Abstract
The cadherin superfamily comprises a large, diverse collection of cell surface receptors that are expressed in the nervous system throughout development and have been shown to be essential for the proper assembly of the vertebrate nervous system. As our knowledge of each family member has grown, it has become increasingly clear that the functions of various cadherin subfamilies are intertwined: they can be present in the same protein complexes, impinge on the same developmental processes, and influence the same signaling pathways. This interconnectedness may illustrate a central way in which core developmental events are controlled to bring about the robust and precise assembly of neural circuitry.
The underlying organization of the nervous system determines its function; the topology and connectivity of neural networks governs how these neuronal ensembles process and propagate information. The centrality of this structure–function relationship is recognized in ongoing efforts to unravel the “connectome” (Denk and Horstmann 2004; Briggman and Denk 2006; Lichtman and Denk 2011). Strikingly, this complex architecture self-assembles during development, and a major intellectual challenge is to understand how developmental processes are coordinated to generate a functional nervous system.
There are two remarkable, yet seemingly incongruent, aspects of neural development: the remarkable degree of “specificity” in neural wiring, and its robustness. As is often remarked on, the mature human brain contains ∼1011 neurons connected by ∼1014 synapses. These connections are not random, but segregate the nervous system into distinct functional systems and parallel neuronal modules that perform specific functions. Moreover, nervous system organization is highly stereotyped, showing reproducible patterns among individuals within a species, as well as between species. The issue of “specificity” is essentially to understand how reproducibility can be attained (“sterotypy”), despite the astronomical numbers of neurons and synapses. Standing in contrast to the apparent improbability of wiring the nervous system is the fact that development is robust. Brain development is able to tolerate genetic diversity, broad ranges of developmental and environmental variability, and microheterogeneities in the number, position, and morphology of neurons. In addition, the broad function of brain regions like the cerebellum, for example, is largely the same, despite species-specific variations in genes and structure.
So, how can robust development of brain architecture be reconciled with specificity? In some ways, this dilemma is analogous to Levinthal’s paradox in the area of protein folding (Levinthal 1969), which imagines that it would take a modest-sized polypeptide longer than the lifetime of the universe to search all of conformational space for its stable, folded conformation; yet, typical proteins routinely perform this task in milliseconds. The simple answer to this paradox is that proteins do not systematically search conformational space. Proteins are guided along kinetically favorable pathways (Dill and Chan 1997), making the vast majority of possible states inaccessible. Similarly, neurons do not choose among all possible connections, and each has access to only a tiny fraction of the 1011 potential synaptic partners. Moreover, there are large populations of neural configurations that are consistent with normal function, so that development is not burdened with the requirement for an overly precise outcome. Just as in the folding of a protein, understanding neural wiring will require determining the cellular and developmental mechanisms that simplify and streamline development by restricting the number of possible outcomes and making patterns of connectivity both specific and robust.
In his “chemoaffinity hypothesis,” Roger Sperry imagined that the topographic mapping of the frog retinotectal system resulted from growth cones responding to two orthogonal gradients (Sperry 1963). In this scenario, thousands of axons can be targeted to their correct positions on a Cartesian coordinate system without the aid of a large number of proteins providing highly refined molecular addresses. Sperry’s view was validated with the discovery of the axon guidance cues, Ephrins, and their receptors, Eph kinases (Cheng et al. 1995; Drescher et al. 1995). Reciprocal gradients of Eph kinases, expressed in retinal ganglion cell axons, and Ephrins, expressed in tectal neurons, are responsible for mapping the nasal-temporal axis of the retina onto the antero–posterior axis of the tectum (Triplett and Feldheim 2012). A comparable set of gradients is responsible for the dorsoventral to medio–lateral mapping. In this and related examples, neither retinal ganglion cell axons nor tectal neurons have specified identities, in that neurons are searching for their “correct” partners on the basis of cell-specific synaptic cues. Retinal axons begin to arborize and form synapses in the area where they are forced to stop growing by guidance cues; tectal neurons form synapses with those axons that terminate in their vicinity. Moreover, these maps can be systematically altered, as occurs when the visual field of barn owls is shifted with prism glasses (Knudsen and Brainard 1991), or in the case of three-eyed frogs (Constantine-Paton and Law 1978). Synaptic rearrangement is consistent with the idea that synapse formation is promiscuous and not rigidly controlled by specific cell surface cues (Jontes and Phillips 2006). This view is not meant to denigrate the role of molecules in guiding neural circuit assembly, but has two primary points: (1) each molecule simply fulfills a flexible role within a larger developmental and molecular context, and (2) a strict molecular hardwiring of connectivity is incompatible with the malleability and robustness of development and the evolution of neural circuit function. Thus, although molecules collaborate to sculpt an evolutionarily conserved gross architecture, the flexibility required for nervous system function suggests that neither is the fine scale architecture overly determined, nor must a molecular code exist that specifies this fine scale organization.
The cadherin superfamily is a large and diverse collection of molecules, defined and related by the presence of multiple repeats of a ∼110 amino acid cadherin motif in their extracellular domain (Hirano and Takeichi 2012). Originally discovered by Takeichi and colleagues (Takeichi 1977; Urushihara and Takeichi 1980) and independently by Kemler (Vestweber and Kemler 1984), the cadherins defined a small family of calcium-dependent cell adhesion molecules with homophilic preferences (Takeichi et al. 1981; Atsumi et al. 1983; Shirayoshi et al. 1986). Subsequently, the cadherin superfamily has expanded to include protocadherins (both clustered and nonclustered), and the atypical cadherins Celsr, Fat, and Dachsous, among others (Hulpiau and van Roy 2011; Hirano and Takeichi 2012). Although the classical cadherins and protocadherins, in particular, have been proposed to regulate circuit assembly (Hirano and Takeichi 2012), accumulating evidence reveals that each of the cadherin subfamilies are heavily involved with multiple aspects of neural development. Nearly every element of neural development can reasonably be tied to circuit formation, including neurogenesis, neuronal polarity, neuronal migration, axon outgrowth, dendrite morphogenesis, synaptogenesis, synaptic rearrangement, and synaptic plasticity. Although cadherin family members are intimately involved in each of these processes, I will focus here on their roles in regulating neurogenesis and in axon development. As further detailed below, two themes are emerging, which illuminate how cadherins contribute to neural circuit assembly that is both robust and specific. First, the developmental events directed by cadherins help generate a coarse neural scaffold of local microcircuits and interregional connections; this restricts the possible outcomes of development and promotes reproducible patterns of connectivity. Second, multiple cadherin families act collaboratively, interacting with each other physically and functionally to guide robust development.
NEUROGENESIS AND NEURONAL MIGRATION
An essential step in the assembly of neural circuitry is the production of the correct number and types of neurons. Neurons are born along the apical surface of the neuroepithelium (Kriegstein and Alvarez-Buylla 2009), derived initially from neuroepithelial cells (NECs) and, at later stages, from radial glial cells (RGCs) (Fig. 1). These progenitor cells must both proliferate to expand the progenitor pool, and give rise to differentiated neurons (Gotz and Huttner 2005). RGCs are elongated cells that remain attached both to the ventricular surface through an apical endfoot and to the external surface of the brain through a long, thin basal process. In the mammalian cortex, RGCs act both as progenitors, and a substrate along which nascent neurons migrate (Rakic 2009). As a neuronal progenitor, each RGC gives rise to multiple progeny that comprise radial clones of sibling neurons. Ubiquitously expressed in the early nervous system, N-cadherin is a major component of apical adherens junctions (AJs) linking adjacent NECs or RGCs (Chenn et al. 1998), and is essential for maintaining the structural integrity of the neuroepithelium (Ganzler-Odenthal and Redies 1998; Lele et al. 2002; Kadowaki et al. 2007). Injection of N-cadherin interfering antibodies into the chick neural tube disrupts the neuroepithelium (Ganzler-Odenthal and Redies 1998), compromising the ventricular surface and giving rise to cellular rosettes. Similarly, the laminar organization of the optic tectum is degraded, with mislocalization of both neurons and proliferating cells. Comparable results are obtained in the zebrafish; mutants lacking N-cadherin show a disorganized neuroepithelium, as well as mislocalization of both mitotic cells and differentiated neurons (Lele et al. 2002; Zigman et al. 2011). Conditional deletion of N-cadherin in the mouse cortex disrupts the apical AJs of NECs and RGCs (Kadowaki et al. 2007). As a consequence, both mitotic cells and differentiated cells scatter randomly in the developing cortex and the pattern of cortical lamination is disrupted. Loss of αE-catenin in neuronal progenitors has comparable effects on both proliferation and cortical architecture (Lien et al. 2006; Stocker and Chenn 2009), suggesting that adhesion by the cadherin–catenin complex is required to maintain epithelial organization and to allow the orderly production of neurons for the assembly of brain architecture.
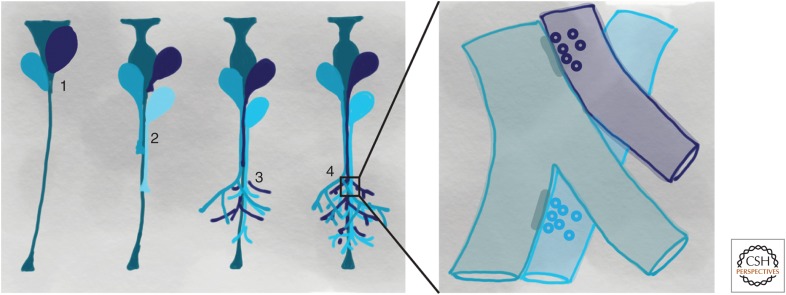
Figure 1.
Potential roles for cadherins in lineage-dependent circuit assembly. Shown here is a model of how protocadherins, cadherins, or other adhesion molecules could guide the formation of local circuits through lineage-dependent mechanisms. (1) Cell–cell adhesion can regulate the production of neurons, either directly from neuronal progenitors or intermediate progenitors. (2) Homophilic adhesion can promote fasciculation or collective extension of axonal or dendritic projections. (3) Axonal and dendritic arborization are promoted by cell–cell interactions. (4) Contact-dependent growth of axons and dendrites promotes contact among sibling neurons. The increased contact among siblings introduces a bias during synaptogenesis, promoting connectivity within a lineage.
The role of the cadherin complex at neuroepithelial AJs is important for more than structural stability, and appears to play an essential role in regulating neuronal differentiation. β-catenin serves a dual role, both as a core component of the adhesive cadherin complex and acting as the primary effector in canonical Wnt signaling. In this latter role, β-catenin regulates proliferation and differentiation in a variety of developmental contexts. Like N-cadherin, elimination of β-catenin in the developing cortex compromises epithelial structure and results in mislocalization of NECs and neurons (Brault et al. 2001; Junghans et al. 2005). Mosaically disrupting β-catenin also induces differentiation of progenitors, and in vitro data suggest that β-catenin signaling is important for maintaining the progenitor pool and inhibiting neuronal differentiation (Woodhead et al. 2006). Supporting this idea, overexpression of β-catenin in cortical progenitor cells expands the progenitor pool (Chenn and Walsh 2002, 2003). In addition to linking adjacent progenitors, AJs also appear to link nascent neurons with their RGC siblings. Many newborn neurons retain an apical process that maintains contact with the ventricular surface, as well as AJs with neighboring cells, and disassembly of these AJs increases neurogenesis through the misregulation of Notch signaling (Hatakeyama et al. 2014).
The transcriptional networks regulating N-cadherin expression also underscore its role in controlling proliferation and neuronal differentiation. The SoxB1 transcription factors are markers of neural progenitor cells (NPCs) and are important for maintaining progenitor identity, as overexpression of Sox2 inhibits neuronal differentiation, while inhibiting Sox2 promotes cell-cycle exit and premature differentiation (Graham et al. 2003; Bani-Yaghoub et al. 2006). Genomic analysis and expression studies reveal several Sox2 binding sites in the cdh2 genomic locus, which positively regulate N-cadherin expression (Matsumata et al. 2005). Conversely, N-cadherin expression is repressed by Foxp2, which is turned on in newly differentiated neurons (Rousso et al. 2012). Reduced levels of N-cadherin promote detachment of neurons from the ventricular surface and migration. Interfering with Foxp2 inhibits the differentiation of neuroepithelial progenitors and compromises the organization of the neuroepithelium. Thus, regulation of the cadherin-catenin complex plays a fundamental role in both maintaining the structural stability of the neuroepithelium and in regulating neuronal differentiation.
In addition to N-cadherin, a growing list of other cadherin superfamily members participates in neuronal differentiation. Expression studies have shown that many protocadherins and atypical cadherins are expressed in the neuroepithelium (Emond et al. 2009; Ishiuchi et al. 2009; Wang et al. 2016). Recently, it was shown in mice that the δ1-pcdh, pcdh11, is expressed in neural progenitors and regulates neuronal differentiation (Zhang et al. 2014). Knockdown of pcdh11 in progenitors, using short interfering RNA (siRNA), increases the number of neurons, while forced expression expands the population of progenitors at the expense of new neurons—similar to what is observed for N-cadherin and β-catenin. Together, these results suggest that pcdh11 expression in neural stem cells inhibits their differentiation into neurons. Similar results have been obtained in the zebrafish, in which loss of the δ2-Pcdh, pcdh19, increases proliferation and the number of pcdh19-expressing neurons in the optic tectum (Cooper et al. 2015). The micro-RNA, miR-484, affects neurogenesis by regulating pcdh19 expression (Fujitani et al. 2016). Increased levels of miR-484 cause hyperactivity in mice, which could be related to neurodevelopmental disorders associated with microduplications of this locus in humans. Forced expression also down-regulates pcdh19 and increases neuronal differentiation. This effect on neurogenesis was rescued by elevating expression of pcdh19. Together, the studies on Pcdh11 and Pcdh19 reveal a role for delta-protocadherins (δ-Pcdhs) in controlling neuronal differentiation. Moreover, evidence from nonneuronal systems suggests that this role involves links to Wnt signaling pathways, as both Pcdh10 and Pcdh17 have been shown to act as tumor suppressors by inhibiting β-catenin and Wnt signaling (Xu et al. 2015b; Yin et al. 2016). Additionally, several δ-Pcdhs interact with the Wnt receptor, Ryk (Berndt et al., 2011).
As was found for N-cadherin, each of the nonclustered δ-pcdhs is a target of the SoxB1 proteins, Sox2 and Sox3, suggesting that they are expressed in neural progenitors (Bergsland et al. 2011; McAninch and Thomas 2014). Similarly, in zebrafish, pcdh18a and pcdh18b were shown to be direct targets of SoxB1 proteins (Okuda et al. 2010). The involvement of cadherins in neurogenesis also extends beyond the δ-Pcdhs, as the atypical cadherins Fat, Dachsous, and Celsr have all been shown to regulate neuronal differentiation. In embryonic cortex, Fat4 interacts heterophilically with Dachsous1 to help organize the apical membrane at contacts between adjacent NECs (Ishiuchi et al. 2009). When levels of Dachsous1 or Fat4 are reduced in the neuroepithelium of developing mice, neuronal differentiation is reduced, with a commensurate increase in neuronal progenitors. Similarly, loss of Fat1 also alters apical junctions in mice, resulting in increased proliferation of neuronal progenitors. Some evidence also suggests a role for Celsr3 in regulating progenitor fates, as Celsr3-Fzd3 present on nascent neurons facilitates Notch signaling in NPCs; loss of Celsr3-Fzd3 results in premature neuronal differentiation and depletion of NPCs. It is clear that multiple cadherin family members (classical cadherins, δ-Pcdhs, Fats, Celsrs, and Dachsous) all act to control the proliferation and differentiation of neuronal progenitors, creating a network of cell surface interactions that sculpts the production of neurons.
As RGCs function both as neuronal progenitors and a substrate for radial migration, they produce over time a radial clone of sibling neurons (Fig. 1). Studies in both mouse visual cortex and Xenopus optic tectum reveal that sibling neurons respond to stimuli with similar features (Li et al. 2012; Muldal et al. 2014). Further, connectomic approaches show that neurons with similar feature selectivity tend to form synaptic subnetworks (Lee et al. 2016), which is consistent with one report that shows that sibling neurons are preferentially synaptically coupled (Yu et al. 2009). Collectively, these studies suggest that lineage plays a major role in organizing the development of local circuitry: developmental modules may largely coincide with functional modules. However, it is not known how synaptogenesis is biased to favor sibling neurons. Observations in the zebrafish optic tectum suggest a possible mechanism (Cooper et al. 2015). The optic tectum of more ancient vertebrates is homologous to the superior colliculus of mammals, which is a midbrain structure responsible for visual attention. Within the zebrafish optic tectum, the expression of δ-pcdhs, such as pcdh19, define radial columns of neurons; radial stripes of neurons express a common δ-Pcdh, each being a clone of sibling neurons derived from a common protocadherin-expressing progenitor (Cooper et al. 2015). Homophilic adhesion by δ-Pcdhs within a clone could promote ongoing contact between the growing axonal and dendritic arbors of siblings, biasing toward synaptogenesis among these neurons (Fig. 1). Such a mechanism could apply to the superior colliculus, which also has a columnar functional organization. Additionally, a similar radial organization has recently been shown in the developing mouse cortex, in which a LacZ knockin to the PCDH19 locus reveals radial stripes (Pederick et al. 2016). This mechanism does not appear to be limited to δ-Pcdhs, as retinal progenitor cells that express Cdh6 have been shown to generate cdh6-positive clones that include retinal ganglion cells, amacrine cells, bipolar cells, horizontal cells, and photoreceptors (De la Huerta et al. 2012).
AXON EXTENSION, TARGETING, AND ARBORIZATION
Although lineage contributes to the formation of local subnetworks, other mechanisms are required to connect distinct regions of the nervous system. This is largely accomplished, although the control of axon outgrowth and guidance, including guidance of axons to the correct region and arborization within the correct target areas (Katz and Shatz 1996; Benson et al. 2001). In general, the formation of axon tracts has two stages. First, the growth cones of pioneer axons respond to guidance cues, both contact-dependent and diffusible gradients. Second, follower growth cones, less affected by guidance cues, migrate along the previously established pathways. Although not traditional axon guidance molecules, the classical cadherins are intimately involved in axon outgrowth and targeting. In particular, N-cadherin appears to participate in multiple aspects of axon development and morphogenesis, including elongation, fasciculation, and target selection. Expression of dominant-interfering truncated forms of N-cadherin in Xenopus retinal ganglion cells impairs axon outgrowth (Riehl et al. 1996). Moreover, recent evidence suggests that N-cadherin helps establish neuronal polarity in the developing cortex and the initiation of axon outgrowth (Xu et al. 2015a). After the initiation of the axonal growth cone, most axons fasciculate as they join stereotyped axon tracts (Fig. 2). Both in vitro and in vivo studies support a role for N-cadherin in axon fasciculation (Masai et al. 2003; Sakai et al. 2012). Application of function-blocking N-cadherin antibodies to cultured dorsal root ganglion axons causes an increase in defasciculated growth cones (Honig et al. 1998). Zebrafish embryos harboring mutations in ncad show both fasciculation defects, as well as aberrant patterns of retinal ganglion axonal arborization (Masai et al. 2003). Thus, N-cadherin function appears to be important for the ability of follower growth cones to join existing axon tracts. Once axons grow into their target regions, they stop extending and begin to arborize. Work in the chick retinotectal system has suggested that N-cadherin is also important for laminar targeting and arbor elaboration of retinal ganglion cell axons (Inoue and Sanes 1997). In coculture experiments, chick retinal ganglion cell axons project to adjacent optic tectum slices, arborizing in the retinorecipient layer, the “stratum griseum et fibrosum superficiale.” When treated with function blocking antibodies against N-cadherin, these retinal ganglion cell axons overshoot their target laminae and have less elaborate arbors. A similar role in laminar targeting has been shown for mouse thalamocortical axons (Poskanzer et al. 2003).
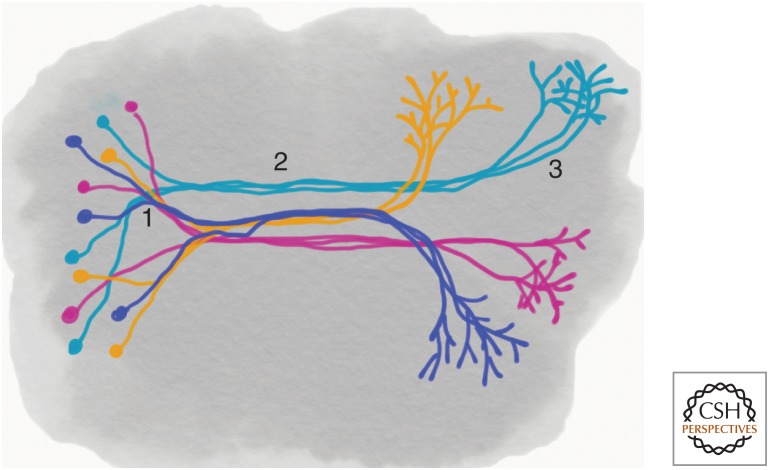
Figure 2.
Potential roles for cadherins in long range neural connectivity. Protocadherins, cadherins, and other adhesion molecules participate in selective fasciculation. (1) Axons expressing a common cadherin or protocadherin sort during elongation through contact-dependent promotion of growth cone motility. (2) Within tracts, axons segregate, as a result of homophilic interactions. This provides an underlying organization to fascicles that are densely populated with a large diversity of axons. Much of axon guidance is mediated by pioneer growth cones responding to arrays of both short- and long-range cues, and occurs when the nervous system is relatively simple. Follower growth cones extend along existing axons, relying heavily on selective adhesion. (3) Homophilic interactions promote target recognition and axonal arborization within the target area. This coarse targeting can be further refined by Ephrin gradients or other guidance cues.
Although N-cadherin is broadly expressed in the developing nervous system, other classical cadherins have more restricted expression patterns, which could suggest involvement in the assembly of specific neural circuits. Several of the type II classical cadherins are present in functionally related brain regions, as well as the axon tracts that connect them (Redies 2000). Like N-cadherin, the type II classical cadherins are implicated in multiple aspects of axon outgrowth and patterning. In particular, these proteins appear to be important for fasciculation and target recognition. Within the chick optic tectum, axons of tectal neurons form a stereotyped bundle that exits the caudal tectum; within this bundle, axons segregate to form distinct tectofugal fascicles. Several classical cadherins are expressed in the tectum, and distinguish among these subsets of tracts (Wohrn et al. 1999; Treubert-Zimmermann et al. 2002). Forced expression of individual cadherins by electroporation was able to redirect axons to inappropriate tracts, indicating that these classical cadherins play an instructive role in selective fasciculation to organize the tectofugal tracts (Treubert-Zimmermann et al. 2002). In organotypic hippocampal slice cultures, both N-cadherin and Cadherin-8 appear to be important for fasciculation of the mossy fiber projection from the dentate gyrus (DG) to CA3 (Bekirov et al. 2008), and blockade of adhesion by Cadherin-8 resulted in exuberant growth and arborization.
Once growth cones arrive at the appropriate target, they defasciculate and begin to arborize and initiate synaptogenesis (Fig. 2). Evidence supports a role for type II classical cadherins in target selection. Pontine neurons from the hindbrain project to the cerebellar cortex where they form synapses with granule cells (Kuwako et al. 2014). Cadherin-7 is expressed both by pontine neurons and granule cells. The presence of Cadherin-7 on the granule cells appears to inhibit the growth of pontine neuron axons and to promote synaptogenesis. Cadherin-6 may play a similar role in the visual system, as loss of Cadherin-6 in a subset of retinal ganglion cell axons causes targeting defects in their projection to thalamic targets (Osterhout et al. 2011). In each case, homophilic recognition could provide a stop signal, indicating that the appropriate target has been reached and promoting the transition to arborization and synaptogenesis. Thus, the cellular roles of cadherins are conditional, as they appear to promote fasciculation and growth in some circumstances and may act as stop signals in other contexts. This could reflect their integration with other axon guidance networks, as well as their interactions with other cadherin family members. Deciphering the precise roles of type II cadherins may not always be straightforward, as revealed by work in the mouse retina. Cadherins-8 and -9 are expressed in distinct subsets of bipolar cells, BC2, and BC5, respectively, which project to distinct sublaminae of the inner plexiform layer (IPL) (Duan et al. 2014). Conditional targeting of Cadherin-8 in BC2 cells or Cadherin-9 in BC5 cells results in imperfect targeting of their axonal arbors within the sublaminae. However, cell-autonomous rescue showed that homophilic adhesion is not required for correct targeting, leaving the mechanism of targeting unresolved.
Like the classical cadherins, the δ-protocadherins are also heavily involved in axon development. As was shown for classical cadherins, expression of a dominant interfering form of Pcdh7/NF-Pcdh disrupts the outgrowth of retinal ganglion cell axons in Xenopus (Piper et al. 2008). Pcdh7 is further involved in guidance at intermediate targets in the retinotectal projection (Leung et al. 2013) and appears to intersect both with the Semaphorin and the Netrin axon guidance pathways (Leung et al. 2013, 2015). Also like the classical cadherins, Pcdh10/OL-protocadherin shows regionalized expression in the brain, and in subsets of axonal pathways (Hirano et al. 1999; Aoki et al. 2003). In the chick spinal cord, Pcdh10 is found on motor axons, but not sensory axons, which segregate in the spinal nerve, suggesting a potential role in axon fasciculation (Nakao et al. 2005). Both Pcdh10 and Pcdh17 have been shown to promote contact-dependent cell motility by recruiting the WAVE complex (Nakao et al. 2008; Hayashi et al. 2014), an activator of actin-assembly. Among axons that express it, Pcdh17 clusters at interaxonal contacts. Similarly, Pcdh17 promotes growth cone recognition of axons and migration along other Pcdh17-expressing axons, and loss of Pcdh17 impairs growth cone migration. In each case, cell–cell contact recruits the WAVE complex, along with Lamellipodin and Ena/VASP to promote actin assembly and axon elongation, allowing axons to sort from one another by recognizing a preferred growth substrate (Hayashi et al. 2014). As all δ2-Pcdhs, as well as some other cadherin subfamilies, interact with the WAVE complex through a conserved motif, the WAVE interacting receptor sequence (WIRS) site (Chen et al. 2014), they could promote differential fasciculation and tract formation through their distinct homophilic preferences (Fig. 2). Their homophilic interactions could be important for establishing the initial scaffold of axon tracts or for targeting axons to their correct pathways. The δ-Pcdhs may also play a role in axon arborization, as knockdown of zebrafish Pcdh18b reduces the rate of branch initiation and overall arbor complexity (Biswas et al. 2014). In addition to the δ-Pcdhs, the α-Pcdhs are also involved in axon growth and targeting. Mice lacking the exons that encode the constant cytoplasmic domain of the α-Pcdhs show defects in the sorting of olfactory axons, as well as their targeting to glomeruli (Hasegawa et al. 2008). In these mice, serotonergic neurons also show abnormal terminal arborization patterns (Katori et al. 2009). The atypical cadherins Celsr and Fat also play important roles in the development of axon tracts. Both Celsr2 and Celsr3 participate in axon tract development in vertebrates (Zhou et al. 2008; Qu et al. 2014; Feng et al. 2016). Targeted inactivation of mouse celsr3 disrupts several major axon tracts, with a reduction of the cortical intermediate zone and a reduction or absence of other tracts, including the anterior commissure and the internal capsule (Zhou et al. 2008). A similar phenotype is observed in fzd3 knockouts, suggesting that the Celsr3-Fzd3 complex functions together in axon tract development (Feng et al. 2016). Less is known about the roles of the atypical Fat cadherins, although Fat3 labels axon fascicles in the developing mouse brain (Nagae et al. 2007).
SYNAPTIC DEVELOPMENT
Although Sperry’s chemoaffinity hypothesis imagined that topographic mapping was the result of growth cones responding to gradients of guidance molecules (Sperry 1963), this idea has often been permuted or extended to postulate the existence of recognition molecules responsible for point-to-point matching of pre- and postsynaptic partners. As classical cadherins show homophilic binding preferences, mediate cell sorting in vitro, and show differential patterns of expression, they are plausible candidates to guide selective synapse formation or stabilization. Although there is a dearth of evidence to support this hypothesis, cadherins do appear to play important roles in synaptic development.
Cadherins are prominent components of synapses, tending to be localized perisynaptically in more mature synapses (Fannon and Colman 1996). Studies in cultured hippocampal neurons show that N-cadherin is a very early marker of synapses (Benson and Tanaka 1998). Similarly, in vivo timelapse imaging in zebrafish spinal cord reveal that N-cadherin-GFP in enriched in the central domain of axonal growth cones and that puncta of N-cadherin-GFP are left in the wake of the migrating growth cone (Jontes et al. 2004). Moreover, the timing of punctum formation matches that of synaptic vesicle accumulation, indicating that N-cadherin may be recruited to nascent synapses shortly after initial axon–dendritic contact. N-cadherin appears to help organize presynaptic boutons, as expressing dominant-interfering mutants impairs vesicle clustering (Togashi et al. 2002; Bozdagi et al. 2004). In addition, N-cadherin can modulate voltage-gated calcium channels through RhoA and Rho-associated kinase (Marrs et al. 2009). Postsynaptically, dominant interfering forms of N-cadherin disrupt the accumulation of PSD-95 (Togashi et al. 2002; Bozdagi et al. 2004), a marker of glutamatergic synapses, and destabilizes dendritic spines. Deletion of N-cadherin, αN-catenin or δ-catenin delays the maturation of dendritic spines, with mutants showing a higher proportion of dynamic filopodia and fewer stable spines (Togashi et al. 2002; Abe et al. 2004; Arikkath et al. 2009). Thus, during synapse assembly, the role of the cadherin-catenin complex may partially be to balance the dynamics and motility of axons and dendrites, which is required to initiate cell–cell contacts, and the stability to allow these contacts to mature and assemble synaptic machinery.
Type II cadherins, which are expressed in more specific patterns, have also been shown to influence synaptic development. Mice lacking Cadherin-11 show enhanced long-term potentiation in the CA1 region of the hippocampus (Manabe et al. 2000). Additionally, in the hippocampus, Cadherin-9 is expressed both in pyramidal cells of both the DG and their targets in CA3 (Williams et al. 2011). The mossy fiber synapses formed between DG and CA3 neurons have a complex and distinctive structure, with multiple active zones interdigitating with an elaborate, multiheaded spine. Knockdown of Cadherin-9 in DG neurons alters the morphology of the presynaptic varicosity, which appears less elaborate with fewer filopodia extending from the bouton. In contrast, Cadherin-9 knockdown in the CA3 neurons causes an increase in dendritic filopodia, similar to what occurs in αN-catenin knockouts (Abe et al. 2004) or expression of dominant-interfering cadherins (Togashi et al. 2002). Thus, Cadherin-9 appears to contribute to synaptic assembly and maturation in a manner similar to what has been observed for N-cadherin in cultured neurons: presynaptically, loss of Cadherin-9 impairs bouton assembly, and, postsynaptically, loss of Cadherin-9 promotes filopodia dynamics.
Protocadherins and atypical cadherins also appear to play important roles in synapse stability. Pcdh8/Arcadlin was originally identified in a screen for genes up-regulated in the hippocampus after electroconvulsive shock (Yamagata et al. 1999). Under baseline conditions, Pcdh8 is weakly expressed in cultured hippocampal neurons, but is dramatically up-regulated after electrical stimulation. On expression, Pcdh8 interacts with N-cadherin and promotes its internalization by receptor-mediated endocytosis (Yasuda et al. 2007). These results suggest that Pcdh8 promotes synaptic elimination and that it does so by directly interacting with N-cadherin and antagonizing N-cadherin adhesion. A study of the transcription factor, myocyte enhancer factor 2 (MEF2), suggests that Pcdh10 also plays a role in synapse elimination (Tsai et al. 2012). Pcdh10 is a target for MEF2, which is activated by neuronal activity and leads to synapse elimination (Pfeiffer et al. 2010). MEF2-induced synapse elimination is proposed to be caused by ubiquitin-dependent degradation of the postsynaptic scaffolding protein, PSD-95. On activation of MEF2, PSD-95 becomes ubiquitinated by the E3 ubiquitin ligase Mdm2, and is then targeted to the proteasome for degradation. Knockdown of Pcdh10, using shRNA, inhibits MEF2-induced synapse elimination and PSD-95 degradation, as does overexpression of the Pcdh10 intracellular domain. Although several questions about this process remain unresolved, these results parallel those found for Pcdh8: up-regulation of a δ2-Pcdh by neural activity removes an important structural protein from the synapse to initiate synapse disassembly. Analysis of mice lacking Pcdh17 suggests a role in regulating active zone size (Hoshina et al. 2013). Pcdh17 is perisynaptic in the anterior striatum and lateral globus pallidus. This perisynaptic localization is similar to that observed for classical cadherins. Although there was no difference in the number or size of synaptic contacts in mice lacking pcdh17, there was an increase in both the total number of synaptic vesicles and in the number of docked vesicles. This was accompanied by an increase in paired-pulse facilitation, a result opposite to what is observed in β-catenin knockouts (Bamji et al. 2003). Thus, δ2-Pcdhs can negatively regulate synapse function and stability and, in at least one case, act by destabilizing N-cadherin adhesion at the synaptic membrane.
CONCLUDING REMARKS
After their original discovery, classical cadherins were revealed to participate in a broad range of developmental processes. Over time, this view has been enriched and extended to incorporate the broader cadherin superfamily. In the context of the nervous system, N-cadherin contributes to neurulation, neurogenesis, neuronal migration, axon extension and targeting, dendrite morphogenesis and dynamics, synapse assembly, and synaptic plasticity. It provides a continuous thread, linking the earliest events in nervous system formation with the molecular events of experience-dependent synapse modification. As the diversity of the cadherin superfamily expanded, it was reasonable to expect that there would be a commensurate increase in functional diversity: different molecules would do different things. Increasingly, it appears that, in contrast, their functions are linked and that they reinforce each other to sculpt developmental events. Their activities each represent continuous threads and these threads are woven together to provide robust developmental outcomes. They can modify, regulate and contextualize each other’s behaviors throughout development, rather than pursuing independent and/or parallel activities. In addition to their integrated roles in developmental processes, cadherins are linked in more intimate ways. There are multiple examples of distinct subfamilies associating as part of larger macromolecular assemblies. Classical cadherins associate with δ-protocadherins (Yasuda et al. 2007; Biswas et al. 2010; Emond et al. 2011), α-Pcdhs associate with β-Pcdhs and γ-Pcdhs (Thu et al. 2014), γ-Pcdhs associate with Ret kinase (an atypical cadherin) (Schalm et al. 2010), and N-cadherin, δ-Pcdhs, γ-Pcdhs, Celsrs, Fats, and Dachsous are all found in complex with the Wnt receptor, Ryk (Berndt et al. 2011). In several cases, these physical interactions reflect functional relationships. Finally, nearly all of the cadherin subfamilies intersect with Wnt signaling pathways, which also contribute to their interleaving functions.
As is the case in folding a protein, wiring the nervous system uses mechanisms that channel development toward a restricted subset of acceptable configurations, and the cadherins play essential roles in this process. In the formation of local circuitry, this may be through lineage-based mechanisms that establish functional modules. In this case, homophilic adhesion promotes persistent contact within a clone and biases the inherent promiscuity of synaptogenesis to favor connectivity among sibling neurons. This modular assembly confers robustness to the development of local circuitry. In the case of regional projections, selective axon fasciculation and target selection provide constraints that limit the number of potential synaptic partners available to each neuron. Again, these broad mechanisms bias the inherently nonspecific process of synapse formation to generate a coarse map of connections, providing a substrate for experience-dependent plasticity to integrate local with regional networks.
A rich understanding of how the cadherin superfamily contributes to neural circuit assembly will require a far more comprehensive survey than is currently available. While studies understandably focus on an individual molecule in a specific context, it will be necessary to determine how these examples extrapolate to other family members and contexts. Do all δ-Pcdhs promote collective axon extension, like Pcdh17? Do all type II classical cadherins mediate target recognition, like Cadherin-6? Comparable studies of other cadherins and protocadherins will be required to verify their function and to construct a developmental map of their activities during circuit assembly. Similarly, further work is required to determine if all δ-Pcdhs act like Pcdh19 to regulate lineage-based assembly of local circuits, and whether similar mechanisms are used broadly in the nervous system—beyond the optic tectum or superior colliculus. Finally, a full accounting of cadherin function will entail revealing the functional interactions between the cadherin subfamilies. For example, if N-cadherin, type II classical cadherins and δ-cdhs are all important for axon outgrowth, extension, and fasciculation, how are their respective roles integrated? Similarly, if classical cadherins, δ-Pcdhs, Fat/Dachsous and Celsr all influence Wnt signaling to control neuronal differentiation, what are the interaction and signaling networks that govern cell decisions? While there has been much progress, it remains unclear how molecular networks, including cadherins, collaborate to assemble neuronal networks during development.
ACKNOWLEDGMENTS
I thank Michelle Emond and Sarah Light for critical reading of the manuscript. Our work is supported by Grants from National Institutes of Health (NIH) (R01EY027003) and National Science Foundation (NSF) (IOS-1457126).
Footnotes
Editors: Carien M. Niessen and Alpha S. Yap
Additional Perspectives on Cell–Cell Junctions available at www.cshperspectives.org
REFERENCES
- Abe K, Chisaka O, Van Roy F, Takeichi M. 2004. Stability of dendritic spines and synaptic contacts is controlled by α N-catenin. Nat Neurosci 7: 357–363. [DOI] [PubMed] [Google Scholar]
- Aoki E, Kimura R, Suzuki ST, Hirano S. 2003. Distribution of OL-protocadherin protein in correlation with specific neural compartments and local circuits in the postnatal mouse brain. Neuroscience 117: 593–614. [DOI] [PubMed] [Google Scholar]
- Arikkath J, Peng IF, Ng YG, Israely I, Liu X, Ullian EM, Reichardt LF. 2009. δ-Catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J Neurosci 29: 5435–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T, Takeichi M, Okada TS. 1983. Selective expression of cell type specific cell–cell adhesion molecules in mouse hybrid cells. Differentiation 24: 140–144. [DOI] [PubMed] [Google Scholar]
- Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, Lu B, Reichardt LF. 2003. Role of β-catenin in synaptic vesicle localization and presynaptic assembly. Neuron 40: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, et al. 2006. Role of Sox2 in the development of the mouse neocortex. Dev Biol 295: 52–66. [DOI] [PubMed] [Google Scholar]
- Bekirov IH, Nagy V, Svoronos A, Huntley GW, Benson DL. 2008. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus 18: 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DL, Tanaka H. 1998. N-Cadherin redistribution during synaptogenesis in hippocampal neurons. J Neurosci 18: 6892–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DL, Colman DR, Huntley GW. 2001. Molecules, maps and synapse specificity. Nat Rev Neurosci 2: 899–909. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J. 2011. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev 25: 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. 2011. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling. J Cell Biol 194: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Emond MR, Jontes JD. 2010. Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. J Cell Biol 191: 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Emond MR, Duy PQ, Hao le T, Beattie CE, Jontes JD. 2014. Protocadherin-18b interacts with Nap1 to control motor axon growth and arborization in zebrafish. Mol Biol Cell 25: 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. 2004. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol Cell Neurosci 27: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. 2001. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253–1264. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Denk W. 2006. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol 16: 562–570. [DOI] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. 2014. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HJ, Nakamoto M, Bergemann AD, Flanagan JG. 1995. Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell 82: 371–381. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. 2002. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297: 365–369. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. 2003. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in β-catenin overexpressing transgenic mice. Cereb Cortex 13: 599–606. [DOI] [PubMed] [Google Scholar]
- Chenn A, Zhang YA, Chang BT, McConnell SK. 1998. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci 11: 183–193. [DOI] [PubMed] [Google Scholar]
- Constantine-Paton M, Law MI. 1978. Eye-specific termination bands in tecta of three-eyed frogs. Science 202: 639–641. [DOI] [PubMed] [Google Scholar]
- Cooper SR, Emond MR, Duy PQ, Liebau BG, Wolman MA, Jontes JD. 2015. Protocadherins control the modular assembly of neuronal columns in the zebrafish optic tectum. J Cell Biol 211: 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Huerta I, Kim IJ, Voinescu PE, Sanes JR. 2012. Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci 109: 17663–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Horstmann H. 2004. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol 2: e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA, Chan HS. 1997. From Levinthal to pathways to funnels. Nat Struct Biol 4: 10–19. [DOI] [PubMed] [Google Scholar]
- Drescher U, Kremoser C, Handwerker C, Loschinger J, Noda M, Bonhoeffer F. 1995. In vitro guidance of retinal ganglion cell axons by RAGS, a 25 kDa tectal protein related to ligands for Eph receptor tyrosine kinases. Cell 82: 359–370. [DOI] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. 2014. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158: 793–807. [DOI] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Jontes JD. 2009. Protocadherin-19 is essential for early steps in brain morphogenesis. Dev Biol 334: 72–83. [DOI] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Blevins CJ, Jontes JD. 2011. A complex of Protocadherin-19 and N-cadherin mediates a novel mechanism of cell adhesion. J Cell Biol 195: 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannon AM, Colman DR. 1996. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron 17: 423–434. [DOI] [PubMed] [Google Scholar]
- Feng J, Xian Q, Guan T, Hu J, Wang M, Huang Y, So KF, Evans SM, Chai G, Goffinet AM, et al. 2016. Celsr3 and Fzd3 organize a pioneer neuron scaffold to steer growing thalamocortical axons. Cereb Cortex 26: 3323–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani M, Zhang S, Fujiki R, Fujihara Y, Yamashita T. 2016. A chromosome 16p13.11 microduplication causes hyperactivity through dysregulation of miR-484/protocadherin-19 signaling. Mol Psychiatry 22: 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzler-Odenthal SI, Redies C. 1998. Blocking N-cadherin function disrupts the epithelial structure of differentiating neural tissue in the embryonic chicken brain. J Neurosci 18: 5415–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. 2005. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39: 749–765. [DOI] [PubMed] [Google Scholar]
- Hasegawa S, Hamada S, Kumode Y, Esumi S, Katori S, Fukuda E, Uchiyama Y, Hirabayashi T, Mombaerts P, Yagi T. 2008. The protocadherin- α family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol Cell Neurosci 38: 66–79. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Wakamatsu Y, Nagafuchi A, Kageyama R, Shigemoto R, Shimamura K. 2014. Cadherin-based adhesions in the apical endfoot are required for active Notch signaling to control neurogenesis in vertebrates. Development 141: 1671–1682. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Inoue Y, Kiyonari H, Abe T, Misaki K, Moriguchi H, Tanaka Y, Takeichi M. 2014. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev Cell 30: 673–687. [DOI] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. 2012. Cadherins in brain morphogenesis and wiring. Physiol Rev 92: 597–634. [DOI] [PubMed] [Google Scholar]
- Hirano S, Yan Q, Suzuki ST. 1999. Expression of a novel protocadherin, OL-protocadherin, in a subset of functional systems of the developing mouse brain. J Neurosci 19: 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig MG, Petersen GG, Rutishauser US, Camilli SJ. 1998. In vitro studies of growth cone behavior support a role for fasciculation mediated by cell adhesion molecules in sensory axon guidance during development. Dev Biol 204: 317–326. [DOI] [PubMed] [Google Scholar]
- Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T, Yokoyama K, Tezuka T, Sagara H, Hirano S, et al. 2013. Protocadherin 17 regulates presynaptic assembly in topographic corticobasal ganglia circuits. Neuron 78: 839–854. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. 2011. New insights into the evolution of metazoan cadherins. Mol Biol Evol 28: 647–657. [DOI] [PubMed] [Google Scholar]
- Inoue A, Sanes JR. 1997. Lamina-specific connectivity in the brain: Regulation by N-cadherin, neurotrophins, and glycoconjugates. Science 276: 1428–1431. [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T. 2009. Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol 185: 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jontes JD, Phillips GR. 2006. Selective stabilization and synaptic specificity: A new cell-biological model. Trends Neurosci 29: 186–191. [DOI] [PubMed] [Google Scholar]
- Jontes JD, Emond MR, Smith SJ. 2004. In vivo trafficking and targeting of N-cadherin to nascent presynaptic terminals. J Neurosci 24: 9027–9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans D, Hack I, Frotscher M, Taylor V, Kemler R. 2005. β-Catenin-mediated cell-adhesion is vital for embryonic forebrain development. Dev Dyn 233: 528–539. [DOI] [PubMed] [Google Scholar]
- Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M. 2007. N-Cadherin mediates cortical organization in the mouse brain. Dev Biol 304: 22–33. [DOI] [PubMed] [Google Scholar]
- Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. 2009. Protocadherin-α family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci 29: 9137–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. 1996. Synaptic activity and the construction of cortical circuits. Science 274: 1133–1138. [DOI] [PubMed] [Google Scholar]
- Knudsen EI, Brainard MS. 1991. Visual instruction of the neural map of auditory space in the developing optic tectum. Science 253: 85–87. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. 2009. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32: 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwako K, Nishimoto Y, Kawase S, Okano HJ, Okano H. 2014. Cadherin-7 regulates mossy fiber connectivity in the cerebellum. Cell Rep 9: 311–323. [DOI] [PubMed] [Google Scholar]
- Lee WC, Bonin V, Reed M, Graham BJ, Hood G, Glattfelder K, Reid RC. 2016. Anatomy and function of an excitatory network in the visual cortex. Nature 532: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele Z, Folchert A, Concha M, Rauch GJ, Geisler R, Rosa F, Wilson SW, Hammerschmidt M, Bally-Cuif L. 2002. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129: 3281–3294. [DOI] [PubMed] [Google Scholar]
- Leung LC, Urbancic V, Baudet ML, Dwivedy A, Bayley TG, Lee AC, Harris WA, Holt CE. 2013. Coupling of NF-protocadherin signaling to axon guidance by cue-induced translation. Nat Neurosci 16: 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung LC, Harris WA, Holt CE, Piper M. 2015. NF-protocadherin regulates retinal ganglion cell axon behaviour in the developing visual system. PLoS ONE 10: e0141290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal C. 1969. How to fold graciously. In Mossbauer spectroscopy in biological systems (ed. Debrunner P, Tsibris JCM, Münck E), pp. 22–24. University of Illinois Press, Urbana, IL. [Google Scholar]
- Li Y, Lu H, Cheng PL, Ge S, Xu H, Shi SH, Dan Y. 2012. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature 486: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Denk W. 2011. The big and the small: Challenges of imaging the brain’s circuits. Science 334: 618–623. [DOI] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. 2006. αE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science 311: 1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe T, Togashi H, Uchida N, Suzuki SC, Hayakawa Y, Yamamoto M, Yoda H, Miyakawa T, Takeichi M, Chisaka O. 2000. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol Cell Neurosci 15: 534–546. [DOI] [PubMed] [Google Scholar]
- Marrs GS, Theisen CS, Bruses JL. 2009. N-Cadherin modulates voltage activated calcium influx via RhoA, p120–catenin, and myosin–actin interaction. Mol Cell Neurosci 40: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I, Lele Z, Yamaguchi M, Komori A, Nakata A, Nishiwaki Y, Wada H, Tanaka H, Nojima Y, Hammerschmidt M, et al. 2003. N-Cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development 130: 2479–2494. [DOI] [PubMed] [Google Scholar]
- Matsumata M, Uchikawa M, Kamachi Y, Kondoh H. 2005. Multiple N-cadherin enhancers identified by systematic functional screening indicate its group B1 SOX-dependent regulation in neural and placodal development. Dev Biol 286: 601–617. [DOI] [PubMed] [Google Scholar]
- McAninch D, Thomas P. 2014. Identification of highly conserved putative developmental enhancers bound by SOX3 in neural progenitors using ChIP-Seq. PLoS ONE 9: e113361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldal AM, Lillicrap TP, Richards BA, Akerman CJ. 2014. Clonal relationships impact neuronal tuning within a phylogenetically ancient vertebrate brain structure. Curr Biol 24: 1929–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae S, Tanoue T, Takeichi M. 2007. Temporal and spatial expression profiles of the Fat3 protein, a giant cadherin molecule, during mouse development. Dev Dyn 236: 534–543. [DOI] [PubMed] [Google Scholar]
- Nakao S, Uemura M, Aoki E, Suzuki ST, Takeichi M, Hirano S. 2005. Distribution of OL-protocadherin in axon fibers in the developing chick nervous system. Brain Res Mol Brain Res 134: 294–308. [DOI] [PubMed] [Google Scholar]
- Nakao S, Platek A, Hirano S, Takeichi M. 2008. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol 182: 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y, Ogura E, Kondoh H, Kamachi Y. 2010. B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet 6: e1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu SW, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, et al. 2011. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron 71: 632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederick DT, Homan CC, Jaehne EJ, Piltz SG, Haines BP, Baune BT, Jolly LA, Hughes JN, Gecz J, Thomas PQ. 2016. Pcdh19 loss-of-function increases neuronal migration in vitro but is dispensable for brain development in mice. Sci Rep 6: 26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM. 2010. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron 66: 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Dwivedy A, Leung L, Bradley RS, Holt CE. 2008. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J Neurosci 28: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poskanzer K, Needleman LA, Bozdagi O, Huntley GW. 2003. N-Cadherin regulates ingrowth and laminar targeting of thalamocortical axons. J Neurosci 23: 2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Huang Y, Feng J, Alvarez-Bolado G, Grove EA, Yang Y, Tissir F, Zhou L, Goffinet AM. 2014. Genetic evidence that Celsr3 and Celsr2, together with Fzd3, regulate forebrain wiring in a Vangl-independent manner. Proc Natl Acad Sci 111: E2996–E3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 2009. Evolution of the neocortex: A perspective from developmental biology. Nat Rev Neurosci 10: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C. 2000. Cadherins in the central nervous system. Prog Neurobiol 61: 611–648. [DOI] [PubMed] [Google Scholar]
- Riehl R, Johnson K, Bradley R, Grunwald GB, Cornel E, Lilienbaum A, Holt CE. 1996. Cadherin function is required for axon outgrowth in retinal ganglion cells in vivo. Neuron 17: 837–848. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, Morrisey EE, Novitch BG. 2012. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron 74: 314–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai N, Insolera R, Sillitoe RV, Shi SH, Kaprielian Z. 2012. Axon sorting within the spinal cord marginal zone via Robo-mediated inhibition of N-cadherin controls spinocerebellar tract formation. J Neurosci 32: 15377–15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SS, Ballif BA, Buchanan SM, Phillips GR, Maniatis T. 2010. Phosphorylation of protocadherin proteins by the receptor tyrosine kinase Ret. Proc Natl Acad Sci 107: 13894–13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayoshi Y, Hatta K, Hosoda M, Tsunasawa S, Sakiyama F, Takeichi M. 1986. Cadherin cell adhesion molecules with distinct binding specificities share a common structure. EMBO J 5: 2485–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry RW. 1963. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci 50: 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker AM, Chenn A. 2009. Focal reduction of αE-catenin causes premature differentiation and reduction of β-catenin signaling during cortical development. Dev Biol 328: 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 1977. Functional correlation between cell adhesive properties and some cell surface proteins. J Cell Biol 75: 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M, Atsumi T, Yoshida C, Uno K, Okada TS. 1981. Selective adhesion of embryonal carcinoma cells and differentiated cells by Ca2+-dependent sites. Dev Biol 87: 340–350. [DOI] [PubMed] [Google Scholar]
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T. 2014. Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell 158: 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. 2002. Cadherin regulates dendritic spine morphogenesis. Neuron 35: 77–89. [DOI] [PubMed] [Google Scholar]
- Treubert-Zimmermann U, Heyers D, Redies C. 2002. Targeting axons to specific fiber tracts in vivo by altering cadherin expression. J Neurosci 22: 7617–7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JW, Feldheim DA. 2012. Eph and ephrin signaling in the formation of topographic maps. Semin Cell Dev Biol 23: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N-P, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM. 2012. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 151: 1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushihara H, Takeichi M. 1980. Cell–cell adhesion molecule: Identification of a glycoprotein relevant to the Ca2+-independent aggregation of Chinese hamster fibroblasts. Cell 20: 363–371. [DOI] [PubMed] [Google Scholar]
- Vestweber D, Kemler R. 1984. Rabbit antiserum against a purified surface glycoprotein decompacts mouse preimplantation embryos and reacts with specific adult tissues. Exp Cell Res 152: 169–178. [DOI] [PubMed] [Google Scholar]
- Wang W, Jossin Y, Chai G, Lien WH, Tissir F, Goffinet AM. 2016. Feedback regulation of apical progenitor fate by immature neurons through Wnt7–Celsr3–Fzd3 signalling. Nat Commun 7: 10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, et al. 2011. Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron 71: 640–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohrn JC, Nakagawa S, Ast M, Takeichi M, Redies C. 1999. Combinatorial expression of cadherins in the tectum and the sorting of neurites in the tectofugal pathways of the chicken embryo. Neuroscience 90: 985–1000. [DOI] [PubMed] [Google Scholar]
- Woodhead GJ, Mutch CA, Olson EC, Chenn A. 2006. Cell-autonomous β-catenin signaling regulates cortical precursor proliferation. J Neurosci 26: 12620–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Funahashi Y, Watanabe T, Takano T, Nakamuta S, Namba T, Kaibuchi K. 2015a. Radial glial cell–neuron interaction directs axon formation at the opposite side of the neuron from the contact site. J Neurosci 35: 14517–14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Yang Z, Yuan H, Li Z, Li Y, Liu Q, Chen J. 2015b. PCDH10 inhibits cell proliferation of multiple myeloma via the negative regulation of the Wnt/β-catenin/BCL-9 signaling pathway. Oncol Rep 34: 747–754. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, Miki N, Hayashi Y, Yoshioka M, Kaneko K, et al. 1999. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J Biol Chem 274: 19473–11979. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, et al. 2007. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2β and p38 MAP kinases. Neuron 56: 456–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Xiang T, Mu J, Mao H, Li L, Huang X, Li C, Feng Y, Luo X, Wei Y, et al. 2016. Protocadherin 17 functions as a tumor suppressor suppressing Wnt/β-catenin signaling and cell metastasis and is frequently methylated in breast cancer. Oncotarget 9: 51720–51732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YC, Bultje RS, Wang X, Shi SH. 2009. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458: 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wu C, Liu N, Niu L, Yan Z, Feng Y, Xu R. 2014. Protocadherin 11 x regulates differentiation and proliferation of neural stem cell in vitro and in vivo. J Mol Neurosci 54: 199–210. [DOI] [PubMed] [Google Scholar]
- Zhou L, Bar I, Achouri Y, Campbell K, De Backer O, Hebert JM, Jones K, Kessaris N, de Rouvroit CL, O’Leary D, et al. 2008. Early forebrain wiring: Genetic dissection using conditional Celsr3 mutant mice. Science 320: 946–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman M, Trinh le A, Fraser SE, Moens CB. 2011. Zebrafish neural tube morphogenesis requires Scribble-dependent oriented cell divisions. Curr Biol 21: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]