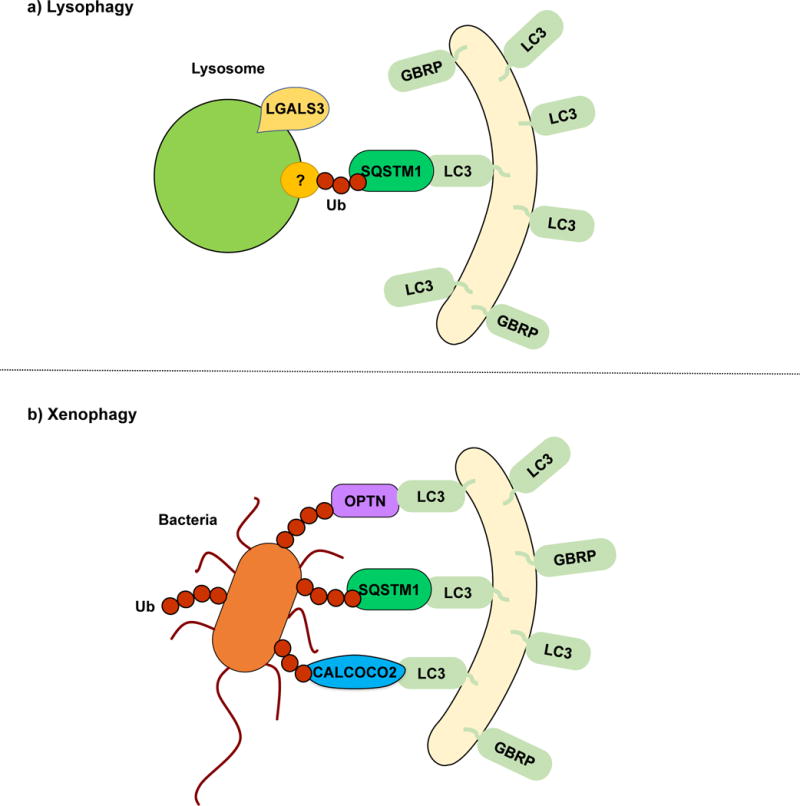
Lysophagy and xenophagy. (a) During lysophagy, unknown lysosomal proteins are ubiquitinated and recognized by SQSTM1, which functions as a receptor, linking the damaged lysosomes with the LC3/GABARAP-conjugated sequestering membranes. LGALS3 binds to exposed lysosomal glycoproteins upon membrane rupture. A specific lysophagy mechanism remains to be elucidated. (b) In xenophagy, intracellular pathogens such as viruses and bacteria are recognized and ubiquitinated. SQSTM1, OPTN, CALCOCO2 and NBR1 have been described as receptor proteins.
Xenophagy
Xenophagy is the collective term used for the selective autophagic degradation of intracellular pathogens including viruses, bacteria and fungi, which constitutes an important part of the immune response120, 121. Once again, ubiquitination and cargo receptor binding play an important role in xenophagy. Following Salmonella typhimurium infection and release into the cytosol, bacterial proteins are rapidly ubiquitinated and recognized by the cargo receptors SQSTM1122, CALCOCO2123 and OPTN123. CALCOCO2 binding to invading bacteria depends on lectin LGALS8 recruitment to damaged bacteria-containing vesicles124. All three receptors possess ubiquitin binding domains and LIRs, thus mediating the interaction between the ubiquitinated bacteria and LC3/GABARAP family members for phagophore sequestration122, 123, 125 (Fig. 3b). Wild et al. showed that these three cargo receptors can bind to the same bacterium125. However, individual silencing of SQSTM1, CALCOCO2 or OPTN is sufficient to increase S. typhimurium replication125. This finding suggests that all three cargo receptors have individual roles in xenophagy that cannot be compensated by the other two. Although probably linked to their individual abilities to recruit other autophagy-inducing factors, further studies will be necessary to determine the specific contributions of each cargo receptor. Additionally, finding the specific pathogen proteins that are ubiquitinated will prove indispensable to therapeutically counter the strategies that pathogens have evolved to avoid autophagy.
Lipophagy
Initially discovered in hepatocytes and later in other cell types, lipophagy describes the selective degradation of lipid droplets (LD) by autophagy. In vivo and in vitro experiments have shown that lipophagy occurs during basal and starvation conditions regulating cellular triglyceride content126. Chaperone-mediated autophagy (CMA) has been proposed as a regulator of lipophagy. In this model, CMA would degrade the LD-associated PLIN (perilipin) proteins leading to lipophagy activation127. Although specific receptors for lipophagy have not been found, the metabolic implications associated with this process have highlighted important insights into energy utilization and possible therapeutic strategies for high-fat diet-induced pathologies. In S. cerevisiae lipid droplets are degraded in a process termed microlipophagy that depends on the core autophagy machinery, but not Atg11128.
Ferritinophagy
Ferritinophagy involves the degradation of the iron-sequestering protein ferritin129. Iron is an essential component of various enzymes and proteins, making it indispensable for several cellular processes. However, free iron promotes ROS generation and is detrimental to the cell130. Ferritin, consisting of multiple heavy chain (FTH1) and light chain (FTL) subunits, acts as a sink for iron when cellular iron levels are high. Conversely, when bioavailable iron levels are low, ferritin is mobilized by ferritinophagy to release iron129.
Ferritinophagy was initially identified in atg5−/− MEF cells, which fail to degrade ferritin upon iron depletion131. Selectivity during ferritinophagy is mediated by the receptor NCOA4 which specifically binds FTH1 and marks ferritin as an autophagic cargo132, 133. The level of NCOA4 is kept low in iron-replete conditions by the iron-dependent interaction between the HECT E3 ligase HERC2 and NCOA4, followed by the ubiquitination and proteasomal degradation of NCOA4132. In response to iron depletion NCOA4 is stabilized, allowing ferritin to be selectively degraded. NCOA4 does not contain a conventional LIR motif in contrast to other autophagy receptors129. Therefore, how NCOA4 links its cargo to phagophores promises to be an intriguing question for the field.
Glycophagy
Glycophagy refers to the selective autophagy-mediated degradation of glycogen, the storage form of glucose in animal cells, by acid α-glucosidase within the lysosome134. Glycophagy is distinct from cytosolic glycogen breakdown via glycogen phosphorylase, and these pathways likely have complementary roles in glycogen catabolism because they preferentially act on slightly different glycogen substrates135. The putative receptor for glycophagy is STBD1 (starch binding domain 1), which possesses a CBN20 glycan-binding domain136 as well as a LIR135. STBD1 localizes to glycogen particles and binds GABARAPL1134 but not LC3B134. Current evidence indicates an important role for glycophagy in cardiac and hepatic pathophysiology, and further mechanistic investigation of this process will be crucial for realizing the full scope of this pathway in carbohydrate metabolism.
Conclusion
Whereas selective autophagy occurs in different forms corresponding to various targets, there is a unifying principle: a receptor, which binds the cargo or which may be an integral part of the cargo (for example as observed with Atg32), links the cargo to the autophagy machinery. Recent years have shown tremendous progress in understanding the mechanisms behind each of these selective processes resulting in a wealth of knowledge on how distinct subsets of the cell’s autophagic machinery are employed to eliminate different cellular components and organelles. As we have highlighted here, there are still important unanswered questions, including the mechanism of mitophagy under physiological (as opposed to experimental) conditions, the post-translational and structural modifications that occur to temporally control receptor-ligand interactions, and the regulatory pathways that integrate stress and developmental signals to coordinate the mode of selective autophagy with the precise cellular needs. Exploring these queries will further our understanding of selective autophagy and may provide important clues for therapeutic strategies.
Acknowledgments
This work was supported by NIH grant GM053396 to DJK.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin M, Liu X, Klionsky DJ. SnapShot: Selective autophagy. Cell. 2013;152:368–368 e362. doi: 10.1016/j.cell.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112(Pt 22):4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 7.Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 8.Nakatogawa H, Mochida K. Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy. 2015;11:2377–2378. doi: 10.1080/15548627.2015.1106665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar S, Ravikumar B, Rubinsztein DC. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 2009;453:83–110. doi: 10.1016/S0076-6879(08)04005-6. [DOI] [PubMed] [Google Scholar]
- 10.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leber R, Silles E, Sandoval IV, Mazon MJ. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J Biol Chem. 2001;276:29210–29217. doi: 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- 12.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffenwimmer T, et al. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 2014;15:862–870. doi: 10.15252/embr.201438932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 17.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 23.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 24.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogov VV, et al. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci Rep. 2017;7:1131. doi: 10.1038/s41598-017-01258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 28.Winslow AR, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 30.Lu K, Psakhye I, Jentsch S. A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy. 2014;10:2381–2382. doi: 10.4161/15548627.2014.981919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Shen Z, Li Y, Gasparski AN, Abeliovich H, Greenberg ML. Cardiolipin Regulates Mitophagy through the Protein Kinase C Pathway. J Biol Chem. 2017;292:2916–2923. doi: 10.1074/jbc.M116.753574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 35.Filimonenko M, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clausen TH, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 37.Lystad AH, et al. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 2014;15:557–565. doi: 10.1002/embr.201338003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonsen A, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 39.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 40.Verhoef LG, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 41.Lu K, den Brave F, Jentsch S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol. 2017;19:732–739. doi: 10.1038/ncb3531. [DOI] [PubMed] [Google Scholar]
- 42.Wurzer B, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4:e08941. doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 44.Nazarko TY, Farre JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–3839. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazarko TY, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014;204:541–557. doi: 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deosaran E, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita S, Abe K, Tatemichi Y, Fujiki Y. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy. 2014;10:1549–1564. doi: 10.4161/auto.29329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nordgren M, et al. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy. 2015;11:1326–1340. doi: 10.1080/15548627.2015.1061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grou CP, et al. The peroxisomal protein import machinery–a case report of transient ubiquitination with a new flavor. Cell Mol Life Sci. 2009;66:254–262. doi: 10.1007/s00018-008-8415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sargent G, et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol. 2016;214:677–690. doi: 10.1083/jcb.201511034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 55.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Jin M, Liu X, Klionsky DJ. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy. 2013;9:1828–1836. doi: 10.4161/auto.26281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redmann M, Dodson M, Boyer-Guittaut M, Darley-Usmar V, Zhang J. Mitophagy mechanisms and role in human diseases. Int J Biochem Cell Biol. 2014;53:127–133. doi: 10.1016/j.biocel.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurihara Y, et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287:3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stotland A, Gottlieb RA. Mitochondrial quality control: Easy come, easy go. Biochim Biophys Acta. 2015;1853:2802–2811. doi: 10.1016/j.bbamcr.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Von Stockum S, Nardin A, Schrepfer E, Ziviani E. Mitochondrial dynamics and mitophagy in Parkinson’s disease: A fly point of view. Neurobiol Dis. 2016;90:58–67. doi: 10.1016/j.nbd.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Aihara M, et al. Tor and the Sin3-Rpd3 complex regulate expression of the mitophagy receptor protein Atg32 in yeast. J Cell Sci. 2014;127:3184–3196. doi: 10.1242/jcs.153254. [DOI] [PubMed] [Google Scholar]
- 62.Kanki T, Furukawa K, Yamashita S. Mitophagy in yeast: Molecular mechanisms and physiological role. Biochim Biophys Acta. 2015;1853:2756–2765. doi: 10.1016/j.bbamcr.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 65.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kondo-Okamoto N, et al. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J Biol Chem. 2012;287:10631–10638. doi: 10.1074/jbc.M111.299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aoki Y, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanki T, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–794. doi: 10.1038/embor.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193:755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abeliovich H, Zarei M, Rigbolt KT, Youle RJ, Dengjel J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun. 2013;4:2789. doi: 10.1038/ncomms3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Georgakopoulos ND, Wells G, Campanella M. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017;13:136–146. doi: 10.1038/nchembio.2287. [DOI] [PubMed] [Google Scholar]
- 72.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei Y, Chiang WC, Sumpter R, Jr, Mishra P, Levine B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell. 2017;168:224–238 e210. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Durcan TM, Fon EA. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 78.Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aerts L, Craessaerts K, De Strooper B, Morais VA. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J Biol Chem. 2015;290:2798–2811. doi: 10.1074/jbc.M114.620906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 83.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 86.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bingol B, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 88.Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McLelland GL, Lee SA, McBride HM, Fon EA. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016;214:275–291. doi: 10.1083/jcb.201603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matsushima M, et al. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- 92.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sheng ZH, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsieh CH, et al. Functional Impairment in Miro Degradation and Mitophagy Is a Shared Feature in Familial and Sporadic Parkinson’s Disease. Cell Stem Cell. 2016;19:709–724. doi: 10.1016/j.stem.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novak I, Dikic I. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011;7:301–303. doi: 10.4161/auto.7.3.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang J, et al. A short linear motif in BNIP3L (NIX) mediates mitochondrial clearance in reticulocytes. Autophagy. 2012;8:1325–1332. doi: 10.4161/auto.20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ni HM, Williams JA, Ding WX. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015;4:6–13. doi: 10.1016/j.redox.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu L, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 101.Bordi M, Nazio F, Campello S. The Close Interconnection between Mitochondrial Dynamics and Mitophagy in Cancer. Front Oncol. 2017;7:81. doi: 10.3389/fonc.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen G, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 103.Palikaras K, Lionaki E, Tavernarakis N. Mitophagy: In sickness and in health. Mol Cell Oncol. 2016;3:e1056332. doi: 10.1080/23723556.2015.1056332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Shirihai OS, Song M, Dorn GW., 2nd How mitochondrial dynamism orchestrates mitophagy. Circ Res. 2015;116:1835–1849. doi: 10.1161/CIRCRESAHA.116.306374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen M, et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. 2016;12:689–702. doi: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu W, et al. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 2016;35:1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chu CT, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sentelle RD, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. 2014;127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 114.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 115.Changou CA, et al. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc Natl Acad Sci U S A. 2014;111:14147–14152. doi: 10.1073/pnas.1404171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dou Z, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mijaljica D, Prescott M, Devenish RJ. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS One. 2012;7:e40013. doi: 10.1371/journal.pone.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kvam E, Goldfarb DS. Structure and function of nucleus-vacuole junctions: outer-nuclear-membrane targeting of Nvj1p and a role in tryptophan uptake. J Cell Sci. 2006;119:3622–3633. doi: 10.1242/jcs.03093. [DOI] [PubMed] [Google Scholar]
- 119.Maejima I, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Colombo MI, Gutierrez MG, Romano PS. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy. 2006;2:162–164. doi: 10.4161/auto.2827. [DOI] [PubMed] [Google Scholar]
- 121.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 122.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 123.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 124.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Zutphen T, et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 130.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Asano T, et al. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol. 2011;31:2040–2052. doi: 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mancias JD, et al. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife. 2015;4 doi: 10.7554/eLife.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Delbridge LM, Mellor KM, Taylor DJ, Gottlieb RA. Myocardial autophagic energy stress responses–macroautophagy, mitophagy, and glycophagy. Am J Physiol Heart Circ Physiol. 2015;308:H1194–1204. doi: 10.1152/ajpheart.00002.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170–184. doi: 10.1038/nrgastro.2016.185. [DOI] [PubMed] [Google Scholar]
- 136.Zhu Y, Zhang M, Kelly AR, Cheng A. The carbohydrate-binding domain of overexpressed STBD1 is important for its stability and protein-protein interactions. Biosci Rep. 2014;34 doi: 10.1042/BSR20140053. [DOI] [PMC free article] [PubMed] [Google Scholar]


