SUMMARY
Organisms in the three domains of life depend on protein polymers to form a cytoskeleton that helps to establish their shapes, maintain their mechanical integrity, divide, and, in many cases, move. Eukaryotes have the most complex cytoskeletons, comprising three cytoskeletal polymers—actin filaments, intermediate filaments, and microtubules—acted on by three families of motor proteins (myosin, kinesin, and dynein). Prokaryotes have polymers of proteins homologous to actin and tubulin but no motors, and a few bacteria have a protein related to intermediate filament proteins.
1. INTRODUCTION—OVERVIEW OF CELLULAR FUNCTIONS
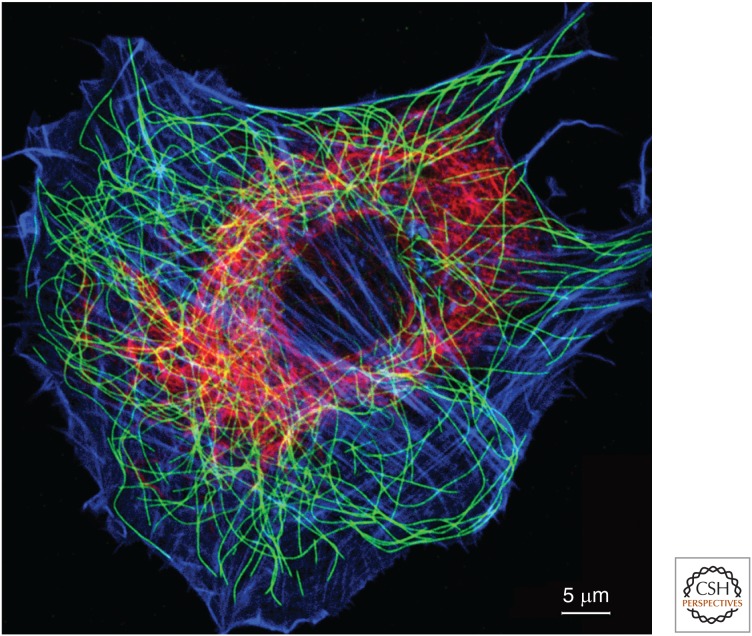
Actin filaments, intermediate filaments, and microtubules form distinctive networks in the cytoplasm of eukaryotic cells. The fibroblast in Figure 1 illustrates their distributions. Actin filaments concentrate around the periphery of the cell in the cortex underlying the plasma membrane and in bundles called stress fibers anchored to plasma membrane adhesions sites (reviewed by Svitkina 2016). Intermediate filaments form a complex network, concentrated in the perinuclear region, from which filaments radiate toward the cell surface, where they are anchored to intercellular junctions called desmosomes and cell–extracellular-matrix junctions called hemidesmosomes (for review, see Jones et al. 2016). Individual microtubules are large enough to image by light microscopy. These long, stiff polymers form a network radiating from the juxtanuclear centrosome or microtubule-organizing center (MTOC) to the edge of the cell.
Figure 1.
Fluorescence micrograph of a cultured fibroblast stained with fluorescent phalloidin (blue) and antibodies to microtubules (green) and intermediate filaments (red). Individual microtubules are large enough to be resolved by light microscopy, but actin filaments (blue) and intermediate filaments are too small and too densely packed to be resolved. In cells with desmosomes or hemidesmosomes, intermediate filaments extend to the periphery of the cell and anchor these adhesive structures. (Courtesy of Harald Herrmann, University of Heidelberg, Germany.)
All three cytoskeletal polymers serve as mechanical elements (reviewed by Pegoraro et al. 2016), and microtubules and actin filaments provide tracks for molecular motors (see Sweeney and Holzbaur 2016 for review). In contrast, intermediate filaments do not serve as tracks for molecular motors (reviewed by Herrmann and Aebi 2016) but, rather, are transported by these motors. Microtubules are tracks for the motors dynein and kinesin that move membrane-bound vesicles, ribonucleoprotein particles, and intermediate filaments over long distances through the crowded cytoplasm (see Barlan and Gelfand 2016 for review). These movements are responsible for the characteristic distributions of the endoplasm reticulum, the Golgi apparatus, and other organelles in the cytoplasm. Microtubules also form the scaffold of the axonemes of cilia and flagella (for review, see Viswanadha et al. 2016) and the mitotic apparatus (reviewed by McIntosh 2016). In both cases, kinesins and dyneins act on the microtubule scaffold to bend axonemes or move chromosomes during mitosis. Polymerization of actin filaments produces forces for cellular movements and membrane traffic. Actin filaments are also tracks for short-range movements of vesicles by myosins (see review by Titus 2016) and cables for contraction by myosin motors during locomotion, cytokinesis, and other cell shape changes (Svitkina 2016). Intermediate filaments provide the mechanical support for the internal (noncortical) cytoplasm and cell surface (Pegoraro et al. 2016), and also participate in shaping the cell and regulating signal transduction (for reviews, see Herrmann and Aebi 2016; Cheng and Eriksson 2016).
2. STRUCTURES OF THE CYTOSKELETAL POLYMERS
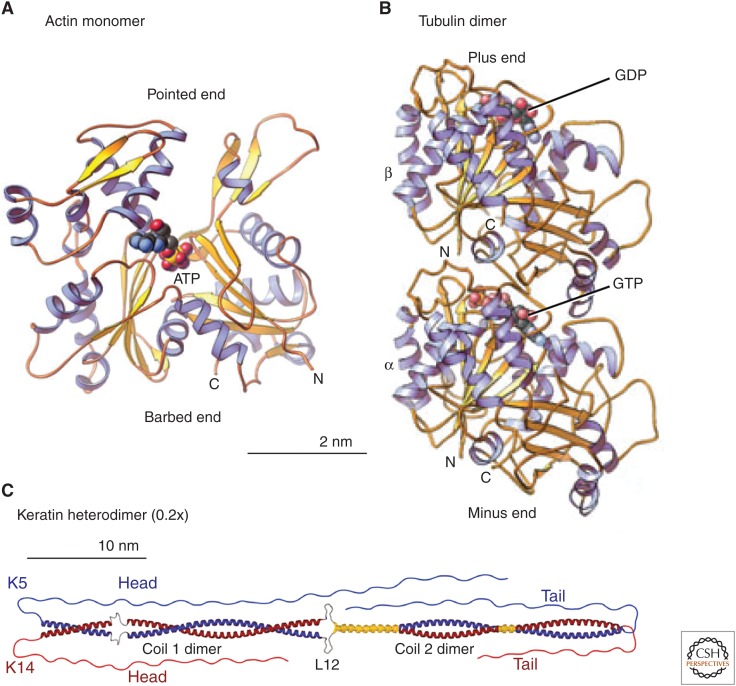
The protein subunits of the three cytoskeletal polymers have strikingly different structures (Fig. 2). The flat actin molecule has a deep cleft to bind ATP (reviewed by Pollard 2016). The tubulin subunit is a dimer of homologous α- and β-subunits, each with a binding site for a guanine nucleotide on one end. GTP is bound at the stable interface between the two subunits and does not exchange with GTP in the cytoplasm (for review, see Goodson and Jonasson 2016). The exposed site on the β-subunit binds either GTP or GDP. The GTP bound to the β-subunit is hydrolyzed after assembly into a microtubule. After depolymerization, this GDP is exchanged for GTP. The main structural feature of intermediate filament proteins is a long, rod-shaped, α-helical, coiled coil flanked by additional residues at both ends (Herrmann and Aebi 2016). These head and tail domains vary in size in the various isoforms. No nucleotides are known to bind to intermediate filament proteins.
Figure 2.
Structures of (A) an actin monomer, (B) a tubulin dimer, and (C) a keratin heterodimer. Ribbon diagrams compare the building blocks of the three cytoskeletal polymers. The scale of the keratin is 20% that of actin and tubulin to fit this long molecule into the figure. (A,B, Reprinted from Pollard and Earnshaw 2008; C, reprinted from Herrmann and Aebi 2016.)
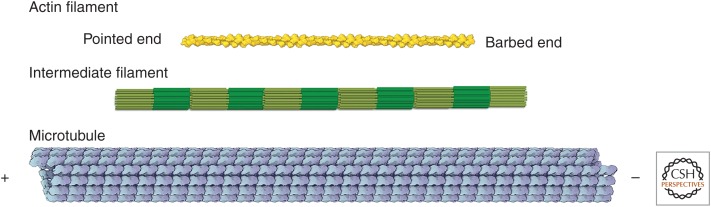
Each cytoskeletal polymer has a different arrangement of its protein subunits (Fig. 3). The subunits in actin filaments form a helix that can be described as two, parallel, right-handed strands offset by half a subunit or as a short-pitch, left-handed helix with a single strand going from each subunit to the next in the opposite strand. All of the asymmetric actin subunits are oriented in the same direction along the polymer, and so the polymer is polar. The two ends of actin filaments are called “barbed” and “pointed” ends, names coming from the polarized arrowhead pattern created when they are saturated with myosin (see Pollard 2016). Both ends can grow or shrink, but the barbed end grows faster than the pointed end. Most myosins move toward the barbed end (see Sweeney and Holzbaur 2016).
Figure 3.
Scale drawings of the actin filament, intermediate filament, and microtubule. The actin filament and microtubule models are based on reconstructions of cryo-electron micrographs. The model of the intermediate filament is based on measurements of mass per unit length and characterization of the oligomeric intermediates that anneal to form the filament. Actin filaments and microtubules are polar; the names of their ends are indicated. Intermediate filaments are not polar because they assemble from bipolar tetramers of helical polypeptides. The dark green represents zones where pairs of coiled coils overlap to make antiparallel tetramers. (The actin filament and microtubule are courtesy of Graham Johnson. The intermediate filament model is reprinted from Herrmann and Aebi 2016.)
Tubulin dimers associate end-to-end to form protofilaments that run along the length of cylindrical microtubules. α-subunits form lateral bonds with neighboring α-subunits and β-subunits are next to β-subunits. The protofilaments are offset slightly from each other, making what seems to be a helix. However, most cytoplasmic microtubules have 13 protofilaments, so the lattice of subunits has a seam in which α-subunits are next to β-subunits. The end of the microtubule with exposed β-subunits is called the plus end, and the other end is called the minus end (Fig. 3). Dimers only associate or dissociate at the ends, generally faster at plus ends than minus ends (Goodson and Jonasson 2016). Dyneins move toward the minus ends of microtubules, and most kinesins move toward plus ends.
The internal structure of intermediate filaments is not yet known, but they consist of subunits comprising four polypeptides. These “tetramers” are formed from two dimeric molecules with their rod domains interacting side by side with opposite polarity (Herrmann and Aebi 2016). Owing to the opposite orientation of dimers within the tetrameric subunits, mature intermediate filaments lack the polarity inherent in the structure of actin and microtubules, which might explain why no motors are known to move on intermediate filaments.
The three cytoskeletal polymers have distinctive mechanical properties. Persistence length is the commonly used measure of their stiffness. (Persistence length scales with stiffness and, formally, is the distance in which the correlations between the tangents to a polymer are lost.) With a persistence length of >1000 µm, microtubules are far stiffer than the other two polymers. If enlarged a millionfold, a microtubule would have physical properties similar to a 25-mm diameter tube of a stiff plastic such as Plexiglas. In contrast, intermediate filaments are the most flexible of the three cytoskeletal polymers, with a persistence length of 0.3–1.0 µm. They would feel like a braided, plastic rope 10 mm in diameter, if enlarged a millionfold. Like a rope, they have great tensile strength, so they resist stretching in spite of being flexible. Because of their flexibility, intermediate filaments show strain stiffening, where they become very stiff after stretching beyond 3.5 times their resting length (see Pegoraro et al. 2016). Actin filaments are as stiff as steel, but they are very thin, and so their persistence length is ∼10 µm, 100 times more flexible than a microtubule. These differences in stiffness are apparent in the micrograph in Figure 1.
3. ASSEMBLY OF CYTOSKELETAL POLYMERS
All three cytoskeletal polymers assemble spontaneously under physiological conditions. For microtubules and actin, nucleation of polymerization is unfavorable, and so cells use regulatory proteins to specify when and where new polymers form. Once started, actin filaments and microtubules elongate at both ends by addition of ATP–actin monomers or GTP–tubulin dimers. The barbed ends of actin filaments, and plus ends of microtubules, grow faster than the other ends.
The nucleotides bound to actin and tubulin impact their polymerization properties. These effects differ in magnitude but are conceptually similar for the two polymers in spite of having evolved independently.
ATP bound to actin is hydrolyzed rapidly to ADP during polymerization, followed by slow dissociation of the γ-phosphate. ADP–Pi actin is much like ATP–actin, but ADP–actin dissociates faster from filaments than ATP–actin. Therefore, the crucial reaction is dissociation of phosphate rather than ATP hydrolysis (see Pollard 2016 for details).
Similarly, assembly of GTP–tubulin dimers into a microtubule promotes the hydrolysis of the GTP on the β-subunit. Contacts with an α-subunit along a protofilament complete the active site for hydrolysis. After dissociation of the γ-phosphate, GDP–tubulin dissociates very rapidly if exposed on the end of a microtubule. However, a cap of GTP–tubulin subunits can stabilize the end. Loss of the GTP cap destabilizes the end of a microtubule and results in rapid depolymerization by dissociation of short pieces of protofilaments. At steady state, many microtubules alternate between phases of steady elongation and catastrophic disassembly—behavior called dynamic instability (see Goodson and Jonasson 2016 for details). After a GDP–dimer dissociates from a microtubule, the GDP is exchanged for GTP in the cytoplasm in preparation for another round of assembly.
Intermediate filaments self-assemble into complex polymers under environments of physiological ionic conditions and pH without a requirement for nucleotides such as GTP and ATP. The polymerization process is of a hierarchical nature, beginning with the formation of tetramers. On average eight tetramers interact to form a unit-length filament comprising 32 polypeptides. These unit-length filaments interact end-to-end in series to form mature intermediate filaments, followed by a radial compaction. There are eight tetramers in a cross section of the typical 10-nm diameter intermediate filament (Herrmann and Aebi 2016). This assembly mechanism allows intermediate filaments to incorporate subunits all along their lengths.
4. OVERVIEW OF THE EVOLUTION OF THE CYTOSKELETON
The actin and tubulin genes arose in the common ancestor of life on Earth and each diverged in fascinating ways during the past three billion years. Today, the homologous proteins form different polymers with different functions in contemporary prokaryotes and eukaryotes. The genes for intermediate filaments arose in early eukaryotes and diversified by gene duplication and divergence, especially in animals during the past five hundred million years.
Genes for the proteins that regulate the assembly of actin and tubulin seem to have arisen separately in prokaryotes and eukaryotes because they are not homologous. Furthermore, eukaryotes evolved proteins with similar functions in the actin and tubulin systems, including proteins that bind nonpolymerized subunits, nucleate polymers, promote elongation, cap polymer ends, sever polymers, stabilize polymers by lateral association, and cross-link polymers. Remarkably, the genes for these proteins with parallel functions evolved independently, and so the genes for proteins with similar functions are not homologous.
4.1. Evolution of the Actin System
Most contemporary organisms have genes for actin or closely related proteins (Gunning et al. 2015), and so the common ancestor of life on Earth must have had an actin gene about three billion years ago. This ancestral organism and its primordial actin gene have disappeared, but contemporary actin genes provide some clues about its evolution.
A likely precursor to the actin gene encoded an ATP-binding protein half as large as actin (one of the two domains flanking the central cleft of actin). An early gene duplication created a protein with two homologous domains with an ATP-binding cleft in between (Fig. 2). Hexokinase has the same fold as actin and binds ATP in a cleft between two homologous domains, and so this glycolytic enzyme likely shares a common ancestor with actin.
Genes encoding actin are widespread in bacteria and archaea, as well as their plasmids and bacteriophages. These genes and proteins were hard to identify as their sequences have diverged very far from the conserved actins in eukaryotes. Bacteria have three broad families of actin genes, the FtsA, MreB, and ParM families, each encoding a diversity of proteins with distinct functions. FtsA participates in cell division, MreB has a role in cell wall synthesis, and ParM separates plasmids during mitosis. Bacteriophages and plasmids also encode genes for approximately 30 different divergent actins, many of which participate in DNA segregation (Derman et al. 2009). Structures of a sample of these proteins show that they share the basic actin fold, with ATP bound in a central cleft, but vary in the architecture of their surfaces (Ghoshdastider et al. 2015). Some of these prokaryotic actins assemble filaments with two strands of subunits, similar to eukaryotic actin filaments, but others are quite different, including filaments comprising a single strand of subunits and another with two strands running in opposite directions (Ghoshdastider et al. 2015).
Eukaryotes inherited the gene encoding actin from an archaeal cell related to contemporary Lokiarchaeota (Spang et al. 2015). This organism is known from a nearly complete genome sequence obtained from a deep-sea environmental sample, but the organism itself has not yet been seen. Lokiarchaeota is related to the archaeal cell that gave rise to eukaryotes, having in addition to actin a number of other genes typical of eukaryotes but not found in other prokaryotes.
During the divergence of eukaryotes from a common ancestor, many species duplicated their actin genes, but these actins are among the most conserved eukaryotic genes and proteins, sharing similar sequences, molecular structures, biochemical properties, and filament structures. In contrast, duplication and divergence of actin genes gave rise to eukaryotic actin-related proteins (Arps) with sequences and functions that diverged to become quite different from those of actin. For example, Arp2 and Arp3 are part of the Arp2/3 complex that nucleates actin filament branches (Pollard 2016), whereas Arp1 forms a filament in the dynactin complex (Barlan and Gelfand 2016; Sweeney and Holzbaur 2016). Furthermore, other Arp proteins are parts of nucleosome-remodeling complexes in the nucleus.
Eukaryotes and prokaryotes use different strategies to deploy actins for physiological purposes. Eukaryotes have single actins (yeast) or multiple closely related actins (plants, animals) that all form double-stranded filaments. Numerous regulatory proteins allow actin filaments to make many different structures for cellular motility, cytokinesis, membrane traffic, and cellular integrity (for reviews, see Glotzer 2016; Pollard 2016; Svitkina 2016). In contrast, prokaryotes used multiple specialized actins to make a range of structures and have relatively few proteins that regulate these actins.
4.2. Evolution of the Microtubule System
The common ancestor of life on Earth had a gene for a tubulin-like protein in addition to a gene for actin. It is not clear how the α- and β-tubulin genes arose in eukaryotes. However, they were present in the very early eukaryotes because microtubule-based axonemes have been a characteristic feature of single-celled eukaryotes for more than one billion years (Viswanadha et al. 2016).
The tubulin gene in contemporary Archaea and bacteria encodes the protein FtsZ. This GTP-binding protein has the same fold as tubulin, but the sequences of FtsZ and tubulin have diverged nearly beyond recognition. Monomers of GTP–FtsZ assemble protofilaments rather than cylindrical microtubules, hydrolyze the bound GTP, and disassemble. FtsZ polymers concentrate at the site of cell division in prokaryotes, and FtsZ reconstituted in liposomes in vitro can constrict the lipids. However, the main function of FtsZ is to organize the assembly of transmembrane proteins that synthesize the cell wall in the furrow that divides the cell. Eukaryotes acquired genes for FtsZ from the symbiotic bacteria that evolved into mitochondria and chloroplasts. Today, FtsZ contributes to the division of chloroplasts, but not mitochondria, in most species. Few prokaryotes have genes encoding true tubulins. However, one group has genes for both α- and β-tubulin that they seem to have acquired by lateral gene transfer from a eukaryote.
Genes for α- and β-tubulin are found in all eukaryotes and encode the subunits of microtubules. With the exception of fungi and most plants, eukaryotes generally have genes for δ-, ɛ-, ζ-, and η-tubulin, components of centrioles and basal bodies. The sequences of these genes are highly conserved so that >75% of amino acids are identical in the tubulins of animals and plants. The ciliate Tetrahymena makes many different structures from single genes for α- and β-tubulin, whereas vertebrates have six to eight genes for α- and β-tubulin that are expressed in different tissues.
4.3. Evolution of the Intermediate Filament System
The genes for intermediate filament proteins are distributed much differently across the phylogenetic tree than the universal presence of genes for actins and tubulins. Lamin genes are found in many metazoans (reviewed by Adam 2016) and diverse species on branches that arose from the common eukaryotic ancestor more than one billion years ago. However, genes for intermediate filament proteins have not been identified in plants or fungi or in most prokaryotes. Therefore, the original gene for lamins likely arose before the common eukaryotic ancestor, but these lamin genes have apparently been lost multiple times during evolution (Peter and Stick 2015). The features of the lamin gene, including the introns, nuclear localization signals, and a carboxy-terminal immunoglobulin domain have generally been conserved within the metazoan radiation (Adam 2016).
A duplication of the lamin gene in metazoan species on the path to chordates created the first gene for a cytoplasmic intermediate filament protein. As the two genes diverged during subsequent evolution, the nuclear localization sequence and the CAAX box—a carboxy-terminal prenylation site—were lost from the gene for the cytoplasmic intermediate filament. Later deletions in early chordates removed the sequence for 42 residues in the rod domain and the immunoglobulin domain from the intermediate protein gene. Later, during vertebrate evolution, further rounds of gene duplication and divergence created the contemporary families of genes for cytoplasmic intermediate filament proteins (Herrmann and Aebi 2016). During this divergence, orthologs have been much more highly conserved than paralogs, suggesting that each type of intermediate protein has unique functions.
Most prokaryotes lack genes for proteins related to intermediate filaments, with the exception of the α-proteobacterium Caulobacter crescentus, which has a protein with a coiled-coil rod similar in some ways to intermediate filament proteins. The bacterium depends on this crescentin protein for its curved shape, but the protein lacks several features that are important for the assembly of intermediate filaments. Convergent evolution is possible, but given the isolated distribution of crescentin, Caulobacter might have acquired the gene encoding crescentin by lateral transfer from a eukaryote.
4.4. Evolution of Motor Proteins
Three families of motor proteins with ATPase activities move on cytoskeletal polymers in eukaryotic organisms. A large family of myosin proteins uses actin filaments as tracks. Another large family of kinesin proteins and a modest number of dynein proteins walk on microtubules (Sweeney and Holzbaur 2016).
The common ancestor of the genes for myosin and kinesin was a prokaryotic gene for a small nucleotide-binding protein—probably a GTPase—which gave rise to contemporary GTPases, myosins, and kinesins. Dynein is a member of the ancient AAA ATPase family. Although precursors of all three of these motor families originated in prokaryotes, none of these motors have been detected in any contemporary prokaryote.
The time of appearance of functional myosins and kinesins during the evolution of eukaryotes is unclear. Phylogenetic analysis suggests that the first myosin appeared before the last common ancestor of eukaryotes about 1 billion years ago. It was a myosin-I, with a single head and short tail. A gene encoding myosin-V appeared shortly after the crown group of eukaryotes began to branch from the common ancestor. It gave rise to myosins VIII and XI in plants. Myosin-II appeared on the branch leading to amoebas, fungi, and animals, in which it contributed to cytokinesis (Glotzer 2016) and, eventually, to the evolution of muscle cells (reviewed by Sweeney and Hammers 2016).
The last common eukaryotic ancestor had approximately 11 genes for kinesins. Over the past billion years, the kinesin gene family has expanded by duplication and divergence, so that eukaryotes now have 17 families of kinesins, including multiple isoforms in many families. Some organisms have lost one or more of these ancient kinesin genes over time.
Dynein is the most ancient of the three motor proteins. This is remarkable as it is the largest and most complicated of the three. The multiple genes required to make dynein were present in very early eukaryotes, which are believed to have had motile axonemes. As for the kinesins and myosin, genes encoding dynein have been lost multiple times during evolution, so neither red algae nor flowering plants have dyneins or motile axonemes.
5. CONCLUSION
The reviews in this collection explain the structures and functions of the cytoskeleton, starting with the proteins that comprise the three systems—actin filaments, intermediate filaments, and microtubules. Additional associated reviews, a subset of which is cited above, describe how cells assemble these proteins into functional supramolecular structures and how these assemblies give cells their mechanical integrity, contribute to adhesion to extracellular molecules and other cells, transport materials inside cells, move entire cells, move their cilia, separate chromosomes during mitosis, and divide cells in two during cytokinesis.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- *.Adam SA. The nucleoskeleton. 2016. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Barlan K, Gelfand VI. 2016. Microtubule-based transport and distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cheng F, Eriksson JE. 2016. Intermediate filaments and the regulation of cell motility during regeneration and wound healing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. 2009. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: Regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 73: 534–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshdastider U, Jiang S, Popp D, Robinson RC. 2015. In search of the primordial actin filament. Proc Natl Acad Sci 112: 9150–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Glotzer M. 2016. Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Goodson HV, Jonasson EM. 2016. Microtubules and microtubule-associated proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. 2015. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci 128: 2009–2019. [DOI] [PubMed] [Google Scholar]
- *.Herrmann H, Aebi U. 2016. Intermediate filaments: Structure and assembly. Cold Spring Harb Perspect Biol 8: a018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jones JCR, Kam CY, Harmon RM, Woychek AV, Hopkinson SB, Green KJ. 2016. Intermediate filaments and the plasma membrane. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.McIntosh JR. 2016. Mitosis. Cold Spring Harb Perspect Biol 8: a023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pegoraro AF, Janmey J, Weitz DA. 2016. Mechanical properties of the cytoskeleton and cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A, Stick R. 2015. Evolutionary aspects in intermediate filament proteins. Curr Opin Cell Biol 32: 48–55. [DOI] [PubMed] [Google Scholar]
- *.Pollard TD. 2016. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8: a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Earnshaw WC. 2008. Cell biology, 2nd ed Saunders/Elsevier, Philadelphia. [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Svitkina T. 2016. The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney HL, Hammers DW. 2016. Muscle contraction. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney HL, Holzbaur ELF. 2016. Motor proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Titus MA. 2016. Myosin-driven intracellular transport. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a21972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Viswanadha R, Sale WS, Porter ME. 2016. Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]